Abstract
The insulin-like growth factor type 1 (IGF-I) plays an important role in neuronal physiology. Reduced IGF-I levels are observed during aging and this decrease may be important to age-related changes in the brain. We studied the effects of IGF-I on total protein oxidation in brain tissues and in cell cultures. Our results indicate that in frontal cortex the level of oxidized proteins is significantly reduced in transgenic mice designed to overproduce IGF-I compared with wild-type animals. The frontal cortex of IGF-I-overproducing mice exhibited high chymotrypsin-like activity of the 20S and 26S proteasomes. The proteasome can also be activated in response to IGF-I in cell cultures. Kinetic studies revealed peak activation of the proteasome within 15 min following IGF-I stimulation. The effects of IGF-I on proteasome were not observed in R- cells lacking the IGF-I receptor. Experiments using specific kinase inhibitors suggested that activation of proteasome by IGF-I involves phosphatidyl inositol 3-kinase and mammalian target of rapamycin signaling. IGF-I also attenuated the increase in protein carbonyl content induced by proteasome inhibition. Thus, appropriate levels of IGF-I may be important for the elimination of oxidized proteins in the brain in a process mediated by activation of the proteasome.
Keywords: Insulin-like growth factor-I, Proteasome, Aging, Brain, Protein oxidation
1. Introduction
Insulin-like growth factor-I (IGF-I) and its IGF I type receptor are widely expressed in neural cells, including astrocytes and neurons, during development and in the adult. IGF-I acts locally near its sites of expression and on distant targets in an autocrine or paracrine fashion (D’Ercole and Ye, 2008), and its actions are primarily, if not entirely, mediated by IGF1R (Baker et al., 1993; Efstratiadis, 1998; Liu et al., 1993). The phosphatidyl inositol (PI) 3-kinase-Akt and MAP (mitogen-activated protein) kinase intracellular signaling pathways mediate many effects of IGF-I signaling and IGF-mediated responses in neural cells (Ye and D’Ercole, 2006). While global reductions in growth hormone or IGF-I seem to be involved in increased longevity, IGF-I is a critical growth factor in most if not all organs, perhaps especially in the brain. For example, IGF-I expression is essential for brain development and both IGF-I and its cognate receptor are widely expressed in the central nervous system (Popken et al., 2005; Russo et al., 2005). Disruption of the Igf1 gene in mice results in profound retardation of brain growth (Baker et al., 1993; Beck et al., 1995; Ye et al., 2002). Increased expression of IGF-I has been accomplished using a number of approaches such as placing the complementary DNA (cDNA) under the control of myelin basic protein promoter or the nestin promoter (D’Ercole and Ye, 2008; Luzi et al., 2004; Popken et al., 2004). Multiple studies suggest that increased expression of IGF-I in the brain leads to increased brain size through both increased proliferation of neural precursors and decreased apoptosis in neurons and oligodendrocytes and their precursors (D’Ercole and Ye, 2008).
In the adult brain, emergent evidence indicates that IGF-I is a neurotrophic and neuroprotective factor vital to the preservation of homeostasis. IGF-I is important for the maintenance of cognitive status, the prevention of tissue atrophy, the avoidance of vascular dysfunction, and the clearance of deleterious substances (Fernandez et al., 2007). Replacement studies to increase growth hormone or IGF-I in aged animals demonstrate cognitive improvements (Markowska et al., 1998; Ramsey et al., 2004) and IGF-I expression or administration attenuates damage in the brain following demyelinating insult and ischemia reperfusion injury (Guan et al., 2001; Mason et al., 2000). In aging long-lived Ames dwarf mice, IGF-I is increased in the dentate gyrus, resulting in an increased cell proliferation. In addition, several lines of evidence suggest that IGF-I may play a neuroprotective role in the clearance of amyloid, glycated products (Carro et al., 2002), and oxidized proteins (Li and Ren, 2007). In this report we examine the role and mechanisms of IGF-I in the control of oxidized protein abundance in vivo and in vitro. In order to examine the influence of IGF-I on the clearance of oxidized proteins in the brain, we examined protein carbonyl content and proteasome activity in mice that either overproduce or underproduce IGF-I.
In mammalian cells, the ubiquitin/proteasome pathway constitutes the major non-lysosomal proteolytic pathway. The 26S proteasome, responsible for the degradation of the majority of intracellular proteins, is a large multicatalytic protease composed of the 20S catalytic core and two 19S (PA700) multisubunit regulatory complexes that confer ubiquitin specificity and ATP-dependence (Goldberg and St John, 1976; Hershko and Ciechanover, 1998). In general, to be processed by the 26S proteasome pathway, proteins must be targeted for recognition and subsequent degradation by covalent attachment of monomers of the 76 amino acid polypeptide ubiquitin. The 20S proteasome in turn, works independently of ATP and ubiquitin and has been implicated in the degradation of damaged or unfolded proteins (Grune et al., 1997). 26S proteasome activity can be identified as the ATP-associated activity. ATP stabilizes the 26S complex and allows the opening of channels in the rings of the 20S core by ATPases located in the 19S regulatory particle (Köhler et al., 2001). The 20S proteasome, can be identified as the SDS-associated activity. Low concentrations of SDS cause gate opening and activation of the 20S particles (Coux et al., 1996). Our results suggest that IGF-I stimulates proteasome activity, in a process mediated by the IGF-I receptor, PI3 kinase/mTOR (mammalian target of rapamycin) signaling.
2. Materials and methods
2.1.1 Materials
Media and serum were purchased from Life Technologies, Inc. (Carlsbad, CA). Epoxomicin, MG115, N-succinyl-LLVY-AMC, Boc-LRR-AMC, Z-LLE-AMC, U0126, LY29402, rapamycin, antibodies against α (PW 8195), core α/β (PW 8155), β5 (PW 8895), and β5i (PW 8355), subunits were purchased from Biomol Research Laboratories, Inc. (Plymouth Meeting, PA). Oxyblot kit was purchased from Chemicon International (Serologicals Corp., Norcross, GA). Human IGF-I was purchased from Novozymes GroPep, Ltd. (Thebarton, Australia). Unless otherwise indicated, all other reagents were purchased from Sigma (St. Louis, MO).
2.1.2. Animals
We used a unique transgenic mouse line that produces IGF-I from an independent promoter that is not growth hormone dependent (cardiac myosin) (Reiss et al., 1996). This line (referred as cm-Igf1) has been used in several studies and is known to be in excellent health until at least 24 months (Li et al., 2007). A second transgenic mouse line that harbors a hypomorphic allele of the Igf1 gene (Igf1TM2s/IM) was obtained from Jackson Laboratories (Bar Harbor, ME) (Lembo et al., 1996). Mice homozygous for this allele are referred to as midi mice for the purposes of this work.
