Abstract
Ubiquitin-like proteins (Ubls) are conjugated by dynamic E1-E2-E3 enzyme cascades. E1s activate Ubls by catalyzing Ubl C-terminal adenylation, forming a covalent E1~Ubl thioester intermediate, and generating a thioester-linked E2~Ubl product, which must be released for subsequent reactions. We report structural analysis of a trapped Ubl activation complex containing NEDD8’s heterodimeric E1 (APPBP1-UBA3), two NEDD8s (one thioester-linked to E1, one noncovalently-associated for adenylation), a catalytically-inactive E2 (Ubc12), and MgATP. The results suggest that a thioester switch toggles E1-E2 affinities. Two E2 binding sites depend on NEDD8 being thioester-linked to E1. One is unmasked by a striking E1 conformational change. The other comes directly from the thioester-bound NEDD8. After NEDD8 transfer to E2, reversion to an alternate E1 conformation would facilitate release of the E2~NEDD8 thioester product. Thus, transferring the Ubl’s thioester linkage between successive conjugation enzymes can induce conformational changes and alter interaction networks to drive consecutive steps in Ubl cascades.
Post-translational modification with ubiquitin-like proteins (Ubls), such as ubiquitin, NEDD8, and SUMO, is an essential eukaryotic regulatory mechanism1. Ubiquitin, NEDD8, and other Ubls are covalently conjugated via their C-termini to targets by related, but distinct cascades that involve the sequential actions of E1, E2, and E3 enzymes2-12 (Fig. 1a). For clarity, we designate covalent complexes with a tilde (~), and noncovalent complexes with a hyphen (-). E1s activate Ubls via multiple steps. First, E1 binds ATP, Mg2+, and the Ubl, and catalyzes adenylation of the Ubl’s C-terminus. Second, E1’s catalytic cysteine attacks the Ubl~adenylate, producing a covalent thioester-linkage between the E1’s catalytic cysteine and the Ubl’s C-terminus. The thioester-linked Ubl will be transferred directly to an E2. However, before this transfer, the E1 binds another Ubl molecule at the adenylation active site. Thus, during the activation cycle, the E1 binds two Ubl molecules, each at a distinct site: Ubl(T) is linked to the E1’s catalytic cysteine via a thioester, and Ubl(A) is bound noncovalently at the adenylation active site. Next, this doubly-Ubl-loaded E1 associates with an E2. Finally, a transthiolation reaction ensues whereby Ubl(T) is transferred from the E1’s catalytic cysteine to the E2’s catalytic cysteine, the E2~Ubl thioester product is released from E1, and the activation cycle continues for the noncovalently-associated Ubl(A) molecule. Consequently, the E1 cycles back and forth between doubly-Ubl-loaded and singly-Ubl(A)-loaded forms as it binds each free E2 substrate and releases each E2~Ubl product. Following the activation process, the E2~Ubl complex typically associates with an E3, which facilitates Ubl transfer to the target.
Figure 1. Structure of APPBP1–UBA3~NEDD8(T)–NEDD8(A)–MgATP–Ubc12(C111A), a trapped Ubl activation complex.

a, Ubl conjugation2-12. b, APPBP1–UBA3~NEDD8(T)–NEDD8(A)–MgATP–Ubc12(C111A) structure. Surfaces, APPBP1–UBA3’s adenylation and cysteine domains; secondary structures, UBA3’s ufd, NEDD8s, and Ubc12. Blue, APPBP1; pink/red, UBA3; yellow, NEDD8(T); lime, NEDD8(A); cyan, Ubc12; green, UBA3’s catalytic Cys216 and Ubc12’s residue 111 (here Ala; Cys in wild-type Ubc12). c, APPBP1–UBA3–NEDD8(A)20, colored/oriented as left APPBP1–UBA3~NEDD8(T)–NEDD8(A)–MgATP–Ubc12(C111A) in b. d, APPBP1–UBA3~NEDD8(T)–NEDD8(A)–MgATP–Ubc12(C111A)’s NEDD8(T) overlaid with APPBP1–UBA3–NEDD8(A)’s ufd20, colored/oriented as in b, c. X, structural overlap. e, Close-up of cryptic Ubc12-binding surface from APPBP1–UBA3~NEDD8(T)–NEDD8(A)–MgATP–Ubc12(C111A), oriented as right view in b. f, Close-up of region corresponding to e from APPBP1–UBA3–NEDD8(A)20.
