Abstract
Alzheimer’s disease (AD) is the most common form of progressive dementia in the elderly. It is a neurodegenerative disorder characterized by the neuropathologic findings of intracellular neurofibrillary tangles and extracellular amyloid plaques that accumulate in vulnerable brain regions. AD etiology has been studied by many groups, but since the discovery of the APOE ε4 allele, no further genes have been mapped conclusively to the late-onset form of the disease. In this study, we examined genetic association with late-onset Alzheimer’s susceptibility in 738 Caucasian families with 4704 individuals and an independent case-control dataset with 296 unrelated cases and 566 unrelated controls exploring 11 candidate genes with 47 SNPs common to both samples. In addition to tests for main effects and haplotype analyses, the Multifactor Dimensionality Reduction Pedigree Disequilibrium Test (MDR-PDT) was used to search for single-locus effects as well as 2-locus and 3-locus gene-gene interactions associated with AD in the family data. We observed significant haplotype effects in ACE in both family and case-control samples using standard and cladistic haplotype models. ACE was also part of significant 2-locus and 3-locus MDR-PDT joint effects models with Alpha-2-Macroglobulin (A2M), which mediates the clearance of Aβ, and Leucine-Rich Repeat Transmembrane 3 (LRRTM3), a nested gene in Alpha-3 Catenin (CTNNA3) which binds Presenilin 1. This result did not replicate in the case-control sample, and may not be a true positive. These genes are related to amyloid beta clearance; thus this constellation of effects might constitute an axis of susceptibility for late-onset AD. The consistent ACE haplotype result between independent data sets of families and unrelated cases and controls is strong evidence in favor of ACE as a susceptibility locus for AD, and replicates results from several other studies in a very large sample.
Keywords: Alzheimer’s disease, complex disease, epistasis, Multifactor Dimensionality Reduction, MDR-PDT
INTRODUCTION
Alzheimer’s disease (AD) (OMIM 104300, 104310) is the most common form of progressive dementia in the elderly. More than 4 million Americans are afflicted with this debilitating disorder and many studies have been conducted to elucidate an etiology [Martin et al., 2005]. The discovery of the APOE ε4 risk factor demonstrated that genetic analysis can be successful in complex disease research. However, between 42% and 68% of cases do not carry the ε4 allele [Henderson et al., 1995; Lucotte et al., 1994; Ritchie et al., 1996; Hardy et al., 2004]. Additional environmental and genetic factors likely play a role in Alzheimer’s susceptibility. Some of these putative factors are explored in the current study. Genes known to interact with presenilins, amyloid beta (Aβ) clearance, and cardiovascular disease are evaluated due to their known biological relevance.
Amyloid precursor proteins and presenilins influence autosomal dominant, early-onset disease due to altered Amyloid Protein Precursor processing, leading to Aβ deposition [Goate, 2006; Hardy, 1997; Levy-Lahad et al., 1995; Rogaev et al., 1995; Sherrington et al., 1995]. These mutations have not been shown to influence late-onset susceptibility, which is far more prevalent. They do, however, provide potential insight into the pathophysiology of the disorder.
Fourteen years after the discovery of APOE, single-locus approaches by many groups have not discovered any additional candidates consistently associating with late-onset AD (LOAD). The ε4 allele of APOE causes increased risk for AD, while the ε2 allele is protective [Chartier-Harlin et al., 1994; Corder et al., 1993]. The mechanism by which APOE ε4 influences risk of AD is unknown, but is likely related to Aβ processing [Bales et al., 1999]. This inability to unravel the mechanism underlying the trait, given steadily increasing ascertainment and genotyping capability, illustrates the difficulty of finding LOAD genes.
A gene that has been carefully studied for association with LOAD with inconsistent results is angiotensin converting enzyme (ACE) [Alvarez et al., 1999a; Kehoe et al., 1999; Scacchi et al., 1998; Chapman et al., 1998]. Large meta-analyses of ACE markers support the hypothesis that ACE is a susceptibility locus for AD [Bertram et al., 2007a; Lehmann et al., 2005]. ACE functions in several biological systems that may be related to AD, such as the renin-angiotensin system regulating salt homeostasis [Reid, 1992] and Aβ degradation pathways [Hemming and Selkoe, 2005; Hu et al., 2001].
Increased ACE activity and expression has also been associated with AD patients and ACE isoform I accumulates perivascularly in cases of severe cerebral amyloid angiopathy [Miners et al., 2008a]. The N-teminal catalytic domain of ACE has recently been indicated as primarily responsible for Aβ degradation [Oba et al., 2005]. Other studies have suggested the C-terminal domain also participates in Aβ catabolism [Sun et al., 2008]. Additionally, species of Aβ that are associated with aging are selectively degraded by ACE [Toropygin et al., 2008], and ACE has been demonstrated to cleave the putatively pathogenic form of Aβ-42 to the more benign Aβ-40 [Zou et al., 2007]. The statistical and molecular evidence for ACE as an AD locus has motivated the discussion of ACE inhibitors as therapeutics for AD in the literature [Kehoe and Wilcock, 2007; Miners et al., 2008b; Nalivaeva et al., 2008], as well as studies evaluating ACE inhibitors in vivo in both animal models of Aβ deposition [Eckman et al., 2006; Hemming and Selkoe, 2005; Hemming et al., 2007] and clinical trials for the effect of ACE inhibitors on dementia and AD [Hanon and Forette, 2004; Khachaturian et al., 2006; Ohrui et al., 2004; Tzourio et al., 2003].
Some other genes where previous associations have been observed but inconsistently replicated are alpha-2-macroglobulin (A2M) [Blacker et al., 1998; Rogaeva et al., 1999; Alvarez et al., 1999b; Blennow et al., 2000] and alpha-T-catenin (CTNNA3) [Bertram et al., 2007b; Busby et al., 2004; Li et al., 2008; Martin et al., 2005], and a nested gene in CTNNA3 leucine-rich repeat transmembrane protein 3 (LRRTM3) [Ertekin-Taner et al., 2003; Martin et al., 2005]. A2M has protease inhibitor activity [Bergqvist and Nilsson, 1979], and mediates the clearance of Aβ deposits. CTNNA3 binds to beta catenin which then interacts with presenilin 1, which has been associated with early-onset AD [Sherrington et al., 1995]. There have also been previous reports of linkage to late-onset AD at the CTNNA3/LRRTM3 locus in addition to CTNNA3 association with Aβ-42 levels in late-onset AD families [Ertekin-Taner et al., 2000; Ertekin-Taner et al., 2003].
ACE, A2M and LRRTM3 were also found as the best multilocus model by Multifactor Dimensionality Reduction Pedigree Disequilibrium Test (MDR-PDT) analysis of our family sample. Previous analyses of the family samples presented here restricted to only to CTNNA3/LRRTM3 and APOE detected a multilocus model with a synergistic effect [Martin et al., 2005; Martin et al., 2006]. Here we expand this analysis to several more candidate genes, including some previously studied by our group for main effects in samples with various degrees of overlap with these, such as A2M [Rogaeva et al., 1999], LRRTM3/CTNNA3 [Liang et al., 2007; Martin et al., 2005; Martin et al., 2006], and LRP1 [Scott et al., 1998].
Undiscovered gene-gene interactions associating with LOAD could explain why the search for LOAD loci since the APOE discovery has been relatively fruitless [Ioannidis, 2007]. Such interactive effects can exist without the presence of substantial main effects, making detection with single-locus analysis unlikely [Hirschhorn et al., 2002]. Methods for the analyses of large interaction search spaces are now available and were applied here for family and case-control data [Martin et al., 2006; Ritchie et al., 2001; Ritchie et al., 2003]. Due to strong biological and epidemiological evidence of a genetic etiology for LOAD but lack of consistent single-locus findings, LOAD would appear to be an ideal trait to begin a search for epistasis among past candidates.
The goal of this study is to explore effects explaining LOAD through single-locus analysis, haplotypes, and epistatic gene-gene interactions among candidate gene SNPs.
METHODS
STUDY POPULATION
FAMILY DATA
The data for this study consisted of genotypes in both a family sample and an independent case-control sample. The family sample contains 738 families consisting of 4704 indivduals collected through three ascertainment groups: the Collaborative Alzheimer Project (CAP: The Joseph and Kathleen Bryan ADRC and the Center for Human Genetics at Duke University, the Center for Human Genetics Research at Vanderbilt University Medical Center, and the University of California at Los Angeles Neuro-psychiatric Institute); National Institutes of Mental Health (NIMH); and the National Cell Repository for AD at Indiana University Medical Center (IU). The family sample is described in Table 1. The singleton dataset contains 158 families with one sampled affected family member and any number of unaffected siblings. The multiplex dataset contains 580 families with at least two sampled affected family members.
Table 1.
Family data details and ascertainment
| Family Type | Total Families | CAP Families | NIMH Families | IU Families | Discordant sibling pairs | Affected Relative Pairs |
|---|---|---|---|---|---|---|
| Multiplex | 580 | 87 | 349 | 124 | 1111 | 1153 |
| Singleton | 158 | 78 | 3 | 29 | 161 | 0 |
|
| ||||||
All affected individuals met the NINDS/ADRDA criteria for probable or definite AD. Unaffected relatives from the CAP and NIMH sites showed no signs of dementia upon examination. Unaffected individuals from IU were classified based on self report. The mean (SD) age at onset (AAO) in affected individuals was 72.31 (9.09) years, and the mean (SD) age at examination (AAE) was 74.82 (11.02) years. For more information on the family sample, see [Martin et al., 2005]
CASE-CONTROL DATA
The case-control dataset consisted of 296 unrelated cases and 566 unrelated controls independent of the family data. The average age of exam (standard deviation) for cases was 79.02(6.76), and controls was 73.63(6.30). The average age of onset (SD) for cases was 71.78(7.82). The ages of onset and controls were not significantly different. Unrelated cases were determined to be affected by examination based on the same criteria as the cases in the family data. Priority for selection was given to cases where age of onset was known, Parkinson’s disease (PD) was not present, depression status was known, and documentation proving AD was available. Unaffected controls required unaffected status confirmed by examination, no first-degree relatives with AD, no PD, otherwise no dementia, and adequate DNA for genotyping. Unrelated cases and controls were collected at the Center for Human Genetics at Duke University and the Center for Human Genetics Research at Vanderbilt University Medical Center. Also ascertained in the case control data was hypertension status, which was measured by survey as having ever being diagnosed with hypertension.
GENOTYPING METHODS
The list of SNPs selected for this study is shown in Table 2. The rationale for including each gene in the list of candidates is detailed in Table 3. The SNPs were designed to be genotyped on the Applied Biosystems, Taqman 7900HT allelic discrimination system and were either custom (Assay by Design) or inventoried (Assay on Demand) assays. All genotyping reactions were run according to the standard genotyping methods as outlined by Applied Biosystems protocols and were performed on 3ng of genomic DNA per reaction. All SNPs were held to a minimum genotyping efficiency of 95%. Quality control was performed on the SNPs by using matched pairs of quality control samples placed within and between the 384 well plates. Laboratory technicians were blinded to the matching pattern, affection status, and pedigree information. In the family sample 48 SNPs were genotyped, including A2M SNP rs1800433, which was not genotyped in the case-control data. In the case-control sample, 55 SNPs were genotyped, including SNPs not genotyped in the family data in AGT, NCSTN and A2MP. These differences are detailed in Table 2.
Table 2.