2.2 Methods
2.2.1. Cell culture
Cell lines were grown at 37°C, 5% CO2 in minimum Eagle medium (MEM) (cell lines WI38, U118MG) or Dulbecco modified Eagle medium (DMEM) (cell lines LN229, U87MG, C6, IGFIR-), supplemented with 2 mM L-glutamine and 10% fetal bovine serum (FBS). Cells were seeded at 1 × 104 cells/cm2 and grown until they reached 70% to 80% confluence. G0-arrested cultures were obtained by 2 days incubation in serum-free MCDB 104 medium or medium containing 0.5% FBS. WI38 fibroblasts were considered to be at early passage when less than 50% of their replicative lifespan was completed. Cultures at the end of the replicative lifespan (senescent cells) were those at greater than 90% of lifespan completed when cultures were unable to complete one population doubling during a 4-week period with weekly refeeding.
2.2.2. IGF-I determination
Quantitative determination of IGF-I in brain tissues was performed by using a mouse IGF-I high-sensitivity enzyme-linked immunosorbent assay (ELISA) kit (Immunodiagnostic Systems, Ltd., Boldon, Tyne and Wear, UK), following the instructions provided by the manufacturer.
2.2.3. Proteasome activity
Proteasome activity was assayed as previously described (Torres et al., 2006) with the following modifications. Control cells or after treatments were washed with cold phosphate-buffered saline (PBS), collected and resuspended in ice-cold proteasome buffer (10 mM Tris, pH 7.8, 1 mM EDTA, 5 mM MgCl2, 0.5 mM DTT). After 10 min on ice cells were lysed by 30 strokes in a glass-polytetrafluoroethylene Dounce homogenizer. Cell debris was removed by centrifugation for 1 h at 10,000 × g at 4°C and the supernatant was used for the assay. Frontal cortex tissue was homogenized in a glass-polytetrafluoroethylene homogenizer in cold proteasome buffer, debris was removed by centrifugation for 1 h at 10,000 × g at 4°C, and the supernatant was used for the assay. Proteasome activity was assayed in proteasome buffer at 37°C, using 10 μg of protein extracts in the presence of 100 μM of the specific fluorogenic peptide substrates, Suc-LLVY-AMC (chymotrypsin-like activity); Boc-LRR-AMC (trypsin-like activity; and Z-LLE-AMC (caspase-like activity). The reaction was monitored by measuring 7-amino-4-methylcoumarin released over a period of 1 h by fluorimetric measurements every 10 min (ex. 350 nm, em. 460 nm) in a Synergy HT Multi-Detection microplate reader and software KC4 v.3.0 (Bio-Tek Instruments, Inc, Winooski, VT). Proteasome activity corresponds to the remaining activity after subtraction of the activity determined in the presence of 20 μM epoxomicin.
2.2.4. Metabolic labeling
Cells were labeled for 30 min by adding 70 μCi/mL of Pro-mix (Pro-mix L-35S, Amersham Bioscience, Fairfield, CT) to the media (1 mL) in 12-well plates. After labeling, cycloheximide was added to 100 μg/mL and incubated for 5 min (to block further protein synthesis). Cells were then washed twice with PBS, lysed in 1 mL of RIPA buffer, transferred to microcentrifuge tubes, and placed at −20°C. For the trichloroacetic acid (TCA; Fisher Scientific) precipitation (ppt), 75 μL of extract was added to 1 mL of ice cold 10% TCA and incubated on ice for 30 min. The mixture was then passed through a glass filter (GF/C Millipore, St. Charles, MO) and washed using a vacuum manifold, according to the manufacturer’s instructions. The dried filters were placed into 5 mL of scintillation fluid and read in a scintillation counter. For total cpm, 20 μL of total extract was placed onto a glass filter, dried, and read as above. Counts were normalized to microliters of extract and %TCA incorporation was determined by dividing the TCA ppt cpm/μL extract by total cpm/μL extract. All experiments were performed a minimum of 2 times with duplicate wells.
2.2.5. Protein carbonyl content
Protein carbonyl groups were determined by using Oxyblot, a commercially available kit from Chemicon International (Serologicals Corp.). In brief, 15 μg of total proteins extracted from cells or brain tissues were derivatized at room temperature with 2,4 dinitrophenylhydrazine (DNPH) for 15 min in the presence of 5% 2-mercaptoethanol and 6% sodium dodecyl sulfate. Protein carbonyl groups were analyzed by Western blot using an anti-DNP polyclonal antibody.
2.2.6. ROS production
Intracellular reactive oxygen species (ROS) production was determined by using 2′,7′-dichlorofluorescein diacetate (DCFH-DA) as previously described (Wang and Joseph, 1999) with the following modifications. Cells were seeded at 1 × 104 cells/cm2 into 10-cm plates 2 days before the experiment. After 6 h of incubation with 10 ng/mL IGF-I, the cells were washed twice with KRPG and incubated with 10 μM DCFH-DA in MEM 1% FBS for 30 min, at 37°C in the incubator. The cells were washed twice with KRPG, harvested with a Teflon scrapper, and sonicated. An aliquot of the homogenates were placed in 96-well μClear Bottom plates (Greiner Bio-One, Inc., Longwood, FL) and the fluorescence at 530 nm was measured after excitation at 485 nm. Fluorometric determinations were performed in a Synergy HT Multi-Detection microplate reader, using the software KC4 v.3.0 (Bio-Tek Instruments, Inc.). Fluorescence values are expressed as fluorescence unit per micrograms of total protein.
2.2.7. Statistics
Unless otherwise specified, all experiments were done at least in triplicate. Data are expressed as mean ± SEM and were analyzed by two-sample Student t test. Differences were considered statistically significant at p < 0.05.
3. Results
3.1. Effects of IGF-I on levels of oxidized proteins
We initially studied the effects of IGF-I on total levels of oxidized proteins in brain tissue and in cell cultures by examining protein carbonyl content using mice that produce varying levels of IGF-I. Mice that harbor the IGF-I cDNA under the control of the cardiac myosin promoter have previously been shown to produce elevated levels of serum IGF-I (Reiss et al., 1996). We examined the level of IGF-I in the brains of these animals and found that compared with wild-type controls (FVB+/+), IGF-I heterozygous (FVB+/cm-Igf1) and homozygous (FVBcm-Igf1/cm-Igf1) contain significantly more IGF-I (Fig. 1A). A second mouse line, known as the midi mouse, produces lower levels of IGF-I due to an insertion in exon 3 of the IGF-I gene (Lembo et al., 1996). In mice homozygous for the midi allele, serum levels of IGF-I are reduced by approximately 60%, (data not shown) and we find a similar reduction in IGF-I levels in the brain (Fig. 1B).
