Summary
Membrane rafts are ordered microdomains of the plasma membrane consisting of cholesterol, sphingolipids, and saturated fatty acids which appear to regulate many cellular signaling pathways. One such type of membrane raft is caveolae, which are cave-like invaginations of the plasma membrane. Interestingly, changes in the acyl composition of cellular membranes have been shown to alter the specific localization of membrane raft associated proteins and their function. This is noteworthy because modification of membrane acyl composition is readily accomplished through changes in dietary fat composition. Here we describe a common approach used to fractionate cell membranes to obtain an enriched preparation of caveolae and gas chromatographic techniques to determine fatty acyl composition. In addition, methods used to visualize and quantify lipid rafts using a fluorescent probe Laurdan in living cells will also be described.
Keywords: Caveolae, Fatty acids, Laurdan, Lipid rafts, Membrane rafts, Nutrition
1. Introduction
Alteration of cell membrane structure, specifically the acyl fatty acids of phospholipids, is readily achieved through simple changes in dietary fat composition. This alteration is very rapid and can be readily detected within hours in cell culture or days in humans. The influence and impact of dietary fat composition on membrane structure has gained increasing interest due to advancements in techniques and refinements in our understanding of membrane rafts (1). Membrane rafts include structures known as caveolae, which are cave-like structures enriched in cholesterol, sphingolipids, and phospholipids containing saturated fatty acids. These membrane rafts are now implicated in a diversity of biological functions which impact on human health and disease (2–4). Studies have shown that alterations in dietary fat composition to include omega-3 fatty acids from marine oils can impact on the structure and function of colonocyte, immune, and cancer cell membranes (2, 5–7). Collectively, this body of work demonstrates that simple nutritional interventions can have significant impact on cell signaling in part mediated through membrane rafts.
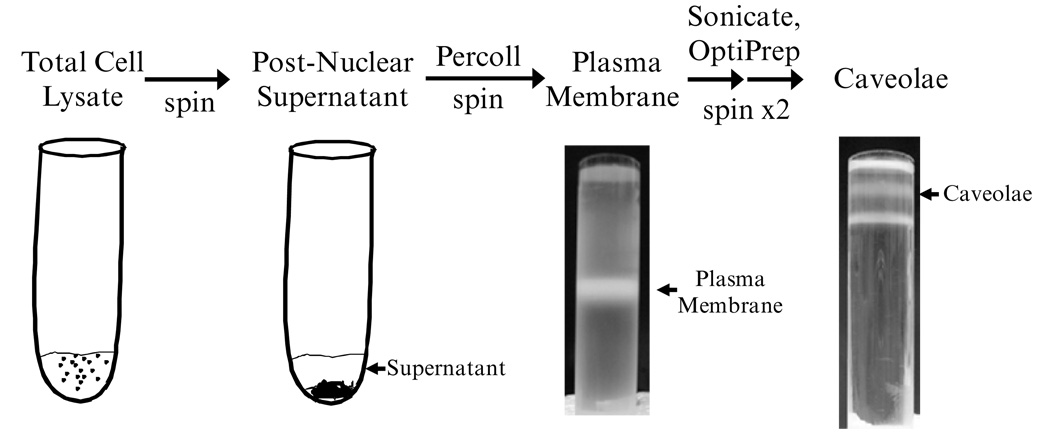
Concentrated caveolae can be isolated using a nondetergent approach following the methodology developed by Smart et al. (2, 8, 9). This protocol works well with isolated cells from cell culture and single cell populations from tissue. The procedure below describes the isolation of caveolae from cell culture. Depending upon the cell type and expression of caveolae, preliminary work is required to determine the optimum number of cells required to obtain sufficient caveolae. Start with 100 × 106 cells, which may be the equivalent of several T-175 flasks. An overview of the procedure and resultant fractions are shown in Fig. 1.
Fig. 1.
Shown in this figure are the bands obtained during the caveolae isolation procedure from cultured cells.
2. Materials
2.1. Equipment
Ultracentrifuge at 4°C.
Benchtop centrifuge.
Water bath at 37°C.
Gradient maker.
Stirrer plate.
2.2. Reagents
Optiprep (60% v/v solution of iodixanol).
Percoll.
Sucrose.
Na4EDTA (monohydrate, 380.2 MW).
Tricine.
