Abstract
Survivin, an antiapoptotic protein highly expressed in cancer, regulates multiple cellular network associated with cancer cell viability and drug resistance. Inhibition of survivin expression has been pursued as a valid cancer therapeutic target. In this study, we showed that selenium, an effective chemopreventive agent for many types of cancers, down-regulated survivin expression. Selenium inhibited survivin expression in both mRNA and protein levels in a dose- and time-dependent manner. Using a series of survivin promoter–luciferase constructs, a 37-bp DNA element in the survivin core promoter region that mediates the ability of selenium to inhibit survivin transcription was identified. Gel mobility shift assays and chromatin immunoprecipitation analyses revealed that selenium prevents the binding of Sp1 or Sp1-like proteins to the 37-bp cis-acting DNA element in the survivin promoter. Furthermore, inhibition of survivin expression by small interfering RNA enhanced selenium’s inhibitory effects on cell growth, whereas overexpression of survivin in LNCaP human prostate cancer cells desensitized cancer cells to selenium effect, suggesting that the expression of survivin plays an important role in determining the response of cancer cells to selenium. Taken together, these results suggest that selenium down-regulated survivin expression by preventing the binding of Sp1 or Sp1-like proteins to the promoter of survivin, which contributes at least in part to the inhibitory effect of selenium on survivin gene transcription. In addition, down-regulation of survivin expression may account for one of the molecular mechanisms of the anticancer effects of selenium.
Introduction
Selenium is an essential nutrient that has a chemopreventive effect against a variety of malignancies including prostate cancer. A number of case-controlled epidemiologic studies have shown an inverse relationship between selenium status and prostate cancer risk (1–5). The biological activity of selenium is dependent on its chemical form. Methylseleninic acid (CH3SeO2H; abbreviated as MSA) was developed specifically for in vitro studies (6) because cultured cells respond poorly to selenomethionine (a commonly used selenium reagent) due to very low levels of β-lyase activity, which is required for the conversion of selenomethionine to the active methylselenol (7). The effect of physiologic concentrations of MSA on cultured cells has been documented in several studies (6, 8–14).
Accumulating in vitro studies showed that selenium inhibited the growth of prostate cancer cell lines, including androgen-sensitive LNCaP and androgen-insensitive DU145 and PC3 cells (9, 10, 15, 16). In vivo studies also support the antitumorigenic role of selenium in prostate cancer. Administration of selenium resulted in a reduction of tumor growth in PC3 and LNCaP tumors in mice (14, 17). There are a number of potential mechanisms proposed for the antiproliferative effects of selenium, including anti-oxidant effects, enhancement of immune function, stimulation of apoptosis, induction of cell cycle arrest (15), and disrupting nuclear receptor signaling (8, 9, 11, 13, 18, 19).
Survivin is a unique member of the inhibitor of apoptosis protein family involved in both control of cell division and inhibition of apoptosis (20–24). It is highly expressed in embryonic and fetal organs but is undetectable in most normal adult tissues (25). Survivin is overexpressed in virtually every human cancer, making survivin as the top 4 “transcriptome” expressed in cancer cells compared with normal tissues in genome-wide searches (26). Accumulating data indicate that altered expression of survivin in cancer cells is associated with cancer progression, drug and radiation resistance, and poor disease-free or overall survival (27, 28). Due to its differential expression in cancer compared with normal tissues and functionally control of apoptosis and regulation of cell division, survivin seems to be an important cancer drug target. Different approaches aimed to target survivin, including antisense oligonucleotides (29), ribozymes (30, 31), small interfering RNAs (32), dominant negative mutants (33, 34), triplex DNA formation (35), and pharmacologic inhibitors of cyclin-dependent kinase, have been used for cancer therapeutics (36, 37). However, none of these studies focus on inhibition of survivin transcription as a potential therapeutic approach. It seems that, due to the multiple function of survivin, inhibition of survivin transcription could be an important approach to inhibit survivin expression for cancer treatment (38). Previous report indicated that the constitutive expression of survivin in cancer cells is largely resulted from the multiple Sp1 sites in the survivin core promoter region (39, 40). Thus, inhibition of Sp1 function and/or abrogation of Sp1 binding to its DNA motif could be an effective way to inhibit survivin transcription/expression.
In this study, we investigate the effect of selenium on survivin expression. We found that selenium inhibits survivin expression by preventing the binding of Sp1 or Sp1-like proteins to its specific site, thereby suppressing survivin transcription expression. Down-regulation of survivin expression resulted in an increase of cellular sensitivity to selenium-induced cell death, whereas overexpression of survivin desensitized cancer cells to selenium effect.
Materials and Methods
Reagents and Cell Culture
MSA was synthesized as described previously (6). Human PC3, LNCaP, and C4-2 prostate cancer cells were maintained in RPMI 1640 supplemented with 10% fetal bovine serum. Human MCF-7 breast cancer cells were cultured in DMEM medium supplemented with 10% fetal bovine serum, 2 mmol/L glutamine, 100 units/mL of penicillin, and 100 μg/mL of streptomycin. The cells were grown at 37°C in 5% CO2 and 95% air.
Vector Transfection and Luciferase Reporter Assay
The survivin promoter–luciferase constructs pLuc-6270, pLuc-1430, pLuc-230, pLuc-178, pLuc-123, pLuc-108, pLuc-86, and pLuc-74, representing different contiguous deletions of survivin promoter, were generated and described previously (41). For luciferase reporter assays, cells were seeded in 24-well plates (5 × 104 cells per well) and grown to about 50% to 60% confluence. Each of the relevant survivin promoter–luciferase constructs was cotransfected with pRL-TK (TK promoter–Renilla luciferase construct as internal control) into C4-2 cells. Briefly, 490 ng of pLuc-survivin construct and 10 ng of pRL-TK were added in 50 μL of serum-free RPMI 1640 in a 1.5-mL tube for each well of 24-well plates. After incubation at room temperature for 30 min, the DNA-LipofectAMINE mixture was added to each well containing 200 μL of serum-free RPMI 1640. The DNA-LipofectAMINE complex was replaced by complete medium containing 10% fetal bovine serum after incubation for 2 to 3 h. Cells were treated with MSA and processed for luciferase assays 8 to 24 h after MSA treatment. For the luciferase assay, a dual luciferase reporter assay system (Promega) was used. Cell lysates (20 μL/well) was used for measurement of luciferase activity in a luminometer by first mixing the cell lysates (20 μL) with 20 μL of luciferase assay reagent for measuring firefly luciferase activity and subsequently adding 20 μL of Stop-Glo reagent for measuring Renilla luciferase activity. Data were normalized to Renilla luciferase activity (internal control) as arbitrary units.