Fig. 1.
IGF-I levels in brain frontal cortex. IGF-I levels were determined by ELISA in brain frontal cortex of mice as indicated in Materials and methods. (A) Wild-type: FVB+/+. Heterozygous: FVB+/cm-Igf1. Homozygous: FVBcm-Igf1/cm-Igf1. (B) C57 wild-type and hypomorphic midi mice (n = 5).
We next examined the level of protein oxidation in the brains of these animals. Our results indicate that in frontal cortex the levels of protein carbonyls are significantly reduced in animals overexpressing IGF-I (p < 00.5). Compared with wild-type animals, heterozygous and homozygous cm-Igf1 mice show a decrease of about 60% and 80% in protein carbonyl content, respectively (Fig. 2, left panel). In concordance with these results, the frontal cortex of midi mice showed a significant increase in protein carbonyls compared to controls (p < 0.05) (Fig. 2, right panel).
Fig. 2.

Effect of IGF-I on protein carbonyl content brain tissue. Fifteen micrograms of total proteins from frontal cortex were DNPH-derivatized and carbonyl groups were analyzed by Western blot using an anti-DNP polyclonal antibody. Left panel, protein carbonyls in wild-type (wt), heterozygous (het), and homozygous (hom) transgenic cm-IgfI mice. Right panel, protein carbonyls in wild-type C57 (wt) and midi mice. Bottom gels show Ponceau S staining. Graphs show carbonyl levels relative to respective controls, (average quantitation of 3-4 immunoblots). * p < 0.05 compared to wild type animals.
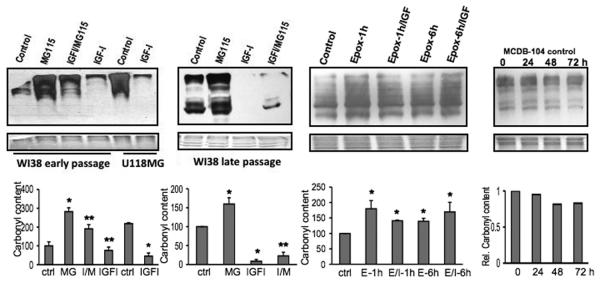
In order to examine the effect of direct stimulation with IGF-I on levels of oxidized proteins, we examined several cell types including human brain cells (U118MG cells) and human fibroblasts (WI38 cells). In both early-passage WI38 human fibroblasts and U118MG human glioblastoma (Fig. 3, left panel), IGF-I stimulation led to a reduction in protein carbonyl content. Because senescent cells are known to increase with age in vivo (Herbig et al., 2006) and it has been suggested that these cells can influence the local tissue environment (Coppe et al., 2008), we examined both the level of oxidized proteins and the response to IGF-I in late-passage WI38 human fibroblasts. There is a substantial protein carbonyl content in senescent fibroblasts and the level of carbonyl modification is attenuated by IGF-I stimulation. Thus, the IGF-I-induced reduction in carbonyls is more evident in late-passage cells where controls already show a high level of oxidized proteins (Fig. 3, central-left panel). As expected the reversible proteasome inhibitor MG115 increased carbonyl content. Surprisingly this increase was mitigated by the presence of IGF-I in the incubation medium, but not by the presence of the irreversible inhibitor epoxomicin (Fig. 3, central-right panel). MCDB 104 medium alone had no effect on carbonyl levels, even during incubation periods of up to 72 hours (Figure 3 right panel). These results suggest that although alternative mechanisms could trigger the reduction in protein carbonyl in response to IGF-I, proteasome activation seems to be involved. This result is consistent with our previous studies, in which we established that impairment of proteasome function is associated with a significant increase in total protein carbonyl content (Torres et al., 2006). Taken together these results suggest that IGF-I enhances the clearance of oxidized and damaged proteins in a process mediated by the proteasome.
Fig. 3.
Effect of IGF-I on protein carbonyl content in vitro. Fifteen micrograms of total proteins from cell extracts were DNPH-derivatized and carbonyl groups were analyzed by Western blot using an anti-DNP polyclonal antibody. Cells were cultured in MEM/10% FBS, synchronized in G0 by serum deprivation in MCDB 104 medium, and incubated 6 h with 10 ng/mL of human IGF-I (IGF-I), or 6 h with 5 μM MG115 (MG115) or 1 h and 6 h with 1 μM epoxomicin (Epox). For IGF-I/inhibitor experiments, cells were pretreated 30 min with proteasome inhibitors and then incubated with IGF-I for 6 h (MG115 treatments, I/M) or 1h (E/I-1), 6h (E/I-6) (epoxomicin, E/I treatments). Left panel, early-passage WI38 human fibroblasts and U118MG human glioblastoma cell lines. Center-left panel, late-passage WI38 human fibroblasts. Center-right panel, early-passage WI38 treated with epoxomicin. Right panel shows the effect of MCDB 104 medium at 0, 24, 48, 72 hours. Bottom gels show Ponceau S staining. Graphs show carbonyl levels relative to respective controls, (average quantitation of 3-4 immunoblots, except for MCDB 104 experiment where the graph represent the quantization of 2 blots). * p < 0.05 compared to controls; ** p < 0.05 compared to MG115 treatments.
3. 2. Mechanisms involved in the modulation of the clearance of oxidized proteins by IGF-I
In order to determine whether IGF-I influences proteasome activity, we evaluated the activity of proteasome both in vivo and in vitro. Figure 4A,B shows that, compared with wild-type animals, the frontal cortex in heterozygous and homozygous cm-Igf1 mice has a higher level of chymotrypsin-like proteasome activity. Relative to wild type the chymotrypsin-like activity associated with 20S proteasome shows an increase of 126 % (p < 0.05) and 114% (p < 0.05) in heterozygotes and homozygotes, respectively (Fig. 4A). A lower but statistically significant increase of 40% and 25% was observed for the ATP-dependent 26S chymotrypsin-like activity respectively (Fig. 4B). We also measured proteasome activity in the midi mice, which have reduced IGF-I levels in the brain. Evaluation of proteasome function in the frontal cortex of the midi mice shows a significantly lower chymotrypsin-like proteasome activity associated with 20S, and 26S proteasomes (30 % reduction, p < 0.05) (Fig. 4C,D). These changes in proteasome activity correspond with variations in IGF-I levels determined by ELISA in the brains of the respective animals.
Fig. 4.