3. Methods
3.1. Fatty Acid Treatment Protocol
Adherent cells are first grown for 2 days in basal medium to acclimate cells (see Note 1).
On day 3, media are replaced with media containing supplemental fatty acids.
Stock nonesterified fatty acids (Nu-Chek Prep, Elysian Falls, MN, USA) are conveniently stored in ethanol at 1–10 mg/mL.
Prepare media containing 0–100 µM fatty acids. Note: concentrations exceeding 200 µM are toxic. Stock fatty acids are transferred to a 15-mL sterile Falcon tube and ethanol is evaporated under a gentle stream of nitrogen.
Fatty acids are bound to bovine serum albumin (fraction V) (0.1%w/v) by incubating in fetal bovine serum and basal medium in a shaking water bath at 37°C for 30 min.
Media is replaced.
Cells are grown until ~90% confluency. Media should be replaced every 3 days.
3.2. Caveolae Isolation
3.2.1. Solutions to be Prepared in Advance
1× PBS. 500 mL warm (37°C), 500 mL cold (4°C)
| Buffer A: (stable for 1 week stored at 4°C) | Final Conc: |
|---|---|
| 20 mL 2.5 M Sucrose | 0.25 M |
| 0.076 g Na4EDTA (monohydrate, 380.2 MW) | 1 mM |
| 0.716 g tricine | 20 mM |
Combine with 170 mL water, pH to 7.8, bring to final volume of 200 mL
2XA Buffer: (stable for 1 week stored at 4°C)
| 13.32 mL 2.5 M Sucrose | 0.5 M |
| 0.05 g Na4EDTA | 2 mM |
| 0.48 g tricine | 40 mM |
Combine with 40 mL of water, pH to 7.8, bring to final volume of 66.6 mL
OptiPrep Diluent: (stable for 1 week stored at 4°C)
| 1 mL 2.5 M Sucrose | 0.25 M |
| 0.022 g Na4EDTA | 6 mM |
| 0.216 g tricine | 120 mM |
Combine with 7 mL water, pH to 7.8, bring to final volume of 10 mL
3.2.2. Solutions to be Prepared on Day of Experiment
-
30% Percoll:
30 mL Percoll
50 mL 2XA
20 mL water
-
50% OptiPrep:
36 mL OptiPrep
7.2 mL OptiPrep diluent
-
OptiPrep (60% v/v solution of iodixanol) Solutions:
5% – 0.4mL 50% OptiPrep + 3.6mL Buffer A
10% – 2.94mL 50% OptiPrep + 11.74mL Buffer A
15% – 1.5mL 50% OptiPrep + 3.5mL Buffer A
20% – 5.86mL 50 OptiPrep + 8.8mL Buffer A
-
1.
Harvest cells at 90% confluency then centrifuge at 100 × g for 3 min. After isolation, keep cells at 4°C or on ice.
-
2.
Resuspend cells in 50 mL of cold 1× PBS and centrifuge at 100 × g for 3 min.
-
3.
Discard supernatant and resuspend pellet in 30 mL of Buffer A and centrifuge at 100 × g for 3 min.
-
4.
Discard supernatant and suspend pellet in 2 mL of Buffer A with protease inhibitor cocktail (Sigma) (40 µL/mL) and phosphatase inhibitor (orthovanadate) (10 µL/mL).
-
5.
Homogenize 20 strokes on ice using Teflon /glass dounce homogenizer. Save aliquot (50 µL).
-
6.
Spin at 1,000 × g, 10 min, 4°C.
Meanwhile precool ultracentrifuge and make 30% Percoll.
-
7.
Collect supernatant (postnuclear) and keep on ice.
-
8.
Resuspend pellet in 1.5 mL of Buffer A, therefore final volume is 3.5 mL. Combine supernatants, save an aliquot (50 µL).
-
9.
The combined PNS (3.5 mL) was overlaid on 9 mL of 34.86% (v/v) Percoll solution in 13.2 mL Ultra-Clear Beckman Centrifuge Tubes. The 34.86% Percoll solution is made by diluting Percoll with 46.53% (v/v) 2XA Buffer.
-
10.
Centrifuge in Beckman SW41 Ti rotor at 84,000 × g for 30 min.
Meanwhile make remaining OptiPrep Solutions
-
11.
Collect 2 mL (up to 3 mL) of plasma membrane fraction (visible band 6 cm from bottom of tube). Place in 15 mL Beckman tubes.