It is important to understand how structural properties of different enzyme forms drive successive steps in conjugation. Recent structures of the NEDD8 and SUMO E1s, alone and in noncovalent singly-loaded complexes with NEDD8(A) or SUMO(A), respectively, revealed similar overall domain orientations13-15. However, several previous studies suggest distinct structural properties for E1 and E2 forms involved in latter steps of Ubl activation. First, E1s and E2s display different relative affinities for each other in their free and covalent thioester-linked enzyme~Ubl states: free E1s display low affinity for E2s, doubly-Ubl-loaded E1s bind their free E2 substrates with high affinity, and E2~Ubl thioester products are released from E1s4,6,7. Second, these differential affinities between distinct enzyme forms are required for progression of E1-E2-E3 cascades, because there is structural overlap between the E1 and E3 binding sites on E2s, and E2s cannot bind their E1 and E3 partners simultaneously16-19. Finally, upon docking the structure of a complex between E1 and E2 domains onto full-length structures of apo or singly-Ubl(A)-loaded E1s, an E2 would bind the opposite side and face away from the E1’s catalytic cysteine13,15,17,20,21. Thus, significant conformational changes would be required for the E1 and E2 catalytic cysteines to face each other. This raises the questions of what roles different E1 conformations might play during the activation cycle, and what would drive E1 conformational changes. To understand the molecular switches influencing E1, E2, and Ubl interactions, we determined the structure of a trapped activation complex for the NEDD8 pathway.
Trapped Ubl activation complex structure
Wild-type activation complexes containing an E1, Ubl(T), Ubl(A), and an E2 are not stable, because Ubl(T) is readily passed from the E1’s catalytic cysteine to that of E2. Although an E2’s catalytic cysteine is also essential for long-range allosteric changes in E1 structure22, it was necessary to use a catalytically-inactive E2 harboring a cysteine-to-alanine mutation to trap an activation complex that would provide insights into intermolecular interactions among all the components. Using this approach, we determined the crystal structure of a trapped activation complex containing APPBP1–MBP-UBA3 (NEDD8’s heterodimeric E1, with UBA3 fused to the C-terminus of the MBP crystallization tag), two NEDD8s [NEDD8(T) thioester-bound to UBA3’s catalytic Cys216, NEDD8(A) noncovalently-associated at the adenylation active site], MgATP, and a catalytic cysteine-to-alanine (C111A) mutant of Ubc12 (NEDD8’s E2). All MBP contacts are to regions of APPBP1, UBA3, and NEDD8(A) in conformations identical to previous structures, so MBP is not discussed further in the text. This trapped activation complex is referred to hereafter as APPBP1–UBA3~NEDD8(T)–NEDD8(A)–MgATP–Ubc12(C111A) (Table 1, Supplementary Table 1, Supplementary Fig. 1).
Table 1. X-ray refinement statistics.
| Resolution (Å) | 50–2.8 |
| Rwork/Rfree | 0.241/0.274 |
| Number of atoms | |
| Protein | 13,012 |
| Ligand/ion | 33 |
| Water | 45 |
| B-factors | |
| Protein | 83.8 |
| Ligand/ion | 64.4 |
| Water | 59.4 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 1.44 |
Rwork = Σ|Fo-Fc|/ΣFo. Rfree is the cross-validation of R-factor for the test set of reflections (5% of total) omitted in model refinement. R.m.s., root mean square.