Gene and SNP information for Alzheimer’s candidate genes
| Gene | SNP rs# | Position (bp) | Chromosome Band | Alleles Major/Minor | Role |
|---|---|---|---|---|---|
| PZP | rs10842971 | 9194563 | 12p13.31 | A/T | Coding exon |
| rs3213831 | 9208040 | 12p13.31 | T/C | Coding exon | |
| rs2277413 | 9209051 | 12p13.31 | C/T | Coding exon | |
| rs3213832 | 9212768 | 12p13.31 | C/T | Coding exon | |
| rs12230214 | 9238059 | 12p13.31 | C/G | Coding exon | |
|
| |||||
| A2MP | rs16918212B | 9276225 | 12p13.31 | C/A | Not annotated |
| rs34362B | 9276692 | 12p13.31 | C/T | Not annotated | |
| rs17804080B | 9279277 | 12p13.31 | C/T | Not annotated | |
|
| |||||
| LRP1 | rs1799986 | 55821533 | 12q13.3 | C/T | Coding exon |
| rs1800127 | 55825349 | 12q13.3 | C/T | Coding exon | |
| rs1800174 | 55846076 | 12q13.3 | G/A | Intron (boundary) | |
| rs1800181 | 5864555 | 12q13.3 | C/T | Intron (boundary) | |
| rs2075699 | 55871411 | 12q13.3 | C/T | Coding exon | |
| rs1800154 | 55875926 | 12q13.3 | C/T | Coding exon | |
| rs1800165 | 55877493 | 12q13.3 | T/C | Intron (boundary) | |
| rs11172124 | 55881222 | 12q13.3 | G/A | Intron (boundary) | |
| rs9669595 | 55881333 | 12q13.3 | G/A | Intron | |
| rs7956957 | 55889082 | 12q13.3 | G/C | Promoter | |
|
| |||||
| CTNNA3 | rs1786927 | 67352267 | 10q21.3 | G/A | Intron |
| rs2126750 | 67507709 | 10q21.3 | T/A | Intron | |
| rs7911820 | 67534145 | 10q21.3 | G/T | Intron | |
| rs12357560 | 67534187 | 10q21.3 | T/C | Intron | |
| rs7070570 | 67534610 | 10q21.3 | A/G | Intron | |
| rs7074454 | 67534965 | 10q21.3 | T/C | Intron | |
| rs6480140 | 67538887 | 10q21.3 | A/C | Intron | |
| rs997225 | 67952976 | 10q21.3 | G/A | Intron | |
|
| |||||
| LRRTM3 | rs1925583 | 68349950 | 10q21.3 | G/T | Promoter |
| rs942780 | 68406547 | 10q21.3 | A/G | Intron | |
| rs1925617 | 68434823 | 10q21.3 | T/G | Intron | |
|
| |||||
| NCSTN | rs6668576 | 157130094 | 1q23.2 | T/C | Intron |
| rs10494342 | 157130193 | 1q23.2 | T/G | Intron | |
| rs2038781 | 157130457 | 1q23.2 | G/C | Intron | |
| rs12239747B | 157134138 | 1q23.2 | A/G | Coding exon | |
| rs7528638B | 157136976 | 1q23.2 | C/G | Intron (boundary) | |
| rs6427515 | 157138184 | 1q23.2 | C/T | Intron | |
| rs4656256 | 157144092 | 1q23.2 | A/G | Promoter | |
|
| |||||
| COG2 | rs3789662 | 227135608 | 1q42.2 | A/G | 3' UTR |
| rs7536290 | 227143437 | 1q42.2 | A/G | 3' UTR | |
|
| |||||
| AGT | rs3789670 | 227150449 | 1q42.2 | C/T | Intron |
| rs2478545 | 227150856 | 1q42.2 | C/T | Intron | |
| rs4762B | 227152712 | 1q42.2 | G/A | Coding exon | |
| rs2148582 | 227156534 | 1q42.2 | T/C | Intron (boundary) | |
| rs5051B | 227156607 | 1q42.2 | C/T | Promoter | |
| rs5050B | 227156621 | 1q42.2 | T/G | Promoter | |
| rs1326886 | 27166495 | 1q42.2 | A/G | Promoter | |
|
| |||||
| A2M | rs3832852 | 9137444 | 12p13 | CCATA/del | Splice Site |
| rs1800433A | 9123618 | 12p13 | A/G | Coding Exon | |
|
| |||||
| APOE | ε2, ε3, ε4 | 50101007 | 19q13.31 | ε2, ε3, ε4 | - |
|
| |||||
| ACE | rs4291 | 58907926 | 17q23.3 | A/T | Promoter |
| rs4295 | 58910030 | 17q23.3 | G/C | Intron (boundary) | |
| rs4311 | 58914495 | 17q23.3 | C/T | Intron (boundary) | |
| rs4329 | 58917190 | 17q23.3 | A/G | Intron | |
| rs4646994 | 58919636 | 17q23.3 | del/ins | Intron | |
| rs4343 | 58919763 | 17q23.3 | G/A | Coding exon | |
| rs4353 | 58924154 | 17q23.3 | A/G | Intron | |
| rs4978 | 58927493 | 17q23.3 | T/C | Coding exon | |
genotyped only in family data
genotyped only in case-control data
Table 3.
Names and role of candidate genes in AD
| Gene Symbol | Gene Name | Candidate Rationale |
|---|---|---|
| PZP | Pregnancy Zone Protein | closely related to A2M, maps to the same region |
| A2MP | Alpha 2 Macroglobulins of pregnancy | PZP analog, may participate in cardiovascular remodeling |
| LRP1 | Low density lipoprotein receptor-related protein 1 | APOE receptor related protein |
| CTNNA3 | Catenin, Alpha-3 | alpha-T-catenin binds beta-catenin; beta-catenin interacts with PSEN1 |
| LRRTM3 | Leucine rich repeat transmembrane 3 | nested gene in CTNNA3 |
| NCSTN | Nicastrin | forms complex with PS1 and 2 |
| COG2 | Component of oligomeric golgi complex 2 | essential component of intracellular protein trafficking |
| AGT | Angiotensinogen | enzymatic target of ACE |
| A2M | Alpha 2 Macroglobulin | mediates the clearance of Aβ, |
| APOE | Apolipoprotein Epsilon | validated AD association, VLDL transport |
| ACE | Angiotensinogen converting enzyme | associations found previously between ACE and AD, also mediates Aβ clearance |
FAMILY DATA ANALYSIS
Pedigree Disequilibrium Test (PDT)
To examine association between alleles and genotypes and AD in the family data, the Pedigree Disequilibrium Test (PDT) and genotype Pedigree Disequilibrium Test (genoPDT) sum statistics were used [Martin et al., 2000; Martin et al., 2003a] The PDT statistics measure transmission of alleles or genotypes to affected offspring from informative pedigrees, testing for excessive transmission of particular alleles or genotypes.
An informative pedigree is either a nuclear family with at least one affected child, both parents genotyped at the locus and at least one heterozygous parent, a discordant sibling pair (DSP) with different genotypes at the locus with or without parental genotypes, or an extended pedigree with at least one informative nuclear family or DSP. Most information about association with the trait in these data comes from DSPs due to the late onset of AD.
MDR-PDT
The MDR-PDT is a within-family measure of multilocus association between genotypes and phenotype [Martin et al., 2006]. The genoPDT statistic functions within the framework of the Multifactor Dimensionality Reduction (MDR) algorithm [Hahn et al., 2003; Ritchie et al., 2001; Ritchie et al., 2003] by establishing which multilocus genotypes are positively associated with the outcome of interest. Positive values of the genoPDT test statistic classify multilocus genotypes as high-risk, creating a binary variable useful for summarizing the association at a multilocus model, and retaining the useful property of robustness to population stratification.
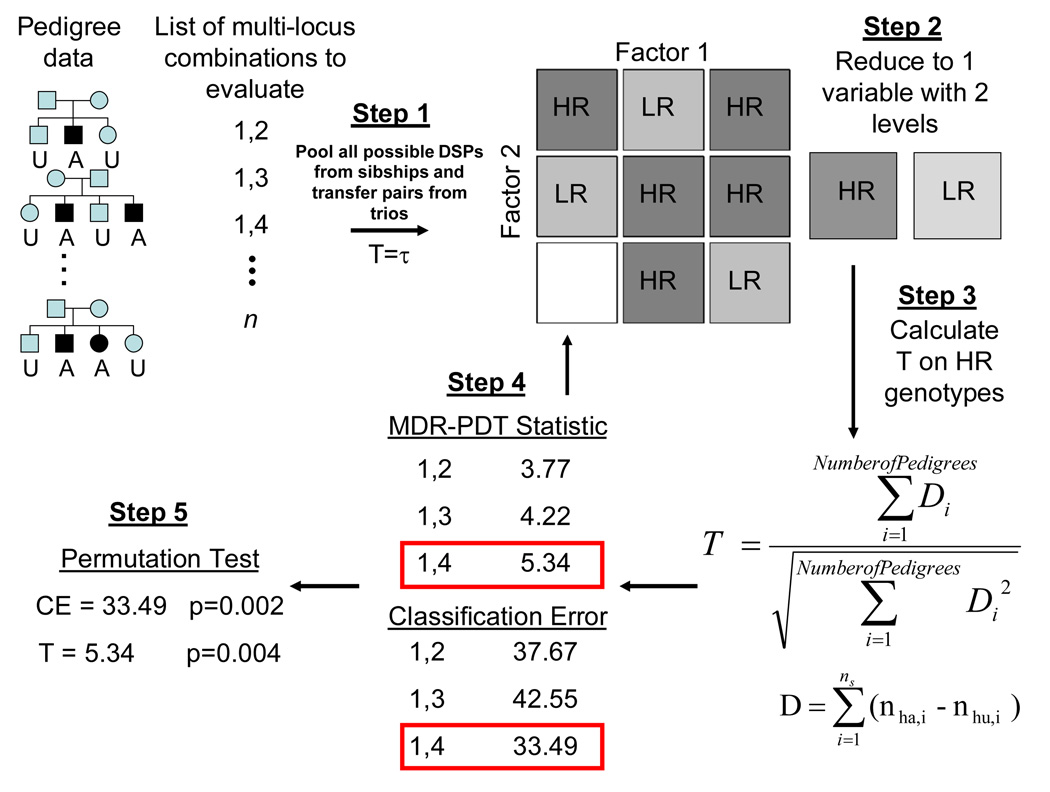
MDR-PDT has been described previously [Martin et al., 2006]. A brief description of the algorithm is shown in Figure 1. The steps of MDR-PDT are:
All possible DSPs are generated within each sibship and pooled.
Each genotype is determined to be high or low risk by comparing the ratio of affecteds to unaffecteds from the pooled DSPs to a threshold τ (τ = 1); whether above or below the threshold indicates positive or negative association with affected status.
Statistics for high-risk genotypes are calculated using the PDT. This is the MDR-PDT statistic for this model. A classification error (CE), calculated as the number of low-risk affecteds plus high-risk unaffecteds divided by the total sample, is also calculated for the model.
Steps 2–3 are repeated for all possible combinations of loci, calculating an MDR-PDT statistic and CE for each, choosing the largest MDR-PDT statistic as the final result.
A permutation test is performed to determine the distribution of the statistic under the null hypothesis, to which the result from step 4 is compared for significance assessment.
Figure 1.
MDR-PDT algorithm
To examine whether the best model of a given order that is observed by MDR-PDT is a real signal or is the result of sampling error, a permutation test is conducted. The permutation test consists of randomizing status for offspring, holding the proportion of affected individuals constant within sibships across permutations, calculating the statistic, and repeating many times to estimate the distribution of the null hypothesis. The test based on the permutation procedure should have the correct type I error, even for sparse data. This validity is due to all contingency table cells from each permutation containing the same number of observations.
Tag SNPs in family data were chosen using tagger, a function within the haploview software package [Barrett et al., 2005; Gabriel et al., 2002] for the MDR-PDT analyses to remove redundant variables from the data, which reduce the power of MDR-PDT. An r2 threshold of 0.8 and LOD of 3 were used to choose tag SNPs in order to eliminate nearby markers with very similar information and maximize power for MDR-PDT analysis. The abridged data contained 32 of the original 47 markers.
