Abstract
Human embryonic stem (hES) cells can differentiate into virtually all somatic cell types. In order to incorporate these derivatives into scientific or clinical applications, efficient methods of directing hES cell differentiation to pure subpopulations are required. Here we describe a robust strategy for generating cytokeratin 14+ (K14+)/p63+ keratinocyte progenitors from hES cells through stage-specific application of retinoic acid (RA) and bone morphogenetic protein-4 (BMP4). Induction of undifferentiated hES cells with RA stimulates expression of epithelial genes such as K18 and p63. Subculture of RA-treated cells in defined keratinocyte medium enables isolation of relatively pure K14+ epithelial populations; these cells also retain the capacity to terminally differentiate. The use of defined media throughout differentiation allows for detailed characterization of keratinocyte lineage specification from hES cells through the use of gene expression and immunofluorescence analyses.
Keywords: Human embryonic stem cells, directed differentiation, ectoderm, retinoic acid, bone morphogenetic protein, p63, epithelial progenitors, keratinocytes
1. Introduction
Human embryonic stem (hES) cells are capable of proliferating extensively and differentiating to form cells of the three embryonic germ layers (1). As such, these pluripotent cells can serve as a model system for studying early events during human development and are a powerful resource of non-transformed human cells for use in diagnostic and potentially therapeutic applications. Successful exploitation of hES cell derivatives requires the ability to direct hES cell differentiation to specific lineages in defined, efficient, and scalable systems (2). While researchers have identified strategies of obtaining endodermal, mesodermal, and neuroectodermal progenitors from hES cells (3-5), efficient methods of deriving non-neural epithelia have only recently been identified (6, 7). The method described here involves treatment of hES cells with retinoic acid (RA) and bone morphogenetic protein-4 (BMP-4) in defined medium at specific stages of differentiation to direct hES cells to form relatively pure populations of epithelial cells and more definitive keratinocytes.
Early induction of undifferentiated hES cell cultures is necessary to ensure efficient differentiation to epithelial lineages by RA and BMP4. This process is marked by significant increases in cytokeratin 18 (K18) and p63 transcription and relative decreases in pluripotency (e.g. Oct-4, SSEA-4, Nanog) and neural gene transcription (7), as measured by quantitative PCR or flow cytometry. These initial findings are supported by a previous study in mice that identified a role for RA and retinoic acid receptor α (RARα) in regulation of p63 (8), which, in turn, is involved in ectodermal fate choices (neural vs. non-neural epithelia) in the zebrafish (9). After early induction with RA and BMP4, these K18+/p63+ cells are subcultured in a defined epithelial medium and ultimately give rise to K14+/p63+ keratinocytes, as demonstrated by immunofluorescent staining and flow cytometry. A consistent epithelial morphology and lack of detectable Brachyury (T) or FOXA2 transcription throughout the differentiation process provide evidence that these K14+ epithelia are of ectodermal origin. However, markers associated with various epithelial tissues are detected in culture, demonstrating multipotent potential of these epithelial progenitors. This highly efficient process enables robust generation of keratinocyte progenitors that may be incorporated into engineered tissues for clinical or analytic use.
2. Materials
2.1. Cell Growth and Differentiation
hES cell growth medium: Dulbecco's Modified Eagle's Medium (DMEM)/F12 (1:1) (Invitrogen, Carlsbad, CA) supplemented with 20% Knockout Serum Replacer (KSR, Invitrogen), 1X MEM non-essential amino acids, (Invitrogen), 1 mM L-glutamine (Invitrogen), 0.1 mM β-mercaptoethanol (Sigma).
Basic fibroblast growth factor (bFGF, Invitrogen): added to conditioned hES cell growth medium at a concentration of 4 ng/ml.
Matrigel (BD Biosciences, San Jose, CA). Store at -80°C in single use aliquots. Thaw at 4°C; all manipulations must be conducted on ice using chilled pipettes to avoid gelation of Matrigel.
Dispase (Invitrogen). Reconstituted in DMEM/F12 at 2 mg/ml; store aliquots at -20°C.