Previous structural studies revealed that APPBP1–UBA3 and other E1s display common modular architectures, with individual domains specifying each activity: an adenylation domain, a catalytic cysteine-containing domain, and a domain structurally resembling ubiquitin (ubiquitin-fold domain, ufd) that binds E213,15,17,23. Ubls, such as ubiquitin and NEDD8, have two regions: an N-terminal globular domain and a flexible C-terminal tail24,25. NEDD8’s E2, Ubc12, also has two regions: a unique N-terminal sequence, and a catalytic core domain conserved among all E2s17,21,26. These features are all visible in APPBP1–UBA3~NEDD8(T)–NEDD8(A)–MgATP–Ubc12(C111A), which adopts a compact overall structure. In this complex, the three APPBP1–UBA3 domains pack to generate a large central groove, which cradles the MgATP, both molecules of NEDD8, and Ubc12 substrates together (Fig. 1b). A crossover loop connecting the adenylation and catalytic-cysteine domains divides the groove into two clefts that are continuous both below and above the loop. As in previous structures, when viewed facing the E1 catalytic cysteine located centrally above the adenylation domain, NEDD8(A)’s globular domain binds in the right cleft (Cleft 2) with its C-terminal tail extending under the crossover loop to approach ATP’s α-phosphate in the left cleft (Cleft 1)13,20. Ubc12’s unique peptide-like extension docks in a groove unique to UBA3’s adenylation domain, and Ubc12’s core domain binds UBA3’s ufd17,21.
NEDD8(T) is in the center of the complex, with its C-terminus tethered within a channel focused on the thioester-bond (Supplementary Fig. 2). A network of charged and polar side-chains contacts UBA3’s catalytic Cys and NEDD8(T)’s C-terminus. Mutational analysis shows these residues contributing to APPBP1–UBA3~NEDD8(T) and Ubc12~NEDD8 complex formation. Conservation of this electrostatic network suggests that common mechanisms underlie E1-catalyzed formation of E1~Ubl and E2~Ubl thioester complexes.
Relative to apo and singly-Ubl(A)-loaded E1 structures13,15,20,21, the APPBP1–UBA3~NEDD8(T)–NEDD8(A)–MgATP–Ubc12(C111A) structure reveals a striking ~120° rotation of the E2-binding ufd (Fig. 1, Supplementary Fig. 3). The ufd rotation results in remodeling of the APPBP1–UBA3 central groove to accommodate NEDD8(T)’s globular domain above the crossover loop in the middle of the groove, and Ubc12’s core domain in Cleft 1. In the alternative conformation found in previous apo and singly-Ubl(A)-loaded E1 structures, the central portion of the E1 groove is partially occupied by the E1’s ufd13,15,20,21 (Fig. 1c). With APPBP1–UBA3 doubly-loaded with two NEDD8 molecules, the thioester-bound NEDD8(T) would clash with UBA3’s ufd in the apo/singly-Ubl(A)-loaded orientation (Fig. 1d), suggesting that the ufd conformational change would accompany doubly-NEDD8-loading of APPBP1-UBA3.
The large-scale ufd rotation reorients Ubc12 as compared to previous models such that (1) Ubc12’s catalytic cysteine faces the direction of UBA3’s catalytic cysteine; (2) Ubc12 is adjacent to the thioester-bound NEDD8(T); and (3) Ubc12 can bind two new E2-binding surfaces not present in apo and singly-Ubl(A)-loaded E1 forms. Interactions between doubly-NEDD8-loaded APPBP1–UBA3 and Ubc12 bury 5600Å2.