Stable Transfection
Transfections with the plasmid expressing the cDNA encoding a full open reading frame of survivin or empty vector were done using Superfect (Qiagen) according to the manufacturer’s protocol. Stable clones were selected in 800 μg/mL G418 and maintained in 300 μg/mL G418.
Nuclear Protein Preparation
C4-2 cells were cultured in the absence or presence of 5 μmol/L MSA for 24 h. Cells were harvested, washed twice with PBS, and resuspended in a hypotonic buffer [10 mmol/L HEPES-KOH (pH 7.9), 1.5 mmol/L MgCl2, 10 mmol/L KCl, and 0.1% NP40] and incubated on ice for 10 min. Nuclei were precipitated by 3000 × g centrifugation at 4°C for 10 min. The supernatant was collected as the cytosolic fraction. After washing once with the hypotonic buffer, the nuclei were lysed in a lysis buffer [50 mmol/L Tris-HCl (pH 8.0), 150 mmol/L NaCl, 1% Triton X-100] and incubated on ice for 30 min. The nuclear lysate was precleared by 10,000 rpm centrifugation at 4°C for 15 min. Protein concentration was determined using the Coomassie Plus protein assay kit (Pierce).
Northern Blot Analysis
Total RNA was extracted from cells with TRIzol reagent (Life Technologies). Twenty micrograms of each sample were electrophoresed on 1.2% denaturing agarose gels and transferred to a nylon membrane (MSI). A 1,600-bp fragment of survivin cDNA was labeled with [α-32P]dCTP (3,000 Ci/mmol; ICN) using the Ready-To-Go DNA labeling beads (Amersham Pharmacia Biotech). Hybridization was carried out for 3 h at 65°C in Rapid-hyb buffer (Amersham). Membranes were washed for 15 min at 65°C in 2× SSC, 0.1% SDS (twice); 0.5× SSC, 0.1% SDS; and 0.1× SSC, 0.1% SDS. Radioactivity in the membranes was analyzed with a Storm Phosphoimager System.
Western Blot Analysis
The protein extracts were resolved on 12.5% SDS-PAGE. Proteins were then transferred to nitrocellulose membrane. After blocking overnight at 4°C in 5% milk in PBS–0.1% Tween 20, membranes were incubated for 1 h at room temperature with antisurvivin rabbit polyclonal antibody (Santa Cruz Biotechnology) or anti–α-actin (Sigma) diluted in 1% milk in PBS–Tween 20. After secondary antibody incubation, immunoreactive proteins were visualized with an enhanced chemiluminescence detection system (Amersham Pharmacia Biotech).
Small Interfering RNA Preparation and Transfection
A human survivin mRNA-specific RNA oligonucleotides with 3′-TT overhangs (5′-GGCUGGCUUCAUCCACUGCTT) and reverse chain (5′-GCAGUGGAUGAAGCCAGCCTT) were chemically synthesized and purified by high performance liquid chromatography (Xeragon). Equal moles of each RNA oligonucleotide were mixed together to a final concentration of 20 μmol/L in annealing buffer [100 mmol/L KAc, 30 mmol/L HEPES-KOH, 2 mmol/L MgAc2 (pH 7.4)]. After denaturation at 90°C for 1 min, the survivin small interfering RNA mixture was annealed at 37°C for 60 min and stored at −80°C for transfection experiments. A scramble sequence (5′-CAGUCGCGUUUGCGACUGGTT) and reverse chain (5′-CCAGUCGCAAACGCGACUGTT) was not present in mammalian cells by BLAST search at National Center for Biotechnology Information. The effectiveness of the survivin small interfering RNA on survivin inhibition was 75% to 90% and confirmed in HeLa cells and MCF-7 cells (42). One day before transfection, 3 × 105 cells were seeded in six-well plate. Cells at 40% to 60% confluence were transfected with the scramble and survivin small interfering RNAs, respectively, as follows. Serum-free media (100 μL) containing 3 μg small interfering RNA were mixed with 100-μL serum-free media containing 9 μL LipofectAMINE reagents and held at room temperature. After the medium in a six-well plate was replaced by serum-free medium (800 μL/well), the small interfering RNA–LipofectAMINE mixture prepared above was added onto each well in the six-well plate within 20 to 45 min after the mixture was prepared. The transfected cells were treated with or without MSA for 72 h posttransfection. Survivin expression was analyzed by Western blots, and cell viability was analyzed by trypan blue exclusion.
Chromatin Immunoprecipitation Assay
C4-2 cells were cultured in RPMI 1640 supplemented with 10% fetal bovine serum and treated with or without 5 μmol/L MSA for 24 h. The survivin and protein complexes were cross-linked inside the cells by the addition of formaldehyde (1% final concentration) to the cells in culture. Whole-cell extracts were prepared using sonication, and an aliquot of the cross-linked receptor protein complexes were immunoprecipitated by incubation with either the Sp1-specific antibody (Santa Cruz Biotechnologies), Sp3-specific antibody (Santa Cruz Biotechnologies), or IgG antibody overnight at 4°C with rotation. Chromatin-antibody complexes were isolated from solution by incubation with protein G–Sepharose beads for 1 h at 4°C with rotation. The Sepharose-bound immune complexes were washed and eluted from beads with elution buffer (1% SDS and 0.1 mol/L NaHCO3) and DNA extracted. DNA samples from chromatin immunoprecipitation preparations were analyzed by PCR using primers spanning survivin gene in the region of promoter (forward, 5′-CGCGTTCTTTGAAAGCAGTC; reverse, 5′-CAAATCTGGCGGTTAATGGC).
Electrophoretic Gel Mobility Shift Assays
The nuclear extracts from C4-2 cells treated with or without MSA were made from the cells using low-salt and high-salt buffers, respectively, as described previously (43). Ten micrograms of nuclear protein were incubated with binding buffer containing 10 mmol/L HEPES (pH 7.9), 400 mmol/L NaCl, 1 mmol/L EDTA, and 40% glycerol and 1 μg/reaction poly (dI-dC) with 105 cpm of the γ-p32-ATP labeled oligonucleotides located in the regions of the survivin promoter between −161 and −147 bp (5′-CTACAACTCCCGGCAC) and between −146 and −124 bp (5′-ACCCCGCGCCGCCCCGCCTCTA, S22), or ′-p32-ATP labeled oligonucleotides of AR promoter (5′-TCGGGTCCCGCCCCCACCGGGC), or Oct-1 oligos (TGTCGAATGCAAATCACTAGAA) for 20 min at room temperature, respectively. The reactions were stopped with the addition of 6× DNA loading buffer and electrophoresed on a 5% nondenaturing polyacrylamide gel. The gel was dried and exposed to a phosphorimager screen. For super shift assays, the reaction mixtures were incubated with the appropriate antibodies for 45 min after the initial 20 min reaction, and the reactions were stopped with loading buffer and run on a 5% nondenaturing polyacrylamide gel.
