Abstract
The action of dopamine on the aggregation of the unstructured alpha-synuclein (α-syn) protein may be linked to the pathogenesis of Parkinson's disease. Dopamine and its oxidation derivatives may inhibit α-syn aggregation by non-covalent binding. Exploiting this fact, we applied an integrated computational and experimental approach to find alternative ligands that might modulate the fibrillization of α-syn. Ligands structurally and electrostatically similar to dopamine were screened from an established library. Five analogs were selected for in vitro experimentation from the similarity ranked list of analogs. Molecular dynamics simulations showed they were, like dopamine, binding non-covalently to α-syn and, although much weaker than dopamine, they shared some of its binding properties. In vitro fibrillization assays were performed on these five dopamine analogs. Consistent with our predictions, analyses by atomic force and transmission electron microscopy revealed that all of the selected ligands affected the aggregation process, albeit to a varying and lesser extent than dopamine, used as the control ligand. The in silico/in vitro approach presented here emerges as a possible strategy for identifying ligands interfering with such a complex process as the fibrillization of an unstructured protein.
Introduction
Parkinson's disease (PD) is a neurodegenerative movement disorder, affecting an estimated four million people worldwide [1], [2]. It is characterized by the loss of the neuromelanin expressing dopamine (DOP) neurons in the substantia nigra pars compacta and the deposition of Lewy bodies in many of the cells remaining in this region [3], [4], [5], [6], [7], [8], [9]. The major components of the Lewy bodies are fibrillar aggregates of the alpha-synuclein protein (α-syn) [6], [10], [11]. Thus, α-syn fibrillization and DOP metabolism are likely to be linked to PD pathogenesis [11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21]. DOP and some derivatives (Figure 1), which may be present in oxidizing conditions, form non-covalent and/or covalent adducts with α-syn [21], [22]. These molecules inhibit the conversion of α-syn to mature fibrils, promoting instead, accumulation of oligomeric (or protofibril) forms [23], [24], [25], [26], [27].
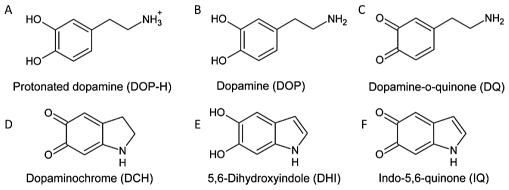
Figure 1. Dopamine and the oxidation derivatives known to interact with α-syn.
(A) Protonated Dopamine (DOP-H), (B) Dopamine (DOP), (C) Dopamine-o-quinone (DQ), (D) Dopaminochrome (DCH), (E) 5,6-Dihydroxyindole (DHI), (F) Indol-5,6-quinone (IQ).
Recently, molecular dynamics (MD) simulations based on α-syn's nuclear magnetic resonance (NMR) structural ensemble [28] in combination with biophysical methods, led some of us to propose a structural basis for DOP non-covalent inhibition of α-syn fibrillization [29]. This may be caused, at least in part, by 1) the formation of nonspecific hydrophobic contacts between DOP and its oxidation derivatives with the C-terminal; this includes the 125YEMPS129 region, as in agreement with experimental evidence [23], [28] and 2) long-range electrostatic interactions with residues in the NAC region which are involved in the fibrillization process [29].
Molecules structurally and electrostatically similar to a given ligand might provide similar structure/activity relationships [30]. We screened ligands structurally and electrostatically similar to DOP (Figure 2) from the ligand.info meta-database [31]. The ability of these ligands to bind to α-syn was then explored by MD simulations. The ligands bound weaker to the protein than DOP. Consistently, high-resolution atomic force microscopy (AFM) and transmission electron microscopy (TEM) data showed that the ligands affected α-syn fibril assembly, but to a lower extent than DOP. Remarkably, the best analogs revealed the most inhibitory effects upon protein aggregation in terms of fibril length and quantity.
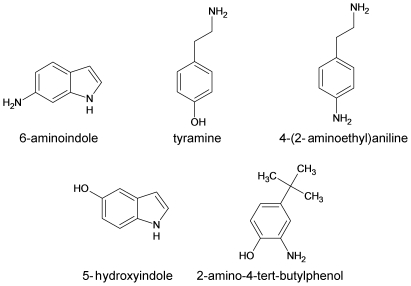
Figure 2. Chemical structures of the five DOP analogs chosen for the in silico/in vitro analysis.