In the present structure, which represents a trapped rather than a functioning activation complex, there is a ~20Å gap between Ubc12’s residue 111 (here an alanine, but in wild-type Ubc12 the catalytic cysteine) and UBA3’s catalytic Cys216. Although this gap is still greater than the distance required for transfer of NEDD8(T) to Ubc12, this finding is consistent with previous kinetic studies indicating that long-range conformational changes in E1 structure are induced by an E2’s catalytic cysteine22, which is absent from our structure. Accordingly, a corresponding ~20Å gap in APPBP1–UBA3’s central groove between NEDD8(T) and NEDD8(A) could accommodate further conformational changes to support the transthiolation reaction.
A cryptic Ubc12-binding site unmasked
One Ubc12-binding surface is a V-shaped groove. One face of the V comes from UBA3’s ufd, and the other comes from the adenylation domain portion of UBA3, immediately adjacent to the ATP binding site (Figs. 1b, 1e, 2a). In apo and singly-Ubl(A)-loaded E1s, this surface is concealed by the ufd in its alternate position13,15,20,21 (Fig. 1f). However, in the structure containing two NEDD8s, the ufd has turned around and peeled away from the adjacent adenylation domain to create a single, continuous Ubc12-binding surface involving both domains. Mutational analysis underscores the importance of this cryptic site for E2 recruitment (Supplementary Fig. 4). Notably, the location of this Ubc12 binding site would allow transmission of subtle structural differences resulting from occupation of the nucleotide-binding site to the Ubc12 binding site.
Figure 2. Two Ubc12 binding sites depend on NEDD8(T) being thioester-linked APPBP1–UBA3.

a, Close-up view of the cryptic Ubc12-binding surface near the nucleotide-binding site, with the adenylation domain portion of UBA3 in pink, NEDD8(A) in lime, and Ubc12 in cyan. b, Close-up view of direct interactions between Ubc12 (cyan) and NEDD8(T) (yellow).
NEDD8(T) – also a binding site for Ubc12
Ubc12 also binds directly to NEDD8(T). A hydrophobic groove formed by Ubc12’s α3 helix wraps around NEDD8(T)’s β1β2-loop, and additional electrostatic interactions further stabilize the complex (Fig. 2b). Other Ubl pathways likely involve parallel interactions recruiting E2s to their corresponding doubly-Ubl-loaded E1 complexes, as structural studies of thioester analog complexes between ubiquitin and SUMO with their E2s also involve interactions between Ubl surfaces corresponding to NEDD8’s β1β2-loop and E2 surfaces corresponding to Ubc12’s α3 helix19,27-29.
Alanine-scanning over Ubc12’s core domain revealed that mutations at each interaction surface significantly decrease formation of the Ubc12~NEDD8 thioester product (Supplementary Fig. 5). Further enzyme kinetic analyses revealed increased Km values for mutants with substitutions for residues involved in binding (1) UBA3’s ufd, (2) UBA3’s adenylation domain adjacent to the ATP binding site, and (3) the thioester-bound NEDD8(T) (Supplementary Table 2). Thus, the structurally observed E2-binding surfaces are critical for Ubc12 recruitment for the transthiolation reaction.
Model for E1-E2 transthiolation
Our structure suggests a model for transthiolation, in which the E1 and E2 cysteine-to-cysteine gap would be closed. Continuing along the same trajectory as between the apo/singly-NEDD8(A)-loaded E1 and the trapped activation complex presented here, an additional ~10° rotation of UBA3’s ufd would bring the E1 and E2 cysteines into close proximity (Fig. 3a). Because E2 core domains have oblong, tower-like structures, a small rotation at the base translates into a large translation for the catalytic cysteine. Several features of our analyses support this model. First, this rotation would also require a corresponding movement of NEDD8(T), and there is an equivalent gap in APPBP1–UBA3’s central groove to accommodate NEDD8(T) in a rotated position without clashes. Second, other than interactions with Ubc12, NEDD8(T) is tethered to APPBP1–UBA3 through its flexible C-terminus. The structural flexibility of Ubl C-terminal tails24,25 would allow NEDD8(T) to rotate along with Ubc12. Finally, our model predicts one additional Ubc12 surface involved in transthiolation: the channel leading to Ubc12’s catalytic Cys111 would secure NEDD8(T)’s C-terminal tail for the reaction. In support of the model, our alanine-scanning revealed that residues lining the channel to Cys111 contribute the only side-chains involved in forming the Ubc12~NEDD8 thioester complex that are outside the structurally-observed interfaces (Supplementary Fig. 5). It seems likely that Ubc12’s catalytic cysteine, absent from our trapped complex, stabilizes the rotated, active conformation22. It is also possible that adenylation of the noncovalently-bound NEDD8(A) may trigger conformational changes in UBA3’s adenylation domain contributing to rotation at the base of Ubc12.