Effect of IGF-I on proteasome activity in vivo. Proteasome activity was assayed using 10 μg of protein extracts from frontal cortex in the presence of the specific fluorogenic peptide substrates, Suc-LLVY-AMC (chymotrypsin-like, CT-like activity); Boc-LRR-AMC (trypsin-like, T-like activity), and Z-LLE-AMC (caspase-like activity). The reaction was monitored by measuring 7-amino-4-methylcoumarin released (ex. 350 nm, em. 460 nm). Top graphs, proteasome activity in frontal cortex from cm-IgfI transgenic mice, wild-type: FVB+/+, Heterozygous: FVB+/cm-Igf1 and Homozygous: FVBcm-Igf1/cm-Igf1; (A) 20S proteasome, SDS-associated, 0.02% SDS, (B) 26S proteasome ATP-associated, 2 mM ATP. Bottom graphs, show chymotrypsin-like proteasome activity in frontal cortex from hypomorphic midi mouse, of wild-type C57 and midi mice; (C) 20S proteasome, SDS-associated, 0.02% SDS, (D) 26S proteasome ATP-associated, 2 mM ATP. AFU (arbitrary fluorescence unit) values correspond to the remaining activity after subtraction of activity determined in the presence of 20 μM epoxomicin (n = 5). * p < 0.05 compared to wild type animals.
3. 3. Mechanisms of the modulation of proteasome by IGF-I
To determine whether IGF-I could directly stimulate the proteasome, we analyzed the effect of IGF-I on proteasomal function in vitro. Stimulation with IGF-I alone for 24 h increases the chymotrypsin-like activity associated with the proteasome significantly (150% activation, p < 0.05) in C6 cells, a rat glial line (Fig. 5A). Kinetic studies in WI38 cells reveal an initial peak of activation of the proteasome after 15 min of incubation with IGF-I. This elevation in proteasome activity is maintained elevated for up to 24 h (Fig. 5B). We sought to determine whether IGF-I increased proteasome activity was associated with changes in the expression of specific proteasome subunits. As shown in figure 5C, no significant variations were observed in the abundance of β5i inducible, “core” α/β (α5/α7, β1, β5, β5i, β7), and α-structural subunits after 24 or 48 h stimulation of G0-arrested cultures with IGF-I. Although IGF-I seems to induce an slight increase in subunit β5, which is responsible for chymotrypsin-like activity, the short-term activation of proteasome activity (15 minutes) is not likely to be due to increased synthesis of proteasome subunits, but rather with a more rapid postranslational modification.
Fig. 5.
Effect of IGF-I on proteasome in vitro. C6 glioblastoma cells (A) and human WI38 fibroblasts (B) were synchronized in Go by 48 h of incubation in the serum-deficient medium MCDB 104 and then stimulated with 10 ng/mL IGF-I by the indicated times. Cell extracts were prepared and the chymotrypsin-like proteasome activity was assayed as described in Materials and methods. (C) Representative immunoblot showing the effect of IGF-I on steady state levels of proteasome subunits. WI38 cells where incubated for 24 h or 48 h in the absence (control) or in the presence of 10 ng/mL IGF-I. Twenty-five g of total proteins were analyzed by Western blot to evaluate proteasome subunit abundance. Core subunits α/β show a range from 22 to 27 kDa, representing total α-structural (α5, α7) and β-catalytic (β1, β5, β5i, β7) proteasome subunits. α subunits show two bands of 29 and 32 kDa representing α1, α2, α3, α5, α6, α7. A, B (n=3). C (n=2) * p < 0.05 compared to time 0.
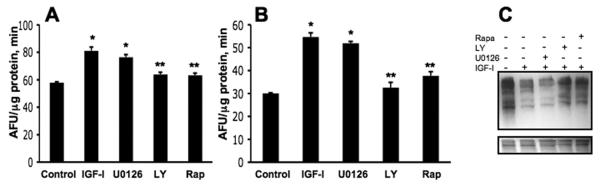
Interestingly, the effects of IGF-I on proteasome activation were not observed in knockout cells for the IGF-I receptor (Fig. 6), indicating there is an absolute requirement for the IGF-I receptor and suggesting that activation of proteasome by IGF-I involves cellular signaling transduction pathways from the IGF-I receptor. To further investigate the signaling pathway involved, WI38 human fibroblasts and a human glioblastoma cell line were preincubated in the presence of kinase inhibitors and then incubated with IGF-I. The results shown in Fig. 7 A, B indicate that the presence of LY29402 (PI3-kinase inhibitor) and rapamycin (mTOR inhibitor) both significantly reduced proteasome activation by IGF-I (p < 0.05). Interestingly inhibition of PI3-kinase and mTOR by these inhibitors block the effect of IGF-I in reducing protein carbonyls, supporting a role for PI3-kinase and mTOR in the regulation proteasome function (Fig. 7C). To further examine the role of PI3-kinase in proteasome function, we determined proteasome activity in U87MG, a phosphatidylinositol phosphate 3′-phosphatase (PTEN)-deficient cell line compared with LN229 with normal PTEN levels. As showed in Fig. 8, the PTEN-deficient U87MG cells show elevated proteasome activity relative to the PTEN-competent LN229 cells. Taken together, these results suggest that activation of proteasome by IGF-I involves initial activation of IGF-I receptor and the Akt/PI3-kinase/mTOR cascade signaling.
Fig. 6.

Effect of IGF-I on proteasome in IGF-I receptor–deficient cells. IGFI-R- cells were incubated 48 h in MEM containing 0.5% FBS (gray bars) or in the serum-deficient medium MCDB 104 (white bars). Then 10 ng/mL IGF-I was added to the cultures by the indicated times. Cells were washed with PBS and lysed and the chymotrypsin-like proteasome activity was assayed in 10 μg of protein extracts as indicated. Values represent proteasome activities relative to time 0. (n=3).
Fig. 7.
Effect of kinase inhibitors on proteasome activity and protein carbonyl content. Human WI38 fibroblasts (A) and human U118 glioblastoma (B) cell lines were incubated for 48 h in serum-free MCDB 104 medium. Cultures were incubated 30 min in the presence of 10 ng/mL IGF-I (IGF-I) or preincubated for 30 min in the presence 10 μM U0126 (MEK inhibitor), or 50 μM LY294002 (LY, PI3-kinase inhibitor) or 100 nM rapamycin (Rap, mTOR inhibitor) and then activated with IGF-I. Cells were washed with PBS, lysed and CT-like proteasome activity was determined as indicated in Material and methods. * p < 0.05 compared to controls; ** p < 0.05 compared to IGF-I treatments. (n=3). (C) Representative immunoblot showing the effects of inhibitors on protein carbonyl. WI38 cells were incubated 48 h in the MEM medium containing 0.5% FBS and then incubated 24 h in the presence of the inhibitors as indicated above, and protein carbonyl analyzed as indicated in Material and methods.