Results
Dopamine Mimics: Identification and Binding to α-Syn
We screened seventy molecules of the ligand.info database [31] with the largest similarity with DOP and its oxidation derivatives (Table S1). The similarity was quantified according to the Tanimoto's equations [32]. The ligands were docked onto the six structural representatives of human α-syn in aqueous solution. The procedure was identical to that of previously reported [29], except that a refined set of structures of α-syn were used (see Material and Methods). The resulting complexes were ranked in terms of the number of contacts with the 125YEMPS129 region known-to-be targeted by DOP [24], [26], [29]. Such ranking was used only as a means to fast screen the ligands, and not to provide structural predictions. In fact, there are severe limitations of this procedure for an unstructured protein, as discussed by Slutzki et al [33] Five commercially available ligands chosen randomly from Table S1 and forming top ranking adducts (Figure 2), underwent 36 ns MD simulations in water solution (Table S2). The same procedure was used for DOP and its derivatives (Figure 1).
DOP and its oxidation derivatives bound for 69% or more of their time to α-syn (the criteria used to identify bound complexes are described in Materials and Methods) and bound preferentially to the target 125YEMPS129 region, similarly to previous MD simulations [29]. As found previously [29], they did not form specific interactions with the target region (Table S2). Moreover, they interacted with one or both negative residues (E83 and E61) of the NAC region (Table 1), a region known to be involved in the fibrillization of the protein. DOP-E83 interaction might play a role in the observed inhibition of fibrillization by DOP, as suggested experimentally [29].
Table 1. α-syn binding of ligands in figures 1, 2 in aqueous solution observed in MD simulations.
DOP and its oxidation derivatives abbreviations are explained in detail in figure 1. Column titles from left to right: (i) ligand name (ii) percentage of time in which the ligands are bound to α-syn and (iii) to the 125YEMPS129 ‘target’ region, (iv-v) stabilizing electrostatic interaction energies between the ligands and E83 and E61, two negatively charged residues of the NAC region. Energy values of the force field are very approximate1 and do not take into account the screening of the solvent. They should be taken here only for qualitative comparisons. Here they are normalized with respect to the most negative interaction energy between the neutral ligands and the two negative residues (-2.8 kcal/mol, relative to the interaction between IQ and E83). The DOPH/E61 energy turns out to be very large in absolute value because the ligand is charged, in contrast to all the others.
| Ligand | % protein | % target | Point Charge Model Av ± Std (E83) | Point Charge Model Av ± Std (E61) |
| DOPH | 70 | 32 | 0.0 | −14.9 |
| DOP | 69 | 46 | 0.0 | −0.7 |
| DQ | 49 | 28 | −0.3 | −0.3 |
| DCH | 72 | 32 | −0.8 | −0.5 |
| DHI | 91 | 16 | 0.0 | 0.0 |
| IQ | 62 | 32 | −1.0 | −0.3 |
| 6-aminoindole | 68 | 23 | 0.0 | 0.0 |
| Tyramine | 48 | 19 | 0.0 | 0.1 |
| 4-(2-aminoethyl) aniline | 39 | 9 | 0.0 | -0.2 |
| 5-hydroxyindole | 70 | 23 | -0.9 | 0.0 |
| 2-amino-4-tert-butylphenol | 62 | 19 | 0.0 | 0.0 |
L. Guidoni, V. Torre and P. Carloni, FEBS Letters 477 (2000) 37-42.
Three of the screened ligands (6-aminoindole, 5-hydroxyindole, 2-amino-4-tert-butylphenol) interacted significantly with α-syn (and in particular with the 125YEMPS129 region), although to a lesser extent than most dopamine derivatives (Table 1). In addition, they formed much weaker stabilizing electrostatic interactions with E61 and/or E83 (Table 1).
We have also studied several binding regions of ligands other than the target region. The results are provided in Table S3, where the contacts are listed for those adducts in which the ligands bind to regions other than the target region for more than 50% of their time.
The other two ligands, tyramine and 4-(2-aminoethylaniline), bound much less to α-syn (albeit still interacting with the 125YEMPS129 region), with almost no stabilizing interactions with the two residues (E61 and E83) of NAC region.
As in the case of DOP, all the ligands did not form specific interactions with any residues in the 125YEMPS129 region (Table S2).
Based on these results, we propose that: (i) the DOP mimics may interfere with the fibrillization of α-syn, although to a lesser extent than DOP and (ii) the ligands, that show the strongest binding to α-syn, i.e 6-aminoindole, and 5-hydroxyindole, and 2-amino-4-tert-butylphenol may have the strongest effect on the fibrillization of α-syn. We next proceeded to perform in vitro assays to test these two predictions.