Figure 3. Model of a Transthiolation Complex.

a, An additional ~10° ufd rotation allows juxtaposition of Ubc12’s catalytic Cys111 with the UBA3~NEDD8(T) thioester, in a putative conformation for the transthiolation reaction. The APPBP1–UBA3~NEDD8(T)–NEDD8(A)–MgATP–Ubc12(C111A) structure is in the left panel, the model is in the right panel, and proteins are colored as in Fig. 1b. b, Superposition of the ufd-Ubc12~NEDD8 thioester model onto the APPBP1–UBA3–NEDD8(A) structure20, generated by least squares alignment of Cα atoms in UBA3’s ufd. Much of NEDD8 in the Ubc12~NEDD8 thioester model is not visible due to clashing with the APPBP1–UBA3 surface.
Following the transthiolation reaction, the Ubc12~NEDD8 thioester product is released. Elimination of the covalent tether between NEDD8(T) and UBA3 would allow rotation of UBA3’s ufd back to the conformation observed in the apo and singly-NEDD8(A)-loaded APPBP1–UBA3 structures. To gain insight into this process, we docked the Ubc12~NEDD8 thioester complex from our transthiolation model onto the previous APPBP1–UBA3–NEDD8(A) structure20. With the ufd in the alternate orientation, substantial clashing between Ubc12~NEDD8 and APPBP1–UBA3 could facilitate product release (Fig. 3b). Similarity in the overall domain orientations in the apo and singly-Ubl(A)-loaded structures of SUMO’s E115 (Supplementary Figure 3) indicates that ufd rotations play similar roles in the activation cycles of other Ubls.
A thioester switch modulates affinities
Our results suggest that a Ubl-dependent thioester switch toggles E1–E2 interactions to drive the activation cycle (Fig. 4). First, E1 is switched into a form that binds E2 (Fig. 4a, b). In the doubly-Ubl-loaded E1, with Ubl(T) thioester-linked to E1, the E1’s conformational change reorients the E2-binding ufd and unmasks a cryptic E2-binding site. A second E2 binding site comes directly from Ubl(T) thioester-linked to E1. Transfer of Ubl(T)’s covalent linkage from E1 to the E2’s cysteine generates the E2~Ubl thioester product. In the absence of Ubl(T)’s covalent tether to E1, the switch flips back (Fig. 4c). Lack of Ubl(T)-dependent E2-binding sites, combined with steric clashing between the E2~Ubl thioester product and the singly-Ubl(A)-loaded E1 likely facilitate product release (Fig. 4d). Thus, E1–E2 interactions depend on whether E1 or E2 is thioester-bound to the Ubl. Formation of the E1~Ubl thioester favors interaction with free E2, whereas formation of the E2~Ubl thioester leads to release.
Figure 4. A thioester-switch toggling E1-E2 interactions.
