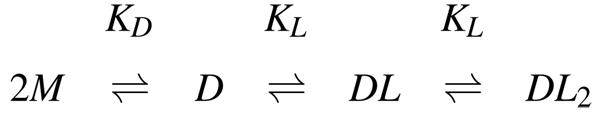Abstract
Dynein light chain LC8 is a small, dimeric, very highly conserved globular protein first identified as an integral part of the dynein and myosin molecular motors but now recognized as a dimerization hub with wider significance. Phosphorylation at Ser88 is thought to be involved in regulating LC8 in the apoptotic pathway. The phosphomimetic Ser88Glu mutation weakens dimerization of LC8 and thus its overall ligand-binding affinity, because only the dimer binds ligands. The 1.9 Å-resolution crystal structure of dimeric LC8S88E bound to a fragment of the ligand Swallow (Swa) presented here shows that the tertiary structure is identical to that of wild-type LC8/Swa, with Glu88 well accommodated sterically at the dimer interface. NMR longitudinal magnetization exchange spectroscopy reveals remarkably slow association kinetics (kon~ 1 sec−1 mM−1) in the monomer-dimer equilibrium of both wild-type LC8 and LC8S88E, possibly due to the strand-swapped architecture of the dimer. The Ser88Glu mutation raises the dimer dissociation constant (KD) through a combination of a higher koff and lower kon. Using a minimal model of titration linked to dimerization, we dissect the thermodynamics of dimerization of wild-type LC8 and LC8S88E in their various protonation states. When both Glu88 residues are protonated, the LC8S88E dimer is nearly as stable as the wild-type dimer, but deprotonation of one Glu88 residue raises KD by a factor of 400. We infer that phosphorylation of one subunit of wild-type LC8 raises KD at least as much, sufficient to prevent dimerization of LC8 at physiological concentration. Some LC8 binding partners may bind tightly enough to promote dimerization even when one subunit is phosphorylated; thus linkage between phosphorylation and dimerization provides a mechanism for differential regulation of binding of LC8 to its diverse partners.
Dynein light chain LC8 (called DYNLL1 in mammals (1)) was first described as an essential component of the dynein (2–4) and Myosin V (5) molecular motors, binding directly to specific sites on the dynein intermediate chain IC74 (6, 7) and the Myosin V heavy chain (8, 9). Because LC8 also binds to some putative dynein cargos, it has been described as a cargo adaptor (10–12); however, a large fraction of LC8 is not associated with any molecular motor (13), and some of its ligands– such as neuronal nitric oxide synthase (nNOS) (14)– are not clearly associated with active transport, leading to the emerging concept that LC8 acts as a dimerization hub, binding to diverse partners with some disordered regions and inducing them to dimerize and to form more ordered structures (6, 15, 16).
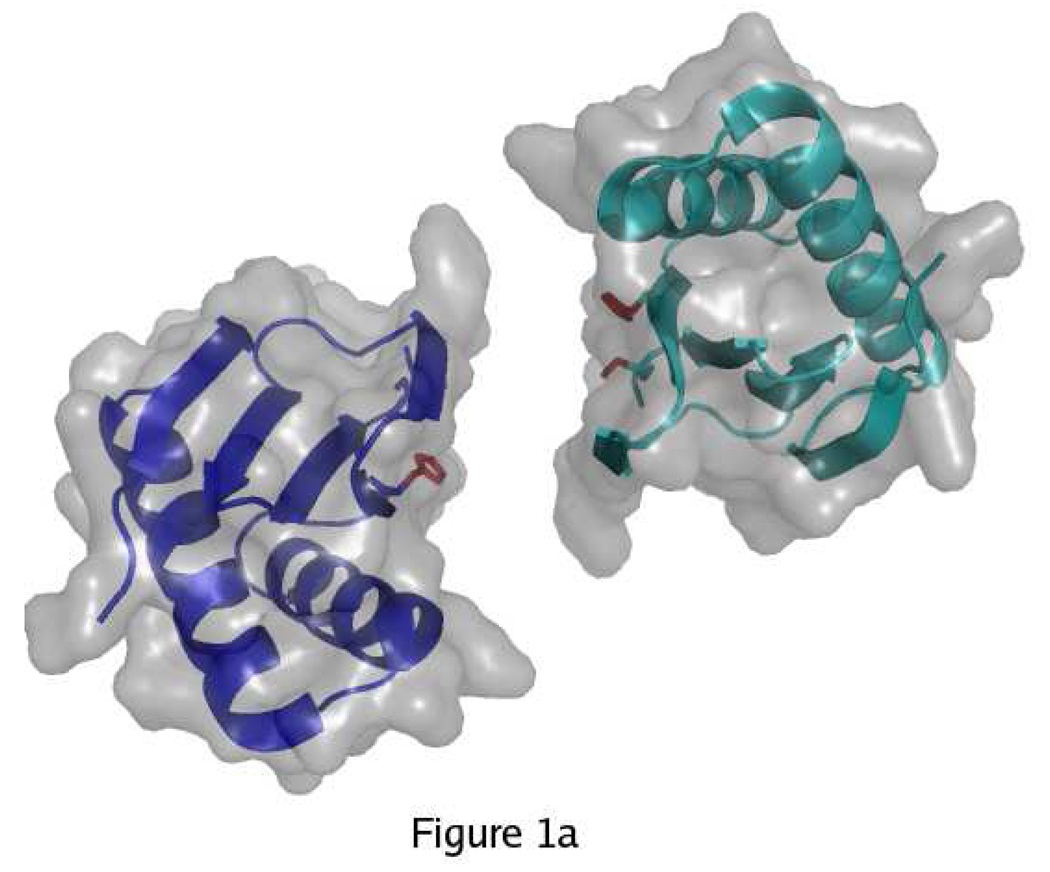
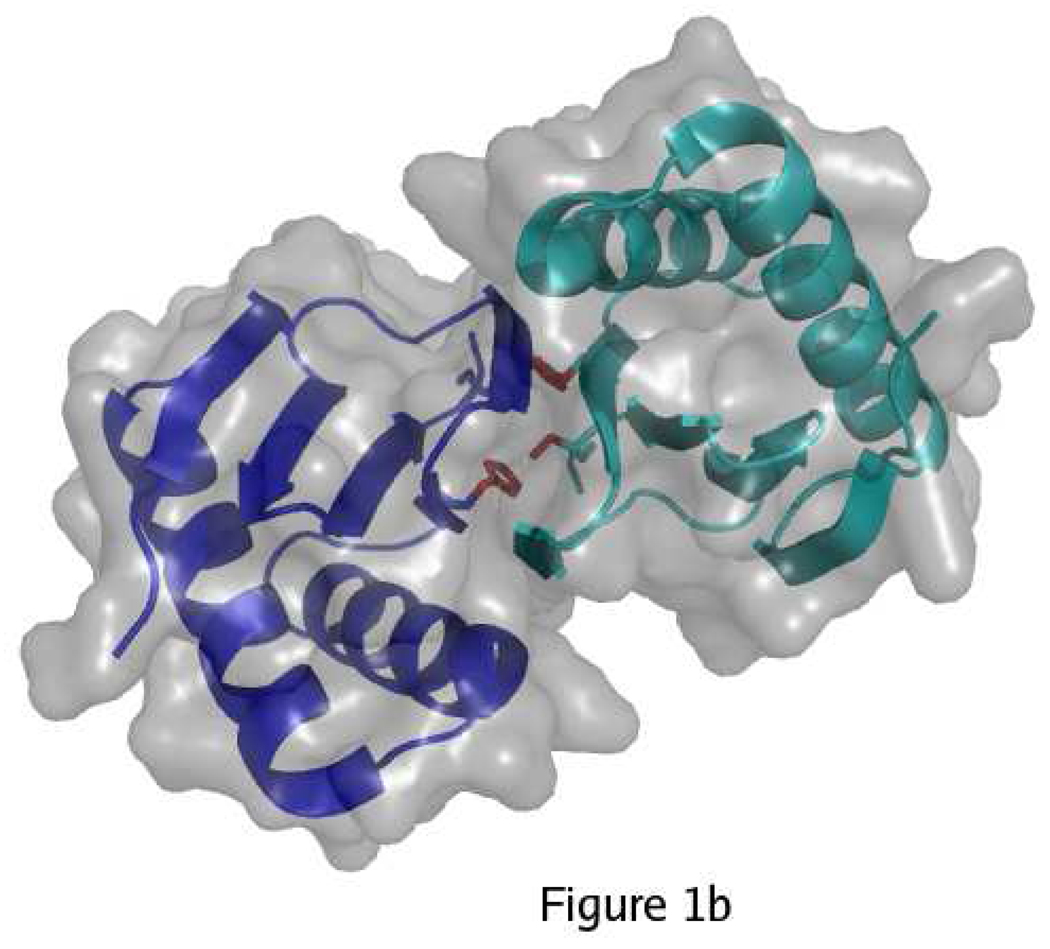
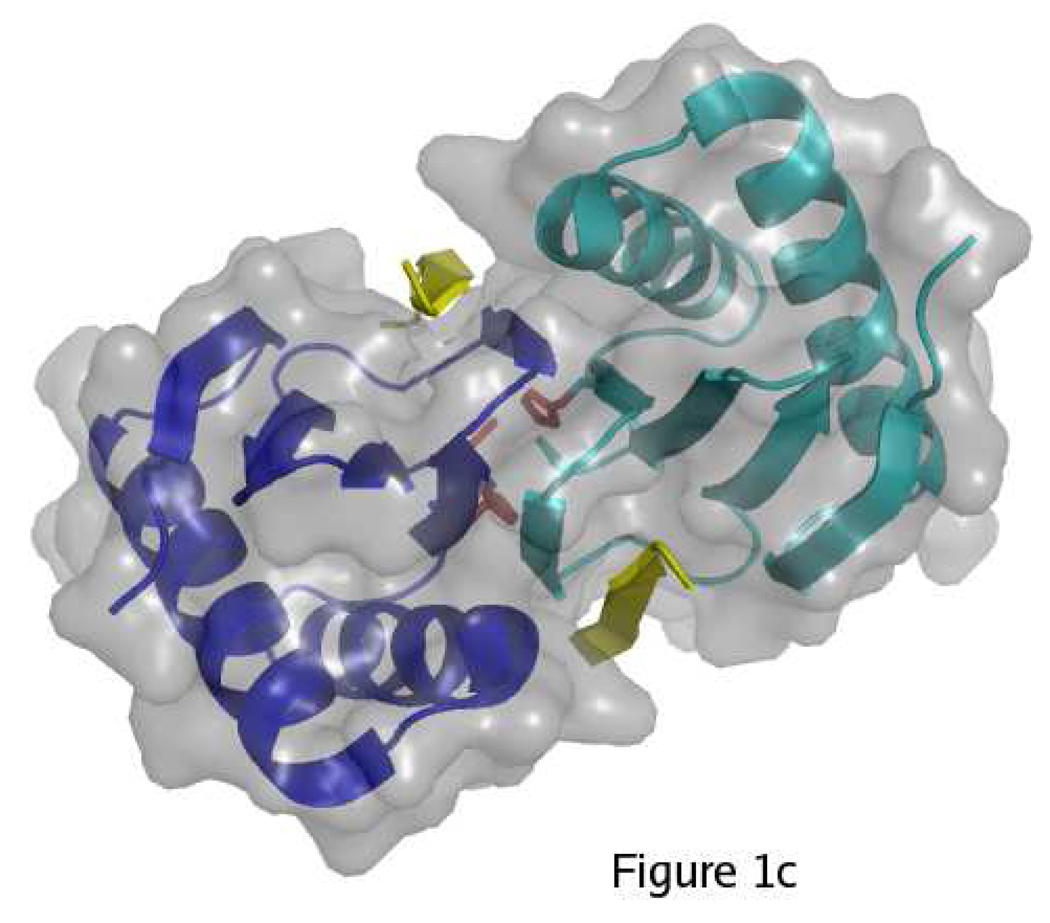
Wild-type LC8 can exist as a folded monomer or a folded dimer, with the dimeric form predominating at neutral pH and the monomeric form predominating at low pH (17). The disruption of the dimer at low pH has been attributed to protonation of His55, which is solvent-exposed in the monomer with pKa ~ 6.5 and becomes buried at the dimer interface with pKa < 4.5 (18). The tertiary structure remains similar in the pH-induced monomer, the apo-dimer, and ligand-bound complexes (19–22). The dimer interface consists of a central β-sheet composed of four strands from one subunit and one strand crossed over from the other subunit (Figure 1). All known ligands bind in a cleft at the dimer interface by appending as a sixth β strand to the central β sheet. Both subunits of the dimer contribute to the ligand-binding site, and thus monomeric LC8 fails to bind ligands (18, 23).
Figure 1.
Overview of LC8 structure. A) In the monomer, His55 and Ser88 (red) are solvent-exposed. The two identical subunits of LC8 are shown in blue and cyan. B) In the dimer, His55 and Ser88 are buried. Strand β3 forms part of the central β-sheet of the opposite subunit. C) Ligands (yellow) bind in a cleft at the dimer interface by appending to the central β-sheet. Figure made with PyMol (41) using PDB structures 3BRI and 3E2B.
Phosphorylation at Ser88 can regulate LC8 in vivo. This was first described in the apoptotic pathway, where phosphorylation disrupts binding to BimL, which in turn inhibits apoptosis (24–26). Initially Pak1, which binds directly to LC8 (27), was implicated as the kinase responsible for phosphorylation (25); however, this has been questioned (27). Despite uncertainty about the kinase involved, the occurrence of LC8 phosphorylation in vivo is clear, and its regulatory role is potentially of much broader importance because LC8 is now recognized as a hub protein and general promoter of dimerization involved in numerous pathways (16).