CONDITIONAL LOGISTIC REGRESSION
For single-locus effect size evaluation, the referent group was the major allele homozygote. The adjustment of Siegmund et al. [Siegmund et al., 2000] was implemented to correct confidence intervals for familial correlation in regions of linkage.
APL
The Association in the Presence of Linkage (APL) statistic [Chung et al., 2006; Martin et al., 2003b] was employed to measure haplotype associations in family data. APL measures the difference in the number of copies of an allele or haplotype in affected offspring from the expected number of copies under the null hypothesis of no association conditional on parental genotypes. APL uses nuclear families with at least one affected offspring. When parental genotypes are missing, they are inferred using the expected probabilities of consistent parental mating types. APL correctly adjusts for correlated transmissions to multiple affected siblings by estimating IBD probabilities. The probability IBD 0, 1, 2 and the haplotype frequency are estimated by EM algorithm [Clark, 1990; Excoffier and Slatkin, 1995; Long et al., 1995].
To estimate the variance of the APL statistic, a bootstrapping approach is used [Chung et al., 2006]. Bootstrap samples are taken with replacement across families, forming same-size pseudosamples consisting of replicates of some families and missing others at random. The variance of the APL statistic calculated for all pseudosamples is the estimated sampling variance for the statistic. This variance can be used to test the null hypothesis of no association allowing for the presence of linkage.
CASE-CONTROL DATA ANALYSIS
CHI-SQUARED AND FISHER’S EXACT TESTS
To test for association of sex with genotypes in the case-control data chi-squared or Fisher’s exact tests of differences between frequencies of alleles and genotypes between sexes were performed in controls at each marker. This test should detect where sampling error has distorted the distribution of alleles or genotypes by sex at autosomal markers. Since there is a difference in prevalence by sex in AD, such a scenario in the data could cause confounding. If the genotype frequency tests were significant at the 0.05 level, then sex-stratified chi-squared or Fisher’s exact tests of Hardy-Weinberg Equilibrium (HWE) and association with disease at alleles and genotypes were performed.
Sex stratification, single site allele and genotype frequency and association, and HWE analyses in controls were performed using Powermarker statistical software [Zaykin et al., 2002]. Where the number of observations for a cell from the 3×2 table stratifying the data by genotype and status was five or less, Fisher’s exact test was used to assess HWE and association with LOAD.
MDR
Multifactor dimensionality reduction (MDR) [Hahn et al., 2003; Ritchie et al., 2001; Ritchie et al., 2003] was used to search for interactions in the case-control data. MDR exhaustively screens all possible interactions and ranks results by the signal detected by balanced accuracy and cross-validation consistency in case-control data to find models with the most potential to be real interactions. MDR has performed well across many genetic simulation scenarios where purely epistatic relationships existed between status and a set of variables with an absence of main effects [Ritchie et al., 2003].
LOGISTIC REGRESSION
To estimate single-locus effect sizes in parallel with the family data, effect sizes in case-control data were estimated using logistic regression using the major allele homozygote as the referent group [Stata Corp, 2005].
HAPLOTYPE ANALYSIS
Haplotype analyses for case-control data were performed using the haplo.cc and haplo.glm functions in Haplo.Stats [Schaid et al., 2002]. A 3-marker sliding window was run to identify associations among correlated sets of markers. Full haplotypes were tested and haplotype exposure odds ratios were estimated using the most frequent haplotype as the referent group.
BIOINFORMATICS TOOLS
The website SNPer [Riva and Kohane, 2004] and Entrez PubMed were used to collect information on candidate genes and genotyped markers. Online Inheritance in Man (OMIM) [OMIM, 2008] was used to collect information about the phenotype and candidate genes. The Alzgene database at www.Alzgene.org [Bertram et al., 2007a] was also used to collect information about AD association studies.
Multiple testing was accounted for depending on the type of analysis. MDR and MDR-PDT both inherently correct for the search conducted with permutation testing. Multiple tests of main effects were corrected using Nyholt’s method [Nyholt, 2004] with the modification of [Li and Ji, 2005]. The effective number of tests for the 47 markers which were in both datasets was 28.9 for the founders from family data and 29.4 for the controls from the case-control data, showing the similarity of correlation among these independent samples. There were 7 additional tests in the case control data than the family sample, and the effective number of independent tests considering all markers for case-control and family samples was 34.6 and 29.9, respectively. These effective numbers of tests for each dataset lead to thresholds for significance of p≤0.0015 for the case-control data and p≤0.0017 for the family data. To reject the null for any test from either sample, the threshold is p≤0.00079.
For purposes of assessing significance where two tests have been performed for the same null hypothesis in independent samples, we used Fisher’s method [Fisher, 1950] to merge p-values from the same SNP in different samples. We then compared the merged p-value to the threshold for significance given the effective number of independent tests established by SNPSpD. This threshold is determined by the Sidak correction for multiple tests [Sidak, 1967] which is slightly more liberal than the Bonferroni correction but provides the exact correction necessary to return the experiment-wise error rate to the desired level.
CLADISTIC HAPLOTYPE MODELLING
Clade-based haplotype analysis was conducted using markers rs4291 and rs4343 as suggested in Katzov et al [Katzov et al., 2004]. These markers denote ancestral haplotype clades A, B and C which have been previously associated with circulating levels of ACE [Keavney et al., 1998; Rieder et al., 1999; Soubrier et al., 2002]. A straightforward cladistic model of the ACE locus was proposed by Farrall et al., in which the variation in the gene could be captured using only a few markers [Farrall et al., 1999]. We analyzed these markers for haplotype association with LOAD.
FAMILY DATA – SINGLE LOCUS RESULTS
For the family dataset, single-locus associations were examined with allele and genotype PDT statistics. These results are presented in Table 4. Seventeen SNPs from the family data yielded a p-value less than 0.05 at tests of either alleles or genotypes. Of these, only APOE is statistically significant after accounting for multiple tests. The PZP SNP rs12230214 (C/G), a nonsynonymous L/V change located in exon 11 was nominally associated with LOAD at genotypes (allele p = 0.18, genotype p = 0.05). Two LRP1 SNPs, rs9669595 (A/G), located in intron 65 (allele p = 0.02, genotype p = 0.04), and rs7956957 (G/C), located in intron 78 (allele p = 0.08, genotype p = 0.02) were nominally associated with LOAD. Two CTNNA3 intron 13 SNPs, rs7911820 (G/T) (allele p = 0.02, genotype p = 0.03) and rs7074454 (C/T) (allele p = 0.01, genotype p = 0.02) and 1 intron 14 SNP, rs12357560 (T/C) (allele p = 0.06, genotype p = 0.03) were nominally associated. One LRRTM3 intron 7 SNP, rs1925617 (T/G) was nominally associated with LOAD (allele p = 0.65, genotype p = 0.01). One NCSTN intron 2 SNP, rs2038781 (G/C) was nominally associated (allele p = 0.05, genotype p = 0.04). rs3832852 (ins/del), a 5-base insertion in A2M that spans the upstream splice site for exon 18 (allele p = 0.01, genotype p = 0.01) showed a nominal association. The APOE allele ε4 was highly significantly associated with AD in both allele and genotype tests (allele p < 0.001, genotype p < 0.001). The remaining seven markers nominally significantly associated with disease at alleles and genotypes were all found in ACE. The ACE markers were: rs4291 (A/T), 239 base pairs upstream of exon 1 (allele p = 0.02, genotype p = 0.01); rs4295 (G/C), an intron 2 marker (allele p = 0.07, genotype p = 0.03); rs4311 (C/T), an intron 9 marker (allele p = 0.1, genotype p = 0.03); rs4646994 (del/ins), a 287bp indel in intron 16 (allele p = 0.02, genotype p = 0.07); rs4343 (A/G), a synonymous coding SNP in exon 16 (allele p = 0.01, genotype p = 0.03); rs4353 (A/G), a marker in intron 19 (allele p = 0.04, genotype p = 0.04); and rs4978 (C/T), a synonymous coding SNP in exon 23 (allele p = 0.01, genotype p = 0.01).
Table 4.
Nominally and statistically significant uncorrected single-locus associations with LOAD
| Gene | rs number | risk allele | family allele | family genotype | CC allele | CC genotype | Fisher's allele | Fisher's genotype |
|---|---|---|---|---|---|---|---|---|
| LRP1 | rs9669595* | A | 0.023 | 0.103 | - | - | - | - |
|
|
||||||||
| CTNNA3 | rs7911820* | G | 0.016 | 0.083 | - | - | - | - |
| CTNNA3 | rs12357560* | C | 0.06 | 0.041 | - | - | - | - |
| CTNNA3 | rs7074454* † | C | 0.005 | 0.048 | - | - | 0.006 | 0.087 |
| CTNNA3 | rs6480140§ † | C | - | - | 0.613 | 0.008 | 0.706 | 0.007 |
| CTNNA3 | rs997225§ | A | - | - | 0.033 | 0.082 | - | - |
|
|
||||||||
| LRRTM3 | rs1925617* ‡ † | T | 0.652 | 0.001 | - | - | 0.618 | 0.004 |
|
|
||||||||
| NCSTN | rs2038781* | G | 0.052 | 0.031 | - | - | - | - |
|
|
||||||||
| A2M | rs1800433‡ | A | - | - | - | - | - | - |
| A2M | rs3832852* † | del | 0.002 | 0.002 | - | - | 0.013 | 0.009 |
|
|
||||||||
| APOE | ε2, ε3, ε4* † | ε4 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
|
|
||||||||
| ACE | rs4291* ‡ | A | 0.015 | 0.038 | - | - | - | - |
| ACE | rs4295* | G | 0.068 | 0.058 | - | - | - | - |
| ACE | rs4311 | T | 0.106 | 0.056 | - | - | - | - |
| ACE | rs4646994* † | ins | 0.017 | 0.116 | - | - | 0.009 | 0.066 |
| ACE | rs4343* § † | A | 0.01 | 0.069 | 0.048 | 0.144 | 0.004 | 0.056 |
| ACE | rs4353* † | G | 0.04 | 0.152 | - | - | 0.022 | 0.132 |
| ACE | rs4978* † | C | 0.008 | 0.048 | - | - | 0.012 | 0.093 |
Nominally significant at alleles or genotypes in family sample
Nominally significant at alleles or genotypes in case-control sample
Part of Significant MDR-PDT models
Significant by Fisher’s merged p-value statistic
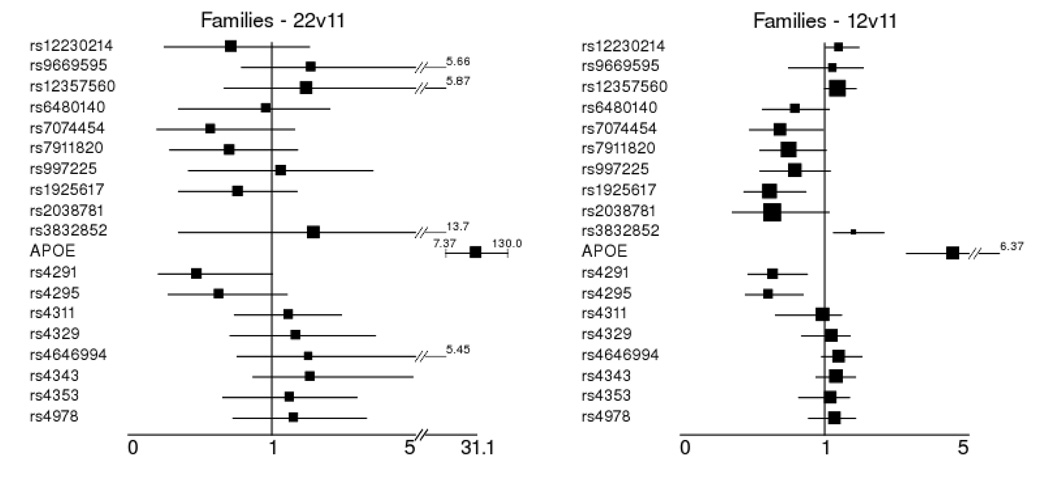
Conditional logistic regression was run to estimate the effect sizes observed in the family sample among those markers that were nominally significant at either alleles or genotypes in families or case-control samples. Of note are effect size estimates for markers that were at least nominally significantly associated in the case-control sample (Table 4, and described below). These estimates attempt to remedy the bias encountered when effect size estimation and association detection are performed on the same data. The major allele homozygote was used as the referent group for these analyses. These results are detailed in Figure 2. APOE had a very strong effect in these data for the ε4 homozygote (OR = 31.1 95% CI = 7.37–130) and the ε4 heterozygote (OR = 4.57 95% CI = 3.28–6.57). Other than APOE, seven nominally significant single-locus genotype effects in five genes were observed in the family data. The PZP marker rs12230214 (OR = 1.42, 95% CI = 1.03–1.95) had a nominally significant effect for the CG heterozygote. Two CTNNA3 markers showed a nominally significant effect: rs12357560 (OR 1.37, 95% CI = 1–1.87) for the TC heterozygote and rs7074454 (OR 0.69, 95% CI = 0.48–0.99) for the TC heterozygote. The LRRTM3 marker rs1925617 (OR 0.619, 95% CI = 0.44–0.87) had a nominally significant effect estimated for the TG heterozygote. The A2M marker rs3832852 (OR = 1.81, 95% CI = 1.25–2.64) had a nominally significant effect estimate for the splice site deletion heterozygote. Two markers in ACE had nominally significant effect estimates. They were rs4291 (OR = 0.48, 95% CI = 0.21–1.0) for the A allele homozygote and (OR = 0.64, 95% CI = 0.47–0.88) for the AT heterozygote, and rs4295 (OR = 0.62, 95% CI = 0.45–0.85) for the GC heterozygote.