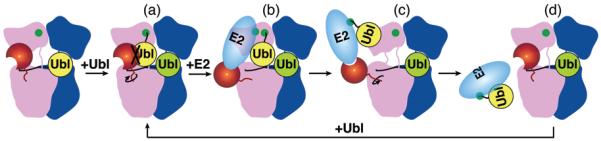
E1 is blue/pink, corresponding to APPBP1/UBA3, with UBA3’s ufd red. E2 is cyan. The first and second Ubls binding E1 are yellow and lime, respectively. Catalytic cysteines are green. a, Ubl(T), thioester-bound to E1, clashes with E1’s ufd in initial conformation. b, E1’s ufd rotation unmasks cryptic E2-binding sites, allowing doubly-Ubl-loaded E1 to bind free E2. c, Ubl transfer to E2’s cysteine eliminates the Ubl’s covalent tether to E1. This removes E2 binding sites, and allows reversion to the alternate E1 conformation. d, Steric-clashing between E1 and E2~Ubl facilitates product-release. Another activation cycle ensues.
Implications for Ubl cascades
A fundamental characteristic of Ubl pathways is the sequential formation of transient complexes during E1-E2-E3 conjugation cascades, and also during handoffs of monoubiquitinated cargo between different ubiquitin-recognition complexes. The same surfaces of enzymes, and of Ubls, are recognized at consecutive steps during many stages of these pathways18,30-35. In order for Ubl pathways to proceed, distinct transient interactions and/or catalytic activities are likely specified by different states of the E1-E2-E3 enzymes, and of Ubl recognition machineries. Our results show how covalent tethering of a Ubl to an enzyme’s active site can allow switching between states, with altered protein-protein interaction abilities, required for progression of the transfer cascade. Notably, NEDD8 and UBA3’s ufd, which occupy overlapping positions in the different E1 states, are structurally similar. Many other proteins in Ubl pathways also contain domains structurally resembling Ubls that could play roles in switching between states. It is likely that the Ubl-handoffs occurring at other stages of conjugation and recognition pathways are likewise driven forward by each reaction inducing conformational changes and altering interaction networks to trigger the next step in the cascade.
Methods
Protein preparation, crystallization, structure determination, and biochemical assays are described in detail in Supplementary Information. APPBP1–UBA3~NEDD8(T)–NEDD8(A)–MgATP–Ubc12(C111A) was generated by mixing purified APPBP1–MBP-Δ11UBA3, Δ5-2Ubc12(Cys111Ala), and NEDD8 at a 1:2:4 molar ratio in 25 mM HEPES, 150 mM NaCl, 5 mM MgCl2, 5 mM ATP (pH 7.0) at 4°C for 18 hours, and purified by gel filtration in 25 mM HEPES, 150 mM NaCl, 0.5 mM MgCl2, 0.5 mM ATP (pH 7.0). Crystals grew immediately upon setting drops at 18°C by the hanging drop vapor diffusion method, in 17% PEG 3350, 0.2 M di-sodium tartrate, 0.1 M HEPES (pH 7.0). The crystals belong to space group P3221 with a=b=156.5 Å, c=190.5 Å, and contain one complex per asymmetric unit. Diffraction data were collected at the APS SERCAT and ALS 8.2.2 and 8.3.1 beamlines. The structure was determined by molecular replacement, with four searchmodels: the APPBP1–UBA3 adenylation domain (APPBP1’s residues 9–167 and 395–534 and UBA3’s residues 12–208 and 293–347), the APPBP1–UBA3 catalytic cysteine domain (APPBP1’s residues 181–374 and UBA3’s residues 213–287), NEDD8 residues 1–73, and the complex between UBA3’s ufd and Ubc12’s core domain13,17,20,21. APPBP1’s residues 1–4, MBP’s residues 1, 299–302, and 312–319, and Ubc12’s residues 1–2 are presumably disordered and are not present in the final model, which was refined to 2.8 Å resolution and R=24.1, Rfree=27.4.
