To the Editor:
We read with interest the recent article in Nature Medicine describing the influence of variation in CCL3L1 copy number and CCR5 genotype on immune recovery during highly active antiretroviral therapy (HAART) in HIV-1 infected individuals1. The chemotactic cytokine CCL3L1 (encoding the MIP-1αP protein) is a potent ligand for the HIV-1 coreceptor CCR5, which is essential for viral entry into human host cells2. The recent study is part of a series that began in 2005 with a paper reporting effects of CCL3L1 copy number variation on HIV-1 acquisiton, viral load, and disease progression3, followed by several publications investigating clinically correlated phenotypes in a largely overlapping set of HIV+ individuals1,4,5.
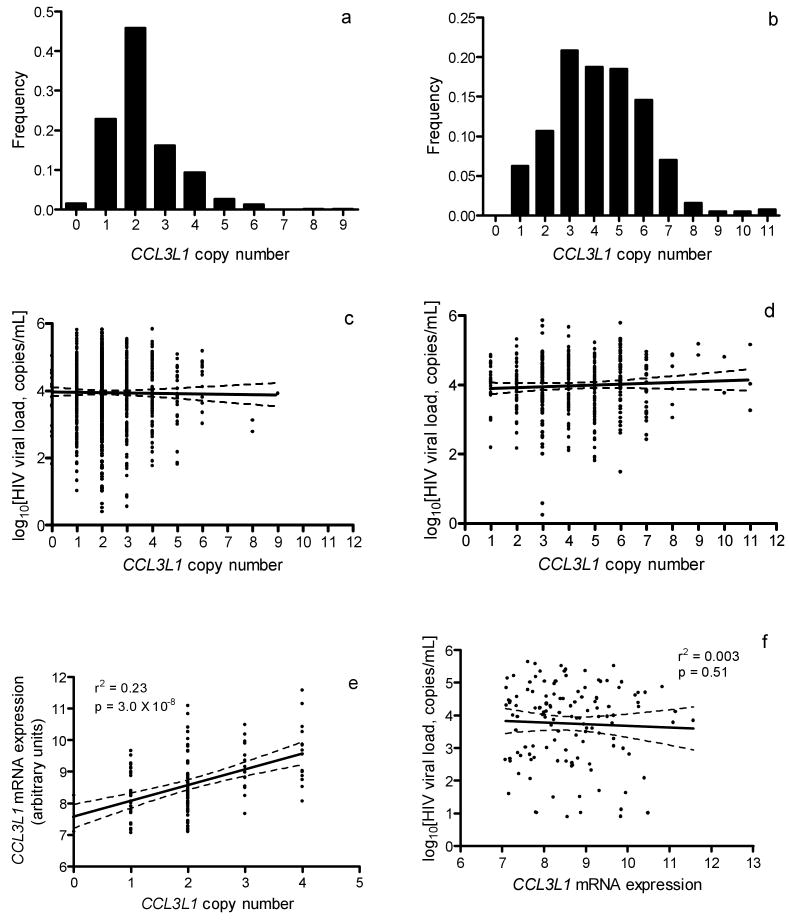
While these studies appear to generate considerable independent support for a role of CCL3L1 in viral control, many of the traits considered are at least partially correlated and the studies include largely overlapping samples and presumably CCL3L1 assay data. For these reasons we sought to re-evaluate a core set of associations related to the effect of CCL3L1 on viral control in a large group of HIV-infected patients with known date of seroconversion enrolled in one of the nine cohorts of the Euro-CHAVI Consortium6 (http://www.chavi.org, n = 1,042), in an African-American cohort from the Tri-Service AIDS Clinical Consortium (TACC) (http://www.idcrp.org/tacc2.html, n = 277) or in the Multicenter AIDS Cohort Study (MACS) (http://www.statepi.jhsph.edu/macs/macs.html) (n = 451 HIV+, n = 195 high-risk seronegative). We assayed for CCL3L1 copy number using the method described by Gonzalez et al. 3 (Supplementary Methods online). A total of 1,855 subjects were successfully genotyped. Distributions of CCL3L1 copy numbers in patients of European or African ancestry were similar to those reported elsewhere, with a median copy number of 2 or 4 in individuals of primarily European (range 0-9) or African (range 1-11) descent, respectively (Fig. 1a,b)1,3,4,7.
Figure 1.