Figure 2.
Odds ratio estimates of effect size from family data using conditional logistic regression and the correction for residual correlation in multiple affected offspring in regions of linkage of Siegmund et al. All estimates are for markers that were significantly associated with AD at alleles or genotypes in either family or case-control data. Estimates are for the homozygote major allele (11) as the referent group versus the other 2 genotypes (221, 122).
1Homozygous minor allele vs. homozygous major allele - 22 genotype compared to 11 (referent group)
2Heterozygote vs. homozygous major allele -12 genotype compared to 11 (referent group)
CASE-CONTROL DATA – SINGLE LOCUS RESULTS
The results of tests at single loci from the case-control data are in Table 4. Three markers in 2 genes significantly deviated from HWE in controls. One was the PZP marker rs12230214, minor allele frequency (MAF): 0.28 (p = 0.01). The AGT markers rs5050, MAF: 0.16 (p = 0.04) and rs4762, MAF: 0.123 (p = 0.01) also significantly deviated from HWE.
Allele and genotype frequency differences among controls between males and females were significant at 4 markers in 4 genes. These tests were conducted to make observations regarding potential confounding by sex where sampling error had caused association of autosomal alleles and genotypes with sex in controls. Such spurious associations in the data might lead to confounding since there is an association between sex and AD.
PZP marker rs12230214 (allele p = 0.01, genotype p = 0.05), LRP1 marker rs1800127 (allele p = 0.03, genotype p = 0.03), LRRTM3 marker rs942780 (allele p = 0.01, genotype p = 0.02), A2M marker rs3832852 (allele p = 0.01, genotype p = 0.02) had significantly different frequencies by sex in controls at both alleles and genotypes. Each of these markers was tested separately in males and females for HWE and allele and genotype frequency differences between cases and controls. Among these tests, significant deviations from HWE were found in control females for PZP marker rs12230214 (p = 0.02).
Nominally significant single-locus differences in allele or genotype frequency between cases and controls were observed at 3 markers in 2 genes. One marker in ACE was nominally significantly associated with disease at alleles. rs4343 (A/G) MAF: 0.46, a synonymous SNP in exon 16 (allele p = 0.05, genotype p = 0.14). Two markers in CTNNA3 were nominally significantly associated with disease. The markers rs6480140 (A/C) MAF: 0.37, a SNP in intron 14 (allele p = 0.61, genotype p = 0.01) and rs997225 (A/G), a SNP in intron 10 (allele p = 0.03, genotype p = 0.08). The APOE marker MAF: 0.09, (allele p < 0.001, genotype p < 0.001) was statistically significantly associated with disease.
Merged p-values for the family and case-control single-locus tests were analyzed using Fisher’s method [Fisher, 1950]. This approach allows for the evidence against the null hypothesis across tests to be combined into a single statistic for each null hypothesis. Global p-values were used from the family-based tests on genotypes. The results of this analysis are presented in Supplementary Table 1. Several markers in ACE were nominally significant at alleles and trending at genotypes. CTNNA3 marker rs7074454 was also nominally significant at alleles and trending at genotypes. A2M SNP rs3832852 was nominally significant at alleles and genotypes. Both of these SNPs had effect size estimates which indicated opposite risk alleles (Figure 2, Figure 3). Again, Only APOE survived a correction for multiple tests.
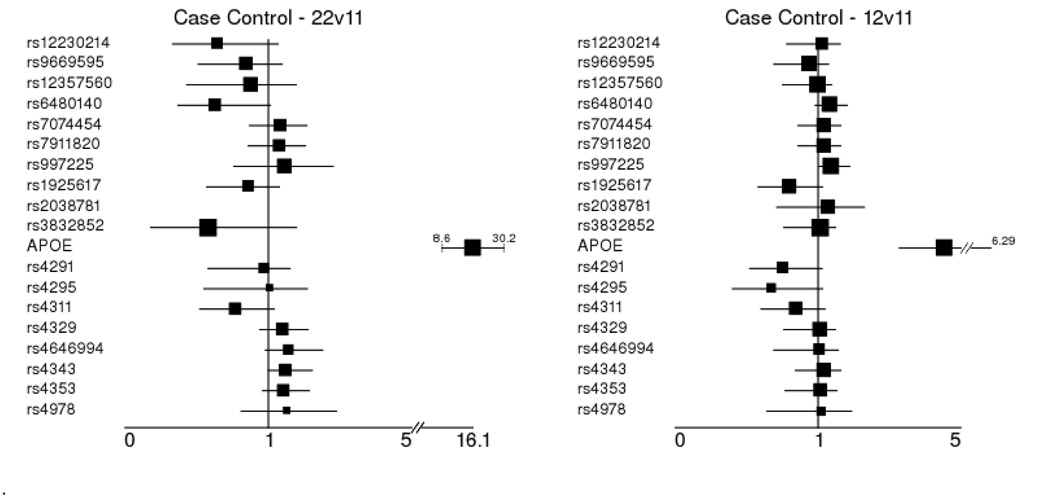
To estimate the effect of each significant finding from family or case-control data, odds ratios and 95% confidence intervals using the major allele homozygote as the referent group were estimated in the case-control data using logistic regression from the STATA statistical software package (STATA). Since the markers associating with LOAD did not significantly differ in genotype frequency by sex, no adjustment for confounding by sex was performed. Also, since no difference was detected between age of onset and controls, no adjustment for age was performed. These results are presented in Figure 3. Of note in these results are those loci which demonstrated nominal association in the family sample. Statistically significant effects were detected at APOE ε4 homozygotes (OR 16.1 95% CI = 8.6–30.2), APOE ε4 heterozygotes (OR 4.55 95% CI = 3.28–6.29). Nominally significant effects were estimated at the CTNNA3 SNP rs997225 GA heterozygote (OR 1.39 95% CI = 1.02–1.89) and ACE SNP rs4343 for the minor allele homozygote, (OR 1.49 95% CI = 1.0–2.23). Markers in ACE were also assessed for effect size adjusted for hypertension status. No regression term for any marker was statistically significant in that analysis, but the OR point estimates did not change, indicating that hypertension was not a confounder for those variables.
Figure 3.
Odds ratio effect size estimates for significant single-locus associations from case-control data. All estimates are for markers that were significantly associated with AD at alleles or genotypes in either family or case-control data. Estimates are for the homozygote major allele (11) as the referent group versus the other 2 genotypes (221, 122).
1Homozygous minor allele vs. homozygous major allele - 22 genotype compared to 11 (referent group)
2Heterozygote vs. homozygous major allele - 12 genotype compared to 11 (referent group)
HAPLOTYPE RESULTS
Haplotype analysis was performed across all candidate markers in pairwise LD as defined by a D’ of 0.95 or greater in the family data with APL using a 3-locus sliding window. These tests identified overlapping 3-locus haplotypes in the ACE gene that were significantly associated with AD in the family data set. Results of this procedure are in Table 5a. These results suggest a consistent signal of association with disease on a common haplotype background throughout these ACE markers. This signal is from a chromosome containing an array of minor alleles at each of these markers. This diffuse association signal is detectable at each individual marker, but this phenomenon is also observed in the case-control data, which makes the family result worthy of note. Also, the p-values observed at these overlapping 3-locus haplotypes are smaller than those for most of the single-locus statistics.
Table 5.
Significant family and case-control data haplotype association in ACE
| a. Family data | |||||
|---|---|---|---|---|---|
| APL haplotypes 3-Marker Scan |
Haplotype | Relative Frequency | P-value |
||
| Gene | Markers | Haplotype | Global | ||
| ACE | rs4291 – rs4295 | AG | 0.495 | 0.383 | 0.672 |
| rs4311 – rs4329 – rs46469941 | TGI | 0.460 | 0.045 | 0.290 | |
| rs4329 – rs46469941 – rs4343 | GIA | 0.450 | 0.013 | 0.020 | |
| rs46469941 – rs4343 – rs4353 | IAG | 0.450 | 0.004 | 0.004 | |
| rs4343 – rs4353 – rs4978 | AGC | 0.450 | 0.003 | 0.020 | |
| b. Case-control data | |||||||
|---|---|---|---|---|---|---|---|
| Haplo.stats 3-Marker Scan |
Haplotype | Relative Frequency |
OR | CI | P-value | ||
| Gene | Markers | Cases | Controls | ||||
| ACE | rs4291 - rs4295 | AG | 0.65 | 0.61 | 1.13 | 0.89–1.45 | 0.338 |
| rs4311 - rs4329 - rs46469941 | TGI | 0.48 | 0.43 | 1.21 | 0.98–1.48 | 0.06 | |
| rs4329 - rs46469941 - rs4343 | GIA | 0.48 | 0.43 | 1.22 | 1–1.49 | 0.046 | |
| rs46469941 - rs4343 - rs4353 | IAG | 0.48 | 0.43 | 1.23 | 1.01–1.51 | 0.048 | |
| rs4343 - rs4353 - rs4978 | AGC | 0.48 | 0.43 | 1.22 | 1.01–1.50 | 0.049 | |
| rs4311 - rs4329 - rs4646994 - rs4343 - rs4353 - rs4978 | TGIAGC | 0.48 | 0.42 | 1.22 | 1.01–1.50 | 0.041 | |
287 base pair Alu repeat: D major allele (289bp absent), I minor allele (289bp present)
289 base pair Alu repeat: D major allele (289bp absent), I minor allele (289bp present)
In the family data, the ACE gene contained several significant markers and overlapping associated haplotypes. No other regions in the family sample contained significant haplotypes. To follow up this observation, and to validate the haplotype findings in ACE from the family data, the Haplo.Stats software package was used to estimate haplotype frequencies and test haplotype associations in ACE in case-control data. Haplo.Stats uses an EM algorithm to estimate haplotype frequencies from unphased genotype data. The results of both sets of tests in family and case-control data are presented in Tables 5a and 5b. A sliding window scan of the markers in ACE, analogous to that performed in the family data, was conducted among markers in strong LD (r2 > 0.9, D’ > 0.95) in both datasets. This scan yielded an odds ratio and 95% confidence interval for all haplotypes versus the most common haplotype, and a chi-squared test for haplotype frequency differences between cases and controls. Every 3-locus haplotype in ACE between rs4311 and rs4978 had a chi-squared p-value < 0.05. The 2-locus haplotype including rs4291 and rs4295 was not significant in either dataset. The 6-locus haplotype including rs4311, rs4329, rs4646994, rs4343, rs4353, and rs4978 had an OR estimate very close to 1.2 and 95% confidence intervals at approximately 1.0–1.5, which was very similar to those estimates for the 3-locus sliding window through that region. This indicates that chromosomes in this area of the gene tend to be either all major or minor alleles with little recombination in two primary haplotypes.