Fig. 8.
Regulation of proteasome activity by PTEN. PTEN-expressing LN229 and PTEN-deficient U87MG cell lines were incubated for 48 h in serum-free MCDB 104 medium. Cells were washed with PBS and lysed and the CT-like proteasome activity was assayed in 10 μg of protein extracts. Gel shows an illustrative Western blot of PTEN protein levels (n=3). * p < 0.05 compared to U87MG cells.
Finally, we examined whether the effects of IGF-I on protein carbonyl content were associated with modulation of ROS production and protein synthesis. Elevated levels of ROS or stimulation of protein synthesis can lead to higher levels of oxidized proteins. In order to determine whether IGF-I influences ROS production, we examined ROS levels following stimulation of cultured cells with IGF-I. As shown in Fig. 9A, IGF-I treatment does not induce significant changes in ROS status compared with control cultures. Similarly, as shown in Fig. 9B, incubation with IGF-I up to a 24-h period does not affect total protein synthesis of G0-arrested human WI38 fibroblasts, indicating that the effects of IGF-I on decreasing the level of oxidized proteins seem not to be associated with variations in the rate of protein synthesis.
Fig. 9.
Effect of IGF-I on ROS production and protein synthesis. (A) ROS production. ROS production was determined in young (PD 30) WI38 quiescent fibroblasts in the absence (control) or after 6 h incubation with 10 ng/μL IGF-I (IGF-I). Cells were incubated 30 min with DCFH-DA and the fluorescence at 530 nm associated with ROS was determined in total cell extracts as indicated in Materials and methods. H2O2 control cells incubated 2 h in the presence of 150 μM H2O2 (n = 3). (B) Protein Synthesis. Young WI38 quiescent fibroblasts were cultured in the absence (control) or presence of IGF-I (20 ng/ul) for 1, 6, and 24 h. Protein Synthesis (TCA/Total) was determined by measuring the amount of 35S –met/cys incorporated into TCA precipitable counts (TCA) normalized to the amount of label found in the total cellular extract (Total) (n = 2). * p < 0.05 compared to controls.
4. Discussion
Numerous studies indicate that ageing (Fujii and Taniguchi, 1999; Lavie et al., 1982; Prasanna and Lane, 1979; Reznick and Gershon, 1979; Stadtman, 1988) and age-associated neurodegenerative diseases such as Alzheimer disease (Oddo, 2008) and Parkinson disease (Lim and Tan, 2007) are accompanied by an accumulation of damaged, misfolded and biologically inactive proteins as a product of oxidative damage. While the proteasome system is known to play a fundamental role in cellular homeostasis by controlling the degradation of normal short-lived proteins, there is also substantial evidence supporting its vital role in the clearance of damaged, oxidized and unfolded proteins (Jung et al., 2009; Poppek and Grune, 2006; Ding et al., 2006; Farout and Friguet, 2006; Squier, 2006). Although the exact mechanism is still unclear, the 20S proteasome has been proposed to play an important role in removal of intracellular oxidized proteins (Jung and Grune, 2008; Grune et al., 2003; Hochstrasser, 1995). However other studies indicate there is a requirement for ATP and ubiquitin in the degradation of oxidized proteins (Goldberg and Boches, 1982; Dudek et al., 2005). More recent work in S. cerevisiae supports a requirement for the ubiquitin proteasome pathway in the degradation of proteins damaged as result of heat shock and oxidation (Medicherla and Goldberg, 2008).
Several lines of evidence suggest that accumulation of oxidized proteins during aging and age-related diseases could be related to a functional impairment in proteasome, an event that is emerging as a common phenomenon in several models of in vitro and in vivo aging and age-related diseases (Bulteau et al., 2000; Ponnappan, 2002; Bulteau et al., 2002; Husom et al., 2004; Louie et al., 2002; Keller et al., 2000; Petropoulos et al., 2000; Sitte et al., 2000; Chondrogianni et al., 2003; Torres et al., 2003;Torres and Perez, 2008). Because the proteasome is involved in a broad range of cellular processes, a functional impairment in this system may have severe consequences not only on protein oxidative status but also on homeostasis and survival during aging. For example, an impairment in proteasome function has been suggested to play a role in neurodegenerative disorders such as Alzheimer’s disease (Oddo, 2008), Parkinson’s disease (Lim, 2007), and Huntington’s disease (Davies et al., 2007; Wang et al., 2008). A role for the proteasome in neurodegenerative disorders is supported by a recent report indicating that the depletion of the proteasome in mouse brain neurons causes neurodegeneration and Lewy-like inclusions resembling human pale bodies (Bedford et al., 2008), This suggests that proteasome dysfunction observed in aged brain could contribute to the accumulation of protein aggregates during neurodegeneration (Oddo, 2008).
The results presented in this study suggest that IGF-I levels may influence the level of oxidized proteins in the frontal cortex of 2 transgenic mouse lines. This reduction in protein carbonyl content agrees with the decreased level of oxidized proteins previously reported in the heart of one of these mouse lines (Li et al., 2007) and with previous work showing that IGF-I treatment reduces accumulation of oxidized proteins in rats (Lo et al., 1997) and in humans (Hussain et al., 1993). Furthermore, in several cell lines the level of oxidized proteins decrease upon exposure to exogenous IGF-I. Our results suggest that the diminished levels of oxidized proteins observed in response to IGF-I could be mediated through activation of proteasome because: 1) proteasome inhibition lead to accumulation of oxidized proteins; 2) the presence of IGF-I significantly attenuates the levels of oxidized proteins induced by inhibition of the proteasome; 3) IGF-I induces a potent activation of the proteasome in vitro; and 4) lack of the IGF-I receptor completely prevents proteasome activation by IGF-I.
Although we have observed an slight increase in β5 subunit where resides chymotrypsin like activity, modulation of proteasome activity by IGF-I seems to be associated with posttranslational events as we observed significant variations proteasomal function in short-term period. In fact, the rapid induction of proteasome activity by IGF-I suggests the activation of signaling cascades. Experiments using specific kinase inhibitors and cell lines with differing PTEN status suggest that activation of proteasome involves phosphorylation events requiring PI3 kinase and mTOR signaling. Although previously proteasome phosphorylation has been described in response to γ-interferon (Bose et al., 2004) our initial experiments did not reveal any changes in phosphorylation status of the proteasome itself (not shown). Additional studies will be needed to determine whether phosphorylation of specific subunits are involved in the activation of the proteasome by IGF-I.