Though phosphorylation at Ser88 disrupts ligand binding, Ser88 is not in direct contact with the ligand in any known structure (21, 28), suggesting that phosphorylation does not act by directly occluding the binding site (23). Phosphorylation can act primarily by altering quaternary structure, as observed in glycogen phosphorylase (29), STAT3 (30), and the Kv channel (31). In LC8, Ser88 is at the dimer interface, and in vitro studies show that the phosphomimetic mutant LC8S88E dimerizes much more weakly than wild-type LC8 (23,26). The stable LC8 monomer is thus of physiological importance, and not just a curiosity of extreme low-pH conditions. Monomeric LC8 cannot bind ligands; therefore, by increasing the monomeric population at the expense of the dimeric population, the Ser88Glu mutation reduces the overall ligand-binding affinity.
The NMR spectra of monomeric, dimeric, and ligand-bound LC8S88E suggest that they are structurally very close to their wild-type counterparts (23). The Ser88Glu mutation therefore introduces a titrable group at the dimer interface, which could disrupt the LC8S88E dimer at neutral pH in the same way that His55 disrupts the wild-type dimer at low pH. To test this model of action we have performed structural, thermodynamic, and kinetic analyses of LC8S88E. The crystal structure at 1.9 Å resolution of dimeric LC8S88E/Swa is essentially unchanged from that of the wild-type complex. A quantitative model linking dimerization with the titration of interface residues, fit to thermodynamic data for dimer association/dissociation, reveals the energetics of dimerization for wild-type LC8 when both, one, or neither His55 residue is protonated, and similary for LC8S88E in its various protonation states.
NMR spectra of LC8 and LC8S88E- recorded at conditions where the dimer dissociation constant is similar to the total protein concentration- contain two distinct sets of signals, corresponding to monomer and dimer. This slow-exchange behavior allows the use of NZ exchange spectroscopy (34) to compare dimerization kinetics in LC8 and LC8S88E. This method offers the advantage of defining both the association and dissociation rates without perturbing the chemical equilibrium (32, 33) and therefore reveals the impact of the phosphomimetic Ser88Glu mutation on the activation energy for dimer dissociation.
Experimental Procedures
X-ray crystallography
Wild-type LC8, LC8S88E and Swa were prepared as described previously (20, 21, 23). Crystals were grown at room temperature in hanging drops made by a 1:1 mixture of the reservoir with a stock solution of 1 mM protein and 2 mM peptide in 20 mM Tris-HCl (pH 8.0). The reservoir solution was 0.2 M calcium acetate, 0.1 M sodium cacodylate, 15% PEG8000, pH 5.5. The crystals grew as hexagonal rods with long axes greater than 300 µm and diameters up to 80 µm, and were flash frozen in liquid nitrogen following transfer to a cryoprotectant consisting of reservoir solution plus 20% (v/v) glycerol.
Diffraction data were collected on beam line 5.0.3 at the Advanced Light Source, Berkeley National Labs (λ=1.0 Å; Δφ = 1°; high-resolution pass: 120 10-second images; low-resolution pass: 100 3-second images). Data sets were processed using the HKL suite of programs (35). Crystals of LC8S88E/Swa belong to space group P6122 with a=b=44.19 Å, c=203.22 Å, with one molecule in the asymmetric unit and a solvent content of 50%.
Before refinement, 10% of the reflection data were set aside for cross-validation. The test set comprised the same reflections used in the test set for the 2 Å LC8/Swa refinement (21), plus new randomly-selected reflections beyond 2 Å resolution. The structure was solved by using the LC8 chain from the previously reported LC8/Swa structure (PDB entry 2p1k (21)) as a search model for molecular replacment at 3.5 Å resolution using MOLREP (36). Rigid body refinement using REFMAC (37) resulted in an initial Rcryst/Rfree of 33%/37%. There was strong electron density present for the bound peptide at the rigid body refinement stage. A model for Swa was built into this density and LC8 residue 88 was changed to Glu. The structure was iteratively refined using REFMAC and coot (38) including TLS refinement (39), to Rcryst/Rfree of 19.0%/25.4%. During refinement, ordered water molecules were added or removed by the criterion of having reasonable hydrogen bonding partners and a peak in the 2Fo-Fc electron density map of at least 1σ. Water molecules were numbered on the basis of final peak electron density from 1 (the highest) to 79 (the weakest).
Per-atom contributions to solvent-accessible surface area were calculated using VOLBL (40). Structure diagrams were produced with PyMol (41).
The coordinates of the LC8S88E/Swa complex have been deposited in the RCSB protein data bank with accession code 3BRL.
