summary
A chromosomal locus for late-onset Alzheimer disease (LOAD) has previously been mapped to 9p21.3. The most significant results were reported in a sample of autopsy-confirmed families. Linkage to this locus has been independently confirmed in AD families from a consanguineous Israeli-Arab community. In the present study we analyzed an expanded clinical sample of 674 late-onset AD families, independently ascertained by three different consortia. Sample subsets were stratified by site and autopsy-confirmation. Linkage analysis of a dense array of SNPs across the chromosomal locus revealed the most significant results in the 166 autopsy-confirmed families of the NIMH sample. Peak HLOD scores of 4.95 at D9S741 and 2.81 at the nearby SNP rs2772677 were obtained in a dominant model. The linked region included the cyclin-dependent kinase inhibitor 2A gene (CDKN2A), which has been suggested as an AD candidate gene. By re-sequencing all exons in the vicinity of CDKN2A in 48 AD cases, we identified and genotyped four novel SNPs, including a non-synonymous, a synonymous, and two variations located in untranslated RNA sequences. Family-based allelic and genotypic association analysis yielded significant results in CDKN2A (rs11515: PDT p=0.003, genotype-PDT p=0.014). We conclude that CDKN2A is a promising new candidate gene potentially contributing to AD susceptibility on chromosome 9p.
Introduction
Alzheimer disease (AD) is the most common form of dementia in the elderly and affects more than 4.5 million Americans (Hebert et al. 2003). AD is characterized by slowly progressive loss of memory, higher intellectual functions, and cognitive abilities (Guttman et al. 1999). A definite diagnosis is only possible retrospectively by brain autopsy. Neuropathological hallmarks include intraneuronal neurofibrillary tangles, deposits of amyloid within senile plaques and cerebral blood vessels, and an overall loss of neurons in the cerebral cortex and hippocampus (Wisniewski et al. 1993).
In a subset of patients, AD manifests with autosomal dominant inheritance. These patients are often characterized by early age at onset (AAO). Three genes have been identified for early-onset AD: amyloid precursor protein (APP) (Goate et al. 1991), and the presenilins 1 and 2 (PS1 and PS2) (Sherrington et al. 1995; Levy-Lahad et al. 1995; Rogaev et al. 1995). While these genes give immense insight into the molecular pathology of AD, they account for less than 2% of AD (Farrer 1997). In contrast, the predominant AD form does not follow obvious Mendelian inheritance patterns and is characterized by late-onset of disease symptoms. Late-onset AD (LOAD) is thought to be a complex disease caused by multiple genetic and environmental risk factors. However, the ε4 allele of apolipoprotein E (APOE) on chromosome 19 is the only repeatedly confirmed genetic risk factor for LOAD. Additional genetic heterogeneity is supported by a number of chromosomal loci, but the identification of the underlying genes has proven to be a difficult task.
The LOAD locus on 9p21.3 has previously been identified by our group in a genome wide microsatellite-based linkage screen on 466 AD families (Pericak-Vance et al. 2000). This chromosomal locus was independently confirmed in a genetic study of a consanguineous Israeli-Arab community (Farrer et al. 2003). Farrer et al. suggested a recessive or additive mechanism primarily because of the observed excess of homozygosity at this locus (Farrer et al. 2003).
In the present study we report continued support for our initial findings in an updated clinical sample of 674 AD families with AAO≥60. This sample consists of three independent ascertainment efforts. 311 families had at least one autopsy-confirmed case. We genotyped 80 SNPs in the chromosomal region of approximately 20 Mb. The analysis included site and autopsy stratification in order to identify a more homogeneous subset that primarily contributes to the significant results. The chromosomal area under the linkage peak contained several potential AD candidate genes. By considering known AD pathways and published expression studies, we decided to study CDKN2A and the adjacent genes CDKN2B, and MTAP. Re-sequencing of these genes revealed several new polymorphisms. Family-based association tests and haplotype analyses were applied to 24 SNPs in the vicinity of these genes in an effort to identify the chromosome 9p21.3 LOAD susceptibility gene.