-
12.
The PM fraction is then sonicated on ice, three times for 2 min each, with 2 min intervals between each sonication (see Note 2)
-
13.
Pour Opti gradient by adding 2.3 mL of 50% OptiPrep to the samples and adjusting the final volume to 5.0 mL using Buffer A as a diluent.
-
14.
A linear 20–10% OptiPrep gradient (3 mL each) is overlaid on the new 23% OptiPrep solution using a gradient maker connected to a peristaltic pump set at a flow rate of 1 mL/min
-
15.
Centrifuge at 52,000 × g (17,000 rpm) 90 min, 4°C in SW41 Ti rotar
-
16.
Collect the top 5 mL into new tube, then prepare ~23% OptiPrep solution by adding 4 mL of 50% OptiPrep to sample and mix.
-
17.
Overlay with a discontinuous gradient with 1 mL of 15% OptiPrep followed by 0.5 mL of 5% OptiPrep
-
18.
Centrifuge at 52,000 × g (17,000 rpm) 90 min, 4°C.
-
19.
Collect top white band (500–700 µL) within 5% OptiPrep layer, just above interface.
3.3. Fast GC Analysis
100 µL aliquots of caveolae are transferred to a 15 mL glass screw cap tube with teflon cap.
Lipids are extracted in chloroform:methanol (2:1), 4 mL and 1 mL of KCl (0.88%w/v), vortex.
Centrifuge at 200 × g to separate phases.
Carefully transfer bottom chloroform fraction to a clean glass tube, with Teflon cap.
Add 2 mL of chloroform to top phase and repeat extraction.
Combine chloroform extracts and dry down under a gentle stream of nitrogen.
Saponify lipids by adding 2 mL of KOH–MeOH, then incubate at 100°C (water bath or oven) for 1 h.
Cool for 5 min in fume hood, then add 2 mL of 14% BF3–MeOH and 2 mL of hexane, then incubate at 100°C (water bath or oven) for 1 h.
Cool for 5 min in fume hood and then add 2 mL of double distilled water.
Centrifuge at 200 × g to separate phases.
Transfer upper hexane phase to clean 2 mL gas chromatography vial.
Dry down hexane under gentle stream of nitrogen.
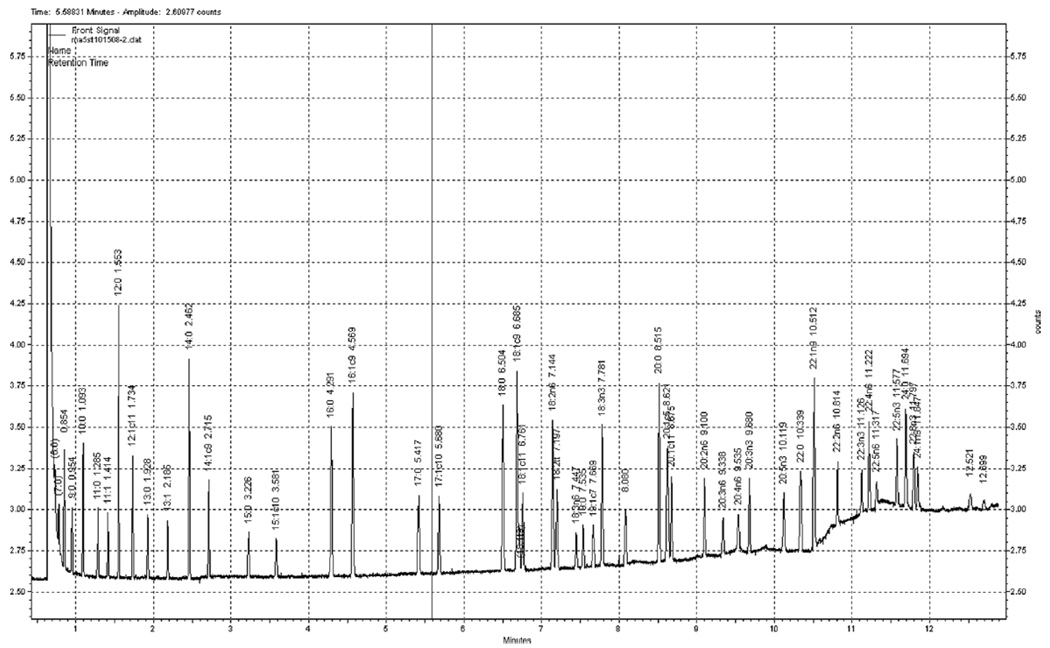
FAME are quantified on an Agilent 7890 flame-ionization detector gas chromatograph (Palo Alto, CA, USA) separated on a DB-FFAP fused silica column, 15 m × 0.10mmID × 0.10 µm (Agilent). Samples were injected in split mode, 200:1. The injector and detector ports should be set at 250°C. FAME are eluted using a temperature program set initially at 150°C for 0.25 min, increased at 35°C/min and held at 170°C for 3 min, increased at 9°C/min and held at 225°C for 0.5 min, increased at 80°C and held at 245°C for 2.2 min. Total run time is 12.9 min. The carrier gas, hydrogen, is set to a 0.5 mL/min constant flow rate. The makeup gas is nitrogen. Fatty acids are identified using authentic standards obtained from Nu-Chek Prep (#GLC-463) (see Note 3) (Fig. 2)
Fig. 2.