NMR spectroscopy
NMR samples of 15N-labeled wild-type LC8 and 15N-labeled LC8S88E were purified as described previously (21, 23). Samples contained 0.5 – 1.0 mM protein, 50 mM sodium citrate, 50 mM sodium chloride, 50 mM sodium phosphate, 3% glycerol, and 10% 2H2O. pH was in the range 3.0 – 7.0 and verified to within ±0.1 using an internal maleic acid standard. All NMR experiments were recorded on a Bruker DRX 600 spectrometer at 30°C. Monomer-dimer exchange rates were measured using NZ-exchange spectroscopy (34). In these experiments, magnetization is first frequency-labeled with the amide nitrogen chemical shift, then after a mixing period is transferred to the amide proton and detected. This leads to four distinct peaks for residues having sufficiently distinct chemical shifts in both the proton and nitrogen dimensions: a monomer-monomer auto-peak; a dimer-dimer auto-peak; and two cross-peaks (monomer-dimer and dimer-monomer) arising from magnetization which has changed environments due to chemical exchange during the mixing period. From the intensities of these four peaks as a function of mixing time, the forward and backward exchange rates can be determined. NZ-exchange spectra were collected in an interleaved fashion, where the mixing time was incremented before the t1 delay. For wild-type LC8, mixing times were 150, 300, and 450 msec. For LC8S88E, mixing times were 35, 70, 105, 140, 175, and 210 msec.
Peak intensities were determined by fitting signals to two-dimensional Lorenzian lineshapes using in-house software. In fitting the cross-peaks of NZ-exchange experiments, we constrained the chemical shifts to match the more easily-measured auto-peaks while allowing the intensity to vary freely. This gives reasonable estimates for the intensities of even very weak cross-peaks, because their position is fixed by the much more intense auto-peaks (33). Uncertainties in peak intensities were estimated using Monte Carlo sampling. Using the best-fit model as a starting point, model parameters (intensity, linewidth, peak position) were randomly varied to generate a set of plausible models, accepting only those meeting a χ2 criterion of p > 0.05. Error estimates for the parameters (e.g. peak intensity) were taken as the standard deviations of the parameters within this model set. KD values were calculated from the intensities of peaks corresponding to monomeric and dimeric populations in 1H-13C CT-HSQC spectra (for wild-type LC8 (18)) or 1H-15N HSQC spectra (for LC8S88E) with a 3-second recycle time. Error estimates for KD were calculated by standard error propagation rules from errors in the peak intensities.
Peak volume as a function of time was modeled by the Bloch equations for two–site exchange between a monomeric environment (M) and a dimeric environment (D):
| (1) |
| (2) |
where Rm and Rd are the T1 relaxation rates for monomer and dimer, respectively. The forward and backward magnetization exchange rates k+ and k− are equivalent to the chemical exchange rates for a unimolecular reaction such as protein unfolding (34). For this bimolecular reaction, they are determined by the relations k+ = konM and k− = koff, where kon and koff are the rate constants of the LC8S88E dimerization equilibrium:
| (3) |
The monomer concentration M is a constant and can be calculated from the (known) total protein concentration and the rate constants kon and koff; therefore kon and koff are sufficient to determine k+ and k−.
An error function was calculated as the sum of squared residuals between measured and predicted peak volumes. For each residue, the fitting procedure included both cross-peaks and both auto-peaks. When too weak to measure, the cross-peak intensity was set to an upper bound of 1 (expressed as a signal-to-noise ratio) for fitting purposes, leading in turn to an upper bound on the kinetic constants. The kinetic parameters kon and koff, when fit on a per-residue basis, clustered within a distribution of ∼ ±50%, suggesting that modeling the data as the result of a single exchange process affecting the entire protein was justified. Therefore we performed a global fit in which monomer and dimer initial peak intensities were independent parameters for each residue, but rate constants were constrained to be equal for all residues. The predicted volume was thus a function of the four independent parameters kon, koff, Rm, and Rd, in addition to two independent parameters for each residue included in the fit.
The best-fit parameters were determined by a gradient-descent search, with error estimates on parameters derived from the gradient at the minimum. The minimization and the solution of the differential equations involved in fitting were both performed using the numerical package R (www.r-project.org).
Minimal model
To explain the pH dependence of kon, koff, and KD, we developed a minimal model based on coupled equilibria of protein dimerization and titrating residues at the dimer interface. Our model is an adaptation of one that has been used to explain the pH dependence of folding kinetics in an SH3 domain (42).
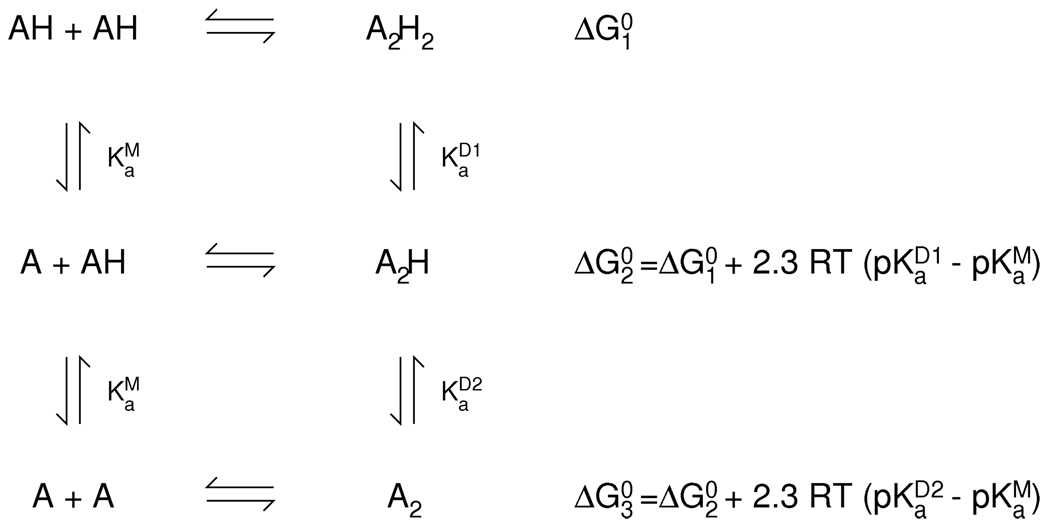
Our model for wild-type LC8 includes His55 as the sole titrable residue at the dimer interface. In such a system, the dimer can form by three distinct pathways: association of two unprotonated monomers; association of one protonated and one unprotonated monomer; or association of two protonated monomers (Scheme 1). The acid dissociation constant in the monomeric state and the first and second acid dissociation constants in the dimeric state are not generally equal, because the titrating group is in a different environment. Because of thermodynamic cycles linking protonation to dimerization, of the six parameters in this model (three acid dissociation constants and three dimer dissociation constants) only four can be varied independently.
Scheme 1.
Model of titration-linked dimerization. A: unprotonated monomer; AH: protonated monomer; A2: unprotonated dimer; A2H: singly-protonated dimer; A2H2: doubly-protonated dimer.
We extended the model to LC8S88E by incorporating titration of Glu88, leading to additional parameters and thermodynamic cycles. In general, the overall free energy of association as a function of pH is given by:
| (4) |
where the sum extends over all titrating residues.
Kinetic predictions can be obtained by considering pKa values in the transition state as well as in the monomeric and dimeric states. For a protein dimerization reaction, the transition state is itself dimeric and resembles the bound state (43, 44). Overall kinetic behavior in the presence of titrating groups can be described in terms of a pH-dependent free energy for the initial state, the final state, and a transition state ensemble (42):
| (5) |
| (6) |
where M, D, and ‡ are the monomeric, dimeric, and transition states, respectively. The sum contains one term for each titrating group at the interface.
From the free energies of the monomeric, dimeric, and transition states, we can calculate dimer dissociation equilibrium and rate constants:
| (7) |
| (8) |
| (9) |
where kb and h are the Boltzmann and Planck constants, respectively, and κ is a transmission coefficient.
For wild-type LC8, His55 and were set initially to the acid dissociation constant of free histidine and then iteratively adjusted to give the best least-squares fit simultaneously to KD, kon, and koff according to equation 7, equation 8, and equation 9. Experimental KD values were taken from (18) and kon and koff were taken from this study. For LC8S88E, titration of His55 and Glu88 were considered. Though the acid dissociation constants for His55 in LC8S88E may differ from those in wild-type LC8, the amount of available data did not justify fitting the pKa values for both residues independently. Thus, to model LC8S88E, we used the acid dissociation constants of His55 from wild-type LC8 and adjusted the acid dissociation constants of Glu88 iteratively to give the best match to the experimentally measured KD, kon, and koff.
