Abstract
Loss-of-function mutations in the nuclear factor erythroid-2 related factor-2 (Nrf2) inhibitor, Kelch-like-ECH-associated protein (Keap1), result in increased Nrf2 activity in non–small-cell lung cancer (NSCLC) and confer therapeutic resistance. We detected point mutations in Keap1 gene leading to non-conservative amino acid substitutions in prostate cancer cells. We found novel transcriptional and post-transcriptional mechanisms of Keap1 inactivation such as promoter CpG island hypermethylation and aberrant splicing of Keap1 in DU-145 cells. Very low levels of Keap1 mRNA were detected in DU-145 cells, which significantly increased by treatment with DNA methyltransferase inhibitor 5-aza-cytidine. The loss of Keap1 function led to an enhanced activity of Nrf2 and its downstream electrophile/drug detoxification pathway. Inhibition of Nrf2 expression in DU-145 cells by RNAi attenuated the expression of glutathione, thioredoxin, and the drug efflux pathways involved in counteracting electrophiles, oxidative stress, and detoxification of a broad spectrum of drugs. DU-145 cells expressing Nrf2-shRNA had lower levels of total glutathione and higher levels of intracellular reactive oxygen species. Attenuation of Nrf2 function in DU-145 cells enhanced sensitivity to chemotherapeutic drugs and radiation-induced cell death. In addition, Inhibition of Nrf2 greatly suppressed in vitro and in vivo tumor growth of DU-145 prostate cancer cells. Thus, targeting Nrf2 pathway in prostate cancer cells may provide a novel strategy to enhance chemo- and radio-therapy responsiveness and ameliorate the growth and tumorigenecity leading to improved clinical outcomes.
Keywords: Nrf2, Keap1, Prostate cancer, mutation, chemo-resistance, radio-resistance, RNAi
Introduction
Prostate cancer is the most common lethal malignancy diagnosed in American men that accounts for 29% of the incident cases and is the second leading cause of cancer mortality in men (1). Current therapies for prostate cancer are prostatectomy, hormonal manipulation, and chemo- and radio-therapy. While most patients with prostate cancer respond to initial androgen ablation, they typically reemerge in an androgen-independent form months to years later. The prognosis of androgen-independent prostate cancer is poor, and it is usually resistant to chemo- and radio-therapy (2). The mechanisms responsible for the acquisition of resistance to chemo- and radio-therapy by hormone-independent prostate cancer are still unclear.
Ionizing radiation kills cancer cells by generation of reactive oxygen species (ROS), mainly superoxide, hydroxyl radicals, and hydrogen peroxide, which causes DNA damage, and up-regulation of antioxidant enzyme expression or addition of free radical scavengers has been reported to protect cells from the damaging effects of radiation (3, 4). Many anticancer agents such as cisplatin, paclitaxel, bleomycin, adriamycin, and etoposide exert their toxic effects on cancer cells by producing free radicals that may exhaust the antioxidant capacity of cancer cells and thereby causing apoptosis. Enzymes involved in xenobiotic metabolism in conjunction with drug efflux proteins act to detoxify cancer drugs, whereas antioxidants confer cyto-protection by attenuating drug-induced oxidative stress and apoptosis. Thus, mode of action of radiation and widely used chemotherapeutic agents includes oxidative insult to cancer cells.
Nuclear factor erythroid-2 related factor-2 (Nrf2), a basic leucine zipper transcription factor, regulates the expression of a battery of genes that maintain cellular redox homeostasis and protects against oxidative stress and apoptosis induced by a variety of stressors including electrophiles, oxidants, radiation, and FAS ligand (5-9). The Nrf2-regulated transcriptional program includes genes that encode for antioxidants, electrophile, and xenobiotic detoxification enzymes and several ATP-dependent multidrug resistant efflux proteins (5-7, 9-11). Keap1 is a cytoplasmic anchor of Nrf2, which maintains steady-state levels of Nrf2 by targeting it for proteasomal degradation (5, 12). Keap1 is located at chromosome 19p13.2 and has three major domains: an N-terminal broad complex, tramtrack, and bric-a-brac (BTB) domain; a central intervening region (IVR); and a series of six C-terminal Kelch repeats (13). The Kelch repeats of Keap1 bind to the Neh2 domain of Nrf2, whereas the IVR and BTB domains are required for the redox-sensitive regulation of Nrf2 through a series of reactive cysteines present throughout this region (14). Recently, we and other investigators have reported point mutations in Keap1 gene that lead to non-conservative amino acid substitutions and nonsense mutations in non–small-cell lung cancer (NSCLC) cell lines and tumors (15-17). Similar mutations have been reported in breast cancer and gall bladder cancer (17-19).
The aim of this study is to provide a proof-of-concept and to determine whether aberrant Nrf2-Keap1 pathway exists in prostate cancer cell lines similar to that in NSCLC, which may confer resistance to chemotherapy and ionizing irradiation (20). Furthermore, we studied whether inhibition of Nrf2 activity in prostate cancer cells affects tumor formation in vivo. The study demonstrates that gain of Nrf2 function in prostate cancer cells promotes in vitro and in vivo cell proliferation and confers chemo- and radio-resistance.
Materials and methods
Cell Lines and Culture
The human prostate cancer cell lines DU-145, PC3, LNCaP, C42B, and CWR22RV1 (CWR22) were obtained from ATCC (Manassas, VA). Transfections were carried out using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Normal prostate epithelial cells (PrEC) were obtained from Lonza Inc. (Allendale, NJ). All chemicals were purchased from Sigma-Aldrich (St. Louis, MO).
PCR and Sequence Analysis
A total of 12 cases of prostate tumor, were chosen in accordance with the Institutional Review Board protocol, and DNA was isolated using DNeasy Kit (Qiagen, Valencia, CA). PCR amplification and sequencing of Keap1 gene was carried out using primer sequences and protocols published by Singh et al (15). All mutations were confirmed by sequencing in both directions. DNA samples harboring mutation were sequenced twice to confirm the mutations. Chromatograms were analyzed by manual review.
Western Blot Analysis
Nrf2 and Keap1 Western blot experiments were carried out using protocols published by Singh et al (15).