In Vitro Fibrillization Assays
Human α-syn was produced and purified as described previously [34], with some modifications. Protein analysis by SDS-PAGE resulted in a single band showing a molecular weight (MW) of ≈14 KDa. The exact MW, as determined by mass spectrometry, was 14459.40±0.43 Da, and far-UV circular dichroism (CD) measurements revealed a randomly coiled secondary structure.
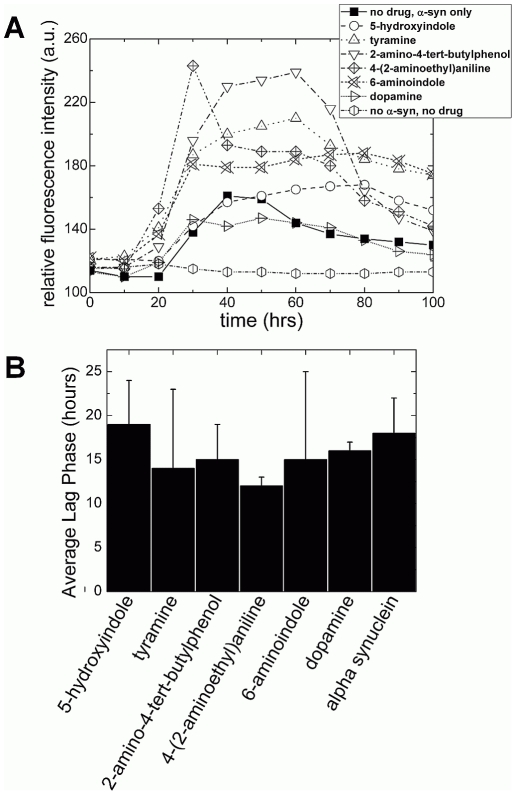
Human α-syn was then used in the amyloid fibrillization experiments in the presence and absence of the test ligands. For each assay run, a concentration of 100 µM of α-syn was used for the synthesis of the fibrils. All ligands were added together with the α-syn in an equimolar concentration at the start of the assay run. Due to its known inhibitory effect on α-synuclein fibrilization, dopamine was used as the control ligand [26]. Fibril formation was achieved using a continuous in vitro fibrillization assay, with amyloid fibril formation monitored by the dye, thioflavin T (ThT). Each assay was always run in triplicate. The assays ran for 100 hours, continuously, under a controlled temperature of 37°C. At the end of the assay, each sample revealed a kinetic curve suggestive of amyloid fibril formation [35], [36], [37] comprising of a lag phase, an exponential growth phase and an end plateau (Figure 3A), which is typical of a nucleated polymerization type process [38]. Moreover, there appeared to be no observable significant differences in the average lag phase time between all ligands (including dopamine) and α-syn alone (Figure 3B).
Figure 3. The kinetics of α-syn fibrillization with the DOP analogs in comparison to α-syn alone.
(A) Kinetics curves of α-syn fibrillization in the presence of the test ligands. The fibrillization buffer (i.e, no protein) was assayed in the absence of both α-syn and the test ligands. All curves represent the mean kinetics output from at least 3 measurements. (B) Bar chart statistics displays no significant difference between the lag phases.
For all in vitro assays, the ligands and the control ligand DOP were dissolved in a concentration of 0.1% dimethylsulfoxide (DMSO). This concentration had been determined empirically by a series of preliminary experiments set out to elucidate the concentration where DMSO had no effect, upon both the lag phase and the assembly of fibrils (see Figure S1, S2 and S3).
The continuous presence of ThT in the assay was assessed by AFM, to determine whether this could have an effect upon the aggregation of the protein and/or the binding of a ligand to the α-syn. As DOP has previously been shown by AFM to inhibit the aggregation of α-syn [26], the reaction of DOP with α-syn protein in equimolar concentration in the assay, was assessed by AFM, both in the presence and absence of the ThT dye. Neither the presence nor the absence of ThT appeared to have any effect on the inhibitory effect of DOP on the aggregation of α-syn (Figure 4).