Shown in this figure is a representative fast gas chromatographic separation of fatty acid methyl esters.
3.4. Protocol for Visualization of Lipid Rafts Using the Fluorescence Probe Laurdan
3.4.1. Poly-l-lysine Precoating on Coverglass
Dilute 0.1% (w/v) poly-l-lysine solution (Sigma) to 0.01% with sterile water.
Fill the chambered coverglass (Nunc) wells with 0.01% polyl-l-lysine solution (1 mL).
Allow solution to sit for 30 min.
Aspirate excess solution and allow the coverglass wells to air dry.
Sterilize the coverglass slides under UV light for 1 h.
3.4.2. Preparation of 10 mM Laurdan (Invitrogen, MW 353.55) in Ethanol
Measure 10 mg of Laurdan in a glass tube covered with aluminum foil (Laurdan is light sensitive).
Dissolve in 2.83 mL of ethanol to make 10 mM of Laurdan stock solution.
Vortex vigorously (~2 min) to dissolve Laurdan.
Keep at 4°C and prewarm at RT prior to use.
3.4.3. Cell Labeling of Laurdan
Cell concentration might differ by type of cells.
Start with 1 × 106 cells/mL, which has been shown to be optimal for CD4+ T cells (10).
Transfer 2 mL of cell suspension into a 15 mL-conical tube.
Add 2 µL of (10 mM) Laurdan solution into 2 mL of cell suspension to dilute to 10 nM of Laurdan (see Note 4).
Vortex for 2 s.
Incubate cells at 37°C for 10 min.
Centrifuge cell suspension at 300× gravity for 5 min.
Resuspend cell pellets in 2 mL of Leibovitz’s medium.
Transfer 1 mL of cell suspension into a poly-l-lysine-precoated chambered glass.
Incubate cells in an appropriate condition, if necessary.
3.4.4. GP-Value Calculation
-
1.
Capture Laurdan-labeled cell images using 2-photon microscopy. (Zeiss LSM 510 META NLO, 40× objective 1.3 NA oil. The coherent Chameleon femtosecond pulsed Ti:Sapphire laser should be set at an excitation wavelength of 770 nm)
-
2.
Save the image files as 8-bit TIFF format.
-
3.
Open the image file in an image analysis software package, such as Adobe Photoshop® CS (v.8.0).
(The following protocol is using Adobe Photoshop®)
-
4.
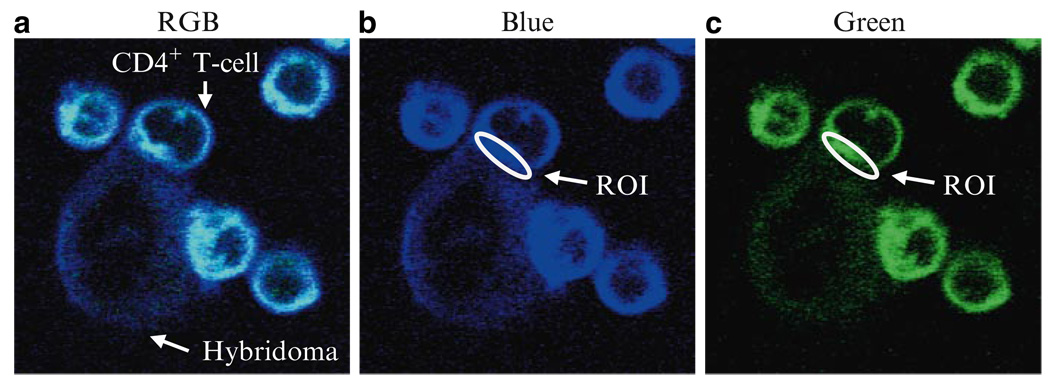
Using a marquee tool, make an oval to select a region of interest (Fig. 3b, c)
-
5.