Real-time RT-PCR
Quantitative real-time reverse transcriptase-polymerase chain reaction (RT-PCR) analyses of human Keap1, Nrf2, GCLc, GCLm, GSR, G6PDH, PRDX1, NQO1, HO-1, TXN1, TXNRD1, ABCC1, and ABCC2 were performed by using assay-on-demand primers and probe sets from Applied Biosystems. β-actin was used for normalization (15).
Luciferase Assay
DU145 cells were seeded onto a 24-well dish at a density of 0.2 × 106 cells/ml for 12 h before transfection. NQO1-ARE luciferase along with WT-KEAP1 cDNA constructs, mutant cDNA constructs (Y255H and T314M) were transfected into the cells along with pRL-TK plasmid expressing Renilla luciferase as a transfection control. Thirty six hours after transfection, cells were lysed and both firefly and Renilla luciferase activities were measured with a Dual-Luciferase Reporter Assay System (Promega). The primers used for generating site-directed mutagenesis were as follows: (1) Y255H, Sense-tgcatcaactgggtcaagcacgactgcgaacagcga and Antisense- tcgctgttcgcagtcgtgcttgacccagttgatgca; (2) T314M, Sense- accctg cacaa gccca tgcag gtgat gccct and Antisense- agggc atcac ctgca tgggc ttgtg cagggt.
Generation of DU-145 cells stably expressing Nrf2 shRNA
Short hairpin RNA (shRNA) targeting the Nrf2 transcript and Keap1 was generated as described previously (15, 20, 21) and transfected into DU-145 cells. A shRNA targeting luciferase gene was used as vector control. Stable cell clones with reduced Nrf2 expression were generated following a protocol published by Singh et al (20).
Cellular Glutathione Measurement
Reduced glutathione was determined using the modified Tietze method (22).
Cell Viability and Proliferation Assays
Chemotherapy drug treatments were performed by using protocols published by Singh et al (20). In vitro drug sensitivity was evaluated by using a cell viability assay kit (Roche, Indianapolis, IN). Cell proliferation assays were conducted using a MTS assay kit from Promega (Madison, WI) and manual trypan blue stained cell counting method.
Measurement of Intracellular ROS levels
Endogenous ROS levels were measured using c-H2DCFDA (Invitrogen, Carlsbad, CA) (20). For N-acetyl-cysteine (NAC) treatment, cells were pretreated with 15mM NAC for 30mins followed by c-H2DCFDA staining.
Clonogenic Assays
A total of 1000 cells were exposed to a high dose rate (0.68Gy/min) radiation using a gamma cell 40 137Cs irradiator (Atomic Energy of Canada, Ontario, Canada) and incubated in complete growth medium at 37°C for 14 days. The cells were stained with 50% methanol-crystal violet solution and only colonies with more than 50 cells were counted.
Drug Accumulation
Tritium (23) labeled paclitaxel accumulation in DU-145 cells was carried out using the protocol by Wu et al (24). The [3H] levels in each sample were measured using a scintillation counter (Beckman Instruments Inc., Fullerton, CA), and accumulation was normalized to total protein content.
Bisulfite Genomic Sequencing
Normal WBC, PrEC, and DU-145 genomic DNA were subjected to sodium bisulfite treatment using the EZ-DNA Methylation Kit (Zymo Research, Orange, CA). One CpG island region upstream of the transcriptional start site (TSS) of the longest known isoform of Keap1 was amplified using primers Keap1-F1 (5′-GGTTAGGAGTTTAAGGTTGTAGTGAGT-3′) and Keap1-R 1 (5′-ACCAAACCCCCCTTCTCACTA-3′). Second CpG island region flanking the TSS of the longest Keap1 isoform was amplified using primers Keap1-F2 (5′-TAGTGAGAAGGGGGGTTTGGT-3′) and Keap1-R2 (5′CCAATCCAAAAAATTCCTACCTTAC-3′). Third CpG island region, located downstream of the longest Keap1 isoform, but upstream of the TSS for a shorter Keap1 isoform was amplified using Keap1-F3 (5′-TAAGGTAGGAATTTTTTGGATTGG-3′) and Keap1-R3 (5′-AAAAATAAAATAAACACCCCTCCC-3′). PCR products were sub-cloned into pCR2.1-TOPO vector (Invitrogen, Carlsbad, CA) and 10 to 15 independent clones of each amplicon were sequenced. Clones showing <95% conversion of non-CpG cytosine to thymine were considered uninformative and discarded. Sequencing data from each clone were then aligned to a virtually bisulfite converted reference sequence corresponding to the amplicon, and schematic maps of methylation status at each CpG dinucleotide in each clone were generated using custom software developed in Visual Basic.
5-aza-cytidine Treatment and Keap1 Gene Expression Analysis
One million DU-145 cells were treated with 5μM of 5-Aza-deoxycytidine (5-AzaC) (Sigma-Aldrich, St. Louis, MO) for 7 days. For trichostatin-A (TSA) co-treatment, cells were incubated with 5-AzaC for 6 days, followed by incubation with 100 nM TSA for 16 hrs, and cells were harvested for RNA isolation.
Alternative Splicing
The protein coding region of Keap1 was amplified using the forward primer: 5′-AGGTGGTGGTGTTGCTTATCTT-3′ and the reverse primer: 5′-ACAATGATACTCCCCATTGGAC-3′ from total RNA using Superscript-one step RT-PCR kit (Invitrogen, Carlsbad, CA). The two differentially spliced transcripts were cloned in pCDNA3.0 expression vector (Invitrogen, Carlsbad, CA) and verified by sequencing.
Xenograft
Five million DU145-LucshRNA and DU145-Nrf2shRNA cells were implanted subcutaneously in the flanks of athymic nude mice (National Cancer Institute, Frederick, MD). Tumor size was measured by caliper once per week. Tumor volumes were calculated by using following formula: [length (mm) × width (mm) × width (mm) × 0.52].
Statistics Analysis
Statistical comparisons were performed by Student's t-tests or Wilcoxon rank-sum test. A P-value<0.05 was considered statistically significant.