In summary, our results indicate that IGF-I can increase proteasome activity through the mTOR pathway in vitro and that IGF-I levels in the brain correlate with proteasome activity. This suggests that the neuroprotective effect of IGF-I may involve the clearance of deleterious substances in the adult brain and favor the idea that maintaining IGF-I levels in brain may be beneficial during aging (Fernandez et al., 2007). In fact, the cm-Igf mice live longer than genetically matched controls (Li and Ren, 2007). This is difficult to reconcile with the fact that the low IGF-I midi mice also live longer than genetically matched controls (Lerner et al., 2009), however, it is the balance of positive and negative influences that ultimately dictate lifespan. The cm-Igf mice seem to derive a benefit from increased levels of IGF-I in the heart (Li et al., 2007) and the brain (results presented in this manuscript) but the Igf1 hypomorphic midi mice derive benefit in other tissues in the form of enhanced autophagy (Lerner et al., 2009). Thus the tissue specific effects of altered IGF-I signaling must be considered when comparing different models. It is the balance of IGF-I levels (along with other related factors such as growth hormone) that may be critical to optimal lifespan and this balance may explain why animals with low levels of IGF-I, such as the midi mice, may show a lifespan extension at the same time that other animals, such as the cm-Igf mice, show a lifespan extension due to elevated levels of IGF-I.
Acknowledgements
This study was supported by USPHS grants AG022443 and AG022443-S1 from the National Institutes of Health. We are grateful to Dr. Peiro Anversa for providing the Cardiac IGF-1 producing mice.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baker J, Liu JP, Robertson EJ, Efstratiadis A. Role of insulin-like growth factors in embryonic and postnatal growth. Cell. 1993;75:73–82. [PubMed] [Google Scholar]
- Beck KD, Powell-Braxton L, Widmer HR, Valverde J, Hefti F. Igf1 gene disruption results in reduced brain size, CNS hypomyelination, and loss of hippocampal granule and striatal parvalbumin-containing neurons. Neuron. 1995;14:717–730. doi: 10.1016/0896-6273(95)90216-3. [DOI] [PubMed] [Google Scholar]
- Bedford L, Hay D, Devoy A, Paine S, Powe DG, Seth R, Gray T, Topham I, Fone K, Rezvani N, Mee M, Soane T, Layfield R, Sheppard PW, Ebendal T, Usoskin D, Lowe J, Mayer RJ. Depletion of 26S proteasomes in mouse brain neurons causes neurodegeneration and Lewy-like inclusions resembling human pale bodies. J. Neurosci. 2008;28:8189–8198. doi: 10.1523/JNEUROSCI.2218-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose S, Stratford FLL, Broadfoot KI, Mason GF, Rivett J. Phosphorylation of 20S proteasome alpha subunit C8 (α7) stabilizes the 26S proteasome and plays a role in the regulation of proteasome complexes by © interferon. Biochem. J. 2004;378:177–184. doi: 10.1042/BJ20031122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulteau AL, Petropoulos I, Friguet B. Age-related alterations of proteasome structure and function in aging epidermis. Exp. Gerontol. 2000;35:767–777. doi: 10.1016/s0531-5565(00)00136-4. [DOI] [PubMed] [Google Scholar]
- Bulteau AL, Szweda LI, Friguet B. Age-dependent declines in proteasome activity in the heart. Arch. Biochem. Biophys. 2002;397:298–304. doi: 10.1006/abbi.2001.2663. [DOI] [PubMed] [Google Scholar]
- Carro E, Trejo JL, Gomez-Isla T, LeRoith D, Torres-Aleman I. Serum insulin-like growth factor I regulates brain amyloid-beta levels. Nat. Med. 2002;8:1390–1397. doi: 10.1038/nm1202-793. [DOI] [PubMed] [Google Scholar]
- Chondrogianni N, Stratford FL, Trougakos IP, Friguet B, Rivett AJ, Gonos ES. Central role of the proteasome in senescence and survival of human fibroblasts: induction of a senescence-like phenotype upon its inhibition and resistance to stress upon its activation. J. Biol. Chem. 2003;278:28026–28037. doi: 10.1074/jbc.M301048200. [DOI] [PubMed] [Google Scholar]
- Coppe JP, Patil CK, Rodier F, Sun Y, Munoz DP, Goldstein J, Nelson PS, Desprez PY, Campisi J. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6:2853–2868. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- Davies JE, Sarkar S, Rubinsztein DC. The ubiquitin proteasome system in Huntington’s disease and the spinocerebellar ataxias. BMC Biochem. 2007;8(Suppl 1):S2. doi: 10.1186/1471-2091-8-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ercole AJ, Ye P. Expanding the mind: insulin-like growth factor I and brain development. Endocrinology. 2008;149:5958–5962. doi: 10.1210/en.2008-0920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Dimayuga E, Keller JN. Proteasome regulation of oxidative stress in aging and age-related diseases of the CNS. Antioxid. Redox Signal. 2006;8:163–172. doi: 10.1089/ars.2006.8.163. [DOI] [PubMed] [Google Scholar]
- Dudek EJ, Shang F, Valverde P, Liu Q, Hobbs M, Taylor A. Selectivity of the ubiquitin pathway for oxidatively modified proteins: relevance to protein precipitation diseases. FASEB J. 2005;19:1707–1709. doi: 10.1096/fj.05-4049fje. [DOI] [PubMed] [Google Scholar]
- Efstratiadis A. Genetics of mouse growth. Int. J. Dev. Biol. 1998;42:955–976. [PubMed] [Google Scholar]
- Farout L, Friguet B. Proteasome function in aging and oxidative stress: implications in protein maintenance failure. Antioxid. Redox Signal. 2006;8:205–216. doi: 10.1089/ars.2006.8.205. [DOI] [PubMed] [Google Scholar]
- Fernandez S, Fernandez AM, Lopez-Lopez C, Torres-Aleman I. Emerging roles of insulin-like growth factor-I in the adult brain. Growth Horm. IGF Res. 2007;17:89–95. doi: 10.1016/j.ghir.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Fujii J, Taniguchi N. Down regulation of superoxide dismutases and glutathione peroxidase by reactive oxygen and nitrogen species. Free Radic. Res. 1999;31:301–308. doi: 10.1080/10715769900300861. [DOI] [PubMed] [Google Scholar]