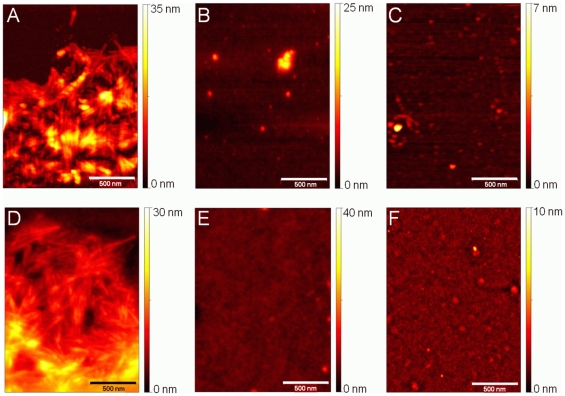
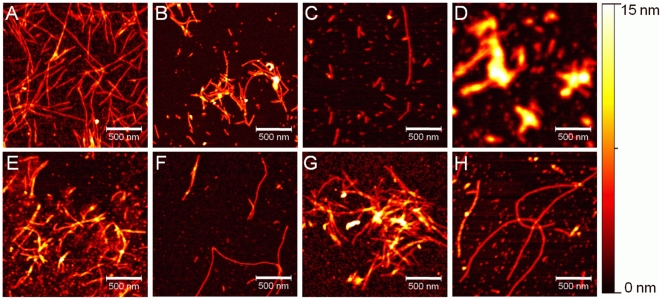
Figure 4. AFM analysis of α-syn aggregation in the presence or absence of both ThT and DOP.
AFM height images were acquired from assay end products, which had been deposited onto freshly cleaved mica surfaces after 100 hours of incubation. The presence or absence of ThT revealed no observable effect upon either the formation or morphology of α-syn fibrils when assay was run with α syn alone, (A) +ThT, (D)–ThT. The inhibitory effect of DOP on the α syn fibrillization was clearly observed when the assay was run in the presence of DOP. (B, C, E, F). Moreover, the presence (B, C) or absence (E, F) of ThT did not show any effect upon the inhibitory action of DOP on α-syn fibrillization.
AFM and TEM Analysis of the Ligands on the Aggregation of α-Syn
Samples were analyzed by both AFM and TEM. In order to detect any distinctive effect upon the assembly of the fibrils, we developed a detailed classification of the fibrillar structures that had been formed. Based on AFM analysis, fibrillar structures were classified as follows: mature fibrils (>0.75 µm in length), intermediate fibrils (0.5–0.75 µm), short fibrils/fragments (or protofibrils) (<0.5 µm). Qualitatively, AFM analysis revealed some differences in terms of α-syn fibril assembly in the presence of the ligands (Figure 5A–G ).
Figure 5. AFM analysis of α-syn aggregation in presence of test ligands as DOP analogs and alone.
AFM height images were acquired from assay end products, which had been deposited onto freshly cleaved mica surfaces after 100 hours of incubation. All images are displayed using the same color scale as shown on the right hand side. (A–B) 5-hydroxyindole; (C) 4-(2-aminoethyl)aniline; (D-E) 6-aminoindole; (F) 2-amino-4-tert-butylphenol; (G) tyramine; (H) α-syn only.
The α-syn fibrils in the absence of any of the ligands consistently revealed predominantly ‘mature’ fibrils (≈>0.75 µm in length) with an average width of ≈9.2 nm ±2.2 nm on TEM micrographs (107 fibrils) (Figure 4A and D , Figure 5H and Figure 6A and B , and Figure S2 and Figure S3), in agreement with previous reports [39], [40], [41]. Moreover, they frequently appeared in clusters (Figure 4A and D , Figure 6A and B , Figure S2 and Figure S3). In addition, round spherical structures (possibly oligomers) were observed (Figure 5H and Figure S2, Figure S3).
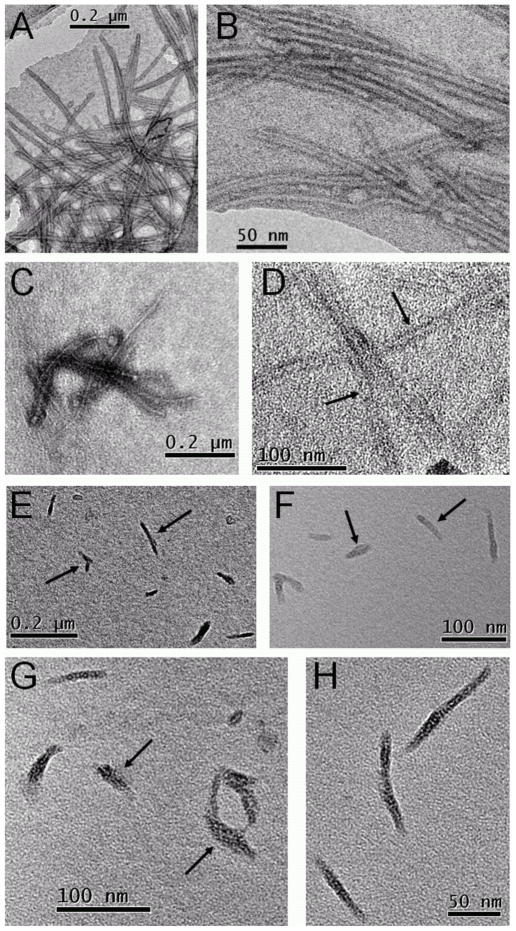
Figure 6. TEM micrographs showing the effect of the presence of 6-aminoindole or 5-hydroxindole on the aggregation of α-syn.