Results
Somatic mutations in Keap1 gene in prostate cancer cell lines and tumors
To determine whether mutations in Keap1 gene exist in prostate cancer cells, we sequenced all five protein-coding exons and intron–exon boundaries of the Keap1 gene in 6-prostate cancer cell lines. Sequencing of Keap1 in CWR22Rv1 revealed a C-to-T transition (Fig. 1A), resulting in threonine to methionine substitution at 314th amino acid in Keap1 protein. As C42B cells are derived from LNCaP cells, both the cell lines showed a T-to-C transition (Fig. 1A), resulting in tyrosine to histidine change at 255th amino acid position. These non-synonymous amino acid changes were present in central intervening region of Keap1 and altered highly conserved amino acids (Fig. 1B). DU-145, LAPC4, and PC3 had a wild type (WT) Keap1 sequence. Sequencing of Keap1 gene in 12 primary prostate tumor samples revealed an A-T transversion mutation in one tumor sample resulting in methionine to leucine substitution at 209 amino acid position in Keap1 protein. To determine the functional consequences of somatic Keap1 mutations on its activity and resultant increases in Nrf2 activity, we generated cDNAs harboring the same mutations seen in tumor cell lines, LNCaP, C42B (Y255H) and CWR22Rv1 (T314M). We transfected the WT and mutant constructs of Keap1 and NQO1 ARE-luciferase reporter plasmid into DU145 cells. Importantly, overexpression of WT-Keap1 completely abolished the Nrf2-mediated ARE reporter activity whereas ectopic expression of mutant Keap1 constructs demonstrated significantly lower repression of Nrf2 dependent ARE reporter activity (Supplementary Fig. S1).
Figure 1. Keap1 mutations in prostate cancer cell lines.
(A) A heterozygous C–T transition mutation was detected in exon-3 of Keap1 in CWR22Rv1 cell line resulting in tyrosine to threonine substitution. In LNCaP and C42B cells, a heterozygous C–T transition mutation in exon-3 of Keap1 was noted resulting in threonine to methionine substitution. Wild type (WT) sequence shown was obtained from PrEC. Keap1 mutations in C42B and LNCaP cells were identical (data not shown). (B) Multiple protein sequence alignment showing location of amino acid changes was detected in LNCaP and CWR22Rv1.
Effect of Keap1 mutation on target antioxidant genes in prostate cancer
We examined Keap1 and Nrf2 mRNA expression in prostate cancer cells by real-time RT-PCR. Nrf2 expression did not change significantly between the prostate cancer cell lines but Keap1 mRNA was dramatically reduced in DU-145 cells (Fig. 2A). To further assess whether Keap1 mutations correlated with Nrf2 protein level changes in these prostate cancer cells, we examined Nrf2 protein levels using western blot. DU-145 cells demonstrated higher accumulation of Nrf2 in the nucleus compared to other prostate cancer cells. On the other hand, LNCaP and C42B cells bearing heterozygous Keap1 mutations showed moderate increase in the level of Nrf2 protein compared to PC3 and LAPC4 prostate cancer cells with WT Keap1 (Fig. 2B). Furthermore, we detected reduced Keap1 protein levels in DU-145 and LNCaP cells.
Figure 2. Dysregulated Keap1-Nrf2 pathway in prostate cancer cells.
(A) Real-time RT-PCR analysis of Keap1 and Nrf2 expression among prostate cancer cells. The expression levels in PC3 cells are arbitrarily valued as 1. Columns, mean (n = 3); bars, standard deviation. (B) Western blot analysis of Keap1-Nrf2 in prostate cancer cells. Immunoblot with nuclear protein showing increased Nrf2 levels in DU-145 cells and other prostate cancer cells. To detect Keap1 expression, equal amount of whole cell extract was loaded. GADPH, and lamin B were used as normalization controls (C) Real-time RT-PCR analysis of Nrf2 target genes, HO1, NQO1, Gclc, and GSR in prostate cancer cells. The expression levels in PC3 are arbitrarily valued as 1. Columns, mean (n = 3); bars, standard deviation.
To investigate whether altered levels of Nrf2 protein leads to up-regulation of Nrf2 dependent pathway in prostate cancer cells, we measured the expression of Nrf2 pathway genes. As shown in Figure 2C, the expression of Nrf2 targets, such as NQO1, HO1 mRNA levels were increased in DU-145, LNCaP and C42B cells. The expression of glutathione reductase (GSR) was increased in DU-145, LNCaP, CWR22Rv1, C42B, and LAPC4 cells but not in PC3 cells (Fig. 2C). We also compared the relative expression of Nrf2 dependent genes between non-transformed normal prostate epithelial cell line (PrEC) and prostate cancer cells. The expression of Nrf2 target genes was found to be significantly higher in prostate cancer cells as compared with normal prostate epithelial cells (Supplementary Fig. S2)
Keap1 core promoter CpG dinucleotides methylation and Keap1 mRNA aberrant splicing in DU-145 cells
We explored the possible mechanisms involved in the establishment of the low levels of Keap1 mRNA expression in DU-145 cells. Previous studies have shown that promoter CpG island hypermethylation is a frequent cause of epigenetic gene silencing in prostate cancer cell lines (25, 26). To test whether Keap1 promoter DNA methylation is associated with the low level expression of Keap1 in DU-145 cells, we isolated genomic DNA from DU-145 cells and carried out bisulfite genomic sequencing of CpG island sequences surrounding the Keap1 TSS. There are dramatic increases in methylated CpG dinucleotides around TSS (-114 bp to +163 bp) and an upstream region (-393 to -39 bp) of the Keap1 gene in DU-145 cells compared to PrEC cells and peripheral WBCs (Fig. 3A). In contrast, in a CpG island region that is downstream of the Keap1 TSS (+138 bp to +337 bp), all three cell types showed complete absence of methylation (Fig. 3A). Furthermore, treatment of DU-145 cells with 5-aza-C+TSA resulted in a significant increase in Keap1 expression and a parallel decrease in the expression of Nrf2 downstream genes (Fig. 3B left panel). In addition to the up-regulation of full-length Keap1 transcript, we detected amplification of additional truncated Keap1 transcripts in 5-aza-C and 5-aza-C+TSA treated DU-145 cells (Fig. 3B right panel). Taken together, these findings suggest that promoter CpG island methylation is involved in ameliorating Keap1 transcription in DU-145 cells.