(a and b) Distribution of CCL3L1 copy number in HIV-infected individuals of recent (a) European or (b) African ancestry. Median CCL3L1 copy number was 2 in patients of recent European descent and 4 in patients of recent African descent. (c and d) Relationship between HIV viral load at setpoint and CCL3L1 copy number among patients of recent European (c) or African (d) ancestry. Linear regression of HIV viral load at setpoint on CCL3L1 copy number showed no significant effect of CCL3L1 dose (European: r2 = 0.0006, P = 0.14; African: r2 = 0.0022, P = 0.27). (e and f) Relationship among CCL3L1 copy number, CCL3L1 mRNA expression, and viral load at set point. Specific expression of CCL3L1 mRNA in CD4+ T cells was determined using the Illumina WG-6 v3 expression array. CCL3L1 expression in CD4+ T lymphocytes showed a strong correlation with copy number, but was not associated with viremia.
We then tested for association of CCL3L1 copy number with HIV viral load at set point by linear regression after stratifying according to ethnicity and correcting for known covariates (gender, age at seroconversion, and ancestry as determined by a principal components method described previously8), and found no evidence of association (European:, P = 0.14; African: P = 0.27) (Fig. 1c,d). Dividing the sample into the previously described “high risk” (CCL3L1low) and “low risk” (CCL3L1high) genotype groups (where high risk vs. low risk is defined as having copy number below vs. equal to or above the population median, respectively)3, we again found no evidence of association, either within each population (European: P = 0.10; African: P = 0.41) or in the combined sample (P = 0.35) (Table 1). Furthermore, a model including known functional polymorphisms in the CCR5 receptor (CCR5Δ32, CCR5*HHE) in a subset of n=820 individuals of European descent for which CCR5 effects had been tested previously (Fellay et al., unpublished data), showed that while the CCR5 polymorphisms were strongly associated with viral load (CCR5Δ32: β = -0.29 +/- 0.08 log RNA copies, P = 0.001; CCR5*HHE: β = 0.14 +/- 0.05 log RNA copies, P = 0.005), there remained no appreciable effect of CCL3L1 copy number (copy number: P = 0.24; genotype risk group: P = 0.12).
Table 1.
Results of statistical tests for association of CCL3L1 copy number or genotype risk group (GRG) status with HIV infection risk or HIV-related outcomes.
| Continuous Traits | n | beta | r2 | p-value |
|---|---|---|---|---|
| Set point vs. copy number (EUR) | 1138 | -.0342 | 0.0006 | 0.14 |
| Set point vs. copy number (AFR) | 366 | .0244 | 0.0022 | 0.27 |
| Set point vs. CCL3L1high/CCL3L1low (EUR) | 1138 | -.1022 | 0.0019 | 0.10 |
| Set point vs. CCL3L1high/CCL3L1low (AFR) | 366 | .0741 | 0.0021 | 0.41 |
| Set point vs. CCL3L1high/CCL3L1low (combined) | 1504 | -.0395 | 0.0006 | 0.42 |
| Time to progression vs. copy number (EUR) | 682 | 6.64 | 0.0002 | 0.90 |
| Time to progression vs. CCL3L1high/CCL3L1low (EUR) | 682 | 62.5 | 0.0001 | 0.65 |
| Binary Traits | n (progressor) | n (nonprogressor) | Odds Ratio | p-value |
| Progressor/Nonprogressor vs. copy number (EUR) | 611 | 71 | 1.14 | 0.33 |
| Progressor/Nonprogressor vs. CCL3L1high/CCL3L1low (EUR) | 611 | 71 | 1.25 | 0.46 |
| Survival Analysis | n | Hazard ratio | 95% CI | p-value |
| Time to progression vs. CCL3L1high/CCL3L1low (EUR) | 744 | 0.95 +/- 0.09 | 0.78 – 1.15 | 0.59 |
| Tests for Association with Infection Status | n (HIV+) | n (HRSN) | Odds Ratio (95% CI) | p-value |
| HIV infection status vs. copy number | 451 | 195 | 0.86 – 1.08 | 0.53 |
| HIV infection status vs. CCL3L1high/CCL3L1low | 451 | 195 | 0.52 – 1.13 | 0.18 |
EUR, individuals of recent European ancestry; AFR, individuals of recent African ancestry (African Europeans and African Americans). Beta, odds ratios, hazard ratios and p-values are reported for the CCL3L1 copy number or GRG term after adjusting for gender, age at seroconversion, and population structure. R2 values represent the fraction of variation explained by the CCL3L1 copy number or GRG term before correction for other covariates.