Methods
Study Samples
We ascertained 674 AD families of European descent. Of these, 311 were autopsyconfirmed families defined as a family in which at least one individual has been diagnosed as Alzheimer disease by autopsy. Criteria for a definite AD diagnosis were based on the neuropathological Braak stages (Braak & Braak 1991). The overall sample fell into three independent sets: 1) CAP (Miami Institute for Human Genomics (MIHG), Center for Human Genetics Research at Vanderbilt University, and Joseph and Kathleen Bryan Alzheimer Disease Research Center at Duke University); 2) the AD Genetics Initiative at the National Institutes of Mental Health (NIMH), and 3) the National Cell Repository for Alzheimer's Disease at Indiana University Medical Center (NCRAD).
Only LOAD families were used, defined as all patients having AAO values ≥ 60 years. We included 674 multiplex families of Caucasian ethnicity. Refer to Table 1 for a detailed description. The number of affecteds sampled per multiplex family ranged from 2 to 9 (mean=2.4, stdev=0.8). 12.3% of multiplex families were extended, defined as having an affected relative pair other than a full-sibling pair. 21% of singleton families reported a positive family history of AD. The dataset contained 558 families with at least one affected relative pair informative for linkage analysis and 379 families with at least one sampled affected family member and at least one sampled unaffected family member (discordant sibling pair) informative for association analysis.
Table 1.
Dataset of LOAD families included in this study.
| Relation Types | Total Number | Families by Ascertainment Center |
||
|---|---|---|---|---|
| CAP | NIMH | NCRAD | ||
| Discordant Sibling Pairs | 1303 | 397 | 636 | 270 |
| Affected Relative Pairs | 894 | 242 | 480 | 172 |
| Total Number of Families | 674 | 189 | 349 | 136 |
There were 1,454 genotyped affected individuals (69% female); AAO ranged from 60 to 98 (mean=72.93, stdev=6.4) and age at exam (AAE) ranged from 60 to 105 (mean=80.23, stdev= 7.1). There were 928 genotyped unaffected individuals (60% female); AAE ranged from 20 to 102 (mean= 69.92, stdev=11.08). 70% of families were APOE-ε4 positive families, where all affected individuals had at least one copy of the APOE- ε4 allele.
All affected individuals and AD cases were classified according to the National Institute of Neurological and Communicative Diseases and Stroke - Alzheimer's Disease and Related Disorders Association (NINCDS-ADRDA) clinical diagnostic criteria (McKhann et al. 1984). Written consent was obtained from all participants in agreement with protocols approved by the institutional review board at each contributing center.
Molecular analyses and genotyping
Genomic DNA was extracted from peripheral blood leucocytes applying standard procedures. For SNP selection we applied the SNPselector software (Xu et al. 2005). A minor allele frequency (MAF) ≥ 0.10 was required. Correlation between SNPs was set at r2 = 0.64 to identify blocks of LD. The selection of tagging SNPs allowed for a reduction of the number of SNPs required to detect available statistical power (Carlson et al. 2004). Taqman assays were ordered from Applied Biosystems (ABI, Foster City, CA) assay-on-demand and assay-by-design genotyping products. PCR amplifications used standard Taqman chemistry with 3ng of DNA and were performed on GeneAmp® PCR 9700 thermal cyclers (ABI). The PCR products were assigned to an ABI Prism 7900 Sequence Detector (ABI). By applying a Biomek Laboratory Automation Workstation (BeckmanCoulter, Fullerton, CA), 3ng DNA aliquots were placed on 384 well plates. Following previously described quality control procedures, two CEPH standards were included on each plate and samples from six individuals were duplicated across all plates (Rimmler et al. 1998). Laboratory personnel were blinded to QC samples, family relations and affection status. All genotypes were analyzed with the SDS software package (ABI) and stored in the PEDIGENE® information management system (Haynes et al. 1995). Mendelian inconsistencies were identified and resolved using PedCheck (O'Connell & Weeks 1998). Genotyping efficiency was greater than 95%.