Figure 3. Both promoter CpG dinucleotides methylation and mRNA differentially splicing contribute to the decreased Keap1 expression in DU-145 cells.
(A) Schematic representation of sodium bisulfite sequencing of 5-mCpG at the Keap1 promoter in DU-145 cells, human peripheral WBCs, and PrEC cells. Each circle represents an individual CpG site. Closed circles, methylated CpG sites; open circles, unmethylated sites. The beginning and end of each amplicon relative to the TSS of the longest Keap1 isoform is indicated. The region from -393 to -93 in the PrEC genomic DNA contained many non-informative CpG dinucleotides that showed deviation from the consensus virtually bisulfite-converted sequence despite having high rates of non-CpG conversion. (B) Left panel, real-time RT-PCR analysis of Keap1, NQO1, and Gclm expression. Right panel, Amplification of full-length Keap1 transcript. PCR products were resolved on agarose gel. (C) Amplification of full-length keap1 transcript in prostate cancer cells (Left panel) and Schematic representation of Keap1 WT transcript and DU-145 derived alternatively spliced products (right panel). E2 to E6 represent relative position of exon-2 to exon-6, open boxes stand for 5′ and 3′ untranslated regions. (D) Aberrantly spliced Keap1 transcripts code for non-functional truncated Keap1 proteins. The WT and differentially spliced Keap1 transcripts cloned in pDNA3.1 expression vector were transfected into DU-145 cells, and Keap1 protein expression was measured by immunoblot assay (Left panel). Truncated Keap1 proteins are unable to suppress Nrf2 depdendent NQO1 reporter activity. (right panel).
Another possible reason for substantially decreased levels of Keap1 mRNA in DU-145 cells may be due to post-transcriptional mechanisms. We designed primers to specifically amplify the full-length Keap1 protein coding region. The amplification of full-length Keap1 transcript was dramatically decreased in DU-145 cells as compared with other prostate cancer cells. As shown in Fig. 3C, two prominent small PCR products positioned below WT Keap1 transcript were detected in DU-145 cells suggesting the presence of differentially spliced forms. These differentially spliced keap1 transcripts (named as DCL1 and DCL2) were cloned and sequence verified. First form corresponded to the wild type Keap1 mRNA, second form (DCL1) with partial deletion of exon-4 and exon-5 resulted in a truncated Keap1 protein lacking c-terminal portion of the Kelch domain missing, and the shortest clone (DCL2) represented the Keap1 mRNA with exon-3 missing plus partial deletion of exon-5 and exon-6. The skipping of exon-3 created a frameshift mutation in the Kelch domain and introduced a stop codon at 457th amino acid (Supplementary Fig. S3). To confirm whether differentially spliced Keap1 transcripts code for non-functional Keap1 protein, we over-expressed full-length and differentially spliced Keap1 cDNA in DU-145 cells. Immunoblot results demonstrated that aberrantly spliced Keap1 transcripts coded for smaller molecular weight Keap1 protein than the WT Keap1 protein in DU-145 cells (Fig. 3D left panel). To examine the functional significance of truncated Keap1 transcripts, we co-transfected expression vectors coding full-length Keap1 truncated Keap1 cDNA (DCL1 and DCL2) along with a NQO1 promoter reporter vector into DU-145 cells, and measured the NQO1 reporter activity. As shown in Fig. 3D right panel, only full-length Keap1 transcript suppressed Nrf2-dependent NQO1 promoter reporter activity indicating that differentially spliced Keap1 transcripts code for non-functional keap1 protein in DU-145 cells.
Generation of DU-145 cells constitutively expressing Nrf2shRNA
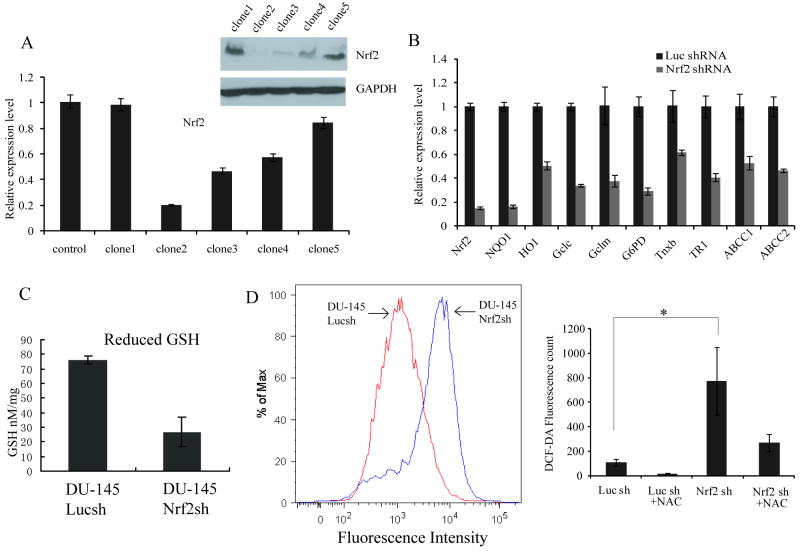
We established DU145 cells stably expressing shRNA targeting Nrf2 transcript (DU145-Nrf2shRNA) and the control DU-145 cells expressing shRNA against luciferase gene (DU145-LucshRNA). We screened several clones of DU-145 by real time RT-PCR and immunoblotting. Among the five clones screened for DU-145 cells expressing Nrf2shRNA, Clone#2 showed maximum reduction in Nrf2 transcript and protein level (Fig. 4A-B). The expression of Nrf2 in DU-145-LucshRNA cells was similar to that of the parent DU-145 cells (Supplementary Fig. S4). We used DU-145-Nrf2shRNA Clone#2 to further study the biological effect of Nrf2 knockdown in DU-145 cells. We also tested whether ectopic expression of WT Keap1 cDNA reconstituted the keap1 activity and inhibited Nrf2 activity. Results showed that over-expression of WT keap1 protein significantly inhibited Nrf2 activity and reduced the expression of classical Nrf2 target genes (Supplementary Fig. S5).