Gelatin powder (Sigma) dissolved in water at 0.1% (w/v).
Differentiation medium: DMEM/F12 supplemented with 1X N2 supplement (Invitrogen), 1 μM all-trans retinoic acid (RA, Sigma), and 25 ng/ml BMP-4 (R&D Systems, Minneapolis, MN) (see Note 1). RA is sensitive to light, heat, and oxidation and should be reconstituted in sterile DMSO at a stock concentration of 10 mM and stored at -20°C for up to 6 months. Prior to use make working stock by diluting stock 10-fold in DMSO (can be stored at 4°C for up to 2 weeks); dilute working stock 1:1000 in culture medium.
Defined Keratinocyte Serum-free Medium and supplement (DSFM, Invitrogen).
Trypsin (0.05%)-ethylenediamine tetraacetic acid (EDTA, 1 mM) from Invitrogen.
Soybean trypsin inhibitor (Invitrogen) dissolved in PBS at 0.5 mg/ml and sterile filtered.
2.2. Quantitative RT-PCR
RNeasy Mini Kit (Qiagen, Valencia, CA).
Omniscript RT reverse transcriptase (Qiagen).
Oligo-dT primers at 50 μM (Invitrogen).
Quantitect SYBR Green qPCR kit (Qiagen).
Fluorescein sodium salt (Sigma) (Fisher).
Primers (Table 1) reconstituted in water to 20 μM and stored at -20°C (IDT DNA, Coralville, IA).
Table 1.
Primers used for quantitative PCR analysis using SYBR Green-based fluorescent quantification.
| Gene | Forward primer | Reverse primer | Amplicon size |
|---|---|---|---|
| GAPDH | 5′-caccgtcaaggctgagaacg-3′ | 5′-gccccacttgattttggagg-3′ | 91 |
| FOXA2 | 5′-gggagcggtgaagatgga-3′ | 5′-tcatgttgctcacggaggagta-3′ | 89 |
| Brachyury (T) | 5′-tgcttccctgagacccagtt-3′ | 5′-gatcacttctttcctttgcatcaag-3′ | 121 |
| Nestin | 5′-tgaagggcaatcacaacagg-3′ | 5′-tgaccccaacatgacctctg-3′ | 136 |
| Sox1 | 5′-caatgcggggaggagaagtc-3′ | 5′-ctctggaccaaactgtggcg-3′ | 464 |
| Pax6 | 5′-ggcaggtattacgagactgg-3′ | 5′-cctcatctgaatcttctccg-3′ | 427 |
| K18 | 5′-ccgtcttgctgctgatgact-3′ | 5′-ggccttttacttcctcttcgtg-3′ | 200 |
| TAp63 | 5′-aagatggtgcgacaaacaag-3′ | 5′-agagagcatcgaaggtggag-3′ | 234 |
| ΔNp63 | 5′-ggaaaacaatgcccagactc-3′ | 5′-gtggaatacgtccaggtggc-3′ | 294 |
| K14 | 5′-gaccattgaggacctgagga-3′ | 5′-attgatgtcggcttccacac-3′ | 157 |
2.3. Immunofluorescent Staining
IF Fixation Buffer: 16% (w/v) paraformaldehyde (PFA, Electron Microscopy Sciences, Hatfield, PA) diluted to 4% (v/v) in PBS prior to use.
Quenching buffer: PBS with 100 mM glycine.
Blocking buffer: PBS with 5% chick serum (Sigma) and 0.4% (v/v) Triton X-100 added.
Primary antibodies (recommended dilution): mouse anti-p63 monoclonal antibody (mAb, 1:100), rabbit anti-K14 polyclonal antibody (pAb, both from Lab Vision, Fremont, CA) (1:200), mouse anti-βcatenin mAb (1:200, BD Biosciences), goat anti-involucrin pAb (1:100), goat anti-filaggrin pAb (1:100, both from Santa Cruz Biotechnology, Santa Cruz, CA).