Statistical Analysis
Student’s t test (two-tailed) was used to determine the significance between control and the treatment groups, and P < 0.05 was considered significant.
Results
Selenium Decreases Survivin Protein and mRNA Expression
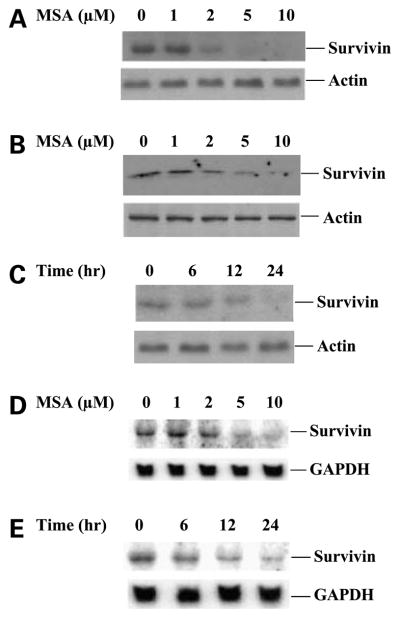
We determined the effect of selenium on the levels of survivin protein and mRNA expression by Western blot and Northern blot analyses, respectively. Human prostate cancer C4-2 and PC3 cells were treated with increasing doses of MSA with different time points. Treatment of MSA resulted in a dose-dependent inhibition of survivin protein expression in both C4-2 and PC3 cells 24 h after MSA treatment (Fig. 1A and B). A marked decrease in survivin protein by 2 μmol/L MSA treatment was detected, and it further dropped to undetectable level by 5 μmol/L MSA treatment. A decrease in survivin protein was detected as early as 6 h after exposure to 5 μmol/L MSA, and almost dropped to undetectable levels after 24 h exposure to 5 μmol/L MSA (Fig. 1C). MSA treatment was also associated with down-regulation of survivin mRNA expression (Fig. 1D and E). The decrease in survivin mRNA levels were not detected at 3 h of treatment but were noticeable after 6 h of treatment (data not shown). The down-regulation of survivin mRNA by MSA, in general, was similar to a decrease of survivin protein. These results suggest a transcriptional mechanism for the modulation of survivin expression by MSA.
Figure 1.
MSA inhibits survivin expression. A and B, Western blot analysis of survivin protein expression in C4-2 and PC3 cells treated with increasing doses of MSA for 24 h. C, time course of inhibition of survivin protein expression by 5 μmol/L MSA. α-Actin expression provided an internal control for the total amount of protein loading. D, Northern blot analysis of survivin mRNA expression in C4-2 cells treated with increasing doses of MSA for 24 h. E, time course of inhibition of survivin mRNA expression by 5 μmol/L MSA. GAPDH mRNA expression was an internal control for total RNA loading.
Selenium Down-regulates Survivin Promoter Activity
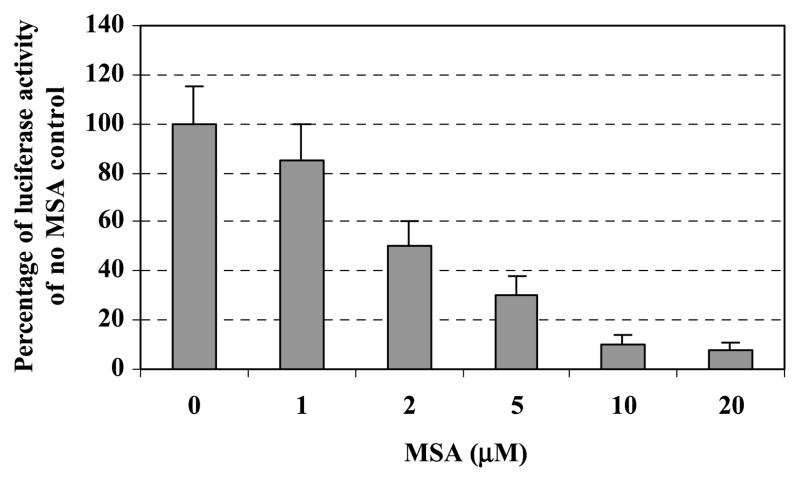
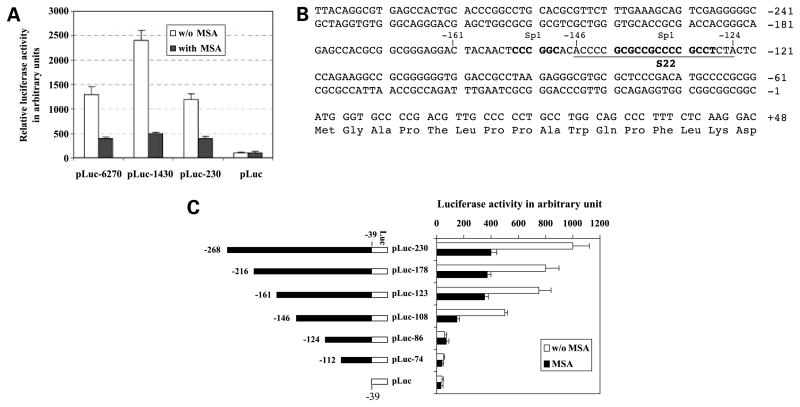
Because MSA down-regulated survivin mRNA expression, we next examined the effects of MSA on survivin promoter activity. C4-2 cells were transfected with a survivin promoter–luciferase construct, pLuc-6270, following increasing doses of MSA treatment for 24 h. MSA considerably decreased survivin promoter activity starting from 2 μmol/L concentrations and further decreased at 5 μmol/L concentration (Fig. 2). We then tried to determine which regions of the survivin promoter were responsible to MSA treatment. For this purpose, a series of truncated survivin promoter–luciferase constructs characterized previously (41) were transfected into C4-2 cells and analyzed for transcriptional activity after MSA treatment for 24 h. A 230-bp survivin core promoter region was sufficient to recapitulate the inhibitory effect of MSA on survivin gene expression (Fig. 3A).