Figure 4. Generation and characterization of DU145 cells constitutively expressing Nrf2shRNA.
(A) Screening of DU-145 cell clones stably expressing Nrf2shRNA by immunoblotting and real time RT-PCR. (B) Gene expression analysis of Nrf2 and its downstream target genes in DU-145 cells stably expressing Nrf2shRNA. The Nrf2shRNA clone#2 was selected for further studies. The expression level in DU-145-Luc-shRNA was arbitrarily valued as 1. (C) Decreased total Glutathione levels in DU-145 cells with reduced Nrf2 expression. (D) Higher endogenous ROS levels in cells expressing Nrf2-shRNA as compared to cells expressing Luc-shRNA (left panel). Pretreatment with NAC (15μM) for 30min decreased the ROS levels (right panel). * P <0.05.
Lowering the expression of Nrf2 in DU-145 cells causes global decrease in the expression of electrophile and drug detoxification system
Since Nrf2 is a master regulator of glutathione and drug detoxification pathway, it is likely that the inhibition of Nrf2 activity would jeopardize glutathione biosynthesis and would lead to accumulation of ROS. Indeed, the inhibition of Nrf2 in DU-145 cells caused a decrease in the expression of all classical Nrf2 target genes such as antioxidants, Phase-II, and Phase-III detoxification genes (Fig. 4B). Ectopic expression of siRNA resistant murine Nrf2 in DU145-Nrf2shNRA cells partially restored the expression of Nrf2 dependent genes (Supplementary Fig. S6)
The level of reduced glutathione in Nrf2 knockdown cells was greatly decreased compared to control luciferase shRNA expressing cells (Fig. 4C). A decrease in antioxidant capacity led to a dramatic increase (∼10 fold) in cellular ROS levels in DU-145-Nrf2shRNA cells compared to control DU154-LucshRNA cells (Fig. 4D). Pre-treatment with NAC dramatically decreased the cellular ROS level in both control and Nrf2shRNA cells (Fig. 4D). These results suggest that the inhibition of Nrf2 activity leading to increased ROS may potentiate the toxicity of therapeutic regimens in DU-145 cells. Attenuation of Nrf2 expression in C42B cells and CWR22v1 by RNAi led to downregulation of Nrf2 dependent gene expression (Supplementary Fig. S7).
RNAi mediated inhibition of Nrf2 expression leads to enhanced chemo- and radio-sensitivity in cancer cells
We have recently shown that high Nrf2 activity in lung cancer cells leads to chemotherapeutic resistance (15). Hence, we tested the hypothesis that enhanced Nrf2 activity in DU-145 cells confers chemo-resistance, and the inhibition of Nrf2 activity will sensitize DU-145 cells to chemotherapeutic drugs.
To address whether targeted inhibition of Nrf2 pathway in DU-145 cells confers chemo-sensitivity, we exposed DU-145-LucshRNA cells and DU-145-Nrf2shRNA to increasing doses of paclitaxel for 48 hrs and cisplatin, etoposide for 72 hrs and measured cell survival by MTT assay. Nrf2 inhibition enhanced the sensitivity of DU-145 cells to paclitaxel, cisplatin, and etoposide, suggesting that Nrf2 pathway is required for detoxification and inactivation of these drugs in DU-145 cells (Fig. 5A). To determine whether enhanced sensitivity to drug in Nrf2 depleted cells results from increased intracellular drug accumulation, we measured [3H]-labeled paclitaxel accumulation in DU-145-LucshRNA and DU-145-Nrf2shRNA cells. We observed significantly higher paclitaxel accumulation in DU-145-Nrf2shRNA cells (Fig. 5B). Increased drug accumulation in cells with low levels of Nrf2 protein suggests that Nrf2 plays an important role in regulating the accumulation/detoxification of drugs in the prostate cancer cells. In PC3 cells, attenuation of Keap1 expression by RNAi approach resulted in upregulation of Nrf2 target genes and enhanced resistance to paclitaxel induced cell death (Supplementary Fig. S8).
Figure 5. Attenuation of Nrf2 expression in DU-145 cells enhances the efficacy of chemotherapeutic drugs and ionizing radiation.
(A) Enhanced sensitivity of DU145-Nrf2shRNA cells to paclitaxel, cisplatin and etoposide. Cells were exposed to drugs for 48 to 72 hrs and viable cells were determined by MTS assay. Cell survival was expressed as absorbance relative to that of untreated controls. Results are presented as means ± SD. (B) Enhanced accumulation of radiolabeled paclitaxel in Nrf2 depleted Du145-Nrf2shRNA cells. ‘*’,P<0.05. (C) Clonogenic survival of DU-145 cells stably expressing NRF2shRNA. Cells expressing non-targeting LucshRNA were used as control.
Next, we determined whether inhibition of Nrf2 expression could also sensitize DU-145 cells to ionizing radiation. DU-145 cells stably expressing luciferase-shRNA and Nrf2-shRNA were exposed to increasing doses of γ-irradiation, and cell survival was measured. (Fig. 5C). Clonogenic survival in DU-145 cells decreased as the dose of radiation increased. At low dose of 3Gy, about 70% of DU-145-LucshRNA cells were able to survive and form colonies compared with only 29% of DU-145-Nrf2shRNA cells. At medium dose of 6Gy, the survival fraction in the DU-145-LucshRNA group was 18%, while it was only 8% in the DU-145-Nrf2shRNA group. Thus, Nrf2shRNA transfectants showed a markedly increased radio-sensitivity that was more pronounced at low and medium doses compared to control LucshRNA cells. These results strongly suggest that increased Nrf2 level in DU-145 cells confers resistance to chemotherapeutic drugs as well as radiation treatment and promotes therapeutic resistance.
Enhanced Nrf2 activity promotes cell proliferation tumor formation in vivo
To determine whether aberrant Nrf2 activity correlated with proliferation in DU-145 cells in vitro, we compared the growth rate of DU-145LucshRNA and DU-145-Nrf2shRNA cells. As shown in Figure 6A, depletion of Nrf2 resulted in a pronounced decrease in cellular proliferation. To study the effect of Nrf2 inhibition on prostate tumor growth in vivo, we injected DU-145-LucshRNA and DU-145-Nrf2shRNA cells into the flank region of nude mice and monitored the tumor growth over a period of 5 weeks. Tumor volume was measured semiweekly and tumor weight was recorded at the termination of the experiment. Suppression of Nrf2 expression in DU-145 cells resulted in complete inhibition of tumor formation in >90% of nude mice recipients (p<0.001). These data indicate that Nrf2 is required for in vitro and in vivo proliferation of prostate tumor cells (Fig. 6B-C).