For re-sequencing, we ranked families by multipoint LOD scores at five markers that spanned the core of the linkage peak and which were not in LD with each other (r2<0.3), rs783562 - rs2805064 - D9S741 - rs1162608 - rs1932420. From each of the 24 highest ranked families two affected individuals were chosen. These individuals 1) shared the same haplotype within the family; 2) were autopsy confirmed; 3) were the youngest AAO in the family in order to avoid phenocopies. DNA samples were placed on 96 well plates as described above. The coding exons and at least 70 bp of flanking intronic DNA sequence of three candidate genes - 5'-methylthioadenosine phosphorylase (MTAP), CDKN2A, and CDKN2B - were amplified in standard PCR reactions. The PCR products were purified over sephadex columns and directly sequenced with the BigDye sequencing kit following the manufacturer’s recommendations (ABI). Sequencing was performed on an ABI3730 automated sequencer (ABI). Sequencing traces were aligned to genomic DNA templates with the Sequencher software (Gene Codes Corporation, Ann Arbor, MI) and analyzed independently by two investigators.
Statistical analyses
Tests for deviations from Hardy-Weinberg equilibrium (HWE) were conducted in unrelated cases, unrelated controls and families (selecting one affected individual and one unaffected individual per family) using the exact test from Genetic Data Analysis (GDA) software (Lewis & Zaykin 2000). Measures of LD were computed with the GOLD program (Abecasis & Cookson 2000). The squared correlation coefficient (r2) and the normalized disequilibrium coefficient (D`) were used as LD measures.
Single-point parametric LOD scores were computed under affecteds-only dominant and recessive models with FASTLINK 4.1P (Cottingham et al. 1993). Disease-allele frequencies were 0.001 and 0.20 for dominant and recessive disease models, respectively. Marker allele frequencies were estimated from all individuals in the family sample. To assess linkage in the presence of genetic heterogeneity, maximum heterogeneity LOD (HLOD) scores were generated from HOMOG 3.35 (Bhat et al. 1999). For single-locus family-based association analysis of AD risk we applied the pedigree disequilibrium test (PDT) version 5.1, the genotype-PDT (Martin et al. 2003). All tests were conducted in the overall sample and several subsets were stratified by the three ascertainment sites and autopsy-confirmation.
Results
Linkage analysis
A total of 80 SNPs and the original peak marker D9S741 were selected to cover the linkage peak according to preexisting AD studies on chromosome 9p21(Pericak-Vance et al. 2000; Farrer et al. 2003). Tests for HWE deviations were calculated for each marker in affected individuals and in unaffected individuals. We found no evidence of deviation from HWE in the markers tested. Significant LD (r2 and D`) for SNP pairs was observed only for short distances.
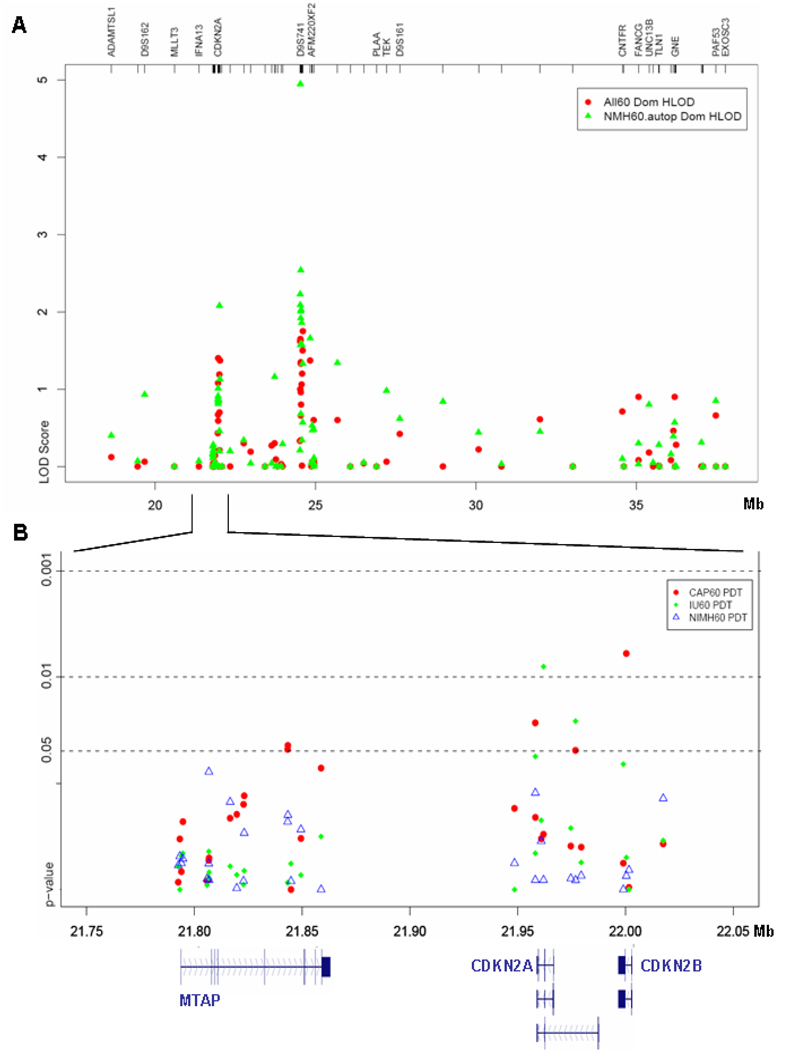