Results
Crystal structure of LC8S88E
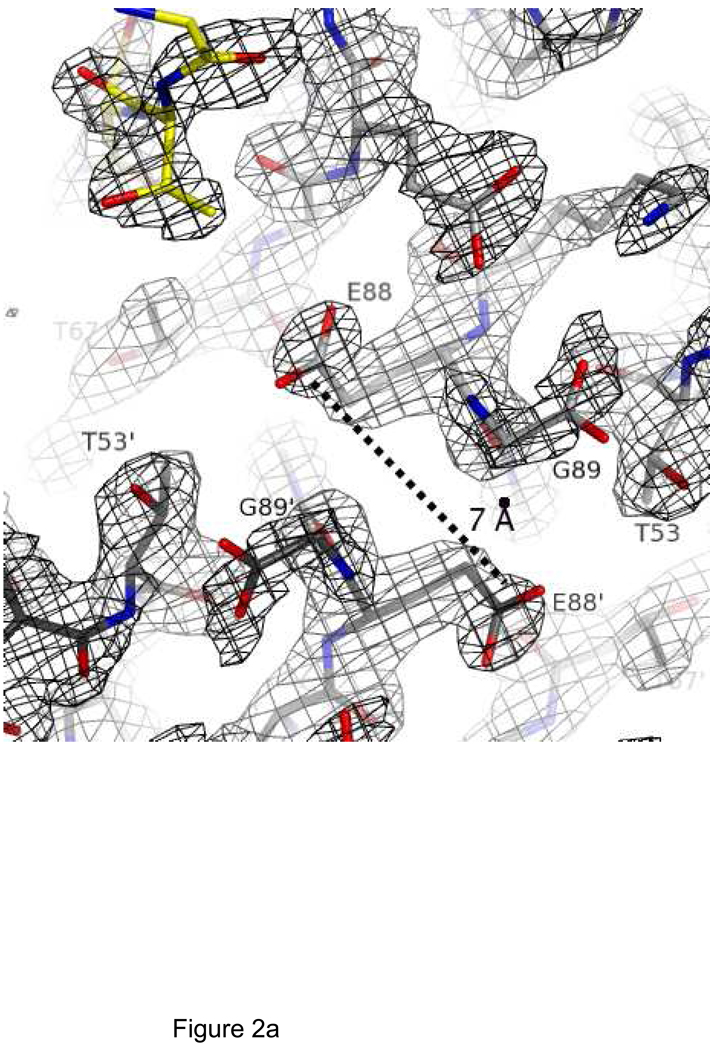
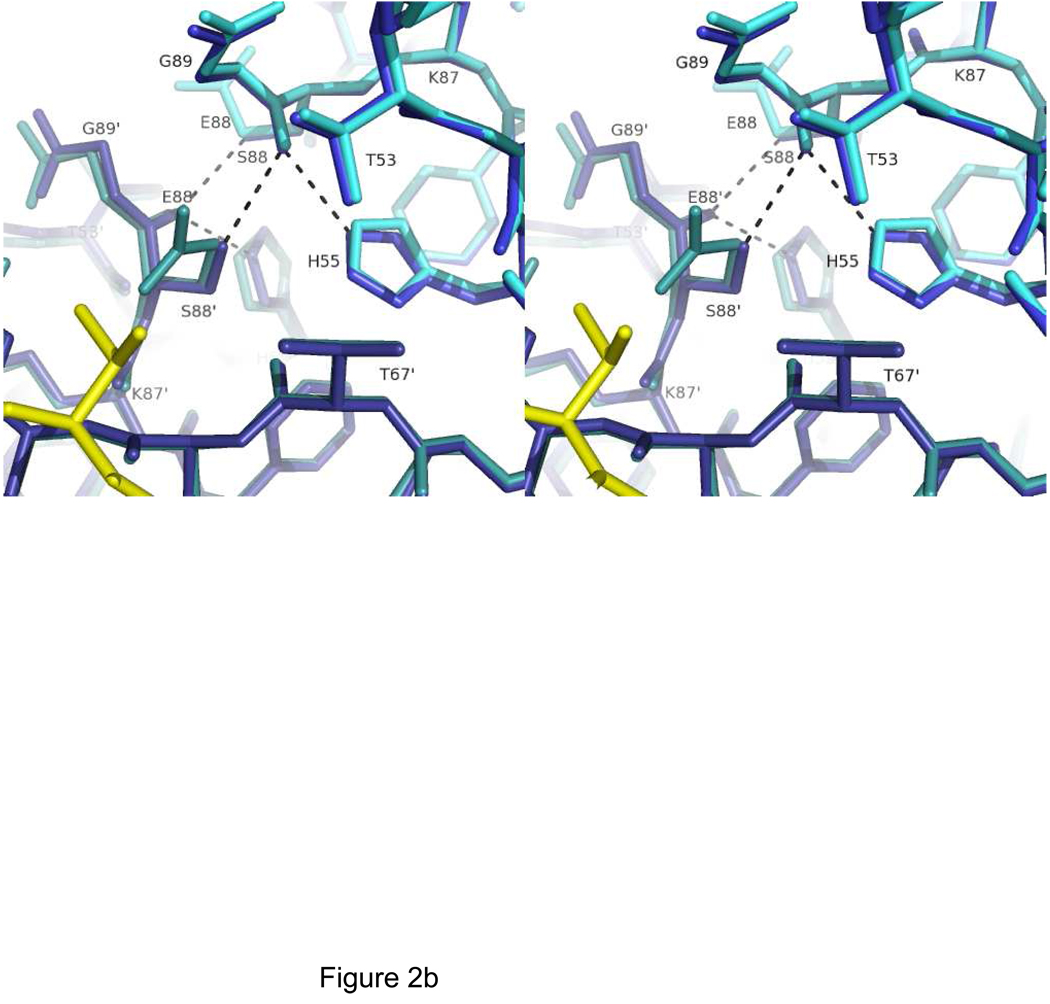
The structure of LC8S88E/Swa was solved by rigid body refinement using the LC8 chain from the wild-type LC8/Swa complex (21) as the initial model, and refined at 1.9 Å resolution to Rcryst=19.0%, Rfree=25.4% (Table 1). All residues were in the most favored Ramachandran region except for Asn51, which as in all LC8 structures has a positive φ angle and is part of a turn which also features a cis-peptide bond between Pro52 and Thr53. Electron density clearly shows the conformation of Glu88 (Figure 2A). The structure of LC8S88E/Swa is nearly identical to that of wild-type LC8/Swa (Cα RMSD of 0.28Å), and even at the site of mutation, the backbone and side chain torsion angles are similar (Figure 2B). In wild-type LC8, Ser88′ O has two hydrogen bond donors: Ser88 Oγ and His55 Nε. In S88E, the hydrogen bond from Ser88 Oγ is lost. Instead, the methylene group of Glu88 is in steric contact with Glu88′ O, at a favorable non-bonded distance (45). Glu88 adopts a rotamer directing the sidechains away from each other, such that the distance between the carboxylates is 7 Å (Figure 2A). The Glu88 side chain is mostly buried in a pocket formed by residues Thr53, His55, and Thr67′, with the carboxylate group only partially exposed (18 Å2 solvent accessible surface compared with 60–80 Å2 for the most exposed carboxylate groups).
Table 1.
Data Collection and Refinement Statistics
| Resolution | ∞ − 1.90 (2.01 − 1.90) |
| Reflections (total / unique) | 195803/10183 |
| Completeness | 100.0 (99.8) |
| Rmeasa | 9.6 (72.4) |
| I/σ | 27.9 (3.8) |
| Refinement | |
|---|---|
| Resolution (Å) | ∞ − 1.90 (1.95 − 1.90) |
| Number of reflections | 9605 (685) |
| Number of amino acids (LC8) | 88 |
| Number of amino acids (ligand) | 10 |
| Number of solvent atoms | 79 |
| Total number of atoms | 880 |
| Average B (all atoms) (Å2) | 34 |
| Rcryst(%) | 19.0 (22.0) |
| Rfree(%) | 25.4 (27.3) |
| RMSD bonds (Å) | 0.009 |
| RMSD angles (degrees) | 1.1 |
| φ, ψ most favoredb | 96/97 |
As defined by Diederichs and Karplus, 1997
As defined in Lovell et. al., 2003
Figure 2.
Crystal structure of LC8S88E, showing the environment of Glu88. A) Electron density for Glu88 in LC8S88E/Swa. View is along the two-fold symmetry axis with 2Fo-Fc density contoured at 1.2σ and showing LC8S88E (cyan) and the peptide (yellow). B) Stereo view of the environment of residue Ser88 in wild-type LC8/Swa (blue) and of residue Glu88 in LC8S88E/Swa (cyan). The peptide is shown in yellow. Chain B is shown in a darker shade than chain A. Figure made with PyMol (41) using PDB structures 3BRL (LC8S88E/Swa) and 3E2B (LC8/Swa).
Thermodynamics of dimerization
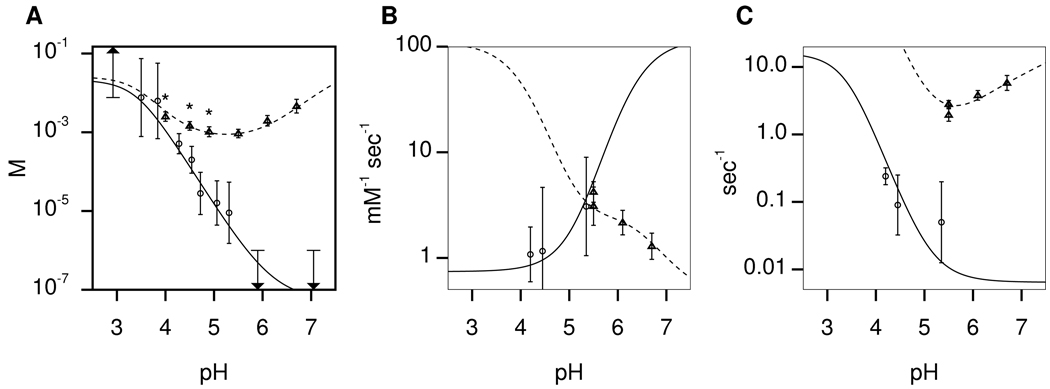
In HSQC spectra of wild-type LC8 in the 4.2–5.3 pH range, and those of LC8S88E in the 4.2–6.7 pH range, certain residues (primarily those at the dimer interface) have two distinct peaks, corresponding to monomer and dimer. Changing the pH, and thus the monomer/dimer populations, alters the intensities but not the positions of these peaks (i.e. they are in the NMR slow-exchange regime), allowing determination of KD through ratios of peak intensities. The KD of wild-type LC8 increases monotonically with decreasing pH with a midpoint at pH 4.5, reaching a limit < 1 µM at high pH (18). The pH dependence of LC8S88E dimerization is remarkably different (Figure 4; supplementary Table 1): KD decreases with increasing pH until reaching a minimum of KD= 930 µM at pH 5.5, and then increases again at higher pH. We attribute these opposing trends to perturbation of titrating residues at the dimer interface- in the dimer, the pKa of His55 is lowered, and that of Glu88 is elevated.