Figure 6. Inhibition of Nrf2 in DU-145 cells decreases cell proliferation in vitro, and reduces tumor growth in vivo.
(A) NRF2 promotes prostate cancer cell proliferation. Fifty thousand cells were seeded in each well of 6-well plates and relative cell number was measured everyday for 5 days. * P<0.001. (B) DU-145-LucshRNA and DU-145-Nrf2shRNA cells were injected into the flanks of male athymic nude mice (n = 13 for LucshRNA cells group; n = 12 for Nrf2sRNA group). Weekly measurements of tumor size were conducted for 5 weeks, and the mean tumor volume was calculated. Weight of the tumor was recorded at the end of the experiment. Data were analyzed using 2-sample Wilcoxon rank-sum test (* U=424-604, P-value<0.001).
Discussion
In the present study, for the first time, we report that Nrf2-Keap1 pathway is dys-regulated in prostate cancer. We found novel mechanisms of Keap1 inactivation in prostate cancer. In addition to mutations in Keap1 gene, we detected promoter methylation and aberrant splicing of Keap1 transcript. In particular, we report that DU-145 cells, which are known to be resistant to some of the chemotherapeutic drugs and apoptotic signals (27), exhibit a dramatic over-expression of Nrf2 protein. Inhibition of Nrf2 pathway re-sensitized DU-145 cells to chemo- and radiation-therapy.
Nrf2-Keap1 interactions are frequently dysfunctional in several cancers. Point mutations in the Keap1 gene leading to non-conservative amino acid substitutions and nonsense mutations have been reported in lung, breast and gall bladder cancer (15, 18, 19). We detected point mutations in Keap1 gene leading to non-conservative amino acid substitutions in CWR22Rv1, C42B, and LNCaP cells in central intervening region of Keap1 (a domain important for redox sensitive regulation of Nrf2). Alteration of the highly conserved amino acids further suggests that these mutations would likely abolish Keap1 repressor activity as shown previously in lung cancer cells (15).
We have demonstrated that both transcriptional and post-transcriptional mechanisms contribute to the down-regulation of Keap1 mRNA levels in DU-145 cells. Compared to other prostate cancer cell lines, DU-145 cells showed very low levels of Keap1 mRNA expression due to alternatively mRNA splicing and promoter methylation and alternatively spliced Keap1 transcript lacking exon-4 is reported in human large cell lung carcinoma (NCBI Human Genome Database); however, there are no reports on existence of alternatively spliced Keap1 transcript lacking exon-4 in prostate and other cancers. We for the first time show that such aberrantly spliced Keap1 transcripts coding for truncated Keap1 protein are also expressed in prostate cancer. Furthermore, the Keap1 promoter region appeared to undergo CpG island hypermethylation. This CpG island hypermethylation also contribute to low levels of Keap1 expression in DU-145 cells as the treatment with DNA methyl-transferase inhibitor 5-aza-cytidine in combination with HDAC inhibitor TSA significantly increased the expression of Keap1 and decreased the expression of Nrf2 target genes. These two mechanisms contribute to the loss of Keap1 activity and lead to aberrant up-regulation of Nrf2 pathway activity in DU-145 cells.
Taxol-based regimen has been one of the commonly used chemotherapeutic option for the treatment of prostate cancer (28). Paclitaxel exerts its cytotoxicity via elevation of intracellular O2•−, H2O2, and NO• levels. Cisplatin forms DNA adducts and generates ROS. Cell lines with higher antioxidant capacity have been shown to be resistant to paclitaxel and platinum drug cytotoxicity (15, 29, 30). Glutathione, thioredoxin and non-protein thiols such as metallothionins are directly linked with platinum and adriamycin resistance (31). Depletion of glutathione with buthionine sulfoximine significantly enhanced the cytotoxicity of chemotherpeutic drugs (32-34). Thus, antioxidant system plays an important role in the development of resistance to cancer therapies (29, 35). There are number of reports to support the involvement of MRP1 (or ABCC1) and glutathione metabolism in clinically relevant drug resistance in prostate cancer. Increased levels of GSH (36), p-glycoprotein, and MRP1 have been recognized in hormone-independent prostate cancer cell lines PC3 and DU-145 (37, 38). Attenuation of Nrf2 expression significantly ameliorated the cytotoxicity of paclitaxel, cisplatin and etoposide in prostate cancer cells. Enhanced paclitaxel accumulation followed by increased cell death in paclitaxel treated DU145-Nrf2shRNA cells group suggests that several mechanisms may be involved in the sensitization of DU-145-Nrf2shRNA cells to paclitaxel. The expression of Nrf2 dependent drug efflux genes such as ABCC1 and ABCC2 were diminished in Nrf2 knockdown cells and may be responsible for increased drug retention and cytotoxicity in Nrf2 depleted cells.
Ionizing radiation triggers the formation of free radicals which interact among themselves and critical biological targets with the formation of a plethora of newer free radicals. It is generally believed that production of these free radicals is the main mechanism through which radiation induces biological damage at lower radiation doses (39). Radio-protective effects by modification of antioxidant enzyme expression or by addition of free radical scavengers have been reported (3, 4, 39). Diminished Nrf2 activity in DU-145 cells led to glutathione depletion and a parallel increase in basal ROS levels. In present studies, we found that alteration of redox status by Nrf2 inhibition in prostate cancer cells enhanced the sensitivity to ionizing radiation through depletion of antioxidants and electrophile detoxification enzymes.