Secondary antibodies: chick anti-mouse IgG Alexa 488 conjugated antibodies, donkey anti-rabbit IgG Alexa 594 conjugated antibodies, donkey anti-goat IgG Alexa 488 conjugated antibodies (all from Invitrogen).
Hoechst 33342 nuclear staining solution (Sigma, 10 mg/ml stock diluted 1:5000 in water).
2.4. Flow Cytometry
FC Fixation Buffer: 1% (v/v) PFA in PBS.
Methanol (Fisher).
FACS Buffer: PBS with 2% (w/v) fetal bovine serum (FBS, Invitrogen), 0.1% (v/v) Triton X-100 (Sigma), and 0.1% (w/v) sodium azide (Sigma) added. Store at 4°C for up to 2 weeks.
Primary antibodies: mouse anti-K18 (DC10) mAb, rabbit anti-K14 pAb (both from Lab Vision), mouse anti-nestin mAb (Santa Cruz Biotechnology).
Secondary antibodies: chick anti-mouse IgG Alexa 647 conjugated antibodies, goat anti-rabbit IgG Alexa 488 conjugated antibodies (both from Invitrogen).
3. Methods
High efficiency directed differentiation of hES cells requires precise manipulation of the cellular microenvironment. Specification of hES cells to epithelial lineages can be effectively accomplished through time- and concentration-dependent application of RA and induction of BMP signaling. Early treatment of undifferentiated hES cells stimulates expression of epithelial genes, including the transcription factor p63, which is involved in maintenance of epithelial progenitors in a variety of tissues (10, 11). Subsequent culture of RA-induced hES cells in defined keratinocyte medium allows for propagation of definitive keratinocytes capable of undergoing terminal differentiation.
The method described here enables production of high purity epithelial populations from hES cells. Initial cultures entering the differentiation process must be pure and undifferentiated, as spontaneously differentiating cultures may not be restricted to epithelial lineages, compromising the ultimate purity of keratinocytes obtained. Procedures for the characterization of differentiating cultures are provided, including quantitative RT-PCR, fluorescence microscopy, and flow cytometry, which provide quantitative and qualitative measures of gene expression. Given the use of defined culture media throughout differentiation, this system provides a unique means of investigating the molecular events involved in early epithelial differentiation during human development.
3.1. Cell Growth and Differentiation
hES cells are cultivated in hES cell growth medium that has been conditioned by irradiated mouse embryonic fibroblasts (MEFs). To condition medium, plate irradiated MEFs (5000-10,000 rads, varies from lot-to-lot) at a density of 5×104 cells/cm2 and incubate with fresh hES cell growth medium for 18 – 24 hours. After conditioning, bFGF is added and complete conditioned medium is sterile filtered. hES cells are routinely passaged every 5 – 6 days (1:3 or 1:4 split) on Matrigel-coated plates using Dispase to remove cell colonies (see Note 2).
Begin treatment with Differentiation Medium after approximately 4 days (prior to cells reaching confluence), adding 2-3 ml/well for 6-well plates. Change medium daily for 7 days. Significant levels of cell death may be observed, though healthy differentiating cells should remain attached.
Aspirate medium and treat cultures with Dispase for 3-5 minutes at 37°C. When epithelial sheets begin to detach, gently aspirate Dispase and resuspend colonies in DSFM by scraping plate with a glass pipette. Centrifuge differentiated cell colonies at 200 × g for 4 minutes, resuspend in DSFM, and centrifuge again. Colonies are then resuspended in DSFM and distributed to gelatin-coated plates at a split ratio of 1:3 (see Note 3). After attachment, culture medium (DSFM) should be changed every other day (2 ml/well for 6-well plate).
After 3-4 weeks of cultivation in DSFM, tightly packed epithelial colonies that express high levels of nuclear p63 and cytoplasmic K14 should be observed. At this point one can subculture the differentiated cells via trypsinization for 5-10 minutes, inactivation with an equal volume of trypsin inhibitor, and 2 subsequent washes via centrifugation. Plate cells at 10,000 cells/cm2 on gelatin-coated tissue culture plates in DSFM (see Note 4).