We confirmed the linkage peak at D9S741 with SNP markers in a now expanded sample of 674 families. In the overall dataset, peak HLOD scores were 1.76 at D9S741 and 1.73 at rs1329853 (6 kb from D9S741) in dominant models. As expected, stratification for autopsy confirmation (311 families) increased the peak HLOD to 2.89 at D9S741 and 2.68 at rs1034168 (75 kb from D9S741). Since the complete sample consisted of three independent ascertainment efforts, we then stratified by site. The highest scores were obtained in a subset of 166 autopsy confirmed NIMH families with HLOD of 4.95 at D9S741 and 2.81 at rs2772577 (13 kb from D9S741). The peak HLOD scores in the other two data sets were 0.85 at rs1162608 in the CAP sample and 1.42 at rs3731249 in the NCRAD sample; however, they were not stratified by autopsy confirmation because of the small number of autopsy cases.
In the NIMH autopsy confirmed sample positive linkage signals (HLOD>0.00) were observed between 21.7 Mb (rs2165408) and 37 Mb (rs4256662). By applying the 1-log-down method based on our SNP data the linkage area would be confined to about 3 Mb between 22 Mb and 25 Mb. Beyond this core linkage region the HLOD signal sharply fell below scores of 1.00.
Resequencing
Under the peak LOD score no genes were mapped and we found only sparse mRNA and EST support for novel transcripts. However, about 2.7Mb upstream of D9S741 resides the gene CDKN2A, which comprises an interesting candidate gene for AD.
Direct sequencing of all exons of CDKN2A and the adjacent genes MTAP and CDKN2B in 48 LOAD patients revealed a number of known and novel genetic variations (Table 2). We identified seven variations in MTAP. Three of these changes, a synonymous, a nonsynonymous, and a 5`-UTR sequence variation, were not reported before. In CDKN2A and 2B we detected six SNPs; one known non-synonymous change and four SNPs in the untranslated regions. One 5’-UTR change has not been described before. Genotyping of the entire LOAD sample revealed the presence of these sequence variants in cases and controls. The novel changes had low allele frequencies (MAF<1%) (Table 2).
Table 2.
Genetic variations detected by re-sequencing in MTAP, CDKN2A, and CDKN2B.
| Gene | Designation | Exon | Variation | Position in bp (NCBIv35) | MAF | Function |
|---|---|---|---|---|---|---|
| MTAP | 9P0427* | 1 | T/C | 21792655 | Not typed | 5’-UTR |
| rs10114559 | T/C | 21806637 | 0.44 | intronic | ||
| rs7023474 | A/G | 21806646 | 0.46 | intronic | ||
| rs7023954 | 3 | GTC>ATC; V56I | 21806758 | 0.42 | non-synonymous | |
| rs10965163 | 6 | CGC>CGT; R187R | 21844740 | 0.0897 | synonymous | |
| 9P0383* | 6 | ATC>GTC; I194V | 21844759 | >0.001 | non-synonymous | |
| 9P0426* | 7 | CCT>CCG; P256P | 21849379 | 0.0094 | synonymous | |
| CDKN2A | rs3814960 | 2 | A/G | 21965017 | Not typed | 5’-UTR |
| rs3731249 | 3 | GCG>ACG; A148T | 21960916 | 0.03 | non-synonymous | |
| rs11515 | 4 | C/G | 21958199 | 0.14 | 3’-UTR | |
| rs3088440 | 4 | C/T | 21958159 | 0.08 | 3’-UTR | |
| CDKN2B | 9P0351* | 1 | G/A | 21999016 | 0.009 | 5’-UTR |
| rs2069426 | C/A | 21996273 | Not typed | intronic | ||
novel variations; MAF – minor allele frequency
Association studies
To further delineate the genetic effect at the MTAP -CDKN2A-CDKN2B locus we performed allelic association tests. We chose 24 SNPs according to the known LD structure that would preferentially ‘tag’ the known haplotypes. In addition, three novel SNPs detected by resequencing were genotyped and analyzed in the overall sample, even though they appeared to be rare. Deviation from HWE was not observed.