Increased ROS generation associated with malignant transformation renders the cancer cell highly dependent on antioxidant systems to maintain redox balance, and thus, susceptible to agents that impair antioxidant capacity (40, 41). Our results demonstrate that inhibition of Nrf2 activity in DU-145 cells suppresses cellular proliferation and attenuates tumor growth in nude mice. In other words, Nrf2 is essential for the growth of prostate cancer cells in vitro and in vivo. We speculate that severely compromised ROS scavenging machinery resulting from decreased antioxidant capacity in Nrf2 depleted cells leads to ROS accumulation and may be responsible for the slow proliferation rate of prostate cancer cells. Recently, Reddy et al (42) reported that type-II epithelial cells isolated from nrf2-/- mice lungs display defects in cell proliferation and GSH supplementation rescues these phenotypic defects (42). Thus, constitutive activation of Nrf2 is indispensable for maintaining the redox balance and growth of prostate cancer cells under homeostatic conditions. These results combined with our previous report showing reduced tumorigenicity of Nrf2 depleted lung cancer cells (20) imply that Nrf2 is crucial for tumorigenicity of different tissue/organ originated cancer cells. These results also suggest that patients with prostate cancers harboring loss of keap1 or high Nrf2 activity could receive enhanced benefit from chemo- or radio-therapy when given in combination with compounds inhibiting the Nrf2 pathway. Further studies are needed to determine the prevalence of Keap1 inactivation and Nrf2 over-activation in human prostate cancer and to test this novel therapeutic strategy in animal and human models.
Supplementary Material
Acknowledgments
We thank Paul Fallon and Jay Bream at Becton Dickinson Immune Function Laboratory, Johns Hopkins University School of Public health for help with flow cytometry. This work was supported by NIH grants P50 CA058184, research grant from the Maryland Cigarette Restitution fund (SB), P30ES03819, developmental grant from prostate spore P50 CA58236, a grant from the Prostate Cancer Foundation (SY), DOD CDMRP/PCRP grant PC073533 (SY), Clinical Innovator Award (SB) and Young Clinical Scientist award to AS from Flight Attendant Medical Research Institute. We thank Dr. David Blake for his help in GSH measurement experiments.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Pienta KJ, Smith DC. Advances in prostate cancer chemotherapy: a new era begins. CA Cancer J Clin. 2005;55:300–18. doi: 10.3322/canjclin.55.5.300. quiz 23-5. [DOI] [PubMed] [Google Scholar]
- 3.Lee HC, Kim DW, Jung KY, et al. Increased expression of antioxidant enzymes in radioresistant variant from U251 human glioblastoma cell line. Int J Mol Med. 2004;13:883–7. [PubMed] [Google Scholar]
- 4.Weiss JF, Landauer MR. Protection against ionizing radiation by antioxidant nutrients and phytochemicals. Toxicology. 2003;189:1–20. doi: 10.1016/s0300-483x(03)00149-5. [DOI] [PubMed] [Google Scholar]
- 5.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 6.Kotlo KU, Yehiely F, Efimova E, et al. Nrf2 is an inhibitor of the Fas pathway as identified by Achilles' Heel Method, a new function-based approach to gene identification in human cells. Oncogene. 2003;22:797–806. doi: 10.1038/sj.onc.1206077. [DOI] [PubMed] [Google Scholar]
- 7.Rangasamy T, Cho CY, Thimmulappa RK, et al. Genetic ablation of Nrf2 enhances susceptibility to cigarette smoke-induced emphysema in mice. J Clin Invest. 2004;114:1248–59. doi: 10.1172/JCI21146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rangasamy T, Guo J, Mitzner WA, et al. Disruption of Nrf2 enhances susceptibility to severe airway inflammation and asthma in mice. J Exp Med. 2005;202:47–59. doi: 10.1084/jem.20050538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thimmulappa RK, Mai KH, Srisuma S, Kensler TW, Yamamoto M, Biswal S. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res. 2002;62:5196–203. [PubMed] [Google Scholar]
- 10.Morito N, Yoh K, Itoh K, et al. Nrf2 regulates the sensitivity of death receptor signals by affecting intracellular glutathione levels. Oncogene. 2003;22:9275–81. doi: 10.1038/sj.onc.1207024. [DOI] [PubMed] [Google Scholar]
- 11.Nguyen T, Sherratt PJ, Pickett CB. Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu Rev Pharmacol Toxicol. 2003;43:233–60. doi: 10.1146/annurev.pharmtox.43.100901.140229. [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi A, Kang MI, Okawa H, et al. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24:7130–9. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adams J, Kelso R, Cooley L. The kelch repeat superfamily of proteins: propellers of cell function. Trends Cell Biol. 2000;10:17–24. doi: 10.1016/s0962-8924(99)01673-6. [DOI] [PubMed] [Google Scholar]
- 14.Zhang DD, Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol. 2003;23:8137–51. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh A, Misra V, Thimmulappa RK, et al. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3:e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Padmanabhan B, Tong KI, Ohta T, et al. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol Cell. 2006;21:689–700. doi: 10.1016/j.molcel.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 17.Sjoblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–74. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 18.Nioi P, Nguyen T. A mutation of Keap1 found in breast cancer impairs its ability to repress Nrf2 activity. Biochem Biophys Res Commun. 2007;362:816–21. doi: 10.1016/j.bbrc.2007.08.051. [DOI] [PubMed] [Google Scholar]