We next tested whether CCL3L1 variation influences disease progression. We used both a quantitative measure of progression introduced by Fellay et al.6 (consisting of measured or estimated time to CD4+ cell count <350/mm3 or initiation of antiretroviral therapy; Supplemental Methods online) and a simple case/control comparison of progressors vs. non-progressors (defined as progression to CD4+ cell count <350/mm3 or antiretroviral therapy within 10 years since seroconversion vs. no progression within 10 years). Finally, we tested for an effect of CCL3L1low vs. CCL3L1high group on these measures as well as progression to AIDS 1987, AIDS 1993, or AIDS-related death using a Cox proportional hazards model. Neither CCL3L1 copy number nor CCL3L1low vs. CCL3L1high genotype group assignment was associated with disease progression under any of these models (P > 0.1 for all tests) (Table 1).
We then tested whether CCL3L1 copy number was associated with risk of HIV infection by comparing the copy number distributions in HIV-infected patients (HIV+) compared with individuals who were judged to be unusually exposed to HIV but remain uninfected (called high-risk seronegative, or HSRN). Using samples from the MACS cohort we compared 451 HIV+ to 195 HRSN individuals. This comparison was well powered to detect effects of CCL3L1 copy number on risk of infection through mucosal exposure (the principal model of transmission in this cohort). No association was found between infection status and either copy number (P = 0.53) or genotype risk group (P = 0.18) (Table 1). In the same sample, CCR5Δ32 homozygosity was strongly associated with reduced risk of infection (CCR5Δ32/CCR5Δ32 genotype frequency: 4.9% in exposed uninfected vs. 0% in infected individuals, P = 3.5 × 10-6). Of note is the enrichment of CCR5Δ32 homozygotes in the HRSN sample (4.9% vs. an estimated 1% in unselected individuals of European descent)9, demonstrating that the effective exposure in the HRSN cohort was very high and therefore that this cohort should provide sufficient power to detect additional genetic risk factors of reasonable effect size. Notably, we also found no effect of CCL3L1 copy number on infection risk after stratifying according to CCR5Δ32 genotype.
We investigated whether CCL3L1 copy number influences CCL3L1 mRNA expression in CD4+ T lymphocytes from 122 HIV+ patients who had not yet initiated antiretroviral therapy, using the Illumina WG-6 v3 expression array (Supplementary Methods online), and found a strong and linear increase of CCL3L1 mRNA levels with copy number (r2 = 0.23, P = 3.0 × 10-8, Fig. 1e). In the same samples, however, CCL3L1 mRNA expression itself shows no correlation with HIV set point (r2 = 0.003, P = 0.51, Fig. 1f).
These observations raise the question of why earlier studies reported positive associations which cannot be replicated here. As a possible explanation we note that measurement of CCL3L1 copy number variation appears highly susceptible to systematic biases related to the preparation and quality of DNA samples. We observed that batch differences in input DNA amounts between cases and controls can lead to biased copy number estimates by the real-time PCR method used here, and in fact found an apparently significant association in the direction opposite to that previously reported (with higher copy number among HIV+ cases compared with controls) before diluting DNA samples into an appropriate range (Supplementary Methods online). Additionally, we compared the results of different assays (the real-time PCR based assay used here and in the previous reports, and a recently published method based on the paralogue ratio test (PRT)10,11) and found that although the results were generally very highly correlated, for one comparison the association statistics from the two assays diverged markedly. Specifically, in a comparison of a small number of HIV+ and HIV- samples from Malawi, copy number estimated by the PRT method showed a strong association with infection status, whereas the real-time PCR-based estimates showed no association; this discrepancy appears to be explained by systematic differences in DNA degradation between case and control samples, in which degradation or shearing of DNA leads to systematic overestimation of copy number by the PRT method specifically (Supplementary Methods online). Among HIV+ individuals, we did not observe either assay method recording a signal of association for any HIV related quantitative trait; these tests are both more statistically powerful than the case/control comparisons and far less sensitive to any “batch effects” on the copy number estimation. Although both of the assays described here are liable to different types of systematic biases, we emphasize that differences between cohorts in the distribution of DNA concentrations are presumed to be far more likely, and perhaps expected, compared with differences in DNA storage or degradation, and thus the real-time PCR method will often be expected to produce a false positive association unless input DNA amounts are carefully considered. We therefore suggest that some of the previously reported associations may reflect differences in DNA quality or concentration which systematically increase or decrease the inferred number of copies of CCL3L1 in cases vs. control samples.