Figure 2.
MSA down-regulates survivin promoter activity. C4-2 cells were transfected with the survivin promoter –luciferase construct pLuc-6270 and the internal control vector pRL-TK. Cells were treated with increasing doses of MSA for 24 h followed by measurement of luciferase activity using a dual luciferase reporter system as described under Materials and Methods section. Columns, percentages of data derived from a representative experiment in triplicate after normalization to Renilla luciferase activity (internal controls); bars, SD.
Figure 3.
Identification of cis-element responsible for inhibition of survivin transcription by selenium. A, the inhibitory effect of MSA on survivin transcription was mapped to the survivin core promoter region (230 bp). C4-2 cells were transfected with various survivin promoter –luciferase constructs and treated with or without MSA (5 μmol/L) for 24 h after transfection, followed by luciferase activity assays. B, DNA sequences of the survivin core promoter region (230 bp). Bold letters, Sp1 sites; underlined letters, S22 probe. C, a 37-bp cis-acting DNA element from −161 to −124 bp was identified playing a major role in MSA-mediated inhibition of survivin promoter activity. Left, the indicated survivin constructs were shown in forward orientation upstream of a luciferase reporter gene in pLuc. All the survivin promoter –luciferase constructs are from −39 bp (the ATG translation start site of survivin as +1). Right, C4-2 cells were transfected with different regions of survivin promoter– luciferase constructs and treated with or without MSA (5 μmol/L) for 24 h after transfection, followed by luciferase activity assays. Luciferase activity was normalized to Renilla luciferase and expressed in arbitrary units. Columns, mean from a representative experiment in triplicate; bars, SD.
We next narrowed the minimal responsive region in the survivin promoter using a series of 5′-deletion constructs within the 230-bp core promoter region (Fig. 3B). Transfection of these survivin promoter–luciferase construct into C4-2 cells following MSA treatment and luciferase activity assay indicated that the inhibition of survivin promoter–luciferase activity by MSA treatment in the pLuc-123 construct is as effective as those in the constructs with longer survivin promoter sequences (Fig. 3C). However, further 5′-end deletion of the survivin promoter to −124 bp resulted in the loss of MSA inhibitory effects with significant survivin promoter activity decrease (Fig. 3C). These results suggest that the minimal responsive region in the survivin promoter should be located within the 37-bp region between −161 and −124 (Fig. 3C).
Identification of Proteins Interacting with Selenium in the Minimal Responsive Regionof the Survivin Promoter
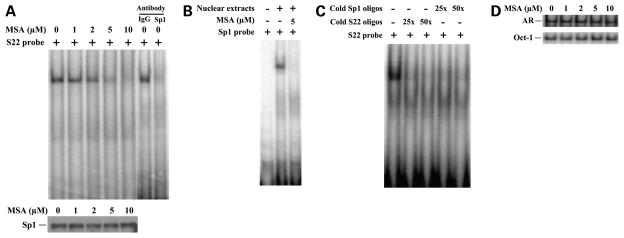
This 37-bp region harbors two consensus-binding sites for the transcription factor Sp1 (Fig. 3B; ref. 40). Sp1 has been suggested to be a major player in regulating survivin gene expression and controlling of apoptosis (40). To identify the transacting factors that interact with the MSA-responsive element, we did EMSA using [32P] oligonucleotides located in the regions between −161 and −147 bp and between −146 and −124 bp (S22) in the survivin promoter containing the two Sp1 sites, respectively. The goal of the EMSA study was to determine whether any of the oligonucleotides are bound by nuclear extracts and whether the degree of binding is altered by MSA treatment. Nuclear extracts were purified from C4-2 cells treated with increasing doses of MSA and incubated with the radiolabeled S22 probes. A DNA-protein binding complex was detected in the region between −146 and −124 bp in the survivin promoter in the absence of MSA treatment, and the intensity of the complex was reduced by 1 μmol/L MSA and disappeared by 5 μmol/L MSA treatment (Fig. 4A). No DNA-protein binding complex was detected in the region between −161 and −147 bp in the survivin promoter with or without MSA treatment in C4-2 cells (data not shown). Because the S22 probe (between −146 and −124 bp) region contains a Sp1 consensus binding site, we next examined whether MSA inhibits any nuclear proteins binding to this site by EMSA using radiolabeled Sp1 oligos as the probe. MSA treatment considerably inhibited the nuclear proteins bound to Sp1 oligos (Fig. 4B). Both cold S22 oligos and cold Sp1 oligos specifically competed with the radiolabeled S22-protein complex (Fig. 4C), suggesting that Sp1 or Sp1-like proteins specifically bind to the S22 oligos, which are inhibited by MSA treatment. To test whether MSA inhibits Sp1 protein expression, C4-2 cells were treated with increasing doses of MSA, and nuclear extracts were isolated and subjected to Western blot analysis using antibody specifically against Sp1. MSA treatment did not affect Sp1 protein expression (Fig. 4A), indicating that MSA prevents Sp1 protein binding to the specific Sp1 site (between −146 and −124 bp) in the survivin promoter rather than inhibits Sp1 protein expression. It has been shown that Sp1 plays a role in the regulation of the transcriptional activity of the AR promoter (44, 45). To test whether the survivin promoter is atypical or whether MSA prevents the binding of Sp1 to other Sp1-targeted promoters, EMSA was done using [32P] oligonucleotides located in the regions of the AR promoter containing a Sp1 site. MSA did not prevent the binding of Sp1 to the AR promoter (Fig. 4D). Taken together, these results suggest that Sp1 or Sp1-like protein binds specifically in the region between −146 and −124 bp in the survivin promoter that may be responsible for the MSA-mediated inhibition of survivin transcription.
Figure 4.
MSA reduces Sp1 or Sp1-like protein binding to the specific sites in the survivin promoter by EMSA experiments. A, MSA reduces nuclear proteins binding to the 22-bp DNA element (S22) between −146 bp and −124 bp in the survivin promoter by EMSA experiments. C4-2 cells were treated with various doses of MSA as indicated. Nuclear proteins were isolated and used for EMSA experiments with radiolabeled S22 probe (top). Specificity of Sp1 DNA binding was confirmed by supershift with antibody against Sp1 or control antibody against IgG. The expression of Sp1 protein by Western blot analysis using Sp1-specific antibody (bottom). B, the nuclear protein extracts from C4-2 cells treated with or without MSA (5 μmol/L) were used for EMSA experiments with radiolabeled Sp1 probe. C, both cold S22 and cold Sp1 oligos specifically competed with radiolabeled S22-protein complex by EMSA experiments. D, the nuclear protein extracts from C4-2 cells treated with various doses of MSA as indicated were used for EMSA experiments with radiolabeled oligos containing Sp1 binding site in the AR promoter and Oct-1 oligos as the controls.