- 19.Shibata T, Kokubu A, Gotoh M, et al. Genetic alteration of Keap1 confers constitutive Nrf2 activation and resistance to chemotherapy in gallbladder cancer. Gastroenterology. 2008;135:1358–68. 68 e1–4. doi: 10.1053/j.gastro.2008.06.082. [DOI] [PubMed] [Google Scholar]
- 20.Singh A, Boldin-Adamsky S, Thimmulappa RK, et al. RNAi-mediated silencing of nuclear factor erythroid-2-related factor 2 gene expression in non-small cell lung cancer inhibits tumor growth and increases efficacy of chemotherapy. Cancer Res. 2008;68:7975–84. doi: 10.1158/0008-5472.CAN-08-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh A, Rangasamy T, Thimmulappa RK, et al. Glutathione peroxidase 2, the major cigarette smoke-inducible isoform of GPX in lungs, is regulated by Nrf2. Am J Respir Cell Mol Biol. 2006;35:639–50. doi: 10.1165/rcmb.2005-0325OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rahman I, Kode A, Biswas SK. Assay for quantitative determination of glutathione and glutathione disulfide levels using enzymatic recycling method. Nat Protoc. 2006;1:3159–65. doi: 10.1038/nprot.2006.378. [DOI] [PubMed] [Google Scholar]
- 23.Katz J, Wals P, Lee WN. Isotopomer studies of gluconeogenesis and the Krebs cycle with 13C-labeled lactate. J Biol Chem. 1993;268:25509–21. [PubMed] [Google Scholar]
- 24.Wu H, Hait WN, Yang JM. Small interfering RNA-induced suppression of MDR1 (P-glycoprotein) restores sensitivity to multidrug-resistant cancer cells. Cancer Res. 2003;63:1515–9. [PubMed] [Google Scholar]
- 25.Yegnasubramanian S, Kowalski J, Gonzalgo ML, et al. Hypermethylation of CpG islands in primary and metastatic human prostate cancer. Cancer Res. 2004;64:1975–86. doi: 10.1158/0008-5472.can-03-3972. [DOI] [PubMed] [Google Scholar]
- 26.Nelson WG, Yegnasubramanian S, Agoston AT, et al. Abnormal DNA methylation, epigenetics, and prostate cancer. Front Biosci. 2007;12:4254–66. doi: 10.2741/2385. [DOI] [PubMed] [Google Scholar]
- 27.Raffo A, Lai JC, Stein CA, et al. Antisense RNA down-regulation of bcl-2 expression in DU145 prostate cancer cells does not diminish the cytostatic effects of G3139 (Oblimersen) Clin Cancer Res. 2004;10:3195–206. doi: 10.1158/1078-0432.ccr-03-0287. [DOI] [PubMed] [Google Scholar]
- 28.Tannock IF, de Wit R, Berry WR, et al. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N Engl J Med. 2004;351:1502–12. doi: 10.1056/NEJMoa040720. [DOI] [PubMed] [Google Scholar]
- 29.Ramanathan B, Jan KY, Chen CH, Hour TC, Yu HJ, Pu YS. Resistance to paclitaxel is proportional to cellular total antioxidant capacity. Cancer Res. 2005;65:8455–60. doi: 10.1158/0008-5472.CAN-05-1162. [DOI] [PubMed] [Google Scholar]
- 30.Varbiro G, Veres B, Gallyas F, Jr, Sumegi B. Direct effect of Taxol on free radical formation and mitochondrial permeability transition. Free Radic Biol Med. 2001;31:548–58. doi: 10.1016/s0891-5849(01)00616-5. [DOI] [PubMed] [Google Scholar]
- 31.Yang P, Ebbert JO, Sun Z, Weinshilboum RM. Role of the glutathione metabolic pathway in lung cancer treatment and prognosis: a review. J Clin Oncol. 2006;24:1761–9. doi: 10.1200/JCO.2005.02.7110. [DOI] [PubMed] [Google Scholar]
- 32.van Brussel JP, Oomen MA, Vossebeld PJ, Wiemer EA, Sonneveld P, Mickisch GH. Identification of multidrug resistance-associated protein 1 and glutathione as multidrug resistance mechanisms in human prostate cancer cells: chemosensitization with leukotriene D4 antagonists and buthionine sulfoximine. BJU Int. 2004;93:1333–8. doi: 10.1111/j.1464-410X.2004.04847.x. [DOI] [PubMed] [Google Scholar]
- 33.Kramer RA, Zakher J, Kim G. Role of the glutathione redox cycle in acquired and de novo multidrug resistance. Science. 1988;241:694–7. doi: 10.1126/science.3399900. [DOI] [PubMed] [Google Scholar]
- 34.Yokomizo A, Ono M, Nanri H, et al. Cellular levels of thioredoxin associated with drug sensitivity to cisplatin, mitomycin C, doxorubicin, and etoposide. Cancer Res. 1995;55:4293–6. [PubMed] [Google Scholar]
- 35.Powis G, Kirkpatrick DL. Thioredoxin signaling as a target for cancer therapy. Curr Opin Pharmacol. 2007;7:392–7. doi: 10.1016/j.coph.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 36.Mulcahy RT, Untawale S, Gipp JJ. Transcriptional up-regulation of gamma-glutamylcysteine synthetase gene expression in melphalan-resistant human prostate carcinoma cells. Mol Pharmacol. 1994;46:909–14. [PubMed] [Google Scholar]
- 37.Zalcberg J, Hu XF, Slater A, et al. MRP1 not MDR1 gene expression is the predominant mechanism of acquired multidrug resistance in two prostate carcinoma cell lines. Prostate Cancer Prostatic Dis. 2000;3:66–75. doi: 10.1038/sj.pcan.4500394. [DOI] [PubMed] [Google Scholar]
- 38.Huisman MT, Chhatta AA, van Tellingen O, Beijnen JH, Schinkel AH. MRP2 (ABCC2) transports taxanes and confers paclitaxel resistance and both processes are stimulated by probenecid. Int J Cancer. 2005;116:824–9. doi: 10.1002/ijc.21013. [DOI] [PubMed] [Google Scholar]
- 39.Weiss JF, Landauer MR. Radioprotection by antioxidants. Ann N Y Acad Sci. 2000;899:44–60. [PubMed] [Google Scholar]
- 40.Petros JA, Baumann AK, Ruiz-Pesini E, et al. mtDNA mutations increase tumorigenicity in prostate cancer. Proc Natl Acad Sci U S A. 2005;102:719–24. doi: 10.1073/pnas.0408894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mirkovic N, Voehringer DW, Story MD, McConkey DJ, McDonnell TJ, Meyn RE. Resistance to radiation-induced apoptosis in Bcl-2-expressing cells is reversed by depleting cellular thiols. Oncogene. 1997;15:1461–70. doi: 10.1038/sj.onc.1201310. [DOI] [PubMed] [Google Scholar]
- 42.Reddy NM, Kleeberger SR, Cho HY, et al. Deficiency in Nrf2-GSH signaling impairs type II cell growth and enhances sensitivity to oxidants. Am J Respir Cell Mol Biol. 2007;37:3–8. doi: 10.1165/rcmb.2007-0004RC. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.