Single-locus association analyses in the entire LOAD dataset were only marginally improved by stratification for ascertainment site and autopsy-confirmation (Table 3). Stratification by ascertainment site did improve the significance level of two markers: rs10118757 and rs598664 (Table 3, Figure 1). The most significant PDT score of 0.003 was measured at rs11515 (Table 3). The significance level after gene-based Bonferoni correction was 0.009. Rs11515 is situated in the 3’-UTR of CDKN2A. However, this SNP is not conserved between human, chimpanzee, and dog. No transcription factor binding site, micro RNA target site, or conserved regulatory potential has been detected at rs11515. Three other SNPs in CDKN2A/B were significant in the PDT test: rs3731246 (PDT, p=0.028; genoPDT, p=0.047), rs3731211 (PDT, p=0.029), and rs598664 (PDT, p=0.015; genoPDT=0.028). Rs11515 is in LD with rs3731246 (r2=0.74), rs3731211 (r2=0.42), and rs598664 (r2=0.72); thus, all significant SNPs fall into a conserved haplo-bin. The three markers with high r2 had similar MAF between 10% and 14% (Table 3)
Table 3.
Significantly associated markers in the vicinity of MTAP – CDKN2A – CDKN2B and their optimal subset.
| Dataset | Gene | Marker | Position (bp) | PDT | genoPDT | Allele Frequencies & Nucleotides | |||
|---|---|---|---|---|---|---|---|---|---|
| Overall | MTAP | rs10118757 | 21843339 | 0.185 | 0.124 | A | 0.87 | G | 0.13 |
| CAP | 0.194 | 0.024 | |||||||
| Overall | CDKN2A | rs11515* | 21958199 | 0.008 | 0.026 | C | 0.14 | G | 0.86 |
| Overall-autopsy | 0.003 | 0.014 | |||||||
| Overall | CDKN2A | rs3731246* | 21961989 | 0.028 | 0.047 | C | 0.91 | G | 0.09 |
| NCRAD | 0.026 | 0.05 | |||||||
| Overall | CDKN2A | rs3731211* | 21976847 | 0.029 | 0.089 | A | 0.73 | T | 0.27 |
| CAP | 0.032 | 0.149 | |||||||
| Overall | CDKN2B | rs598664* | 22017551 | 0.55 | 0.48 | A | 0.91 | G | 0.09 |
| NCRAD | 0.015 | 0.028 | |||||||
All significant results are in bold font. The optimal subsets are in italics.
These markers were in LD with each other.
Figure 1.
Linkage and association test results. (A) Two point linkage results were significantly improved by stratifying the overall dataset (All60) by autopsy confirmation and NIMH site (NMH60.autop). (B) The PDT test revealed significant association in the CDKN2A gene. Stratification by ascertainment site showed the strongest results in the CAP and NCRAD subsets.
Discussion
The present study extended evidence for a LOAD susceptibility gene on chromosome 9p. We confirmed linkage of LOAD to 9p21.3 in an updated and significantly larger sample of LOAD families by applying a dense array of SNP genomic markers. Although positive linkage was observed in all three independent samples (CAP, NIMH, and NCRAD) the contributions to the overall HLOD varied considerably. By stratifying our analysis by ascertainment site we were able to identify a subset of 166 autopsy-confirmed NIMH families as the most important contributor to the linkage signal. Compared to the dominant HLOD of 2.89 at D9S741 in the overall autopsy-confirmed sample, the smaller NIMH sample yielded an HLOD of 4.95. These results improved the findings of the original study, which reported a peak HLOD of 3.04 for an autopsy-confirmed sample (Pericak-Vance et al. 2000). Stratification of the sample by APOE allele status did not change our results (data not shown).