Clade-based haplotype analysis was conducted using markers rs4291 and rs4343 as suggested in Katzov et al [Katzov et al., 2004]. We applied this model to our samples and further evaluated haplotype association to LOAD, observing similar results as the previous study using this approach [Katzov et al., 2004]. For exposure to the 2-marker haplotype corresponding with clade A (frequency: 0.35, A-A) versus clades B (frequency: 0.46, T-G) and C (frequency: 0.18, A-G), the p-value in the family sample was statistically significant at p = 0.0004, and for the case-control data, p = 0.029 (OR = 1.3, 95% CI = 1.06–1.54).
MULTILOCUS RESULTS
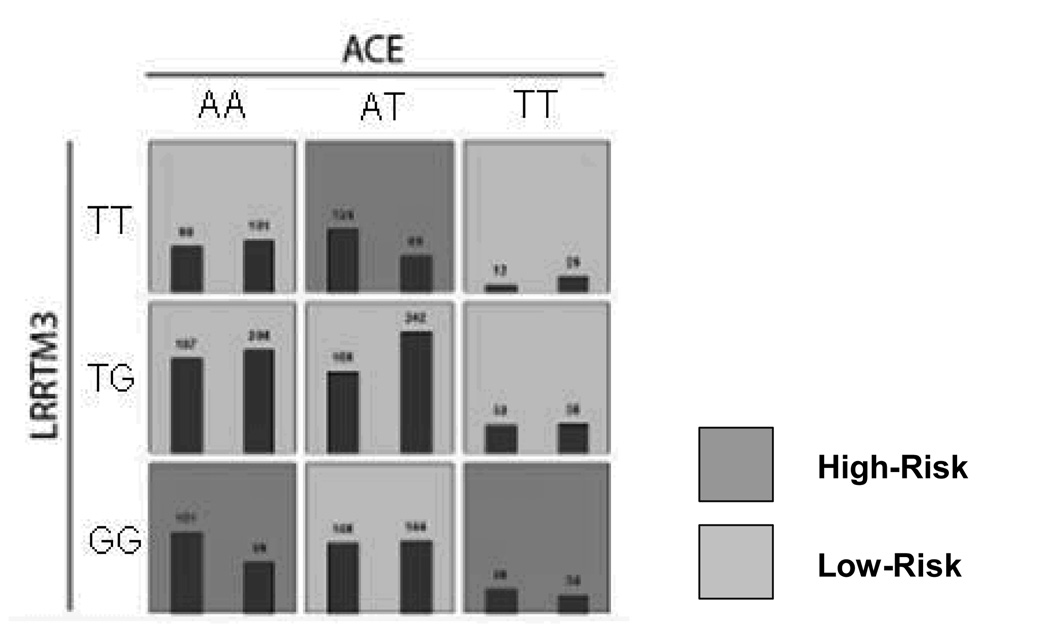
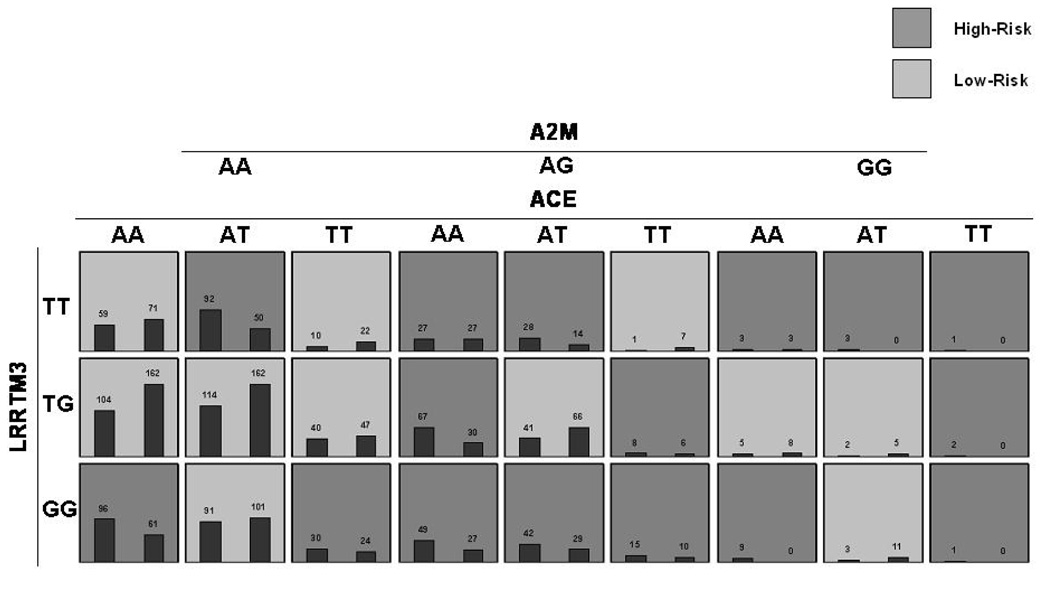
Having explored single-locus main effects at alleles, genotypes, and haplotypes, we began a search for multi-locus signals significantly associated with disease using the MDR-PDT in family data and MDR in case-control data. The MDR-PDT models are presented in Table 6 and Figures 4a and 4b. MDR and MDR-PDT were run with all markers and every model including the APOE marker was highly significant by the permutation test. Since the strength of the APOE signal obfuscated other potentially interesting multilocus models, the APOE locus was excluded from the search and a subset of tag SNPs were chosen from the data. Haplotype tag SNPs were chosen using the haploview software function tagger (r2 = 0.8, LOD = 3), and the best models were found by MDR and MDR-PDT. The best two and 3-locus models from the full data without APOE contained the same markers as those chosen from the tag SNP data for the MDR-PDT. This indicated that the signal observed at these models was detected by MDR-PDT, but the known issue of power loss with increasing numbers of markers caused the failure to reject. No MDR model was significant by the permutation test. The best MDR model was a 3-locus model including LRP1 SNP rs1800165, PZP SNP rs3213831, and PZP SNP rs10842971 (CVC 2/5, PE 43.41, p-value = 0.34). Two significant signals were found by MDR-PDT. The 2-locus model included rs1925617 in LRRTM3 and rs4295 in ACE (MDR-PDT statistic p < 0.001). The 3-locus model included rs1925617 in LRRTM3, rs4291 in ACE, and rs1800433 in A2M (MDR-PDT statistic p < 0.001).
Table 6.
Summary of MDR-PDT results
| # of Loci | Best Model For Each Interaction | SNPs | T Statistic | Classification Accuracy (%) |
|---|---|---|---|---|
| 1 | [ LRRTM3 ] | rs1925617 | 3.32 | 54.32 |
| 2 | [ LRRTM3-ACE ] | rs1925617-rs4291 | 4.49 | 56.89 |
| 3 | [ LRRTM3-ACE-A2M ] | rs1925617-rs4291-rs1800433 | 5.65 | 59.58 |
Figure 4.
Figure 4a. MDR-PDT two locus model. Summary of multilocus interactions between LRRTM and, ACE. Each multifactorial cell is labeled as “high risk” or “low risk”. For each multifactorial combination, empirical distributions of cases (left bar in cell) and controls (right bar in cell) are shown. The classification accuracy for the 2-locus model is 56.89% (permuted p-value <0.001), with a t-statistic of 4.49 (permuted p-value 0.001).
Figure 4b. MDR-PDT three-locus model. Summary of multilocus interactions between LRRTM3, ACE and A2M. Each multifactorial cell is labeled as “high risk” or “low risk”. For each multifactorial combination, empirical distributions of cases (left bar in cell) and controls (right bar in cell) are shown. The classification accuracy for the 3-locus model is 59.58% (permuted p-value = 0.001) with an MDR-PDT-statistic of 5.65 (p-value = 0.001). Of note is the consistent pattern of high-risk cells between ACE and LRRTM3 at the AA genotype of A2M with the 2-locus model, but more high-risk cells at the AT and TT genotypes of A2M, demonstrating the pattern of effect modification on the 2-locus model by A2M.
DISCUSSION
These results highlight the ACE gene as a risk factor in LOAD. In both family and case-control samples, significant associations were observed when considering ACE haplotypes. Notably in the case-control samples, only one single-locus test was marginally significant at rs4343 for the test of allelic frequency differences, but the haplotype tests on specific, overlapping sets of alleles were nominally significant. This result at rs4343 is consistent with previous work in ACE [Katzov et al., 2004; Kehoe et al., 2003]. The p-values from the family data haplotype analysis were also smaller than those from the single-locus analysis, suggesting the variation influencing LOAD was more efficiently captured when considering the entire region. When using markers that denote ancestral clades in European populations, the association signals were even stronger, which is consistent with the results of Katzov et al [Katzov et al., 2004]. This finding strongly supports the existence of a genetic background in the ACE locus in European descent Caucasians that is associated with LOAD.
Evidence for ACE association was found across many studies in recent meta-analyses of association results [Bertram et al., 2007a; Lehmann et al., 2005]. Haplotype associations have also been previously observed in ACE for AD in five independent case-control samples [Kehoe et al., 2003], including a large Swedish sample [Katzov et al., 2004], and in an inbred Israeli Arab sample [Meng et al., 2006]. In the Meng et al paper, the haplotype distribution was quite different from that observed in these samples and the alleles in the associated haplotype were opposite to those reported here. These results agree with the Kehoe et al study, both with regard to approximate effect size and haplotypes, and the Katzov et al study, with regard to A clade association. Additionally, cladistic analysis of haplotypes yielded statistically significant results compared with the nominally significant analysis of larger numbers of SNPs. This result further supports the use of ancestral clades in the ACE locus to evaluate associations to traits in European descent populations. We are unaware of a study evaluating ACE association with LOAD with a larger combined sample not featuring meta-analysis methods in the literature.
ACE plasma concentrations have been shown to be increased in persons bearing a 289bp deletion in intron 16 of the gene [Rigat et al., 1990]. This ACE I/D has also been shown to be a risk locus for cardiovascular disease [Hessner et al., 2001; Malik et al., 1997], which may share some common etiological factors with AD [Breteler et al., 1994; Hofman et al., 1997]. The ACE intron 16 I/D has been previously reported to associate with AD in Caucasians [Alvarez et al., 1999a; Kehoe et al., 1999; Kolsch et al., 2005; Mattila et al., 2000], Japanese [Hu et al., 1999], and Chinese samples [Cheng et al., 2002; Wang et al., 2006; Yang et al., 2000]. This association has a plausible biological explanation, since ACE degrades Aβ peptide in vitro [Hu et al., 2001; Hemming and Selkoe, 2005; Oba et al., 2005; Sun et al., 2008; Toropygin et al., 2008], and the insertion allele, which is on the A clade haplotype [Farrall et al., 1999], is associated with decreased plasma levels of the ACE protein [Hemming and Selkoe, 2005; Rigat et al., 1990; Tiret et al., 1992]. The biological connection between ACE and AD has been explored with clinical trials that provided evidence favoring use of ACE inhibitors to treat AD [Hanon and Forette, 2004] or dementia [Tzourio et al., 2003], and other studies which observed no benefit of ACE inhibition, but did observe significant benefit of hypertension therapy in a large prospective study [Khachaturian et al., 2006]. These studies are reviewed in [Kehoe and Wilcock, 2007]. Additionally, in vivo studies in mouse models have demonstrated that ACE inhibitors can improve cognitive performance and reduce amyloid protein levels [Wang et al., 2007; Zou et al., 2007].