Figure 4.
pH dependence of kinetic and thermodynamic parameters for LC8 dimerization: (A) KD, (B) kon, and (C) koff. Experimental data points are shown as circles for wild-type LC8 and as triangles for LC8S88E. The best-fit model is shown as solid lines for wild-type LC8 and dashed lines for LC8S88E. The experimental data also appears in supplementary table 1. Arrows indicate measurements which are upper or lower bounds. Data points collected after initial model fitting are marked with asterisks (see Results).
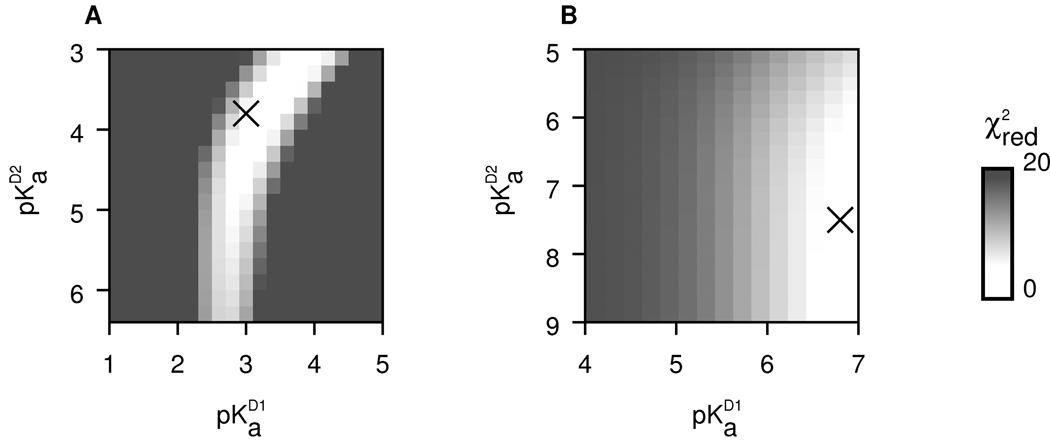
To show that dimerization coupled to titration of residues at the interface can quantitatively account for the different pH dependence of KD for wild-type LC8 and LC8S88E, we developed a minimal model (see Experimental Procedures). We began by constructing a model of wild-type LC8 considering only titration of His55. In fitting KD vs. pH, the freely variable parameters were (the acid dissociation constant in the monomer), and (the acid dissociation constants in the dimer), and either or (Scheme 1). It was not possible to fit all four parameters simultaneously through global fitting procedures, but by iterative refinement from reasonable starting values, we found a local best fit (Table 2). The model based on the best-fit parameters accurately reproduced experimentally-measured KD over the 3–7 pH range (Figure 4). To test the robustness of the reported parameters, we performed systematic searches around the best-fit values (Figure 5). All good fits had the following properties: in the range 6.0 – 6.5, as expected for an exposed histidine; and less than 4.5; and , the free energy of association in the limit of high pH, below 0.3 µM.
Table 2.
Acid dissociation constants in the monomeric, dimeric, and transition states (as defined in Experimental Procedures) derived from the minimal model of titration-linked dimerization. The pKa values for His55 were obtained by fitting the model to the wild-type data and were fixed to be the same in both the wild-type and LC8S88E models (as described in the Methods).
| His55 | Glu88 | ||
|---|---|---|---|
| 6.2 | 4.2 | ||
| 3.0 | 6.8 | ||
| 3.8 | 7.5 | ||
| 4.9 | 6.0 | ||
| 5.3 | 7.0 |
Figure 5.
Exploration of the robustness of the model parameters near the best fit. (A) Grid search of His55 and in wild-type LC8; (B) Grid search of Glu88 and in LC8S88E. The cross symbol marks the location of the best-fit parameters. The goodness of fit, reported as a reduced chi square value, is shown for the surrounding parameter space.
The model of LC8S88E considered titration of His55 and Glu88. We retained the best-fit parameters for His55 from the wild-type model, and iteratively adjusted the parameters of Glu88. As for wild-type LC8, the model accurately reproduced the experimental measurement of KD (Figure 4), including the opposite trends for wild-type LC8 vs. LC8S88E over the 5 – 7 pH range. We used a grid search to investigate the robustness of the pKa values (Figure 5). The best-fit of Glu88 is 4.2, near its unperturbed value in the free amino acid. A good fit required that Glu88 and Glu88 , with best-fit values of 6.8 and 7.5. The initial fit was performed when only three of the six KD measurements for LC8S88E were available (see points marked with an asterisk in Figure 4). The later-collected points, despite their not being included in the initial fit, were in good agreement with the model, serving as a cross-validation of the modeling approach and best-fit parameters. When all of the data were included in the fit, the best-fit pKa values remained unchanged to within 0.1 unit.
Using the relationships in Scheme 1, we calculated separate KD values for LC8 in its distinct protonation states (Table 3). For wild-type LC8, the most tightly-dimerizing form (KD=0.06 µM) is that in which neither His55 is protonated, corresponding to the limiting KD at high pH. The most weakly-dimerizing form (KD=21,000 µM) is that in which both His55 residues are protonated, corresponding to the limiting KD at low pH. Similarly, for LC8S88E, the most tightly-dimerizing form (KD=0.07 µM) is that in which all titrating residues are neutral, and the most weakly-dimerizing form (KD=1.8 × 1010 µM) is that in which all titrating residues are charged. Interestingly, these states- unlike those of wild-type LC8- do not predominate at any pH and thus do not correspond to any observable limit. Over the pH range studied (4.0 – 6.7), the observed macroscopic KD of LC8S88E reflects a mixture of protonation states and reaches a minimum of 0.9 mM at pH 5.2.
Table 3.
Dimer dissociation constants (in µM) for wild-type LC8 (w.t.) and LC8S88E in all possible distinct protonation states, derived from the titration-linked model.
| LC8S88E | ||||||
|---|---|---|---|---|---|---|
| w.t. | Glu88/ Glu88 | |||||
| 2.1 × 104 | 2.5 × 104 | 1.1 × 105 | 1.8 × 1010 | |||
| 14 | 17 | 6800 | 1.2 × 107 | |||
| His55/ His55 | 0.060 | 0.071 | 28 | 5.9 × 104 | ||
Kinetics of dimerization
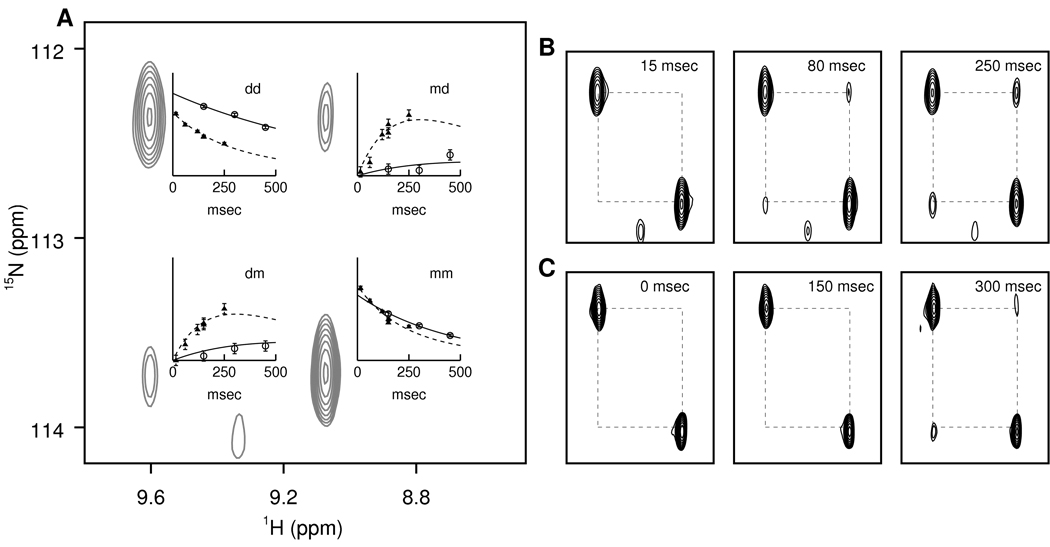
We measured dimer association rate constants (kon) and dimer dissociation rate constants (koff) for wild-type LC8 and LC8S88E using NZ exchange NMR spectroscopy (34) (Figure 3). For four residues (Asp37, Gly59, Gly63, and Ala82) both cross-peaks and both auto-peaks were sufficiently resolved to allow quantitative measurements of exchange rates: for wild-type LC8 between pH 4.2 and pH 5.3, koff = 0.05 – 0.24 sec−1 and kon = 1.0 – 3.0 sec−1 mM−1; and for LC8S88E between pH 5.5 and pH 6.7, koff = 1.9 – 5.7 sec−1 and kon = 1.2 – 3.0 sec−1 mM−1 (Figure 4 and supplementary table 1).
Figure 3.
Monomer-dimer exchange kinetics measured by NMR. (A) Excerpt around residue 59 from an NZ-exchange spectrum of LC8S88E. The relative peak intensity as a function of mixing time (wild-type: circles; LC8S88E: triangles) is plotted beside each of the four component peaks (dd: dimer-dimer auto-peak; mm: monomer-monomer auto-peak; dm: dimer-monomer cross-peak; md: monomer-dimer cross-peak). The vertical scale of the dd and mm plots is four times that of the md and dm plots. The fits to equation 1 are shown as solid lines (wild-type LC8) or dashed lines (LC8S88E). (B) Excerpts of the same region from additional NZ exchange spectra at different mixing times, showing the build-up of the cross-peaks. (C) Similarly for wild-type LC8.