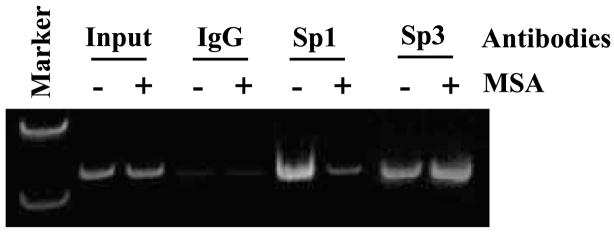
Whether the binding of Sp1 or Sp1-like protein to the specific Sp1 site between −146 and −124 bp was affected by MSA was further examined by chromatin immunoprecipitation. C4-2 cells were cultured in the presence and absence of MSA, and soluble chromatins were prepared and incubated with anti-Sp1 antibody, anti-Sp3 antibody, or normal IgG. The antibody-bound DNAs were analyzed by PCR using primers spanning the regions within the survivin promoter (Materials and Methods). As shown in Fig. 5, with DNA input as the controls, PCR produced comparable amplification for both MSA-treated and untreated samples. In contrast, PCR products amplified with DNA template enriched by anti-Sp1 antibody in the presence of MSA was dramatically reduced compared with that in the absence of MSA. However, equal levels of PCR products were amplified with DNA template enriched by anti-Sp3 antibody in the presence and absence of MSA (Fig. 5). These results further confirm that MSA specifically affect Sp1 or Sp1-like protein binding to its specific site.
Figure 5.
Effects of MSA on the recruitment of Sp1 or Sp1-like proteins and Sp3 protein to the survivin promoter. The effects of MSA in vivo on binding of Sp1 or Sp1-like proteins and Sp3 protein to the survivin promoter were examined by the chromatin immunoprecipitation assay. C4-2 cells were treated in the presence (+) or absence (−) of 5 μmol/L MSA and immunoprecipitated with antibodies against Sp1, Sp3, or IgG. Coprecipitated DNA was amplified by PCR using primers that flank the regions between −161 and −124 bp in the survivin promoter. The presence of total survivin promoter DNA in the soluble chromatin before immunoprecipitation was included as input.
The Levels of Survivin Expression Affect MSA-Induced Cell Growth Inhibition
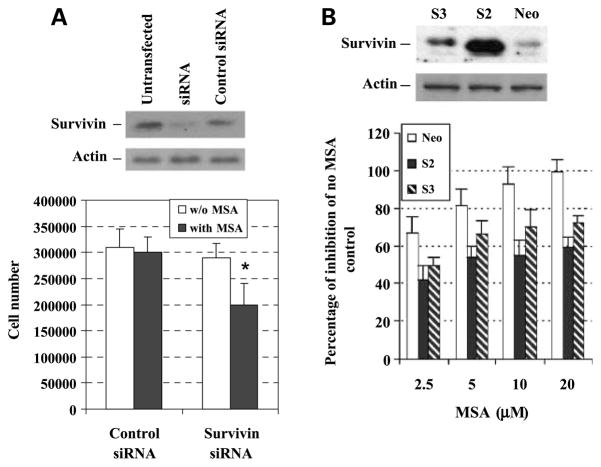
To determine whether survivin expression affects the sensitivity of prostate cancer cells to selenium treatment, C4-2 cells that express endogenous survivin were transiently transfected with a small interfering RNA oligonucleotides against human survivin (Fig. 6A, top) and treated with or without MSA, and the cell number were then determined after 72 h and compared with that of the control small interfering RNA (scramble small interfering RNA) oligonucleotides (Fig. 6A, bottom). Inhibition of survivin expression by survivin small interfering RNA significantly enhanced MSA-mediated growth inhibition compared with that of the control small interfering RNA (Fig. 6A, bottom). Alternatively, LNCaP cells that express very low levels of endogenous survivin were stably transfected with a cDNA, encoding a full-coding region of survivin to increase survivin expression. Two clones, numbers S2 and S3, exhibited overexpression of survivin and were selected and used for further studies (Fig. 6B, top). To determine if overexpression of survivin had an effect on MSA-mediated growth inhibition, we measured in vitro proliferation over 72 h. Overexpression of survivin significantly decreased MSA effects compared with that of neo vector control (Fig. 6B, bottom). These results suggest that the levels of survivin expression directly affect the sensitivity of prostate cancer cells to MSA treatment.
Figure 6.
Effects of survivin expression on the sensitivity of prostate cancer cells to selenium treatment. A, C4-2 cells were sensitized to MSA treatment by inhibition of survivin expression via survivin small interfering RNA. Top, down-regulation of survivin protein expression by small interfering RNA; bottom, C4-2 cells were transfected with control small interfering RNA (scramble small interfering RNA) or with survivin small interfering RNA and treated with MSA (2.5 μmol/L) for 3 d. Cell number was counted by trypan blue exclusion. *, statistical significance compared with control small interfering RNA treated with MSA. B, exogenous expression of survivin in LNCaP cells reduced MSA-mediated growth inhibition. Top, Western blot analysis of survivin expression in vector control (Neo) and survivin stable overexpression clones (S2 and S3) generated by transfecting a cDNA encoding a full-coding region of survivin and selected by G418; bottom, growth comparison between survivin overexpression clones (S2 and S3) and vector neo control cells treated with MSA. The cells were treated with increasing doses of MSA for 3 d; the cells were counted by trypan blue exclusion. Percentage of inhibition of S2 and S3 clones by MSA in all doses were statistically significant compared with the neo control cells treated with MSA.
Discussion
Selenium is an important trace element exhibiting anticancer activity. There are a number of potential mechanisms proposed for the anticancer effects of selenium, including antioxidant effects, enhancement of immune function, stimulation of apoptosis, and induction of cell cycle arrest (15). Selenium disrupts androgen receptor signaling (9), providing additional mechanisms of selenium anticancer action for prostate cancer. In addition, recent studies indicated that selenium down-regulated estrogen receptor signaling in breast cancer cells (11, 18, 19), suggesting that selenium may serve as a chemopreventive agent for breast cancer. In the present study, we examined the effects of selenium on the expression of survivin, one of the most important members of inhibitor of apoptosis family that regulates multiple cellular signaling and differentially expressed in cancer compared with normal tissues. We found that selenium inhibits survivin mRNA, protein, and promoter activity and prevents Sp1 protein binding to the Sp1-specific site, which is responsible, at least in part, for selenium-mediated growth inhibition. These studies provide an important molecular mechanism for selenium chemoprevention and potential therapy in prostate cancer.