Overall, association results in the ACE gene for Caucasians have been inconsistent in past studies. In 31 case-control association studies in Caucasians with markers in ACE reviewed by [Bertram et al., 2007a], there were 12 positive findings, 15 negative findings and 4 trends suggesting association between ACE and AD. The sample sizes were larger in studies where positive associations were observed (1-sided t-test p-value for cases = 0.04, p-value for controls = 0.03, p-value for overall sample size = 0.02), suggesting that differential power might explain some of the previous inconsistency. Additionally, A2M associations have been inconsistent, where in Alzgene.org there have been six positive associations, two trends and 33 negative associations observed in case-control Caucasian samples for markers in that gene. However, in family data there were three positive and one trend associations. Most of those studies were performed on rs3832852, where we observed inconsistent results for allele effects. There was not a significant sample size difference across study outcomes for sample sizes. For CTNNA3, there were seven negative and one positive association in case-control samples, but in family samples three of four studies found variants associating with AD. Evidence exists for associations at each of these genes, and interactions among them may explain some of the previous inconsistency [Ioannidis, 2007].
Interactions likely are relevant to genetic epidemiology of LOAD, and the MDR-PDT provides a capability to search for such effects in family data. MDR, the analogous approach in case-control data, has been used to find several interactive effects for various phenotypes. MDR has been used to detect genetic interactions contributing to risk in several diseases. Some examples are: sporadic breast cancer [Ritchie et al., 2001], essential hypertension [Moore and Williams, 2002; Williams et al., 2004], atrial fibrillation [Tsai et al., 2004], type II diabetes [Cho et al., 2004], coronary artery calcification [Bastone et al., 2004], myocardial infarction [Coffey et al., 2004], schizophrenia [Qin et al., 2005], and amyloid polyneuropathy [Soares et al., 2005]. MDR-PDT has been shown in simulation to have better power than MDR when families are large, as in these data [Martin et al., 2006].
The multilocus model presented here suggests that there may be a functional axis of effects predicting LOAD in these data. It is also possible that this is an artifact arising from the presence of main effects at some of these loci. The non-replication from the case-control dataset casts some further doubt on the strength of this model. There is a chance that this is a type II error, caused by a smaller case-control sample, or by the similar ages of onset of cases to ages of examination in controls, leading to loss of power since controls were not sufficiently aged to be certain not to develop LOAD in the future. We caution that these results should be considered preliminary with regard to the presence of effect modification among these loci. Recent experiments in simulation show that fitting regression models to evaluate effect modification in the same data where MDR or MDR-PDT models are found is extremely unreliable (unpublished results). Independent samples should be obtained to properly test after a search for interactions, or a valid test procedure for the null hypothesis of no interaction should be developed. These candidates also are all related to Aβ clearance, providing a biological rationale for this multilocus model.
Future directions for these investigations into the mechanism underlying late-onset AD should include further investigation of genes in the Aβ degradation pathway as well as functional studies targeting potential molecular etiologies of LOAD involving ACE. Plasma levels of ACE and putative downstream targets relevant to AD should be included in future study designs to make observations regarding coordinate regulation, feedback systems, and continuous measurements further explaining this pattern of association. The presence of A2M and CTNNA3/LRRTM3 SNPs in the MDR-PDT model also point to Aβ accumulation as a factor predicting late-onset AD, as these genes all relate to Aβ clearance. It may be that we have already discovered the main causes of AD in the constellation of weak main effects that have been observed to date. The attributable risk of the ACE haplotype alone explains about 16.6% of late-onset AD cases among those exposed to the haplotype. For the entire Caucasian population, the population attributable risk (PAR) for the haplotype explains about 8%, or 320,000 late-onset Alzheimer’s cases. This contrasts with the single locus PAR of 35% reported by [Kehoe et al., 2003] for rs4343, and the large effect size in ACE reported by [Meng et al., 2006] in an inbred population. When we apply the same dominant model from our case-control data as in the Kehoe paper, the PAR is 21%. For the effect size and exposure rate estimate from our family data, the PAR is 28% for rs4343. The A allele of rs4343 is also reported as associated in the Kehoe et al haplotype, in the Katzov cladistic model, and in our data. The Alzgene meta-analysis for this SNP also shows a significant effect at this locus [Bertram et al., 2007a]. This concordance of statistical and laboratory results implicates ACE variation as not only contributing modest risk, but also supports the biological hypothesis regarding plasma concentrations of ACE and the relationship to Aβ concentrations.
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health grant AG20135. We would also like to thank Dr. Chun Li for providing feedback. We would also like to thank Dana Hancock for providing the code for the conditional logistic regression in SAS.
REFERENCES
- Alvarez R, Alvarez V, Lahoz CH, Martinez C, Pena J, Sanchez JM, Guisasola LM, Salas-Puig J, Moris G, Vidal JA, et al. Angiotensin converting enzyme and endothelial nitric oxide synthase DNA polymorphisms and late onset Alzheimer's disease. J Neurol Neurosurg Psychiatry. 1999a;67(6):733–736. doi: 10.1136/jnnp.67.6.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez V, Alvarez R, Lahoz CH, Martinez C, Pena J, Guisasola LM, Salas-Puig J, Moris G, Uria D, Menes BB, et al. Association between an alpha(2) macroglobulin DNA polymorphism and late-onset Alzheimer's disease. Biochem Biophys Res Commun. 1999b;264(1):48–50. doi: 10.1006/bbrc.1999.1295. [DOI] [PubMed] [Google Scholar]
- Bales KR, Verina T, Cummins DJ, Du Y, Dodel RC, Saura J, Fishman CE, DeLong CA, Piccardo P, Petegnief V, et al. Apolipoprotein E is essential for amyloid deposition in the APP(V717F) transgenic mouse model of Alzheimer's disease. Proc Natl Acad Sci U S A. 1999;96(26):15233–15238. doi: 10.1073/pnas.96.26.15233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bastone L, Reilly M, Rader DJ, Foulkes AS. MDR and PRP: a comparison of methods for high-order genotype-phenotype associations. Hum Hered. 2004;58(2):82–92. doi: 10.1159/000083029. [DOI] [PubMed] [Google Scholar]
- Bergqvist D, Nilsson IM. Hereditary alpha 2-macroglobulin deficiency. Scand J Haematol. 1979;23(5):433–436. [PubMed] [Google Scholar]
- Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007a;39(1):17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- Bertram L, Mullin K, Parkinson M, Hsiao M, Moscarillo TJ, Wagner SL, Becker KD, Velicelebi G, Blacker D, Tanzi RE. Is alpha-T catenin (VR22) an Alzheimer's disease risk gene? J Med Genet. 2007b;44(1):e63. doi: 10.1136/jmg.2005.039263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacker D, Wilcox MA, Laird NM, Rodes L, Horvath SM, Go RC, Perry R, Watson B, Jr, Bassett SS, McInnis MG, et al. Alpha-2 macroglobulin is genetically associated with Alzheimer disease. Nat Genet. 1998;19(4):357–360. doi: 10.1038/1243. [DOI] [PubMed] [Google Scholar]
- Blennow K, Ricksten A, Prince JA, Brookes AJ, Emahazion T, Wasslavik C, Bogdanovic N, Andreasen N, Batsman S, Marcusson J, et al. No association between the alpha2-macroglobulin (A2M) deletion and Alzheimer's disease, and no change in A2M mRNA, protein, or protein expression. J Neural Transm. 2000;107(8–9):1065–1079. doi: 10.1007/s007020070052. [DOI] [PubMed] [Google Scholar]
- Breteler MM, Claus JJ, Grobbee DE, Hofman A. Cardiovascular disease and distribution of cognitive function in elderly people: the Rotterdam Study. BMJ. 1994;308(6944):1604–1608. doi: 10.1136/bmj.308.6944.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby V, Goossens S, Nowotny P, Hamilton G, Smemo S, Harold D, Turic D, Jehu L, Myers A, Womick M, et al. Alpha-T-catenin is expressed in human brain and interacts with the Wnt signaling pathway but is not responsible for linkage to chromosome 10 in Alzheimer's disease. Neuromolecular Med. 2004;5(2):133–146. doi: 10.1385/NMM:5:2:133. [DOI] [PubMed] [Google Scholar]
- Chapman J, Wang N, Treves TA, Korczyn AD, Bornstein NM. ACE, MTHFR, factor V Leiden, and APOE polymorphisms in patients with vascular and Alzheimer's dementia. Stroke. 1998;29(7):1401–1404. doi: 10.1161/01.str.29.7.1401. [DOI] [PubMed] [Google Scholar]
- Chartier-Harlin MC, Parfitt M, Legrain S, Perez-Tur J, Brousseau T, Evans A, Berr C, Vidal O, Roques P, Gourlet V, et al. Apolipoprotein E, epsilon 4 allele as a major risk factor for sporadic early and late-onset forms of Alzheimer's disease: analysis of the 19q13.2 chromosomal region. Hum Mol Genet. 1994;3(4):569–574. doi: 10.1093/hmg/3.4.569. [DOI] [PubMed] [Google Scholar]
- Cheng CY, Hong CJ, Liu HC, Liu TY, Tsai SJ. Study of the association between Alzheimer's disease and angiotensin-converting enzyme gene polymorphism using DNA from lymphocytes. Eur Neurol. 2002;47(1):26–29. doi: 10.1159/000047943. [DOI] [PubMed] [Google Scholar]
- Cho YM, Ritchie MD, Moore JH, Park JY, Lee KU, Shin HD, Lee HK, Park KS. Multifactor-dimensionality reduction shows a two-locus interaction associated with Type 2 diabetes mellitus. Diabetologia. 2004;47(3):549–554. doi: 10.1007/s00125-003-1321-3. [DOI] [PubMed] [Google Scholar]
- Chung RH, Hauser ER, Martin ER. The APL test: extension to general nuclear families and haplotypes and examination of its robustness. Hum Hered. 2006;61(4):189–199. doi: 10.1159/000094774. [DOI] [PubMed] [Google Scholar]
- Clark AG. Inference of haplotypes from PCR-amplified samples of diploid populations. Mol Biol Evol. 1990;7(2):111–122. doi: 10.1093/oxfordjournals.molbev.a040591. [DOI] [PubMed] [Google Scholar]