Epithelial monolayers obtained at this stage express K14 at high purity (>90%) and can be passaged several times before undergoing senescence. These cells can also be incorporated into a variety of epithelial growth and differentiation assays.
3.2. Quantitative RT-PCR
Cells at all stages of differentiation are lysed directly on the culture plate (or in trypsinized cell pellets) for total RNA extraction using the RNeasy kit according to the manufacturer's instructions (see Note 5). On-column DNase treatment is recommended to reduce amplification of genomic DNA in subsequent PCR reactions.
cDNA is generated immediately using the Omniscript RT kit. Briefly, 1 μg total RNA is mixed with 10X RT buffer, oligo-dT primers (1 μM final concentration), RNase inhibitor (0.5 U/μL final concentration), 1 μL RT, and water to 20 μL and incubated for 1 hour at 37°C. cDNA can be stored for at least 1 year at -20°C.
-
For quantitative analysis of RNA levels duplicate qPCR reactions are run for each experimental gene and sample along with triplicate reactions for the reference gene GAPDH using the Quantitect SYBR Green kit. Combine SYBR Green qPCR Master Mix with gene specific primers (0.4 μM), 1 μL cDNA, and water to 50 μL for each reaction and initiate reactions on an iCycler or equivalent thermocycler. When using the iCycler, 10 nM fluorescein must be added to the reaction mixture for well-factor normalization. Gene specific primers of interest and their associated amplicon sizes are listed in Table 1. The annealing temperature for all reactions is 54°C, and 40 cycles are sufficient to detect signal. The iCycler is used to monitor fluorescence throughout the reaction and conduct melt curve analysis of all samples to verify specificity. Thermocycler program is listed below.
Hot start incubation – 15 minutes at 95°C.
Denaturation – 15 seconds at 94°C.
Annealing – 30 seconds at 54°C.
Extension and data acquisition – 30 seconds at 72°C.
Repeat steps b) – d) 39 times to complete reaction.
The iCycler iQ software generates cycle threshold (CT) values for all samples; be sure to manually set the threshold level to the exponential phase of background subtracted data. Relative gene expression (to GAPDH) is subsequently calculated via the following equation (see Note 6):
3.3. Immunofluorescent Staining
Aspirate culture medium from cells and rinse 1X with PBS.
Fix cells for 15-20 minutes at room temperature (RT) in IF Fixation Buffer.
Rinse cells 1X with PBS and incubate in Quenching Buffer for 10 minutes at RT.
Rinse cells 1X with PBS and incubate culture in Blocking Buffer for 2 hours at RT or up to 72 hours at 4°C.
Incubate cells with primary antibodies in fresh Blocking Buffer at the appropriate dilution (see Materials) overnight at 4°C.
Rinse cells 5X with PBS.
Incubate cells with secondary antibodies at a dilution of 1:500 in Blocking Buffer for 30-60 minutes at RT.
Rinse cells 2X with PBS then incubate for 5 minutes with nuclear staining solution.
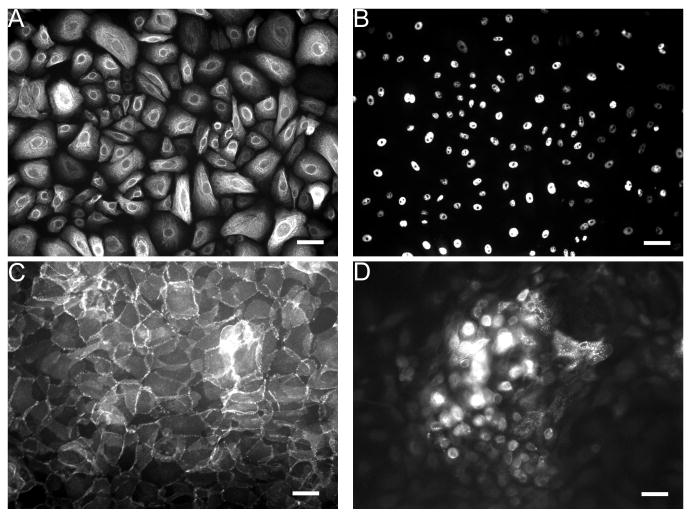
Rinse cells 2X with PBS and view on an epifluorescent microscope, using a mercury lamp and dichroic filters to excite Alexa 488 and 594 dyes as well as Hoechst stain. See Figure 1 for representative images of immunofluorescent stains.