The rate constants for dimerization are pH-dependent, which we explained by extending the model to include acid dissociation constants in the transition state ( and ). These were iteratively adjusted to match the kinetic data, resulting in best-fit values intermediate between those in the monomeric and dimeric states. The resulting model accurately reproduced the experimentally-measured rate constants for wild-type LC8 and LC8S88E (Figure 4).
Discussion
Phosphorylation of Ser88 regulates LC8 by weakening its dimerization, which in turn diminishes overall affinity for ligands which can bind only to the dimer (23, 25, 26). Our biophysical studies of LC8S88E as a phosphomimetic mutant are probing the origin of this weakened dimerization. NMR spectra first suggested that LC8S88E/Swa is structurally very close to its wild-type counterpart (23), and this is now confirmed by the crystal structure (Figure 2B). We have measured dimer stability and dimerization kinetics as a function of pH for both wild-type LC8 and LC8S88E. Using a minimal model we dissect the thermodynamics and kinetics of association for wild-type LC8 and LC8S88E in their various protonation states.
Dimer disruption by titrating residues at the interface
Previously, we showed that titration of His55, uniquely among the histidine residues of LC8, is linked thermodynamically to dimerization (18). When His55 is protonated, formation of the dimer would result in the unfavorable burial of a charge at the interface. His55 and His55′ are in close proximity in the dimer, leading to an additional energetic penalty from like-charge repulsion when both are protonated. Glu88, like His55, is buried at the dimer interface near its symmetry mate. To demonstrate that titration of these two residues sufficiently explains the pH dependence of dimerization, we constructed a minimal model incorporating only His55 (for wild-type LC8) or His55 and Glu88 (for LC8S88E) that accurately reproduces KD over a wide pH range (Figure 4A).
From the best-fit model parameters, we calculated the energetics of dimerization in all possible protonation states (Table 3), allowing us to assess the relative importance of burial of a single charge vs. like-charge repulsion between symmetry mates. For wild-type LC8 in the limit of high pH (conditions where His55 is entirely deprotonated in both the monomer and dimer), the calculated KD is 0.06 µM, which is consistent with a recently reported experimental upper bound of 0.5 µM at pH 7.4 (27). The burial of one charged His55 residue destabilizes the dimer by 3.3 kcal/mol, corresponding to a 250-fold rise in KD (Table 3). Protonation of the second histidine residue incurs an additional energetic penalty of 4.4 kcal/mol, equivalent to a 1500-fold rise in KD. We attribute the greater energetic penalty (by 1.1 kcal/mol) of the second deprotonation to like-charge repulsion between His55 and His55′. This is in reasonable agreement with an expected penalty of 1.6 kcal/mol calculated by considering His55 and His55′ as point charges separated by 6 Å (the distance between the imidazole rings in the crystal structure) and estimating an effective dielectric constant of 40 (taken from a variety of mutagenesis and titration studies of charged residues at or near the protein surface (46–48)).
For states of LC8S88E in which both Glu88 and Glu88′ are protonated, KD is only slightly higher than that for the corresponding states of wild-type LC8 (Table 3). Therefore, steric effects and/or loss of intersubunit hydrogen bonds involving Ser88, which have been proposed as mechanisms for the disruption of the LC8S88E dimer (23,26), appear to play a minor role. This is consistent with the crystal structure of LC8S88E/Swa, which shows Glu88 easily accommodated sterically at the dimer interface, and with the thermodynamics of LC8S88A, which also lacks the hydrogen bonds involving Ser88 yet remains strongly dimeric (49). It is only when the Glu88 side chain is deprotonated that the Ser88Glu mutation becomes disruptive: the first deprotonation destabilizes the dimer by 3.6 kcal/mol, raising KD 400-fold. The second deprotonation destabilizes the dimer by 4.5 kcal/mol and raises KD an additional 2000-fold. We attribute the 0.9-kcal/mol higher penalty to like-charge repulsion between Glu88 and Glu88′, in reasonable agreement with the expected penalty of 1.4 kcal/mol calculated for two point charges separated by 7 Å (the distance between the carboxylates in the crystal structure).
Impact of the Ser88Glu mutation on dimerization kinetics
NZ exchange spectroscopy has been used to measure the kinetics of the folding-unfolding transition of an SH3 domain (34) and of protein dimerization in type II Cadherin (50). The method requires that both states are similarly populated and that exchange rates are in the appropriate range of 0.5 – 10 sec−1 (32, 34, 51). These conditions are satisfied for monomer-dimer exchange in wild-type LC8 over the 4.2 – 5.3 pH range, and for LC8S88E over the 5.5 – 6.7 pH range. Dimer association of both wild-type LC8 and LC8S88E is remarkably slow– the observed kon values of 1.0 – 2.0 sec−1mM−1 are at the low end of the range seen for protein-protein interactions, an order of magnitude below typical values and a million times lower than the fastest, diffusion-limited protein–protein association rates (52). The slow kinetics may reflect the complexity of the dimerization process for LC8, which is not a simple association of complementary surfaces; rather, a disordered strand in each monomer is incorporated into a β sheet, requiring formation of several new intersubunit hydrogen bonds (53).
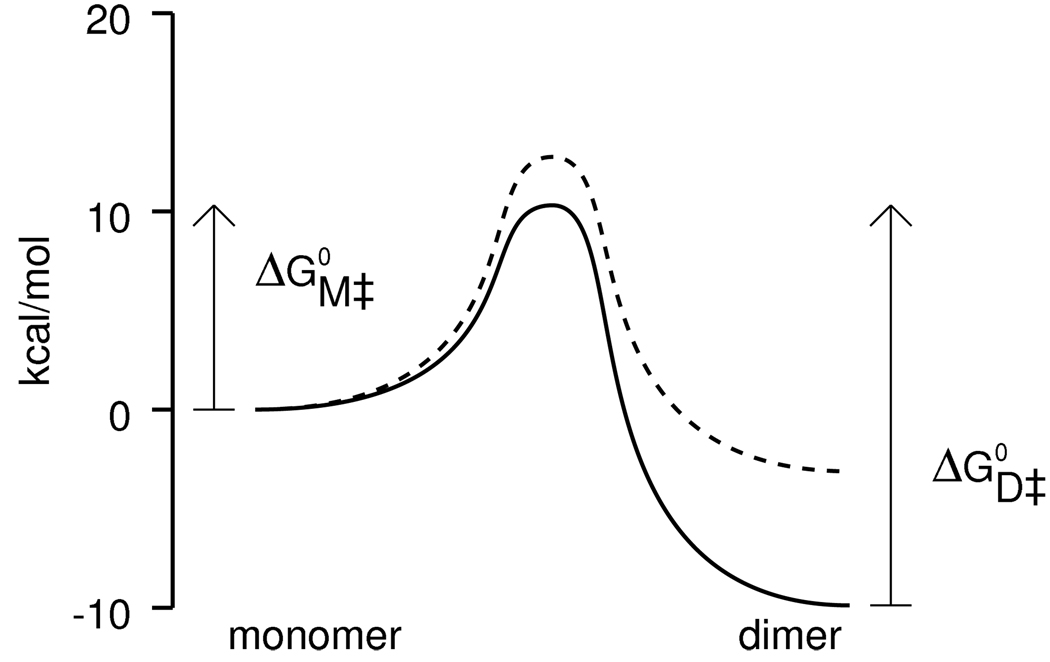
We measured the rates for LC8S88E at near-neutral pH (6.7), but for wild-type LC8 measurements are not possible above pH 5.3 because the monomer population is too low to detect by NMR. Using the model, however, we can extrapolate to neutral pH, obtaining kon = 89 sec−1mM−1 for wild-type LC8 and kon = 1 sec−1mM−1 for LC8S88E (Figure 4B). Thus at physiological pH the Ser88Glu mutation not only disrupts the dimer but also greatly slows association, and hence raises the free energy of the transition state as well as that of the dimeric state (Figure 6).
Figure 6.
Energetic consequences of the Ser88Glu mutation at neutral pH. Free energies for monomer, dimer, and transition state were calculated from the model for wild-type LC8 (solid line) and for LC8S88E (dashed line).
Dimerization coupled to ligand binding
Dissociation of LC8 to monomers, by decreasing the amount of dimer available to bind ligands, decreases the apparent ligand affinity. The effective ligand-binding constant , taking into account the ligand-binding sites that become unavailable due to dimer dissociation (see Scheme 2), is:
| (10) |
For example, consider a 1:1 mixture at 10 µM of LC8 and the dynein intermediate chain (IC), which binds with an affinity of 3 µM (53). KD of wild-type LC8 at neutral pH is less than 0.1 µM, so dimer dissociation lowers the effective ligand affinity by only 1%. In contrast, KD for LC8S88E is 7 mM, leading to a four-fold decrease in effective ligand affinity. Thus dimer dissociation explains the results of GST pulldown experiments in which LC8S88E bound IC with only about half the efficiency of wild-type LC8 (23), even if the intrinsic ligand affinity (KL) is the same for both wild-type LC8 and LC8S88E. Similarly, in a fluorescence assay at micromolar concentration a complex of wild-type LC8 and the ligand Bim formed to completion yet only a small amount of LC8S88E/Bim was detected even when LC8S88E was present in 50-fold excess (26).