Supplementary Material
Acknowledgements
We dedicate this manuscript to the memory of Cecile Pickart. We thank C. Ralston, B. Sankaran, and A. Howard for assistance with data collection at the 8.2.2 beamline at ALS and at the SERCAT beamline at APS. We thank P. Murray, D. Minor, B. Dye, and D. Scott for critical reading of the manuscript, members of the Schulman lab for helpful discussions, and C. Ross for X-ray support. This work was supported in part by ALSAC and grants from the NIH (B.A.S.) and the Charles A. King Medical Foundation (M.O.). B.A.S. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no competing financial interests.
References
- 1.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell. Dev. Biol. 2006;22:159–80. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 2.Haas AL, Warms JV, Hershko A, Rose IA. Ubiquitin-activating enzyme. Mechanism and role in protein-ubiquitin conjugation. J. Biol. Chem. 1982;257:2543–8. [PubMed] [Google Scholar]
- 3.Haas AL, Rose IA. The mechanism of ubiquitin activating enzyme. A kinetic and equilibrium analysis. J. Biol. Chem. 1982;257:10329–37. [PubMed] [Google Scholar]
- 4.Haas AL, Bright PM, Jackson VE. Functional diversity among putative E2 isozymes in the mechanism of ubiquitin-histone ligation. J. Biol. Chem. 1988;263:13268–75. [PubMed] [Google Scholar]
- 5.Ciechanover A, Elias S, Heller H, Hershko A. “Covalent affinity” purification of ubiquitin-activating enzyme. J. Biol. Chem. 1982;257:2537–42. [PubMed] [Google Scholar]
- 6.Hershko A, Heller H, Elias S, Ciechanover A. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J. Biol. Chem. 1983;258:8206–14. [PubMed] [Google Scholar]
- 7.Pickart CM, Rose IA. Functional heterogeneity of ubiquitin carrier proteins. J. Biol. Chem. 1985;260:1573–81. [PubMed] [Google Scholar]
- 8.Pickart CM, Kasperek EM, Beal R, Kim A. Substrate properties of site-specific mutant ubiquitin protein (G76A) reveal unexpected mechanistic features of ubiquitin-activating enzyme (E1) J. Biol. Chem. 1994;269:7115–23. [PubMed] [Google Scholar]
- 9.Hershko A, Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–79. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 10.Hochstrasser M. Biochemistry. All in the ubiquitin family. Science. 2000;289:563–4. doi: 10.1126/science.289.5479.563. [DOI] [PubMed] [Google Scholar]
- 11.Bohnsack RN, Haas AL. Conservation in the mechanism of Nedd8 activation by the human AppBp1-Uba3 heterodimer. J. Biol. Chem. 2003;278:26823–30. doi: 10.1074/jbc.M303177200. [DOI] [PubMed] [Google Scholar]
- 12.Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 13.Walden H, Podgorski MS, Schulman BA. Insights into the ubiquitin transfer cascade from the structure of the activating enzyme for NEDD8. Nature. 2003;422:330–4. doi: 10.1038/nature01456. [DOI] [PubMed] [Google Scholar]
- 14.Wang M, Cheng D, Peng J, Pickart CM. Molecular determinants of polyubiquitin linkage selection by an HECT ubiquitin ligase. EMBO J. 2006;25:1710–9. doi: 10.1038/sj.emboj.7601061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lois LM, Lima CD. Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. EMBO J. 2005;24:439–51. doi: 10.1038/sj.emboj.7600552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bencsath KP, Podgorski MS, Pagala VR, Slaughter CA, Schulman BA. Identification of a multifunctional binding site on Ubc9p required for Smt3p conjugation. J. Biol. Chem. 2002;277:47938–45. doi: 10.1074/jbc.M207442200. [DOI] [PubMed] [Google Scholar]