- Goldberg AL, St John AC. Intracellular protein degradation in mammalian and bacterial cells. Annu Rev Biochem. 1976;45:747–803. doi: 10.1146/annurev.bi.45.070176.003531. [DOI] [PubMed] [Google Scholar]
- Goldberg AL, Boches FS. Oxidized proteins in erythrocytes are rapidly degraded by the adenosine triphosphate-dependent proteolytic system. Science. 1982;215:1107–1109. doi: 10.1126/science.7038874. [DOI] [PubMed] [Google Scholar]
- Grune T, Reinheckel T, Davies KJ. Degradation of oxidized proteins in mammalian cells. FASEB J. 1997;11:526–534. [PubMed] [Google Scholar]
- Grune T, Merker K, Sandig G, Davies KJ. Selective degradation of oxidatively modified protein substrates by the proteasome. Biochem. Biophycs. Res. Commun. 2003;305:709–718. doi: 10.1016/s0006-291x(03)00809-x. [DOI] [PubMed] [Google Scholar]
- Guan J, Bennet L, George S, Wu D, Waldvogel HJ, Gluckman PD, Faull RL, Crosier PS, Gunn AJ. Insulin-like growth factor-1 reduces postischemic white matter injury in fetal sheep. J. Cereb. Blood Flow Metab. 2001;21:493–502. doi: 10.1097/00004647-200105000-00003. [DOI] [PubMed] [Google Scholar]
- Herbig U, Ferreira M, Condel L, Carey D, Sedivy JM. Cellular senescence in aging primates. Science. 2006;311:1257. doi: 10.1126/science.1122446. [DOI] [PubMed] [Google Scholar]
- Hershko M, Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr. Opin. Cell Biol. 1995;7:215–223. doi: 10.1016/0955-0674(95)80031-x. [DOI] [PubMed] [Google Scholar]
- Husom AD, Peters EA, Kolling EA, Fugere NA, Thompson LV, Ferrington DA. Altered proteasome function and subunit composition in aged muscle. Arch. Biochem. Biophys. 2004;421:67–76. doi: 10.1016/j.abb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Hussain MA, Schmitz O, Mengel A, Keller A, Christiansen JS, Zapf J, Froesch ER. Insulin-like growth factor I stimulates lipid oxidation, reduces protein oxidation, and enhances insulin sensitivity in humans. J. Clin. Invest. 1993;92:2249–2256. doi: 10.1172/JCI116828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung T, Grune T. The proteasome and its role in the degradation of oxidized proteins. IUBMB Life. 2008;60:743–752. doi: 10.1002/iub.114. [DOI] [PubMed] [Google Scholar]
- Jung T, Catalgol B, Grune T. The proteasomal system. Mol Aspects Med. 2009;30:191–296. doi: 10.1016/j.mam.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Keller JN, Hanni KB, Markesbery WR. Possible involvement of proteasome inhibition in aging: implications for oxidative stress. Mech. Ageing Dev. 2000;113:61–70. doi: 10.1016/s0047-6374(99)00101-3. [DOI] [PubMed] [Google Scholar]
- Köhler A, Cascio P, Leggett DS, Woo KM, Goldberg AL, Finley D. The axial channel of the proteasome core particle is gated by the Rpt2 ATPase and controls both substrate entry and product release. Mol. Cell. 2001;7:1143–1152. doi: 10.1016/s1097-2765(01)00274-x. [DOI] [PubMed] [Google Scholar]
- Lavie L, Reznick AZ, Gershon D. Decreased protein and puromycinylpeptide degradation in livers of senescent mice. Biochem. J. 1982;202:47–51. doi: 10.1042/bj2020047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lembo G, Rockman HA, Hunter JJ, Steinmetz H, Koch WJ, Ma L, Prinz MP, Ross J, Jr., Chien KR, Powell-Braxton L. Elevated blood pressure and enhanced myocardial contractility in mice with severe IGF-1 deficiency. J. Clin. Invest. 1996;98:2648–2655. doi: 10.1172/JCI119086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner C, Torres C, Lorenzini A, Malaguti M, Roel M, Hrelia S, Ikeno Y, McCarter R, Sell C. Increased lfe span and enhanced autophagy in mice with reduced levels of IGF-I. (in preparation)
- Li Q, Ren J. Influence of cardiac-specific overexpression of insulin-like growth factor 1 on lifespan and aging-associated changes in cardiac intracellular Ca2+ homeostasis, protein damage and apoptotic protein expression. Aging Cell. 2007;6:799–806. doi: 10.1111/j.1474-9726.2007.00343.x. [DOI] [PubMed] [Google Scholar]
- Li Q, Wu S, Li SY, Lopez FL, Du M, Kajstura J, Anversa P, Ren J. Cardiac-specific overexpression of insulin-like growth factor 1 attenuates aging-associated cardiac diastolic contractile dysfunction and protein damage. Am. J. Physiol. Heart Circ. Physiol. 2007;292:H1398–1403. doi: 10.1152/ajpheart.01036.2006. [DOI] [PubMed] [Google Scholar]
- Lim KL. Ubiquitin-proteasome system dysfunction in Parkinson’s disease: current evidence and controversies. Expert Rev. Proteomics. 2007;4:769–781. doi: 10.1586/14789450.4.6.769. [DOI] [PubMed] [Google Scholar]
- Lim KL, Tan JM. Role of the ubiquitin proteasome system in Parkinson’s disease. BMC Biochem. 2007;8(Suppl 1):S13. doi: 10.1186/1471-2091-8-S1-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JP, Baker J, Perkins AS, Robertson EJ, Efstratiadis A. Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r) Cell. 1993;75:59–72. [PubMed] [Google Scholar]
- Lo HC, Hirvonen MD, Kritsch KR, Keesey RE, Ney DM. Growth hormone or insulin-like growth factor I increases fat oxidation and decreases protein oxidation without altering energy expenditure in parenterally fed rats. Am. J. Clin. Nutr. 1997;65:1384–1390. doi: 10.1093/ajcn/65.5.1384. [DOI] [PubMed] [Google Scholar]
- Louie JL, Kapphahn RJ, Ferrington DA. Proteasome function and protein oxidation in the aged retina. Exp. Eye Res. 2002;75:271–284. [PubMed] [Google Scholar]
- Luzi P, Zaka M, Rao HZ, Curtis M, Rafi MA, Wenger DA. Generation of transgenic mice expressing insulin-like growth factor-1 under the control of the myelin basic protein promoter: increased myelination and potential for studies on the effects of increased IGF-1 on experimentally and genetically induced demyelination. Neurochem. Res. 2004;29:881–889. doi: 10.1023/b:nere.0000021233.79076.72. [DOI] [PubMed] [Google Scholar]