Scheme 2.
LC8 dimerization coupled to ligand binding. M: LC8 monomer; D: LC8 dimer; L: ligand; KD: dimer dissociation constant; KL: ligand binding constant.
Binding of ligands to the dimer also decreases the apparent dimer dissociation constant KD′ in the presence of ligands (Scheme 2):
| (11) |
This explains the observation that LC8S88E at 0.2 mM is mostly monomeric, but in the presence of one equivalent of Swa (KL= 0.6 µM; (53)) is 90% bound dimer (23).
Ser88Glu as a phosphomimetic
Because of similarities of charge and side-chain length, glutamic acid is commonly used to mimic phosphoserine, and there is at least one example of a natural “phosphomimetic” mutation: in phosphorylase kinase, a glutamic acid residue in the activation loop replaces a conserved threonine that serves as a phosphorylation target in homologous kinases (54). LC8S88E has been used extensively in experiments both in vitro and in vivo to mimic phosphorylated wild-type LC8 (26). How appropriate is Glu88 as a model for LC8 phosphoserine-88? Glu88 is partially buried at the dimer interface and incurs only a small steric penalty. Phosphoserine is comprised of heavier atoms and one additional atom, and therefore may incur a larger steric penalty. Phosphoserine can have up to two negative charges, and the first acid dissociation constant (pKa1 = 2) ensures that, even if its pKa is perturbed by several pH units in the dimer, phosphoserine always has at least one negative charge at neutral pH (55). Thus we conclude that the KD of wild-type LC8 with one subunit phosphorylated is at least as high as that of LC8S88E with one Glu88 residue deprotonated (28 µM; Table 3), and possibly higher due to steric effects and/or greater charge. By the same argument, we expect the KD of doubly-phosphorylated LC8 at neutral pH to be at least as high as that of LC8S88E when both Glu88 residues are deprotonated (59 mM; Table 3), and we expect the dimerization transition state energy in phosphorylated LC8 to be at least as high as that in LC8S88E, leading to similarly slower association kinetics compared to wild-type LC8.
Functional implications
The LC8 dimer has two identical phosphorylation sites, one on each subunit. Does regulation require phosphorylation of both sites, or is phosphorylation of just one sufficient? Phosphorylation of multiple sites is a way to achieve cooperativity in response to kinase activity, and has been demonstrated in other systems such as the MAPK signaling protein Ste5 (56). Based on our studies with LC8S88E, we conclude that for a heterodimer of phosphorylated and unphosphorylated LC8 KD is 28 µM or higher (Table 3); therefore, phosphorylation of one subunit is sufficient to prevent formation of the apo-LC8 dimer at physiological concentration. Different behavior may occur in the presence of ligands. Swa and the apoptotic factor Bim both bind with affinities better than 1 µM (53), and therefore when present at 10 µM decrease the apparent dimer dissociation constant by a factor of 100 (Equation 11); therefore, tightly-binding ligands such as these may overcome the energetic penalty to dimerization due to phosphorylation of one subunit and allow formation of singly-phosphorylated heterodimers of LC8, whereas more weakly-binding ligands such as the dynein intermediate chain (IC) may not. Thus the presence of two identical phosphorylation sites on LC8 provides this hub protein with a mechanism to regulate its diverse ligands differently in response to phosphorylation.
Supplementary Material
Acknowledgements
We acknowledge the support of the nucleic acid and protein core and the mass spectrometry core in the OSU Environmental Health Sciences Center. We thank Yujuan Song and Jean Yau for protein preparation and Jessica Morgan for assistance collecting NMR data.
Abbreviations
- LC8
10 kDa dynein light chain
- Swa
peptide consisting of residues 281–297 of Swallow
- LC8S88E
LC8 Ser88Glu mutant
Footnotes
This work was supported by NSF grant MCB-0818896 to E.J.B. and an American Heart Association Fellowship (Award #0720019Z) to G.C.B. M.C. was supported by an NSF REU award and HHMI. The OSU Environmental Health Sciences Center is supported by NIH award NIEHS P30 ES00210.
Supporting Information
KD, kon, and koff measurements for wild-type LC8 and LC8S88E (shown in Figure 4) are also available as a supplementary table, available free of charge online at http://pubs.acs.org.
References
- 1.Pfister K, Fisher E, Gibbons I, Hays T, Holzbaur E, McIntosh J, Porter M, Schroer T, Vaughan K, Witman G, King S, Vallee R. Cytoplasmic dynein nomenclature. J Cell Biol. 2005;171:411–413. doi: 10.1083/jcb.200508078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dick T, Ray K, Salz H, Chia W. Cytoplasmic dynein (ddlc1) mutations cause morphogenetic defects and apoptotic cell death in Drosophila melanogaster. Mol Cell Biol. 1996;16:1966–1977. doi: 10.1128/mcb.16.5.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vallee R, Williams J, Varma D, Barnhart L. Dynein: An ancient motor protein involved in multiple modes of transport. J Neurobiol. 2004;58:189–200. doi: 10.1002/neu.10314. [DOI] [PubMed] [Google Scholar]
- 4.Benashski S, Harrison A, Patel-King R, King S. Dimerization of the highly conserved light chain shared by dynein and myosin V. J Biol Chem. 1997;272:20929–20935. doi: 10.1074/jbc.272.33.20929. [DOI] [PubMed] [Google Scholar]
- 5.Espindola F, Suter D, Partata L, Cao T, Wolenski J, Cheney R, King S, Mooseker M. The light chain composition of chicken brain myosin-Va: calmodulin, myosin-II essential light chains, and 8-kDa dynein light chain/PIN. Cell Motil Cytoskeleton. 2001;47:269–281. doi: 10.1002/1097-0169(200012)47:4<269::AID-CM2>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 6.Makokha M, Hare M, Li M, Hays T, Barbar E. Interactions of cytoplasmic dynein light chains Tctex-1 and LC8 with the intermediate chain IC74. Biochemistry. 2002;41:4302–4311. doi: 10.1021/bi011970h. [DOI] [PubMed] [Google Scholar]
- 7.Lo K, Naisbitt S, Fan J, Sheng M, Zhang M. The 8-kDa dynein light chain binds to its targets via a conserved (K/R)XTQT motif. J Biol Chem. 2001;276:14059–14066. doi: 10.1074/jbc.M010320200. [DOI] [PubMed] [Google Scholar]
- 8.Hódi Z, Németh A, Radnai L, Hetényi C, Schlett K, Bodor A, Perczel A, Nyitray L. Alternatively spliced exon B of myosin Va is essential for binding the tail-associated light chain shared by dynein. Biochemistry. 2006;45:12582–12595. doi: 10.1021/bi060991e. [DOI] [PubMed] [Google Scholar]
- 9.Wagner W, Fodor E, Ginsburg A, Hammer J. The binding of DYNLL2 to myosin Va requires alternatively spliced exon B and stabilizes a portion of the myosin’s coiled-coil domain. Biochemistry. 2006;45:11564–11577. doi: 10.1021/bi061142u. [DOI] [PubMed] [Google Scholar]
- 10.Schnorrer F, Bohmann K, Niesslein-Volhard C. The molecular motor dynein is involved in targeting swallow and bicoid RNA to the anterior pole of Drosophila oocytes. Nat Cell Biol. 2000;2:185–190. doi: 10.1038/35008601. [DOI] [PubMed] [Google Scholar]
- 11.Naisbitt S, Valtschanoff J, Allison D, Sala C, Kim E, Craig A, Weinberg R, Sheng M. Interaction of the postsynaptic density-95/guanylate kinase domain-associated protein complex with a light chain of myosin-V and dynein. J Neurosci. 2000;20:4524–4534. doi: 10.1523/JNEUROSCI.20-12-04524.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lo K, Kan H, Chan L, Xu W, Wang K, Wu Z, Sheng M, Zhang M. The 8-kDa dynein light chain binds to p53-binding protein 1 and mediates DNA damage-induced p53 nuclear accumulation. J Biol Chem. 2005;280:8172–8179. doi: 10.1074/jbc.M411408200. [DOI] [PubMed] [Google Scholar]
- 13.King S, Barbarese E, Dillman J, Patel-King R, Carson J, Pfister K. Brain cytoplasmic and flagellar outer arm dyneins share a highly conserved Mr 8,000 light chain. J Biol Chem. 1996;271:19358–19366. doi: 10.1074/jbc.271.32.19358. [DOI] [PubMed] [Google Scholar]
- 14.Jaffrey S, Snyder S. PIN: an associated protein inhibitor of neuronal nitric oxide synthase. Science. 1996;274:774–777. doi: 10.1126/science.274.5288.774. [DOI] [PubMed] [Google Scholar]
- 15.Wang L, Hare M, Hays T, Barbar E. Dynein light chain LC8 promotes assembly of the coiled-coil domain of swallow protein. Biochemistry. 2004;43:4611–4620. doi: 10.1021/bi036328x. [DOI] [PubMed] [Google Scholar]
- 16.Barbar E. Dynein light chain LC8 is a dimerization hub essential in diverse protein networks. Biochemistry. 2008;47:503–508. doi: 10.1021/bi701995m. [DOI] [PubMed] [Google Scholar]