In “Histogram” panel, choose each channel of RGB.
-
6.
Once blue channel is selected, the panel displays “mean intensity” of marquee-selected region.
-
7.
A histogram will also show the pixels chosen by marquee tool.
-
8.
If you change the channel to green, mean intensity is changed, while pixel is not.
-
9.Record green and blue intensities to calculate GP-values by formula
Fig. 3.
Representative (a) RGB images of Laurdan-labeled CD4+ T cells. Purified CD4+ T cells were labeled with 10 µM Laurdan and stimulated by anti-CD3 mAb expressing hybridoma cells at 37°C for 30 min. GP-values were calculated from values of fluorescence intensity identified using Adobe Photoshop® to draw regions of interest (ROI) in each (b) blue or (c) green channel at the immunological synapse.
3.5. Results
3.5.1. Visualization of Lipid Rafts
There have been several methodologies developed to identify and visualize lipid rafts. For instance, flotation of DRM fractions and immunofluorescence microscopy by antibody patching have been used to identify putative raft association and raft proteins (11, 12). In addition, immunoelectron microscopy and single fluorophore tracking microscopy have been utilized to track the location of raft components (13). Fluorescence resonance energy transfer (FRET) has also been applied to determine whether two raft components are spatially impacted (14). However, due to the specificity of probes used, the visualization of heterogeneous lipid rafts have been difficult to determine.
Recently, Gaus et al. demonstrated that the fluorescent probe Laurdan can align itself parallel with the hydrophobic tails of phospholipids in membranes (15–17). Owing to its ability to emit two different wavelengths according to the fluidity of the microenvironment, Laurdan can be used to measure and visualize membrane fluidity in living cells by two-photon microscopy (16). Generalized polarized values (GP-values) derived by the formula GP = (I400 – 460 − I470 – 530)/(I400 – 460 + I470 – 530)), where I stands for intensities of the blue and green channels, respectively, indicates the fluidity of cell membranes. Given that lipid rafts are highly enriched in cholesterol and sphingolipids, the liquid ordered state of the membrane detected by Laurdan labeling is assumed to be heterogeneous lipid rafts. As an example, representative images of Laurdan-labeled CD4+ T cells co-cultured with anti-CD3 mAb expressing hybridoma cells are presented in Fig. 3a.
Acknowledgments
Supported in part by NIH grants CA59034, CA129444, DK071707, and P30ES09106 to R.S. Chapkin and a Natural Sciences and Engineering Research Council of Canada Discovery Grant to D.W.L. Ma.
Footnotes
Isolation of caveolae is best accomplished using a single cell type from cell culture. In addition, this method is also suitable for purified cells isolated from tissue. Caveolae are found in most cell types, however their function is also cell specific. Therefore, if whole tissue is used, it must be recognized that results represent an aggregate from different cell types.
The sonication step may introduce variability in the procedure dependent on the type of sonicator, power, and duration of cell disruption. Furthermore, interindividual variability contributes to differences in overall yield of caveolae.
Although a fast-GC analysis approach is described, any fatty acid methyl ester method will be suitable.
Laurdan concentration and duration of labeling should be optimized for each cell type used.