We have shown that selenium inhibited survivin mRNA and protein expression in a dose- and time-dependent manner in prostate cancer cells. The levels of survivin mRNA and protein expression were significantly inhibited by 5 μmol/L selenium between 12 and 24 h. However, significant cell death induced by 5 μmol/L selenium occurs at least after 48 h of treatment (9). It is noteworthy that the reduction of survivin expression by selenium occurs at least 24 h before any significant decrease in cell number. The expression of survivin has been found to be cell cycle regulated with a low level of survivin expression in G1 phase, and its expression increases in S phase and comes to the highest level in G2-M phase (24). However, it has been shown that expression of survivin is critical for CD34+ hemotopoitic cells to go into cell cycle from G0-G1 to S and G2-M phases (46, 47). Moreover, down-regulation of survivin by vitamin D compounds has been shown to be essential for vitamin D3–induced G1-G0 cell arrest (42). Additionally, it has been also shown that up-regulation of survivin by Taxol is independent from Taxol-mediated G2-M arrest (48). Thus, down-regulation of survivin transcription by selenium compounds may also be critical for selenium-mediated G1 arrest and apoptosis induction (15, 49). The expression of survivin is also regulated by the ubiquitin-proteasome pathway in posttranslational level (50). It is interesting to test whether the ubiquitin-proteasome pathway is involved in selenium-mediated survivin down-regulation. In addition to prostate cancer cells, selenium also inhibited survivin expression in MCF-7 human breast cancer cells (data not shown), suggesting that the effect of selenium on survivin expression is not a cancer type–specific phenomena. Previous studies showed that selenium induced MCF-7 cell death and suppressed estrogen receptor expression (11, 18). Inhibition of survivin expression in MCF-7 cells may account for another important mechanism of selenium-induced cell death in MCF-7 cells.
The survivin core promoter region contains several Sp1 sites (40). The Sp1 protein may contribute to survivin gene expression and control of apoptosis during early development (40). Within the 37-bp minimal region identified in survivin promoter that is responsible for selenium-mediated survivin inhibition, it contains two Sp1 sites (40): one is between pLuc-123 and pLuc-108, and the other is between pLuc-108 and pLuc-86. Interestingly, EMSA shows that nuclear extracts from C4-2 cells bind only to the Sp1 site between pLuc-108 and pLuc-86 (S22), but not to the one between pLuc-123 and pLuc-108. Selenium treatment abrogated the DNA-protein complexes with the S22 Sp1 site. The DNA-protein complex was competed by Sp1-specific oligonucleotides, suggesting that the proteins in the complex are likely Sp1 or Sp1-like proteins. Chromatin immunoprecipitation assays suggest that MSA only prevents Sp1 binding rather than Sp3 binding to the survivin promoter. Moreover, MSA did not prevent the binding of Sp1 to the AR promoter containing Sp1 binding sites (Fig. 4D), suggesting that MSA specifically prevents Sp1 binding to the survivin promoter rather than to all Sp1-targeted promoters. Taken together, these results suggest that the 37-bp DNA motif is the minimal region that is responsible for selenium-mediated inhibition of survivin transcriptional expression and that preventing Sp1 protein bind to its specific Sp1 site by selenium treatment may contribute at least partially to the effect of selenium on the inhibition of survivin promoter activity.
Inhibition of survivin expression by selenium likely has important chemopreventive or therapeutic significance. In C4-2 human prostate cancer cells, suppression of survivin expression by small interfering RNA oligonucleotides enhanced MSA’s effects on cell growth inhibition. On the other hand, overexpression of survivin in LNCaP cells reduced MSA’s effects on cell growth inhibition. These results suggest that the change in levels of survivin expression might be one of the causes underlying the sensitivity of prostate cancer cells to selenium treatment. Survivin is selectively overexpressed in the most common human cancers and has been implicated in control of cell division, inhibition of apoptosis, and tumor cell resistance to certain anticancer agents and ionizing radiation. Thus, survivin has been proposed as an attractive cancer drug target. Pre-clinical studies, including antisense oligonucleotides, small interfering RNA, and small molecule antagonists, have been shown to inhibit survivin expression to achieve therapeutic ability. Selenium is a trace element that has been used widely as chemopreventive and potential anticancer therapeutic agent. The fact that selenium has the ability to inhibit survivin expression in this report implicates that selenium has the potential as an anticancer agent by targeting survivin signaling networks. Furthermore, targeting survivin expression provides an additional molecular mechanism of selenium anticancer activity.
In summary, we have shown in this report for the first time that selenium compound MSA down-regulates survivin protein, mRNA, and promoter activity, at least in part, through abrogation of the Sp1 or Sp1-like proteins binding to the Sp1 site in the S22 DNA element in the survivin core promoter region. Furthermore, survivin small interfering RNA–mediated down-regulation of survivin increased MSA’s inhibitory effects on prostate cancer cell growth, whereas overexpression of survivin decreased MSA’s inhibitory effects on prostate cancer cell growth, indicating the involvement of survivin modulation in selenium biological function.
Acknowledgments
Grant support: NIH grants CA109441 (A.C. Gao), CA118887 (A.C. Gao), and CA109481 (F. Li) and Roswell Park Alliance Foundation (A.C. Gao).