- 17.Barbar E, Kleinman B, Imhoff D, Li M, Hays T, Hare M. Dimerization and folding of LC8, a highly conserved light chain of cytoplasmic dynein. Biochemistry. 2001;40:1596–1605. doi: 10.1021/bi002278+. [DOI] [PubMed] [Google Scholar]
- 18.Nyarko A, Cochrun L, Norwood S, Pursifull N, Voth A, Barbar E. Ionization of His 55 at the dimer interface of dynein light-chain LC8 is coupled to dimer dissociation. Biochemistry. 2005;44:14248–14255. doi: 10.1021/bi0512694. [DOI] [PubMed] [Google Scholar]
- 19.Wang W, Lo K, Kan H, Fan J, Zhang M. Structure of the monomeric 8-kDa dynein light chain and mechanism of the domain-swapped dimer assembly. J Biol Chem. 2003;278:41491–41499. doi: 10.1074/jbc.M307118200. [DOI] [PubMed] [Google Scholar]
- 20.Makokha M, Huang Y, Montelione G, Edison A, Barbar E. The solution structure of the pH-induced monomer of dynein light-chain LC8 from Drosophila. Protein Sci. 2004;13:727–734. doi: 10.1110/ps.03462204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benison G, Karplus P, Barbar E. Structure and dynamics of LC8 complexes with KXTQT-motif peptides: Swallow and dynein intermediate chain compete for a common site. J Mol Biol. 2007;371:457–468. doi: 10.1016/j.jmb.2007.05.046. [DOI] [PubMed] [Google Scholar]
- 22.Benison G, Karplus P, Barbar E. The interplay of ligand binding and quaternary structure in the diverse interactions of dynein light chain LC8. J Mol Biol. 2008;384:954–966. doi: 10.1016/j.jmb.2008.09.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song Y, Benison G, Nyarko A, Hays T, Barbar E. Potential role for phosphorylation in differential regulation of the assembly of dynein light chains. J Biol Chem. 2007;282:17272–17279. doi: 10.1074/jbc.M610445200. [DOI] [PubMed] [Google Scholar]
- 24.Puthalakath H, Huang D, O’Reilly L, King S, Strasser A. The proapoptotic activity of the Bcl-2 family member Bim is regulated by interaction with the dynein motor complex. Mol Cell. 1999;3:287–296. doi: 10.1016/s1097-2765(00)80456-6. [DOI] [PubMed] [Google Scholar]
- 25.Vadlamudi R, Bagheri-Yarmand R, Yang Z, Balasenthil S, Nguyen D, Sahin A, den Hollander P, Kumar R. Dynein light chain 1, a p21-activated kinase 1-interacting substrate, promotes cancerous phenotypes. Cancer Cell. 2004;5:575–585. doi: 10.1016/j.ccr.2004.05.022. [DOI] [PubMed] [Google Scholar]
- 26.Song C, Wen W, Rayala S, Chen M, Ma J, Zhang M, Kumar R. Serine 88 phosphorylation of the 8-kDa dynein light chain 1 is a molecular switch for its dimerization status and functions. J Biol Chem. 2008;283:4004–4013. doi: 10.1074/jbc.M704512200. [DOI] [PubMed] [Google Scholar]
- 27.Lightcap C, Sun S, Lear J, Rodeck U, Polenova T, Williams J. Biochemical and structural characterization of the Pak1-LC8 interaction. J Biol Chem. 2008;283:27314–27324. doi: 10.1074/jbc.M800758200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang J, Jaffrey S, Guo W, Snyder S, Clardy J. Structure of the PIN/LC8 dimer with a bound peptide. Nat Struct Biol. 1999;6:735–740. doi: 10.1038/11501. [DOI] [PubMed] [Google Scholar]
- 29.Lin K, Rath V, Dai S, Fletterick R, Hwang P. A protein phosphorylation switch at the conserved allosteric site in GP. Science. 1996;273:1539–1542. doi: 10.1126/science.273.5281.1539. [DOI] [PubMed] [Google Scholar]
- 30.Becker S, Groner B, Mller C. Three-dimensional structure of the Stat3β homodimer bound to DNA. Nature. 1998;394:145–151. doi: 10.1038/28101. [DOI] [PubMed] [Google Scholar]
- 31.Antz C, Bauer T, Kalbacher H, Frank R, Covarrubias M, Kalbitzer H, Ruppersberg J, Baukrowitz T, Fakler B. Control of K+ channel gating by protein phosphorylation: structural switches of the inactivation gate. Nat Struct Biol. 1999;6:146–150. doi: 10.1038/5833. [DOI] [PubMed] [Google Scholar]
- 32.Sprangers R, Velyvis A, Kay L. Solution NMR of supramolecular complexes: providing new insights into function. Nat Methods. 2007;4:697–703. doi: 10.1038/nmeth1080. [DOI] [PubMed] [Google Scholar]
- 33.Benison G, Barbar E. NMR analysis of dynein light chain dimerization and interactions with diverse ligands. Methods Enzymol. 2009;455:237–258. doi: 10.1016/S0076-6879(08)04209-2. [DOI] [PubMed] [Google Scholar]
- 34.Farrow NA, Zhang OW, Forman-Kay JD, Kay LE. A heteronuclear correlation experiment for simultaneous determination of 15N longitudinal decay and chemical-exchange rates of systems in slow equilibrium. J. Biomol. NMR. 1994;4:727–734. doi: 10.1007/BF00404280. [DOI] [PubMed] [Google Scholar]
- 35.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 36.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 37.Murshudov G, Vagin A, Dodson E. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 38.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 39.Winn M, Isupov M, Murshudov G. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 40.Edelsbrunner H, Facello M, Fu P, Liang J. Measuring proteins and voids in proteins. Proc. 28th Ann. Hawaii Internat. Conf. System Sciences; pp. 256–264. [Google Scholar]
- 41.The pymol molecular graphics system. http://www.delanoscientific.com/
- 42.Tollinger M, Kay L, Forman-Kay J. Measuring pK(a) values in protein folding transition state ensembles by NMR spectroscopy. J Am Chem Soc. 2005;127:8904–8905. doi: 10.1021/ja051942c. [DOI] [PubMed] [Google Scholar]
- 43.Camacho C, Weng Z, Vajda S, DeLisi C. Free energy landscapes of encounter complexes in protein-protein association. Biophys J. 1999;76:1166–1178. doi: 10.1016/S0006-3495(99)77281-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou H. Disparate ionic-strength dependencies of on and off rates in proteinprotein association. Biopolymers. 2001;59:427–433. doi: 10.1002/1097-0282(200111)59:6<427::AID-BIP1047>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 45.Davis I, Leaver-Fay A, Chen V, Block J, Kapral G, Wang X, Murray L, Arendall W, Snoeyink J, Richardson J, Richardson D. Mol-Probity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rogers N, Moore G, Sternberg M. Electrostatic interactions in globular proteins: calculation of the pH dependence of the redox potential of cytochrome c551. J Mol Biol. 1985;182:613–616. doi: 10.1016/0022-2836(85)90248-7. [DOI] [PubMed] [Google Scholar]
- 47.Russell A, Thomas P, Fersht A. Electrostatic effects on modification of charged groups in the active site cleft of subtilisin by protein engineering. J Mol Biol. 1987;193:803–813. doi: 10.1016/0022-2836(87)90360-3. [DOI] [PubMed] [Google Scholar]
- 48.Alden R, Parson W, Chu Z, Warshel A. Calculations of electrostatic energies in photosynthetic reaction centers. J Am Chem Soc. 1995;117:12284–12298. [Google Scholar]
- 49.Mohan P, Hosur R. NMR characterization of structural and dynamics perturbations due to a single point mutation in drosophila DLC8 dimer: functional implications. Biochemistry. 2008;47:6251–6259. doi: 10.1021/bi800531g. [DOI] [PubMed] [Google Scholar]
- 50.Miloushev V, Bahna F, Ciatto C, Ahlsen G, Honig B, Shapiro L, Palmer A. Dynamic properties of a type II cadherin adhesive domain: implications for the mechanism of strand-swapping of classical cadherins. Structure. 2008;16:1195–1205. doi: 10.1016/j.str.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Montelione G, Wagner G. 2D chemical-exchange NMR spectroscopy by proton-detected heteronuclear correlation. J. Am. Chem. Soc. 1989;111:3096–3098. [Google Scholar]
- 52.Gabdoulline R, Wade R. Biomolecular diffusional association. Curr Opin Struct Biol. 2002;12:204–213. doi: 10.1016/s0959-440x(02)00311-1. [DOI] [PubMed] [Google Scholar]
- 53.Hall J, Hall A, Pursifull N, Barbar E. Differences in dynamic structure of LC8 monomer, dimer, and dimer-peptide complexes. Biochemistry. 2008;47:11940–11952. doi: 10.1021/bi801093k. [DOI] [PubMed] [Google Scholar]
- 54.Owen D, Noble M, Garman E, Papageorgiou A, Johnson L. Two structures of the catalytic domain of phosphorylase kinase: an active protein kinase complexed with substrate analogue and product. Structure. 1995;3:467–482. doi: 10.1016/s0969-2126(01)00180-0. [DOI] [PubMed] [Google Scholar]
- 55.Xie Y, Jiang Y, Ben-Amotz D. Detection of amino acid and peptide phosphate protonation using Raman spectroscopy. Anal Biochem. 2005;343:223–230. doi: 10.1016/j.ab.2005.05.038. [DOI] [PubMed] [Google Scholar]
- 56.Strickfaden S, Winters M, Ben-Ari G, Lamson R, Tyers M, Pryciak P. A mechanism for cell-cycle regulation of MAP kinase signaling in a yeast differentiation pathway. Cell. 2007;128:519–531. doi: 10.1016/j.cell.2006.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.