The identification of genes involved in dementia supports considerable genetic heterogeneity in AD. A number of genomic linkage screens have identified additional putative AD genetic loci notably on chromosomes 9q, 10q, and 12 (Blacker et al. 2003; Pericak-Vance et al. 2000; Farrer et al. 2003; Kehoe et al. 1999; Myers et al. 2002; Pericak-Vance et al. 1997). This locus heterogeneity in the presence of relative uniform clinical appearance, the late onset of disease, and uncertain penetrance has been cited as major reasons for the difficulty in identifying AD genes. Our data show that combined stratification by clinical criteria (age of onset, autopsy confirmation) and ascertainment site are effective means to identify a more homogeneous sample of AD families with linkage to 9p. Although AD comprises the major cause for dementia in the elderly only a brain-autopsy is sufficient to delineate other conditions, such as vascular dementia or fronto-temporal dementia. The successful stratification by ascertainment site might represent a population sample that shares a regional genetic background with susceptibility to AD. An example is the study by Farrer et al., who confirmed the 9p21.3 locus in an inbred Israeli-Arab population (Farrer et al. 2003). Alternative explanations are ascertainment methods (advertisement, ascertainment from local hospitals, certain physicians) or differences of the ascertainment schemes at the participating sites that are not recognized. The more homogeneous NIMH sample with linkage to 9p21.3; however, will improve our ability to identify the underlying gene.
We identified and studied an AD candidate gene, CDKN2A, about 2.7 Mbp upstream of D9S741. Significant association of four SNPs implicated a possible genetic effect for CDKN2A in LOAD. Three markers had similar MAF between 10% and 14% and present a conserved haplo-bin (r2=0.42–.074) that covers 60 kb of genomic sequence, and which might contain the true risk allele for AD at 9p21.3 (Fig. 1B). The most significant SNP rs11515 (PDT=0.003) is located in the 3`-UTR of CDKN2A (21.9 Mb) were it could have a yet unidentified regulatory function. Interestingly, we recently performed a whole genome association study in Alzheimer disease that showed a marker at 20.1 Mb of chromosome 9p among the top 35 SNPs ranked by p-value (p-value = 0.0000348) (Beecham et al., submitted). Cyclin-dependent kinases have been implicated in the process of neurodegeneration in AD. They are activated by mitogen-activated protein kinases (MAPK); and recently, such proteins have been found to be differentially expressed in terminally differentiated neurons in AD brains (Arendt et al. 1996; Ueberham et al. 2003; Ueberham & Arendt 2005). These expression changes activate mitogenic pathways such as the p21/MAPK cascade possibly leading to hyperphosphorylated tau or disturbed APP processing (Ueberham & Arendt 2005). Both neurofibrillary tangles and neuritic components of plaques showed strong CDKN2A (p16) immunoreactivity (Arendt et al. 1996). CDKN2A co-localizes with nNOS and p21ras in pyramidal neurons in AD brain (Luth et al. 2000). It has been speculated that an aborted attempt of terminally differentiated neurons to re-enter the cell cycle might be a critical event in the pathology of Alzheimer's disease (Arendt et al. 1996).
Though we found association in the CDKN2A gene, we realize that the signal is rather weak in the light of the strong linkage signal. It is therefore possible that a variation in another gene or in an additional gene in the chromosomal region might account for the main effect of the linkage. Additional haplo-blocks might be present that we did not cover with the set of genotyped SNPs. An intriguing alternative possibility is that multiple relatively rare changes in the gene could account for the linkage signal but would not be detected in an association study. This consideration was behind our re-sequencing approach, and future more extended studies that take advantage of next-generation sequencing technologies might be able to exhaustively probe this possibility.
In summary, we confirmed linkage of LOAD to chromosome 9p21.3 in an extended AD sample by applying an array of SNPs. Stratification by autopsy and site identified a subset of 166 NIMH families that yielded the yet highest HLOD score at this locus of 4.95 in a dominant model. In depth exploration of CDKN2A generated evidence for this gene as a new candidate gene for LOAD. Further studies are required to draw conclusions on the importance of CDKN2A for Alzheimer disease.
Supplementary Material
The following material is available for this article online:
Table S1. Genomic markers genotyped in this study.
Acknowlegment
This work was supported by NIH grants (R01 AG027944-01A2, R01 AG021547-05, R01 AG019757-06 to MA Pericak-Vance) and the Alzheimer Association (IIRG-05-14147 to MA Pericak-Vance).