References
- 1.Brooks JD, Metter EJ, Chan DW, et al. Plasma selenium level before diagnosis and the risk of prostate cancer development. J Urol. 2001;166:2034–8. [PubMed] [Google Scholar]
- 2.Helzlsouer KJ, Huang HY, Alberg AJ, et al. Association between α-tocopherol, γ-tocopherol, selenium, and subsequent prostate cancer. J Natl Cancer Inst. 2000;92:2018–23. doi: 10.1093/jnci/92.24.2018. [DOI] [PubMed] [Google Scholar]
- 3.Li H, Stampfer MJ, Giovannucci EL, et al. A prospective study of plasma selenium levels and prostate cancer risk. J Natl Cancer Inst. 2004;96:696–703. doi: 10.1093/jnci/djh125. [DOI] [PubMed] [Google Scholar]
- 4.Nomura AM, Lee J, Stemmermann GN, Combs GF., Jr Serum selenium and subsequent risk of prostate cancer. Cancer Epidemiol Biomarkers Prev. 2000;9:883–7. [PubMed] [Google Scholar]
- 5.Yoshizawa K, Willett WC, Morris SJ, et al. Study of prediagnostic selenium level in toenails and the risk of advanced prostate cancer. J Natl Cancer Inst. 1998;90:1219–24. doi: 10.1093/jnci/90.16.1219. [DOI] [PubMed] [Google Scholar]
- 6.Ip C, Thompson HJ, Zhu Z, Ganther HE. In vitro and in vivo studies of methylseleninic acid: evidence that a monomethylated selenium metabolite is critical for cancer chemoprevention. Cancer Res. 2000;60:2882–6. [PubMed] [Google Scholar]
- 7.Ip C. Lessons from basic research in selenium and cancer prevention. J Nutr. 1998;128:1845–54. doi: 10.1093/jn/128.11.1845. [DOI] [PubMed] [Google Scholar]
- 8.Cho SD, Jiang C, Malewicz B, et al. Methyl selenium metabolites decrease prostate-specific antigen expression by inducing protein degradation and suppressing androgen-stimulated transcription. Mol Cancer Ther. 2004;3:605–11. [PubMed] [Google Scholar]
- 9.Dong Y, Lee SO, Zhang H, Marshall J, Gao AC, Ip C. Prostate specific antigen expression is down-regulated by selenium through disruption of androgen receptor signaling. Cancer Res. 2004;64:19–22. doi: 10.1158/0008-5472.can-03-2789. [DOI] [PubMed] [Google Scholar]
- 10.Zhao H, Whitfield ML, Xu T, Botstein D, Brooks JD. Diverse effects of methylseleninic acid on the transcriptional program of human prostate cancer cells. Mol Biol Cell. 2004;15:506–19. doi: 10.1091/mbc.E03-07-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee SO, Nadiminty N, Wu XX, et al. Selenium disrupts estrogen signaling by altering estrogen receptor expression and ligand binding in human breast cancer cells. Cancer Res. 2005;65:3487–92. doi: 10.1158/0008-5472.CAN-04-3267. [DOI] [PubMed] [Google Scholar]
- 12.Hu H, Jiang C, Ip C, Rustum YM, Lu J. Methylseleninic acid potentiates apoptosis induced by chemotherapeutic drugs in androgen-independent prostate cancer cells. Clin Cancer Res. 2005;11:2379–88. doi: 10.1158/1078-0432.CCR-04-2084. [DOI] [PubMed] [Google Scholar]
- 13.Chun JY, Nadiminty N, Lee SO, Onate SA, Lou W, Gao AC. Mechanisms of selenium down-regulation of androgen receptor signaling in prostate cancer. Mol Cancer Ther. 2006;5:913–8. doi: 10.1158/1535-7163.MCT-05-0389. [DOI] [PubMed] [Google Scholar]
- 14.Lee SO, Yeon Chun J, Nadiminty N, et al. Monomethylated selenium inhibits growth of LNCaP human prostate cancer xenograft accompanied by a decrease in the expression of androgen receptor and prostate-specific antigen (PSA) Prostate. 2006;66:1070–5. doi: 10.1002/pros.20329. [DOI] [PubMed] [Google Scholar]
- 15.Dong Y, Zhang H, Hawthorn L, Ganther HE, Ip C. Delineation of the molecular basis for selenium-induced growth arrest in human prostate cancer cells by oligonucleotide array. Cancer Res. 2003;63:52–9. [PubMed] [Google Scholar]
- 16.Jiang C, Ganther H, Lu J. Monomethyl selenium-specific inhibition of MMP-2 and VEGF expression: implications for angiogenic switch regulation. Mol Carcinog. 2000;29:236–50. doi: 10.1002/1098-2744(200012)29:4<236::aid-mc1006>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 17.Corcoran NM, Najdovska M, Costello AJ. Inorganic selenium retards progression of experimental hormone refractory prostate cancer. J Urol. 2004;171:907–10. doi: 10.1097/01.ju.0000092859.16817.8e. [DOI] [PubMed] [Google Scholar]
- 18.Shah YM, Al-Dhaheri M, Dong Y, Ip C, Jones FE, Rowan BG. Selenium disrupts estrogen receptor (α) signaling and potentiates tamoxifen antagonism in endometrial cancer cells and tamoxifen-resistant breast cancer cells. Mol Cancer Ther. 2005;4:1239–49. doi: 10.1158/1535-7163.MCT-05-0046. [DOI] [PubMed] [Google Scholar]
- 19.Shah YM, Kaul A, Dong Y, Ip C, Rowan BG. Attenuation of estrogen receptor α (ERα) signaling by selenium in breast cancer cells via down-regulation of ERα gene expression. Breast Cancer Res Treat. 2005;92:239–50. doi: 10.1007/s10549-005-3203-5. [DOI] [PubMed] [Google Scholar]
- 20.Salvesen GS, Duckett CS. IAP proteins: blocking the road to death’s door. Nat Rev. 2002;3:401–10. doi: 10.1038/nrm830. [DOI] [PubMed] [Google Scholar]
- 21.Liston P, Fong WG, Korneluk RG. The inhibitors of apoptosis: there is more to life than Bcl2. Oncogene. 2003;22:8568–80. doi: 10.1038/sj.onc.1207101. [DOI] [PubMed] [Google Scholar]
- 22.Altieri DC. The case for survivin as a regulator of microtubule dynamics and cell-death decisions. Curr Opin Cell Biol. 2006;18:609–15. doi: 10.1016/j.ceb.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 23.Altieri DC. Survivin, versatile modulation of cell division and apoptosis in cancer. Oncogene. 2003;22:8581–9. doi: 10.1038/sj.onc.1207113. [DOI] [PubMed] [Google Scholar]
- 24.Li F, Ambrosini G, Chu EY, et al. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature. 1998;396:580–4. doi: 10.1038/25141. [DOI] [PubMed] [Google Scholar]
- 25.Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3:917–21. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- 26.Velculescu VE, Madden SL, Zhang L, et al. Analysis of human transcriptomes. Nat Genet. 1999;23:387–8. doi: 10.1038/70487. [DOI] [PubMed] [Google Scholar]