- Coffey CS, Hebert PR, Ritchie MD, Krumholz HM, Gaziano JM, Ridker PM, Brown NJ, Vaughan DE, Moore JH. An application of conditional logistic regression and multifactor dimensionality reduction for detecting gene-gene interactions on risk of myocardial infarction: the importance of model validation. BMC Bioinformatics. 2004:549. doi: 10.1186/1471-2105-5-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261(5123):921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- Eckman EA, Adams SK, Troendle FJ, Stodola BA, Kahn MA, Fauq AH, Xiao HD, Bernstein KE, Eckman CB. Regulation of steady-state beta-amyloid levels in the brain by neprilysin and endothelin-converting enzyme but not angiotensin-converting enzyme. J Biol Chem. 2006;281(41):30471–30478. doi: 10.1074/jbc.M605827200. [DOI] [PubMed] [Google Scholar]
- Ertekin-Taner N, Graff-Radford N, Younkin LH, Eckman C, Baker M, Adamson J, Ronald J, Blangero J, Hutton M, Younkin SG. Linkage of plasma Abeta42 to a quantitative locus on chromosome 10 in late-onset Alzheimer's disease pedigrees. Science. 2000;290(5500):2303–2304. doi: 10.1126/science.290.5500.2303. [DOI] [PubMed] [Google Scholar]
- Ertekin-Taner N, Ronald J, Asahara H, Younkin L, Hella M, Jain S, Gnida E, Younkin S, Fadale D, Ohyagi Y, et al. Fine mapping of the alpha-T catenin gene to a quantitative trait locus on chromosome 10 in late-onset Alzheimer's disease pedigrees. Hum Mol Genet. 2003;12(23):3133–3143. doi: 10.1093/hmg/ddg343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Slatkin M. Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol Biol Evol. 1995;12(5):921–927. doi: 10.1093/oxfordjournals.molbev.a040269. [DOI] [PubMed] [Google Scholar]
- Farrall M, Keavney B, McKenzie C, Delepine M, Matsuda F, Lathrop GM. Fine-mapping of an ancestral recombination breakpoint in DCP1. Nat Genet. 1999;23(3):270–271. doi: 10.1038/15449. [DOI] [PubMed] [Google Scholar]
- Fisher RA. Statistical methods for research workers. 1950 11xv-354. [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, et al. The structure of haplotype blocks in the human genome. Science. 2002;296(5576):2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Goate A. Segregation of a missense mutation in the amyloid beta-protein precursor gene with familial Alzheimer's disease. J Alzheimers Dis. 2006;9(3 Suppl):341–347. doi: 10.3233/jad-2006-9s338. [DOI] [PubMed] [Google Scholar]
- Hahn LW, Ritchie MD, Moore JH. Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics. 2003;19(3):376–382. doi: 10.1093/bioinformatics/btf869. [DOI] [PubMed] [Google Scholar]
- Hanon O, Forette F. Prevention of dementia: lessons from SYST-EUR and PROGRESS. J Neurol Sci. 2004;226(1–2):71–74. doi: 10.1016/j.jns.2004.09.015. [DOI] [PubMed] [Google Scholar]
- Hardy J. Amyloid, the presenilins and Alzheimer's disease. Trends Neurosci. 1997;20(4):154–159. doi: 10.1016/s0166-2236(96)01030-2. [DOI] [PubMed] [Google Scholar]
- Hardy J, Myers A, Wavrant-De VF. Problems and solutions in the genetic analysis of late-onset Alzheimer's disease. Neurodegener Dis. 2004;1(4–5):213–217. doi: 10.1159/000080988. [DOI] [PubMed] [Google Scholar]
- Hemming ML, Selkoe DJ. Amyloid beta-protein is degraded by cellular angiotensin-converting enzyme (ACE) and elevated by an ACE inhibitor. J Biol Chem. 2005;280(45):37644–37650. doi: 10.1074/jbc.M508460200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemming ML, Selkoe DJ, Farris W. Effects of prolonged angiotensin-converting enzyme inhibitor treatment on amyloid beta-protein metabolism in mouse models of Alzheimer disease. Neurobiol Dis. 2007;26(1):273–281. doi: 10.1016/j.nbd.2007.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson AS, Easteal S, Jorm AF, Mackinnon AJ, Korten AE, Christensen H, Croft L, Jacomb PA. Apolipoprotein E allele epsilon 4, dementia, and cognitive decline in a population sample. Lancet. 1995;346(8987):1387–1390. doi: 10.1016/s0140-6736(95)92405-1. [DOI] [PubMed] [Google Scholar]
- Hessner MJ, Dinauer DM, Kwiatkowski R, Neri B, Raife TJ. Age-dependent prevalence of vascular disease-associated polymorphisms among 2689 volunteer blood donors. Clin Chem. 2001;47(10):1879–1884. [PubMed] [Google Scholar]
- Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4(2):45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- Hofman A, Ott A, Breteler MM, Bots ML, Slooter AJ, van HF, van Duijn CN, Van BC, Grobbee DE. Atherosclerosis, apolipoprotein E, and prevalence of dementia and Alzheimer's disease in the Rotterdam Study. Lancet. 1997;349(9046):151–154. doi: 10.1016/S0140-6736(96)09328-2. [DOI] [PubMed] [Google Scholar]
- Hu J, Igarashi A, Kamata M, Nakagawa H. Angiotensin-converting enzyme degrades Alzheimer amyloid beta-peptide (A beta ); retards A beta aggregation, deposition, fibril formation; and inhibits cytotoxicity. J Biol Chem. 2001;276(51):47863–47868. doi: 10.1074/jbc.M104068200. [DOI] [PubMed] [Google Scholar]
- Hu J, Miyatake F, Aizu Y, Nakagawa H, Nakamura S, Tamaoka A, Takahash R, Urakami K, Shoji M. Angiotensin-converting enzyme genotype is associated with Alzheimer disease in the Japanese population. Neurosci Lett. 1999;277(1):65–67. doi: 10.1016/s0304-3940(99)00827-7. [DOI] [PubMed] [Google Scholar]
- Ioannidis JP. Non-replication and inconsistency in the genome-wide association setting. Hum Hered. 2007;64(4):203–213. doi: 10.1159/000103512. [DOI] [PubMed] [Google Scholar]
- Katzov H, Bennet AM, Kehoe P, Wiman B, Gatz M, Blennow K, Lenhard B, Pedersen NL, de FU, Prince JA. A cladistic model of ACE sequence variation with implications for myocardial infarction, Alzheimer disease and obesity. Hum Mol Genet. 2004;13(21):2647–2657. doi: 10.1093/hmg/ddh286. [DOI] [PubMed] [Google Scholar]
- Keavney B, McKenzie CA, Connell JM, Julier C, Ratcliffe PJ, Sobel E, Lathrop M, Farrall M. Measured haplotype analysis of the angiotensin-I converting enzyme gene. Hum Mol Genet. 1998;7(11):1745–1751. doi: 10.1093/hmg/7.11.1745. [DOI] [PubMed] [Google Scholar]
- Kehoe PG, Katzov H, Feuk L, Bennet AM, Johansson B, Wiman B, de FU, Cairns NJ, Wilcock GK, Brookes AJ, et al. Haplotypes extending across ACE are associated with Alzheimer's disease. Hum Mol Genet. 2003;12(8):859–867. doi: 10.1093/hmg/ddg094. [DOI] [PubMed] [Google Scholar]
- Kehoe PG, Russ C, McIlory S, Williams H, Holmans P, Holmes C, Liolitsa D, Vahidassr D, Powell J, McGleenon B, et al. Variation in DCP1, encoding ACE, is associated with susceptibility to Alzheimer disease. Nat Genet. 1999;21(1):71–72. doi: 10.1038/5009. [DOI] [PubMed] [Google Scholar]
- Kehoe PG, Wilcock GK. Is inhibition of the renin-angiotensin system a new treatment option for Alzheimer's disease? Lancet Neurol. 2007;6(4):373–378. doi: 10.1016/S1474-4422(07)70077-7. [DOI] [PubMed] [Google Scholar]
- Khachaturian AS, Zandi PP, Lyketsos CG, Hayden KM, Skoog I, Norton MC, Tschanz JT, Mayer LS, Welsh-Bohmer KA, Breitner JC. Antihypertensive medication use and incident Alzheimer disease: the Cache County Study. Arch Neurol. 2006;63(5):686–692. doi: 10.1001/archneur.63.5.noc60013. [DOI] [PubMed] [Google Scholar]
- Kolsch H, Jessen F, Freymann N, Kreis M, Hentschel F, Maier W, Heun R. ACE I/D polymorphism is a risk factor of Alzheimer's disease but not of vascular dementia. Neurosci Lett. 2005;377(1):37–39. doi: 10.1016/j.neulet.2004.11.062. [DOI] [PubMed] [Google Scholar]
- Lehmann DJ, Cortina-Borja M, Warden DR, Smith AD, Sleegers K, Prince JA, van Duijn CM, Kehoe PG. Large meta-analysis establishes the ACE insertion-deletion polymorphism as a marker of Alzheimer's disease. Am J Epidemiol. 2005;162(4):305–317. doi: 10.1093/aje/kwi202. [DOI] [PubMed] [Google Scholar]
- Levy-Lahad E, Wasco W, Poorkaj P, Romano DM, Oshima J, Pettingell WH, Yu CE, Jondro PD, Schmidt SD, Wang K, et al. Candidate gene for the chromosome 1 familial Alzheimer's disease locus. Science. 1995;269(5226):973–977. doi: 10.1126/science.7638622. [DOI] [PubMed] [Google Scholar]
- Li H, Wetten S, Li L, St Jean PL, Upmanyu R, Surh L, Hosford D, Barnes MR, Briley JD, Borrie M, et al. Candidate single-nucleotide polymorphisms from a genomewide association study of Alzheimer disease. Arch Neurol. 2008;65(1):45–53. doi: 10.1001/archneurol.2007.3. [DOI] [PubMed] [Google Scholar]
- Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95(3):221–227. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- Liang X, Martin ER, Schnetz-Boutaud N, Bartlett J, Anderson B, Zuchner S, Gwirtsman H, Schmechel D, Carney R, Gilbert JR, et al. Effect of heterogeneity on the chromosome 10 risk in late-onset Alzheimer disease. Hum Mutat. 2007;28(11):1065–1073. doi: 10.1002/humu.20567. [DOI] [PubMed] [Google Scholar]
- Long JC, Williams RC, Urbanek M. An E-M algorithm and testing strategy for multiple-locus haplotypes. Am J Hum Genet. 1995;56(3):799–810. [PMC free article] [PubMed] [Google Scholar]
- Lucotte G, Visvikis S, Leininger-Muler B, David F, Berriche S, Reveilleau S, Couderc R, Babron MC, Aguillon D, Siest G. Association of apolipoprotein E allele epsilon 4 with late-onset sporadic Alzheimer's disease. Am J Med Genet. 1994;54(3):286–288. doi: 10.1002/ajmg.1320540319. [DOI] [PubMed] [Google Scholar]
- Malik FS, Lavie CJ, Mehra MR, Milani RV, Re RN. Renin-angiotensin system: genes to bedside. Am Heart J. 1997;134(3):514–526. doi: 10.1016/s0002-8703(97)70089-9. [DOI] [PubMed] [Google Scholar]
- Martin ER, Bass MP, Gilbert JR, Pericak-Vance MA, Hauser ER. Genotype-based association test for general pedigrees: the genotype-PDT. Genet Epidemiol. 2003a;25(3):203–213. doi: 10.1002/gepi.10258. [DOI] [PubMed] [Google Scholar]
- Martin ER, Bass MP, Hauser ER, Kaplan NL. Accounting for linkage in family-based tests of association with missing parental genotypes. Am J Hum Genet. 2003b;73(5):1016–1026. doi: 10.1086/378779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin ER, Bronson PG, Li YJ, Wall N, Chung RH, Schmechel DE, Small G, Xu PT, Bartlett J, Schnetz-Boutaud N, et al. Interaction between the alpha-T catenin gene (VR22) and APOE in Alzheimer's disease. J Med Genet. 2005;42(10):787–792. doi: 10.1136/jmg.2004.029553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin ER, Monks SA, Warren LL, Kaplan NL. A test for linkage and association in general pedigrees: the pedigree disequilibrium test. Am J Hum Genet. 2000;67(1):146–154. doi: 10.1086/302957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin ER, Ritchie MD, Hahn L, Kang S, Moore JH. A novel method to identify gene-gene effects in nuclear families: the MDR-PDT. Genet Epidemiol. 2006;30(2):111–123. doi: 10.1002/gepi.20128. [DOI] [PubMed] [Google Scholar]