Figure 1.
Immunofluorescent staining of hES cell-derived keratinocyte progenitors. A, B) Subcultured keratinocytes (as in step 3.1.5. of protocol) immunostained against the cytoskeletal protein K14 (A) and the nuclear transcription factor p63 (B). C) Differentiated hES cells (after 3 weeks culture in step 3.1.4. of protocol) stained against β-catenin; note punctate staining at membranes. D) Keratinocyte progenitors cultured to confluence and treated with 0.8 mM Ca2+ (after 4 weeks of culture in step 3.1.4. of protocol), then stained against the terminal differentiation marker filaggrin; note localization within suprabasal layers. Scale bar denotes 50 μm.
3.4. Flow Cytometry
Incubate cells with Trypsin-EDTA for 5-10 minutes to detach from plate.
Inactivate trypsin with soybean trypsin inhibitor, disperse cells with vigorous pipetting, and pass them through a 40 μm filter to remove large clumps.
Centrifuge samples at 200 × g for 4 minutes and pour off supernatant.
Resuspend cell pellet in 1 ml FC Fixation Buffer and incubate at 37°C for 10 minutes.
Chill tubes on ice for 1 minute and centrifuge at 200 × g and pour off supernatant.
Resuspend cell pellet in 2 ml ice-cold Methanol and hold on ice for 30 minutes (see Note 7).
Centrifuge cells as above, pour off supernatant, and resuspend pellet in 2 ml FACS Buffer. Cells should be counted and aliquoted to 105 cells per tube.
Centrifuge cells and pour off supernatant, add 2 ml FACS Buffer.
Centrifuge cells and again pour off supernatant, leaving approximately 100 μL in each tube.
Pre-dilute primary antibodies (1 μL per sample) in 50 μL FACS Buffer and add to each tube while resuspending pellet. Incubate overnight at 4°C. Be sure to include controls lacking primary antibodies in each combination for sample compensation.
Add 2 ml FACS Buffer to each sample, centrifuge, and pour off supernatant, leaving 100 μL.
Pre-dilute secondary antibodies (0.2 μL per sample) in 50 μL FACS Buffer and add to all tubes while resuspending pellet. Incubate for 30-45 minutes at RT.
Add 2 ml FACS Buffer to each sample, centrifuge, and pour off supernatant, leaving 100 μL.
Resuspend each sample in approximately 300 μL FACS Buffer and hold on ice until analysis.
Analyze samples on a BD FACSCalibur or equivalent cytometer capable of excitation at 488 nm and 630 nm wavelengths. Fixed cells preclude gating of live/dead populations, though the appropriate populations (Figure 1A) should be gated under forward scatter vs. side scatter plots to eliminate cellular debris from analysis. Use of Alexa 488 and 647 fluorophores limits crossover emission; however, fluorescence compensation may need to be applied after measuring control samples (no primary and single stained samples).
Appropriately gated populations enable quantitative comparisons of marker expression in samples under various treatment conditions or at specified time points.
Acknowledgments
The protocols described here were developed under the support of the NIH through the Biotechnology Training Program (C.M.M.) and grant 1R01 EB007534 (S.P.P.), and the NSF through the University of Wisconsin Materials Research Science and Engineering Center (MRSEC).
Footnotes
The described Differentiation Medium is advantageous to use for investigation of signaling events during differentiation given its chemically defined composition. Alternatively, unconditioned hES cell growth medium lacking bFGF and supplemented with 1 μM RA only may substituted to provide similar results (high purity K14+ populations). The presence of BMP activity in the KSR supplement has been previously demonstrated (12), and the omission of BMP-4 is a cost-effective alternative if use of defined medium is not necessary.