In summary, we find the absence of any significant effect of CCL3L1 copy number variation on HIV-1 infection, viral load, or disease progression. We do, however, show a highly significant association of copy number variation with CCL3L1 mRNA levels, demonstrating that the assays are sufficiently accurate to detect the intermediate biological effects of copy number variation. While there is some evidence that reduced expression of the CCR5 receptor may aid in viral control and delay progression to AIDS, there is less reason to believe that CCR5 inhibition is protective from infection without complete CCR5 blockade. Others have demonstrated that CCL3L1/MIP-1αP is expressed at relatively low levels compared with other CCR5 ligands, with measured serum concentrations well below its estimated EC50 based on ex vivo assays7,12. Indeed, concentration increases to orders of magnitude higher than those reported in both healthy and HIV-infected individuals would appear to be required to approach half-maximal occupancy by CCL3L1, whereas a reduction in receptor concentration would effectively multiply the affinity of all CCR5 ligands, including the much more abundant CCL5 (RANTES) and others in addition to MIP-1αP7. We should note that these arguments do not apply to the postulated effects of CCL3L1 that operate independently of direct CCR5 blockade, for example through effects on the expression of innate defense pathways. Such an explanation, however, might also be expected to drive correlations between CCL3L1 expression and viral control in infected individuals which were not observed (Fig. 1d). We point out that a gold standard for copy number determination in this region is yet lacking, and that the current techniques are likely to be influenced by other sources of error beyond the systematic ones describered here. Despite progress in cataloging sequence and structural variation in the CCL3L1 region13, accurate assessment of the contribution of genetic variation in such a complex region will require the development of more accurate assay methods which provide information not only about gene copy number but also gene content. Finally, we emphasize that these results do not cast any doubt on efforts to develop CCR5 antagonists (i.e. MIP-1αP analogs) as therapeutics for HIV prevention and treatment, but merely argue that natural variation in CCL3L1 gene dose does not appear to have any important effects on the control of HIV-1.
Supplementary Material
Acknowledgments
Funding was provided by the NIH-funded Center for HIV/AIDS Vaccine Immunology. J.F. is supported by the Swiss Foundation for Grants in Biology and Medicine, and A.T. is supported by Infectigen and the Swiss National Science Foundation. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This Research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
References
- 1.Ahuja SK, et al. CCL3L1-CCR5 genotype influences durability of immune recovery during antiretroviral therapy of HIV-1-infected individuals. Nat Med. 2008;14:413–420. doi: 10.1038/nm1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Townson JR, Barcellos LF, Nibbs RJ. Gene copy number regulates the production of the human chemokine CCL3-L1. Eur J Immunol. 2002;32:3016–3026. doi: 10.1002/1521-4141(2002010)32:10<3016::AID-IMMU3016>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 3.Gonzalez E, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 4.Dolan MJ, et al. CCL3L1 and CCR5 influence cell-mediated immunity and affect HIV-AIDS pathogenesis via viral entry-independent mechanisms. Nat Immunol. 2007;8:1324–1336. doi: 10.1038/ni1521. [DOI] [PubMed] [Google Scholar]
- 5.Kulkarni H, et al. CCL3L1-CCR5 genotype improves the assessment of AIDS Risk in HIV-1-infected individuals. PLoS ONE. 2008;3:e3165. doi: 10.1371/journal.pone.0003165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fellay J, et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shao W, et al. CCL3L1 and CCL4L1: variable gene copy number in adolescents with and without human immunodeficiency virus type 1 (HIV-1) infection. Genes Immun. 2007;8:224–231. doi: 10.1038/sj.gene.6364378. [DOI] [PubMed] [Google Scholar]
- 8.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 9.Stephens JC, et al. Dating the origin of the CCR5-Delta32 AIDS-resistance allele by the coalescence of haplotypes. Am J Hum Genet. 1998;62:1507–1515. doi: 10.1086/301867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Armour JA, et al. Accurate, high-throughput typing of copy number variation using paralogue ratios from dispersed repeats. Nucleic Acids Res. 2007;35:e19. doi: 10.1093/nar/gkl1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walker S, Janyakhantikul S, Armour JA. Multiplex Paralogue Ratio Tests for accurate measurement of multiallelic CNVs. Genomics. 2009;93:98–103. doi: 10.1016/j.ygeno.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 12.Menten P, et al. The LD78beta isoform of MIP-1alpha is the most potent CCR5 agonist and HIV-1-inhibiting chemokine. J Clin Invest. 1999;104:R1–5. doi: 10.1172/JCI7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paximadis M, Mohanlal N, Gray GE, Kuhn L, Tiemessen CT. Identification of new variants within the two functional genes CCL3 and CCL3L encoding the CCL3 (MIP-1alpha) chemokine: implications for HIV-1 infection. Int J Immunogenet. 2009;36:21–32. doi: 10.1111/j.1744-313X.2008.00815.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.