- Mattila KM, Rinne JO, Roytta M, Laippala P, Pietila T, Kalimo H, Koivula T, Frey H, Lehtimaki T. Dipeptidyl carboxypeptidase 1 (DCP1) and butyrylcholinesterase (BCHE) gene interactions with the apolipoprotein E epsilon4 allele as risk factors in Alzheimer's disease and in Parkinson's disease with coexisting Alzheimer pathology. J Med Genet. 2000;37(10):766–770. doi: 10.1136/jmg.37.10.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y, Baldwin CT, Bowirrat A, Waraska K, Inzelberg R, Friedland RP, Farrer LA. Association of polymorphisms in the Angiotensin-converting enzyme gene with Alzheimer disease in an Israeli Arab community. Am J Hum Genet. 2006;78(5):871–877. doi: 10.1086/503687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miners JS, Ashby E, Van HZ, Chalmers KA, Palmer LE, Love S, Kehoe PG. Angiotensin-converting enzyme (ACE) levels and activity in Alzheimer's disease, and relationship of perivascular ACE-1 to cerebral amyloid angiopathy. Neuropathol Appl Neurobiol. 2008a;34(2):181–193. doi: 10.1111/j.1365-2990.2007.00885.x. [DOI] [PubMed] [Google Scholar]
- Miners JS, Baig S, Palmer J, Palmer LE, Kehoe PG, Love S. Abeta-degrading enzymes in Alzheimer's disease. Brain Pathol. 2008b;18(2):240–252. doi: 10.1111/j.1750-3639.2008.00132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JH, Williams SM. New strategies for identifying gene-gene interactions in hypertension. Ann Med. 2002;34(2):88–95. doi: 10.1080/07853890252953473. [DOI] [PubMed] [Google Scholar]
- Nalivaeva NN, Fisk LR, Belyaev ND, Turner AJ. Amyloid-degrading enzymes as therapeutic targets in Alzheimer's disease. Curr Alzheimer Res. 2008;5(2):212–224. doi: 10.2174/156720508783954785. [DOI] [PubMed] [Google Scholar]
- Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74(4):765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oba R, Igarashi A, Kamata M, Nagata K, Takano S, Nakagawa H. The N-terminal active centre of human angiotensin-converting enzyme degrades Alzheimer amyloid beta-peptide. Eur J Neurosci. 2005;21(3):733–740. doi: 10.1111/j.1460-9568.2005.03912.x. [DOI] [PubMed] [Google Scholar]
- Ohrui T, Matsui T, Yamaya M, Arai H, Ebihara S, Maruyama M, Sasaki H. Angiotensin-converting enzyme inhibitors and incidence of Alzheimer's disease in Japan. J Am Geriatr Soc. 2004;52(4):649–650. doi: 10.1111/j.1532-5415.2004.52178_7.x. [DOI] [PubMed] [Google Scholar]
- OMIM. Online Mendelian Inheritance in Man. McKusick-Nathans Institute for Genetic Medicine, Johns Hopkins University (Baltimore, MD) and National Center for Biotechnology Information, National Library of Medicine (Bethesda, MD); 2008. [Google Scholar]
- Qin S, Zhao X, Pan Y, Liu J, Feng G, Fu J, Bao J, Zhang Z, He L. An association study of the N-methyl-D-aspartate receptor NR1 subunit gene (GRIN1) and NR2B subunit gene (GRIN2B) in schizophrenia with universal DNA microarray. Eur J Hum Genet. 2005;13(7):807–814. doi: 10.1038/sj.ejhg.5201418. [DOI] [PubMed] [Google Scholar]
- Reid IA. Interactions between ANG II, sympathetic nervous system, and baroreceptor reflexes in regulation of blood pressure. Am J Physiol. 1992;262(6 Pt 1):E763–E778. doi: 10.1152/ajpendo.1992.262.6.E763. [DOI] [PubMed] [Google Scholar]
- Rieder MJ, Taylor SL, Clark AG, Nickerson DA. Sequence variation in the human angiotensin converting enzyme. Nat Genet. 1999;22(1):59–62. doi: 10.1038/8760. [DOI] [PubMed] [Google Scholar]
- Rigat B, Hubert C, henc-Gelas F, Cambien F, Corvol P, Soubrier F. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J Clin Invest. 1990;86(4):1343–1346. doi: 10.1172/JCI114844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie K, Kotzki PO, Touchon J, Cristol JP. Characteristics of Alzheimer's disease patients with and without ApoE4 allele. Lancet. 1996;348(9032):960. doi: 10.1016/S0140-6736(05)65371-8. [DOI] [PubMed] [Google Scholar]
- Ritchie MD, Hahn LW, Moore JH. Power of multifactor dimensionality reduction for detecting gene-gene interactions in the presence of genotyping error, missing data, phenocopy, and genetic heterogeneity. Genet Epidemiol. 2003;24(2):150–157. doi: 10.1002/gepi.10218. [DOI] [PubMed] [Google Scholar]
- Ritchie MD, Hahn LW, Roodi N, Bailey LR, Dupont WD, Parl FF, Moore JH. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am J Hum Genet. 2001;69(1):138–147. doi: 10.1086/321276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riva A, Kohane IS. A SNP-centric database for the investigation of the human genome. BMC Bioinformatics. 2004:533. doi: 10.1186/1471-2105-5-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogaev EI, Sherrington R, Rogaeva EA, Levesque G, Ikeda M, Liang Y, Chi H, Lin C, Holman K, Tsuda T, et al. Familial Alzheimer's disease in kindreds with missense mutations in a gene on chromosome 1 related to the Alzheimer's disease type 3 gene. Nature. 1995;376(6543):775–778. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- Rogaeva EA, Premkumar S, Grubber J, Serneels L, Scott WK, Kawarai T, Song Y, Hill DL, bou-Donia SM, Martin ER, et al. An alpha-2-macroglobulin insertion-deletion polymorphism in Alzheimer disease. Nat Genet. 1999;22(1):19–22. doi: 10.1038/8729. [DOI] [PubMed] [Google Scholar]
- Scacchi R, De BL, Mantuano E, Vilardo T, Donini LM, Ruggeri M, Gemma AT, Pascone R, Corbo RM. DNA polymorphisms of apolipoprotein B and angiotensin I-converting enzyme genes and relationships with lipid levels in Italian patients with vascular dementia or Alzheimer's disease. Dement Geriatr Cogn Disord. 1998;9(4):186–190. doi: 10.1159/000017045. [DOI] [PubMed] [Google Scholar]
- Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70(2):425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott WK, Yamaoka LH, Bass MP, Gaskell PC, Conneally PM, Small GW, Farrer LA, Auerbach SA, Saunders AM, Roses AD, et al. No genetic association between the LRP receptor and sporadic or late-onset familial Alzheimer disease. Neurogenetics. 1998;1(3):179–183. doi: 10.1007/s100480050026. [DOI] [PubMed] [Google Scholar]
- Sherrington R, Rogaev EI, Liang Y, Rogaeva EA, Levesque G, Ikeda M, Chi H, Lin C, Li G, Holman K, et al. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer's disease. Nature. 1995;375(6534):754–760. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- Sidak Z. Rectangular Confidence Regions for Means of Multivariate Normal Distributions. Journal of the American Statistical Association. 1967;62(318) 626-&. [Google Scholar]
- Siegmund KD, Langholz B, Kraft P, Thomas DC. Testing linkage disequilibrium in sibships. Am J Hum Genet. 2000;67(1):244–248. doi: 10.1086/302973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares ML, Coelho T, Sousa A, Batalov S, Conceicao I, Sales-Luis ML, Ritchie MD, Williams SM, Nievergelt CM, Schork NJ, et al. Susceptibility and modifier genes in Portuguese transthyretin V30M amyloid polyneuropathy: complexity in a single-gene disease. Hum Mol Genet. 2005;14(4):543–553. doi: 10.1093/hmg/ddi051. [DOI] [PubMed] [Google Scholar]
- Soubrier F, Martin S, Alonso A, Visvikis S, Tiret L, Matsuda F, Lathrop GM, Farrall M. High-resolution genetic mapping of the ACE-linked QTL influencing circulating ACE activity. Eur J Hum Genet. 2002;10(9):553–561. doi: 10.1038/sj.ejhg.5200847. [DOI] [PubMed] [Google Scholar]
- Stata Corp. Stata Statistical Software: Release 10. 2005 [Google Scholar]
- Sun X, Becker M, Pankow K, Krause E, Ringling M, Beyermann M, Maul B, Walther T, Siems WE. Catabolic attacks of membrane-bound angiotensin-converting enzyme on the N-terminal part of species-specific amyloid-beta peptides. Eur J Pharmacol. 2008;588(1):18–25. doi: 10.1016/j.ejphar.2008.03.058. [DOI] [PubMed] [Google Scholar]
- Tiret L, Rigat B, Visvikis S, Breda C, Corvol P, Cambien F, Soubrier F. Evidence, from combined segregation and linkage analysis, that a variant of the angiotensin I-converting enzyme (ACE) gene controls plasma ACE levels. Am J Hum Genet. 1992;51(1):197–205. [PMC free article] [PubMed] [Google Scholar]
- Toropygin IY, Kugaevskaya EV, Mirgorodskaya OA, Elisseeva YE, Kozmin YP, Popov IA, Nikolaev EN, Makarov AA, Kozin SA. The N-domain of angiotensin-converting enzyme specifically hydrolyzes the Arg-5-His-6 bond of Alzheimer's Abeta-(1–16) peptide and its isoAsp-7 analogue with different efficiency as evidenced by quantitative matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 2008;22(2):231–239. doi: 10.1002/rcm.3357. [DOI] [PubMed] [Google Scholar]
- Tsai CT, Lai LP, Lin JL, Chiang FT, Hwang JJ, Ritchie MD, Moore JH, Hsu KL, Tseng CD, Liau CS, et al. Renin-angiotensin system gene polymorphisms and atrial fibrillation. Circulation. 2004;109(13):1640–1646. doi: 10.1161/01.CIR.0000124487.36586.26. [DOI] [PubMed] [Google Scholar]
- Tzourio C, Anderson C, Chapman N, Woodward M, Neal B, MacMahon S, Chalmers J. Effects of blood pressure lowering with perindopril and indapamide therapy on dementia and cognitive decline in patients with cerebrovascular disease. Arch Intern Med. 2003;163(9):1069–1075. doi: 10.1001/archinte.163.9.1069. [DOI] [PubMed] [Google Scholar]
- Wang B, Jin F, Yang Z, Lu Z, Kan R, Li S, Zheng C, Wang L. The insertion polymorphism in angiotensin-converting enzyme gene associated with the APOE epsilon 4 allele increases the risk of late-onset Alzheimer disease. J Mol Neurosci. 2006;30(3):267–271. doi: 10.1385/JMN:30:3:267. [DOI] [PubMed] [Google Scholar]
- Wang J, Ho L, Chen L, Zhao Z, Zhao W, Qian X, Humala N, Seror I, Bartholomew S, Rosendorff C, et al. Valsartan lowers brain beta-amyloid protein levels and improves spatial learning in a mouse model of Alzheimer disease. J Clin Invest. 2007;117(11):3393–3402. doi: 10.1172/JCI31547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SM, Ritchie MD, Phillips JA, III, Dawson E, Prince M, Dzhura E, Willis A, Semenya A, Summar M, White BC, et al. Multilocus analysis of hypertension: a hierarchical approach. Hum Hered. 2004;57(1):28–38. doi: 10.1159/000077387. [DOI] [PubMed] [Google Scholar]
- Yang JD, Feng G, Zhang J, Lin ZX, Shen T, Breen G, St CD, He L. Association between angiotensin-converting enzyme gene and late onset Alzheimer's disease in Han chinese. Neurosci Lett. 2000;295(1–2):41–44. doi: 10.1016/s0304-3940(00)01591-3. [DOI] [PubMed] [Google Scholar]
- Zaykin DV, Westfall PH, Young SS, Karnoub MA, Wagner MJ, Ehm MG. Testing association of statistically inferred haplotypes with discrete and continuous traits in samples of unrelated individuals. Hum Hered. 2002;53(2):79–91. doi: 10.1159/000057986. [DOI] [PubMed] [Google Scholar]
- Zou K, Yamaguchi H, Akatsu H, Sakamoto T, Ko M, Mizoguchi K, Gong JS, Yu W, Yamamoto T, Kosaka K, et al. Angiotensin-converting enzyme converts amyloid beta-protein 1–42 (Abeta(1–42)) to Abeta(1–40), and its inhibition enhances brain Abeta deposition. J Neurosci. 2007;27(32):8628–8635. doi: 10.1523/JNEUROSCI.1549-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.