References
- Abecasis GR, Cookson WO. GOLD--graphical overview of linkage disequilibrium. BioInformatics. 2000;16:182–183. doi: 10.1093/bioinformatics/16.2.182. [DOI] [PubMed] [Google Scholar]
- Arendt T, Rodel L, Gartner U, Holzer M. Expression of the cyclin-dependent kinase inhibitor p16 in Alzheimer's disease. Neuroreport. 1996;7:3047–3049. doi: 10.1097/00001756-199611250-00050. [DOI] [PubMed] [Google Scholar]
- Bhat A, Heath SC, Ott J. Hetergeneity for multiple disease loci in linkage analysis. Hum Hered. 1999;49:229–231. doi: 10.1159/000022879. [DOI] [PubMed] [Google Scholar]
- Blacker D, Bertram L, Saunders AJ, Moscarillo TJ, Albert MS, Wiener H, Perry RT, Collins JS, Harrell LE, Go RC, Mahoney A, Beaty T, Fallin MD, Avramopoulos D, Chase GA, Folstein MF, McInnis MG, Bassett SS, Doheny KJ, Pugh EW, Tanzi RE. Results of a high-resolution genome screen of 437 Alzheimer's Disease families. Hum Mol Genet. 2003;12:23–32. doi: 10.1093/hmg/ddg007. [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol (Berl) 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottingham RW, Idury RM, Schaffer AA. Faster sequential genetic linkage computations. Am J Hum Genet. 1993;53:252–263. [PMC free article] [PubMed] [Google Scholar]
- Farrer LA. Genetics and the dementia patient. Neurologist. 1997;3:13–30. [Google Scholar]
- Farrer LA, Bowirrat A, Friedland RP, Waraska K, Korczyn AD, Baldwin CT. Identification of multiple loci for Alzheimer disease in a consanguineous Israeli-Arab community. Hum Mol Genet. 2003;12:415–422. doi: 10.1093/hmg/ddg037. [DOI] [PubMed] [Google Scholar]
- Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, Fidani L, Giuffra L, Haynes A, Irving N, James L, Mant R, Newton P, Rooke K, Roques P, Talbot C, Pericak-Vance MA, Roses A, Williamson R, Rossor M, Owen M, Hardy J. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer's disease. Nature. 1991;349:704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- Guttman R, Altman RD, Nielsen NH. Alzheimer disease: report of the Council on Scientific Affairs. Archives of Family Medicine. 1999;8:347–353. doi: 10.1001/archfami.8.4.347. [DOI] [PubMed] [Google Scholar]
- Haynes C, Speer MC, Peedin M, Roses AD, Haines JL, Vance JM, Pericak-Vance MA. PEDIGENE: A comprehensive data management system to facilitate efficient and rapid disease gene mapping. Am J Hum Genet. 1995;57:A193. [Google Scholar]
- Hebert LE, Scherr PA, Bienias JL, Bennett DA, Evans DA. Alzheimer disease in the US population-Prevalence estimates using the 2000 census. Arch Neurol. 2003;60:1119–1122. doi: 10.1001/archneur.60.8.1119. [DOI] [PubMed] [Google Scholar]
- Kehoe P, Wavrant-De Vrieze F, Crook R, Wu WS, Holmans P, Fenton I, Spurlock G, Norton N, Williams H, Williams N, Lovestone S, Perez-Tur J, Hutton M, Chartier-Harlin MC, Shears S, Roehl K, Booth J, Van Voorst W, Ramic D, Williams J, Goate A, Hardy J, Owen MJ. A full genome scan for late onset Alzheimer's disease. Hum Mol Genet. 1999;8:237–245. doi: 10.1093/hmg/8.2.237. [DOI] [PubMed] [Google Scholar]
- Levy-Lahad E, Wasco W, Poorkaj P, Romano DM, Oshima J, Pettingell WH, Yu CE, Jondro PD, Schmidt SD, Wang K, Crowley AC, Fu YH, Guenette SY, Galas D, Nemens E, Wijsman EM, Bird TD, Schellenberg GD, Tanzi RE. Candidate gene for the chromosome 1 familial Alzheimer's disease locus. Science. 1995;269:973–977. doi: 10.1126/science.7638622. [DOI] [PubMed] [Google Scholar]
- Lewis PO, Zaykin D. Genetic Data Analysis: Computer program for analysis of allelic data. 2000 [Google Scholar]