- 27.Li F, Brattain MG. Role of the Survivin gene in pathophysiology. Am J Pathol. 2006;169:1–11. doi: 10.2353/ajpath.2006.060121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li F, Ling X. Survivin study: an update of ‘‘what is the next wave”? J Cell Physiol. 2006;208:476–86. doi: 10.1002/jcp.20634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li F, Ackermann EJ, Bennett CF, et al. Pleiotropic cell-division defects and apoptosis induced by interference with survivin function. Nat Cell Biol. 1999;1:461–6. doi: 10.1038/70242. [DOI] [PubMed] [Google Scholar]
- 30.Pennati M, Colella G, Folini M, Citti L, Daidone MG, Zaffaroni N. Ribozyme-mediated attenuation of survivin expression sensitizes human melanoma cells to cisplatin-induced apoptosis. J Clin Invest. 2002;109:285–6. doi: 10.1172/JCI14891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pennati M, Binda M, Colella G, et al. Ribozyme-mediated inhibition of survivin expression increases spontaneous and drug-induced apoptosis and decreases the tumorigenic potential of human prostate cancer cells. Oncogene. 2004;23:386–94. doi: 10.1038/sj.onc.1207071. [DOI] [PubMed] [Google Scholar]
- 32.Ling X, Li F. Silencing of antiapoptotic survivin gene by multiple approaches of RNA interference technology. Biotechniques. 2004;36:450–4. 6–60. doi: 10.2144/04363RR01. [DOI] [PubMed] [Google Scholar]
- 33.Tu SP, Jiang XH, Lin MC, et al. Suppression of survivin expression inhibits in vivo tumorigenicity and angiogenesis in gastric cancer. Cancer Res. 2003;63:7724–32. [PubMed] [Google Scholar]
- 34.O’Connor DS, Grossman D, Plescia J, et al. Regulation of apoptosis at cell division by p34cdc2 phosphorylation of survivin. Proc Natl Acad Sci U S A. 2000;97:13103–7. doi: 10.1073/pnas.240390697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen C, Buck A, Polat B, et al. Triplex-forming oligodeoxynucleotides targeting survivin inhibit proliferation and induce apoptosis of human lung carcinoma cells. Cancer Gene Ther. 2003;10:403–10. doi: 10.1038/sj.cgt.7700581. [DOI] [PubMed] [Google Scholar]
- 36.O’Connor DS, Wall NR, Porter AC, Altieri DC. A p34(cdc2) survival checkpoint in cancer. Cancer Cell. 2002;2:43–54. doi: 10.1016/s1535-6108(02)00084-3. [DOI] [PubMed] [Google Scholar]
- 37.Wall NR, O’Connor DS, Plescia J, Pommier Y, Altieri DC. Suppression of survivin phosphorylation on Thr34 by flavopiridol enhances tumor cell apoptosis. Cancer Res. 2003;63:230–5. [PubMed] [Google Scholar]
- 38.Zhang M, Yang J, Li F. Transcriptional and post-transcriptional controls of survivin in cancer cells: novel approaches for cancer treatment. J Exp Clin Cancer Res. 2006;25:391–402. [PMC free article] [PubMed] [Google Scholar]
- 39.Li F, Altieri DC. The cancer antiapoptosis mouse survivin gene: characterization of locus and transcriptional requirements of basal and cell cycle-dependent expression. Cancer Res. 1999;59:3143–51. [PubMed] [Google Scholar]
- 40.Li F, Altieri DC. Transcriptional analysis of human survivin gene expression. Biochem J. 1999;344(Pt 2):305–11. doi: 10.1042/0264-6021:3440305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu J, Ling X, Pan D, et al. Molecular mechanism of inhibition of survivin transcription by the GC-rich sequence-selective DNA binding antitumor agent, hedamycin: evidence of survivin down-regulation associated with drug sensitivity. J Biol Chem. 2005;280:9745–51. doi: 10.1074/jbc.M409350200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li F, Ling X, Huang H, et al. Differential regulation of survivin expression and apoptosis by vitamin D3 compounds in two isogenic MCF-7 breast cancer cell sublines. Oncogene. 2005;24:1385–95. doi: 10.1038/sj.onc.1208330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ni Z, Lou W, Leman ES, Gao AC. Inhibition of constitutively activated Stat3 signaling pathway suppresses growth of prostate cancer cells. Cancer Res. 2000;60:1225–8. [PubMed] [Google Scholar]
- 44.Ryu S, Zhou S, Ladurner AG, Tjian R. The transcriptional cofactor complex CRSP is required for activity of the enhancer-binding protein Sp1. Nature. 1999;397:446–50. doi: 10.1038/17141. [DOI] [PubMed] [Google Scholar]
- 45.Yuan H, Gong A, Young CY. Involvement of transcription factor Sp1 in quercetin-mediated inhibitory effect on the androgen receptor in human prostate cancer cells. Carcinogenesis. 2005;26:793–801. doi: 10.1093/carcin/bgi021. [DOI] [PubMed] [Google Scholar]
- 46.Fukuda S, Pelus LM. Regulation of the inhibitor-of-apoptosis family member survivin in normal cord blood and bone marrow CD34(+) cells by hematopoietic growth factors: implication of survivin expression in normal hematopoiesis. Blood. 2001;98:2091–100. doi: 10.1182/blood.v98.7.2091. [DOI] [PubMed] [Google Scholar]
- 47.Fukuda S, Pelus LM. Elevation of Survivin levels by hematopoietic growth factors occurs in quiescent CD34+ hematopoietic stem and progenitor cells before cell cycle entry. Cell Cycle. 2002;1:322–6. [PubMed] [Google Scholar]
- 48.Ling X, Bernacki RJ, Brattain MG, Li F. Induction of survivin expression by Taxol (paclitaxel) is an early event, which is independent of Taxol-mediated G2-M arrest. J Biol Chem. 2004;279:15196–203. doi: 10.1074/jbc.M310947200. [DOI] [PubMed] [Google Scholar]
- 49.Dong Y, Ganther HE, Stewart C, Ip C. Identification of molecular targets associated with selenium-induced growth inhibition in human breast cells using cDNA microarrays. Cancer Res. 2002;62:708–14. [PubMed] [Google Scholar]
- 50.Zhao J, Tenev T, Martins LM, Downward J, Lemoine NR. The ubiquitin-proteasome pathway regulates survivin degradation in a cell cycle-dependent manner. J Cell Sci. 2000;113(Pt 23):4363–71. doi: 10.1242/jcs.113.23.4363. [DOI] [PubMed] [Google Scholar]