- 17.Huang DT, et al. Structural basis for recruitment of Ubc12 by an E2 binding domain in NEDD8’s E1. Mol. Cell. 2005;17:341–50. doi: 10.1016/j.molcel.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 18.Eletr ZM, Huang DT, Duda DM, Schulman BA, Kuhlman B. E2 conjugating enzymes must disengage from their E1 enzymes before E3-dependent ubiquitin and ubiquitin-like transfer. Nat. Struct. Mol. Biol. 2005;12:933–4. doi: 10.1038/nsmb984. [DOI] [PubMed] [Google Scholar]
- 19.Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435:687–92. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walden H, et al. The structure of the APPBP1-UBA3-NEDD8-ATP complex reveals the basis for selective ubiquitin-like protein activation by an E1. Mol. Cell. 2003;12:1427–37. doi: 10.1016/s1097-2765(03)00452-0. [DOI] [PubMed] [Google Scholar]
- 21.Huang DT, et al. A unique E1-E2 interaction required for optimal conjugation of the ubiquitin-like protein NEDD8. Nat. Struct. Mol. Biol. 2004;11:927–35. doi: 10.1038/nsmb826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tokgoz Z, Bohnsack RN, Haas AL. Pleiotropic effects of ATP.Mg2+ binding in the catalytic cycle of ubiquitin-activating enzyme. J. Biol. Chem. 2006;281:14729–37. doi: 10.1074/jbc.M513562200. [DOI] [PubMed] [Google Scholar]
- 23.Szczepanowski RH, Filipek R, Bochtler M. Crystal structure of a fragment of mouse ubiquitin-activating enzyme. J. Biol. Chem. 2005;280:22006–11. doi: 10.1074/jbc.M502583200. [DOI] [PubMed] [Google Scholar]
- 24.Vijay-Kumar S, Bugg CE, Cook WJ. Structure of ubiquitin refined at 1.8 A resolution. J. Mol. Biol. 1987;194:531–44. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 25.Whitby FG, Xia G, Pickart CM, Hill CP. Crystal structure of the human ubiquitin-like protein NEDD8 and interactions with ubiquitin pathway enzymes. J. Biol. Chem. 1998;273:34983–91. doi: 10.1074/jbc.273.52.34983. [DOI] [PubMed] [Google Scholar]
- 26.Osaka F, et al. A new NEDD8-ligating system for cullin-4A. Genes Dev. 1998;12:2263–8. doi: 10.1101/gad.12.15.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miura T, Klaus W, Gsell B, Miyamoto C, Senn H. Characterization of the binding interface between ubiquitin and class I human ubiquitin-conjugating enzyme 2b by multidimensional heteronuclear NMR spectroscopy in solution. J. Mol. Biol. 1999;290:213–28. doi: 10.1006/jmbi.1999.2859. [DOI] [PubMed] [Google Scholar]
- 28.Hamilton KS, et al. Structure of a conjugating enzyme-ubiquitin thiolester intermediate reveals a novel role for the ubiquitin tail. Structure. 2001;9:897–904. doi: 10.1016/s0969-2126(01)00657-8. [DOI] [PubMed] [Google Scholar]
- 29.Brzovic PS, Lissounov A, Christensen DE, Hoyt DW, Klevit RE. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol. Cell. 2006;21:873–80. doi: 10.1016/j.molcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 30.Hicke L, Schubert HL, Hill CP. Ubiquitin-binding domains. Nat. Rev. Mol. Cell. Biol. 2005;6:610–21. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- 31.Harper JW, Schulman BA. Structural complexity in ubiquitin recognition. Cell. 2006;124:1133–6. doi: 10.1016/j.cell.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 32.Hurley JH, Emr SD. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomol. Struct. 2006;35:277–98. doi: 10.1146/annurev.biophys.35.040405.102126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hicke L. A new ticket for entry into budding vesicles-ubiquitin. Cell. 2001;106:527–30. doi: 10.1016/s0092-8674(01)00485-8. [DOI] [PubMed] [Google Scholar]
- 34.Morita E, Sundquist WI. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- 35.Hochstrasser M. Lingering mysteries of ubiquitin-chain assembly. Cell. 2006;124:27–34. doi: 10.1016/j.cell.2005.12.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