- Luth HJ, Holzer M, Gertz HJ, Arendt T. Aberrant expression of nNOS in pyramidal neurons in Alzheimer's disease is highly co-localized with p21ras and p16INK4a. Brain Res. 2000;852:45–55. doi: 10.1016/s0006-8993(99)02178-2. [DOI] [PubMed] [Google Scholar]
- Martin ER, Bass MP, Gilbert JR, Pericak-Vance MA, Hauser ER. Genotype-based association test for general pedigrees: the genotype-PDT. Genet Epidemiol. 2003;25:203–213. doi: 10.1002/gepi.10258. [DOI] [PubMed] [Google Scholar]
- McKhann G, Drachman D, Folstein M. Clinical diagnosis of Alzheimer's disease: Report of the NINCDS-ADRDA Work Group under the auspices of the Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- Myers A, Wavrant De-Vrieze F, Holmans P, Hamshere M, Crook R, Compton D, Marshall H, et al. Full genome screen for Alzheimer disease: stage II analysis. Am J Med Genet. 2002;114:235–244. doi: 10.1002/ajmg.10183. [DOI] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pericak-Vance MA, Bass MP, Yamaoka LH, Gaskell PC, Scott WK, Terwedow HA, Menold MM, Conneally PM, Small GW, Vance JM, Saunders AM, Roses AD, Haines JL. Complete genomic screen in late-onset familial Alzheimer disease. Evidence for a new locus on chromosome 12. JAMA. 1997;278:1237–1241. [PubMed] [Google Scholar]
- Pericak-Vance MA, Grubber J, Bailey LR, Hedges D, West S, Kemmerer B, Hall JL, Saunders AM, Roses AD, Small GW, Scott WK, Conneally PM, Vance JM, Haines JL. Identification of novel genes in late-onset Alzheimer disease. Exp Gerontol. 2000;35:1343–1352. doi: 10.1016/s0531-5565(00)00196-0. [DOI] [PubMed] [Google Scholar]
- Rimmler J, McDowell JG, Slotterback BD, Haynes CS, Menold MM, Rogala A, Speer MC, Gilbert JR, Hauser ER, Vance JM, Pericak-Vance MA. Development of a data coordinating center (DCC): Data quality control for complex disease studies. Am J Hum Genet. 1998;63:240. [Google Scholar]
- Rogaev EI, Sherrington R, Rogaeva EA, Levesque G, Ikeda M, Liang Y, Chi H, Lin C, Holman K, Tsuda T. Familial Alzheimer's disease in kindreds with missense mutations in a gene on chromosome 1 related to the Alzheimer's disease type 3 gene. Nature. 1995;376:775–778. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- Sherrington R, Rogaev EI, Liang Y, Rogaeva EA, Levesque G, Ikeda M, Chi H, et al. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer's disease. Nature. 1995;375:754–760. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- Ueberham U, Arendt T. The expression of cell cycle proteins in neurons and its relevance for Alzheimer's disease. Curr Drug Targets CNS Neurol Disord. 2005;4:293–306. doi: 10.2174/1568007054038175. [DOI] [PubMed] [Google Scholar]
- Ueberham U, Hessel A, Arendt T. Cyclin C expression is involved in the pathogenesis of Alzheimer's disease. Neurobiol Aging. 2003;24:427–435. doi: 10.1016/s0197-4580(02)00132-x. [DOI] [PubMed] [Google Scholar]
- Wisniewski T, Golabek A, Matsubara E, Ghiso J, Frangione B. Apolipoprotein E: binding to soluble Alzheimer's beta-amyloid. Biochem Biophys Res Commun. 1993;192:359–365. doi: 10.1006/bbrc.1993.1423. [DOI] [PubMed] [Google Scholar]
- Xu H, Gregory SG, Hauser ER, Stenger JE, Pericak-Vance MA, Vance JM, Zuchner S, Hauser MA. SNPselector: a web tool for selecting SNPs for genetic association studies. BioInformatics. 2005;21:4181–4186. doi: 10.1093/bioinformatics/bti682. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following material is available for this article online:
Table S1. Genomic markers genotyped in this study.