- Markowska AL, Mooney M, Sonntag WE. Insulin-like growth factor-1 ameliorates age-related behavioral deficits. Neuroscience. 1998;87:559–569. doi: 10.1016/s0306-4522(98)00143-2. [DOI] [PubMed] [Google Scholar]
- Mason JL, Ye P, Suzuki K, D’Ercole AJ, Matsushima GK. Insulin-like growth factor-1 inhibits mature oligodendrocyte apoptosis during primary demyelination. J. Neurosci. 2000;20:5703–5708. doi: 10.1523/JNEUROSCI.20-15-05703.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medicherla B, Goldberg AL. Heat shock and oxygen radicals stimulate ubiquitin-dependent degradation mainly of newly synthesized proteins. J Cell Biol. 2008;182:663–673. doi: 10.1083/jcb.200803022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oddo S. The ubiquitin-proteasome system in Alzheimer’s disease. J. Cell Mol. Med. 2008;12:363–373. doi: 10.1111/j.1582-4934.2008.00276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petropoulos I, Conconi M, Wang X, Hoenel B, Bregegere F, Milner Y, Friguet B. Increase of oxidatively modified protein is associated with a decrease of proteasome activity and content in aging epidermal cells. J. Gerontol. A Biol. Sci. Med. Sci. 2000;55:B220–B227. doi: 10.1093/gerona/55.5.b220. [DOI] [PubMed] [Google Scholar]
- Ponnappan U. Ubiquitin-proteasome pathway is compromised in CD45RO+ and CD45RA+ T lymphocyte subsets during aging. Exp. Gerontol. 2002;37:359–367. doi: 10.1016/s0531-5565(01)00203-0. [DOI] [PubMed] [Google Scholar]
- Popken GJ, Hodge RD, Ye P, Zhang J, Ng W, O’Kusky JR, D’Ercole AJ. In vivo effects of insulin-like growth factor-I (IGF-I) on prenatal and early postnatal development of the central nervous system. Eur. J. Neurosci. 2004;19:2056–2068. doi: 10.1111/j.0953-816X.2004.03320.x. [DOI] [PubMed] [Google Scholar]
- Popken GJ, Dechert-Zeger M, Ye P, D’Ercole AJ. Brain development. Adv. Exp. Med. Biol. 2005;567:187–220. doi: 10.1007/0-387-26274-1_8. [DOI] [PubMed] [Google Scholar]
- Poppek D, Grune T. Proteasomal defense of oxidative protein modifications. Antioxid. Redox Signal. 2006;8:173–184. doi: 10.1089/ars.2006.8.173. [DOI] [PubMed] [Google Scholar]
- Prasanna HR, Lane RS. Protein degradation in aged nematodes (Turbatrix aceti) Biochem. Biophys. Res. Commun. 1979;86:552–559. doi: 10.1016/0006-291x(79)91749-2. [DOI] [PubMed] [Google Scholar]
- Ramsey MM, Weiner JL, Moore TP, Carter CS, Sonntag WE. Growth hormone treatment attenuates age-related changes in hippocampal short-term plasticity and spatial learning. Neuroscience. 2004;129:119–127. doi: 10.1016/j.neuroscience.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Reiss K, Cheng W, Ferber A, Kajstura J, Li P, Li B, Olivetti G, Homcy CJ, Baserga R, Anversa P. Overexpression of insulin-like growth factor-1 in the heart is coupled with myocyte proliferation in transgenic mice. Proc. Natl. Acad. Sci. U. S. A. 1996;93:8630–8635. doi: 10.1073/pnas.93.16.8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznick AZ, Gershon D. The effect of age on the protein degradation system in the nematode Turbatrix aceti. Mech. Ageing Dev. 1979;11:403–415. doi: 10.1016/0047-6374(79)90016-2. [DOI] [PubMed] [Google Scholar]
- Russo VC, Gluckman PD, Feldman EL, Werther GA. The insulin-like growth factor system and its pleiotropic functions in brain. Endocr. Rev. 2005;26:916–943. doi: 10.1210/er.2004-0024. [DOI] [PubMed] [Google Scholar]
- Sitte N, Merker K, von Zglinicki T, Grune T. Protein oxidation and degradation during proliferative senescence of human MRC-5 fibroblasts. Free Radic. Biol. Med. 2000;28:701–708. doi: 10.1016/s0891-5849(99)00279-8. [DOI] [PubMed] [Google Scholar]
- Squier TC. Redox modulation of cellular metabolism through targeted degradation of signaling proteins by the proteasome. Antioxid. Redox Signal. 2006;8:217–228. doi: 10.1089/ars.2006.8.217. [DOI] [PubMed] [Google Scholar]
- Stadtman ER. Protein modification in aging. J. Gerontol. 1988;43:B112–B120. doi: 10.1093/geronj/43.5.b112. [DOI] [PubMed] [Google Scholar]
- Torres C, Francis MK, Lorenzini A, Tresini M, Cristofalo VJ. Metabolic stabilization of MAP kinase phosphatase-2 in senescence of human fibroblasts. Exp. Cell Res. 2003;290:195–206. doi: 10.1016/s0014-4827(03)00309-4. [DOI] [PubMed] [Google Scholar]
- Torres C, Lewis L, Cristofalo VJ. Proteasome inhibitors shorten replicative life span and induce a senescent-like phenotype of human fibroblasts. J. Cell Physiol. 2006;207:845–853. doi: 10.1002/jcp.20630. [DOI] [PubMed] [Google Scholar]
- Torres CA, Perez VI. Proteasome modulates mitochondrial function during cellular senescence. Free Radic. Biol. Med. 2008;44:403–414. doi: 10.1016/j.freeradbiomed.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Joseph JA. Quantifying cellular oxidative stress by dichlorofluorescein assay using microplate reader. Free Radic. Biol. Med. 1999;27:612–616. doi: 10.1016/s0891-5849(99)00107-0. [DOI] [PubMed] [Google Scholar]
- Wang J, Wang CE, Orr A, Tydlacka S, Li SH, Li XJ. Impaired ubiquitin-proteasome system activity in the synapses of Huntington’s disease mice. J. Cell Biol. 2008;180:1177–1189. doi: 10.1083/jcb.200709080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye P, Li L, Richards RG, DiAugustine RP, D’Ercole AJ. Myelination is altered in insulin-like growth factor-I null mutant mice. J. Neurosci. 2002;22:6041–6051. doi: 10.1523/JNEUROSCI.22-14-06041.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye P, D’Ercole AJ. Insulin-like growth factor actions during development of neural stem cells and progenitors in the central nervous system. J. Neurosci. Res. 2006;83:1–6. doi: 10.1002/jnr.20688. [DOI] [PubMed] [Google Scholar]