Cultivation of hES cells in TeSR, a chemically defined medium (13), prior to use of Differentiation Medium yields similar results to those described here. Please note that pure, undifferentiated hES cell cultures must be treated with RA-containing medium; appreciable levels (>10%) of spontaneously differentiated cells significantly reduces the purity of epithelial populations ultimately generated using this protocol.
Attachment and growth of hES cells and their derivatives may vary from passage to passage; as such, the split ratio of differentiated cultures may need to be modified slightly. A variety of matrices are effective in this protocol, including Collagen I and Collagen IV; in some cases use of alternative coatings improves adhesion. Finally, we have successfully differentiated cells by adding DSFM directly to the confluent cell layer without passaging on Day 7.
The extent of cell attachment may vary at this stage. However, attached cells will begin dividing within 24 – 72 hours and proliferate extensively for 3 – 4 passages before undergoing senescence (7).
For time course experiments, cell pellets or lysates can be stored at -70°C for simultaneous RNA extraction of all samples within an experiment.
The equation presented assumes all PCR reactions occur at 100% efficiency. If desired, standard curve reactions can be used to generate efficiency values for each primer set and incorporated into the equation as described by Pfaffl (14).
Cells in methanol may be held at -20°C for several days. Methanol fixation/permeabilization can be omitted when staining immediately with the listed antibodies.
References
- 1.Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–7. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Metallo CM, Azarin SM, Ji L, de Pablo JJ, Palecek SP. Engineering tissue from human embryonic stem cells. J Cell Mol Med. 2008;12:709–29. doi: 10.1111/j.1582-4934.2008.00228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D'Amour KA, Bang AG, Eliazer S, et al. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat Biotechnol. 2006;24:1392–401. doi: 10.1038/nbt1259. [DOI] [PubMed] [Google Scholar]
- 4.Li XJ, Du ZW, Zarnowska ED, et al. Specification of motoneurons from human embryonic stem cells. Nat Biotechnol. 2005;23:215–21. doi: 10.1038/nbt1063. [DOI] [PubMed] [Google Scholar]
- 5.Yang L, Soonpaa MH, Adler ED, et al. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453:524–8. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- 6.Aberdam E, Barak E, Rouleau M, et al. A pure population of ectodermal cells derived from human embryonic stem cells. Stem Cells. 2008;26:440–4. doi: 10.1634/stemcells.2007-0588. [DOI] [PubMed] [Google Scholar]
- 7.Metallo CM, Ji L, de Pablo JJ, Palecek SP. Retinoic acid and bone morphogenetic protein signaling synergize to efficiently direct epithelial differentiation of human embryonic stem cells. Stem Cells. 2008;26:372–80. doi: 10.1634/stemcells.2007-0501. [DOI] [PubMed] [Google Scholar]
- 8.Chen CF, Lohnes D. Dominant-negative retinoic acid receptors elicit epidermal defects through a non-canonical pathway. J Biol Chem. 2005;280:3012–21. doi: 10.1074/jbc.M411522200. [DOI] [PubMed] [Google Scholar]
- 9.Bakkers J, Hild M, Kramer C, Furutani-Seiki M, Hammerschmidt M. Zebrafish DeltaNp63 is a direct target of Bmp signaling and encodes a transcriptional repressor blocking neural specification in the ventral ectoderm. Dev Cell. 2002;2:617–27. doi: 10.1016/s1534-5807(02)00163-6. [DOI] [PubMed] [Google Scholar]
- 10.Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–13. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 11.Yang A, Schweitzer R, Sun D, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–8. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 12.Xu RH, Peck RM, Li DS, Feng X, Ludwig T, Thomson JA. Basic FGF and suppression of BMP signaling sustain undifferentiated proliferation of human ES cells. Nat Methods. 2005;2:185–90. doi: 10.1038/nmeth744. [DOI] [PubMed] [Google Scholar]
- 13.Ludwig TE, Bergendahl V, Levenstein ME, Yu J, Probasco MD, Thomson JA. Feeder-independent culture of human embryonic stem cells. Nat Methods. 2006;3:637–46. doi: 10.1038/nmeth902. [DOI] [PubMed] [Google Scholar]
- 14.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]