References
- 1.Pike LJ. Rafts defined: a report on the Keystone symposium on lipid rafts and cell function. J. Lipid Res. 2006;47:1597–1598. doi: 10.1194/jlr.E600002-JLR200. [DOI] [PubMed] [Google Scholar]
- 2.Ma DW, Seo J, Davidson LA, Callaway ES, Fan YY, Lupton JR, Chapkin RS. n-3 PUFA alter caveolae lipid composition and resident protein localization in mouse colon. FASEB J. 2004;18:1040–1042. doi: 10.1096/fj.03-1430fje. [DOI] [PubMed] [Google Scholar]
- 3.Ma DW. Lipid mediators in membrane rafts are important determinants of human health and disease. Appl. Physiol Nutr. Metab. 2007;32:341–350. doi: 10.1139/H07-036. [DOI] [PubMed] [Google Scholar]
- 4.Ma DWL, Seo J, Switzer KC, Fan YY, McMurray DN, Lupton JR, Chapkin RS. n-3 PUFA and Membrane Micro-domains: A New Frontier in Bioactive Lipid Research. J. Nutr. Biochem. 2004;15:700–706. doi: 10.1016/j.jnutbio.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 5.Fan YY, McMurray DN, Ly LH, Chapkin RS. Dietary (n-3) poly-unsaturated fatty acids remodel mouse T-cell lipid rafts. J. Nutr. 2003;133:1913–1920. doi: 10.1093/jn/133.6.1913. [DOI] [PubMed] [Google Scholar]
- 6.Fan YY, Ly LH, Barhoumi R, McMurray DN, Chapkin RS. Dietary docosahexaenoic acid suppresses T cell protein kinase C theta lipid raft recruitment and IL-2 production. J. Immunol. 2004;173:6151–6160. doi: 10.4049/jimmunol.173.10.6151. [DOI] [PubMed] [Google Scholar]
- 7.Schley PD, Brindley DN, Field CJ. (n-3) PUFA alter raft lipid composition and decrease epidermal growth factor receptor levels in lipid rafts of human breast cancer cells. J. Nutr. 2007;137:548–553. doi: 10.1093/jn/137.3.548. [DOI] [PubMed] [Google Scholar]
- 8.Michaely PA, Mineo C, Ying YS, Anderson RG. Polarized distribution of endogenous Rac1 and RhoA at the cell surface. J. Biol. Chem. 1999;274:21430–21436. doi: 10.1074/jbc.274.30.21430. [DOI] [PubMed] [Google Scholar]
- 9.Smart EJ, Ying YS, Mineo C, Anderson RG. A detergent-free method for purifying caveolae membrane from tissue culture cells. Proc. Natl. Acad. Sci. U S A. 1995;92:10104–10108. doi: 10.1073/pnas.92.22.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim W, Fan YY, Barhoumi R, Smith R, McMurray DN, Chapkin RS. n-3 polyunsaturated fatty acids suppress the localization and activation of signaling proteins at the immunological synapse in murine CD4+ T cells by affecting lipid raft formation. J. Immunol. 2008;181:6236–6243. doi: 10.4049/jimmunol.181.9.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harder T, Scheiffele P, Verkade P, Simons K. Lipid domain structure of the plasma membrane revealed by patching of membrane components. J. Cell Biol. 1998;141:929–942. doi: 10.1083/jcb.141.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson RG. The caveolae membrane system. Annu. Rev. Biochem. 1998;67:199–225. doi: 10.1146/annurev.biochem.67.1.199. [DOI] [PubMed] [Google Scholar]
- 13.Wilson BS, Pfeiffer JR, Oliver JM. Observing FcepsilonRI signaling from the inside of the mast cell membrane. J. Cell Biol. 2000;149:1131–1142. doi: 10.1083/jcb.149.5.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kenworthy AK, Petranova N, Edidin M. High-resolution FRET microscopy of cholera toxin B-subunit and GPI-anchored proteins in cell plasma membranes. Mol. Biol. Cell. 2000;11:1645–1655. doi: 10.1091/mbc.11.5.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaus K, Chklovskaia E, Fazekas de St Groth B, Jessup W, Harder T. Condensation of the plasma membrane at the site of T lymphocyte activation. J. Cell Biol. 2005;171:121–131. doi: 10.1083/jcb.200505047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaus K, Zech T, Harder T. Visualizing membrane microdomains by Laurdan 2-photon microscopy. Mol. Membr. Biol. 2006;23:41–48. doi: 10.1080/09687860500466857. [DOI] [PubMed] [Google Scholar]
- 17.Rentero C, Zech T, Quinn CM, Engelhardt K, Williamson D, Grewal T, Jessup W, Harder T, Gaus K. Functional implications of plasma membrane condensation for T cell activation. PLoS ONE. 2008;3:e2262. doi: 10.1371/journal.pone.0002262. [DOI] [PMC free article] [PubMed] [Google Scholar]