Whilst both samples revealed similar kinetic data, typical of a nucleation/polymerization process, microscopy revealed contrasting data. (A and B) α-syn fibrils formed in the absence of any ligands, (C and D) α-syn fibrils formed in the presence of 5-hydroxyindole and (E–H) 6-aminoindole. Arrows indicate fibrillar structures of varying size.
In the presence of the test ligands, in an equimolar concentration with α-syn, the following observations were made. Overall, whilst all ligands showed no complete inhibition on the assembly of α-syn fibrils (Figure 5) compared to the control ligand DOP (Figure 4), differences did appear in both fibril size and distribution of particle aggregates (Figure 5A–G ) when compared to α-syn alone (Figure 4A and D , Figure 5H , Figure S2), which consistently revealed mostly mature fibrils. DOP revealed no fibrils, and only spherical structures (Figure 4B, C, E and F ), whereas 5-hydroxyindole and 6-aminoindole revealed predominantly fibrils of an intermediate and short size (Figure 5A, B, D, E ). Tyramine and 2-amino-4-tert-butylphenol revealed a mixture of both mature and intermediate fibrils (Figure 5F and G ), whereas 4-(2-aminoethylaniline) revealed a mixture of all fibril sizes (Figure 5C ). The ligand 6-aminoindole appeared overall to show the greatest effect on the assembly of α-syn fibrils. As MD simulations predicted that this ligand and 5-hydroxyindole should have the strongest binding to α-syn (Table 1), these two ligands were chosen for a more detailed analysis by higher resolution with TEM.
By TEM, α-syn fibrils alone were generally observed in clusters and longer than 0.75 µm in length (Figure 6A–B). In comparison, 6-aminoindole revealed individual structures, orientated in a fibrillar form (Figure 6E–H ), which at high magnification suggested that these ‘fibrils’ could still be at an early/intermediate stage of the fibrillization process. The ligand 5-hydroxyindole in comparison showed clusters of α-syn fibrils, albeit shorter than α-syn fibrils in the absence of this ligand (Figure 6C–D and Figure 6A–B , respectively).
Discussion
The loss of dopaminergic neurons in the substantia nigra and the presence of α-syn containing Lewy bodies are the classical diagnostic markers of PD. DOP has been shown to inhibit α-syn aggregation by binding to the α-syn protein [21], [22], [23], [24], [25], [26], [27], [29]. Thus, DOP mimics might have some effect on the aggregation of the protein.
We analyzed this issue using a combined in silico and in vitro approach. Ligand screening and molecular docking allowed us to identify the five commercially available ligands used in this study, which are structurally and electrostatically similar to DOP (Figure 2). Thus, we predicted that these ligands might have a weaker inhibitory effect on fibrillization than DOP.
To test our predictions, we developed a continuous in vitro fibrillization assay. We observed the kinetics of the fibrillization process and analyzed the reaction end products by AFM to obtain spatially resolved information of their morphology. AFM revealed that the ligands caused some inhibitory effect, albeit weaker than that of DOP (Figure 5 and Figure 4 respectively). Moreover, it was clearly apparent that the fibrillar structures varied in terms of their length and in the population distribution of the structures for each test ligand (Figure 5), although the kinetic data were typical of an occurring nucleation/polymerization mechanism (Figure 3A). Likewise, the kinetics curves for each ligand did vary in terms of exponential growth time and plateau phases (Figure 3A). The ligand 6-aminoindole revealed the strongest inhibitory effect upon fibril formation, which was consistent with the fact that it binds most strongly (along with 5-hydroxindole) to the 125YEMPS129 ‘target’ region (Table 1). In comparison, 4-(2-aminoethyalinine) showed the weakest effect (Figure 5), consistent with our MD data (Table 1).
Whilst all the ligands used in our study share the same chemical scaffolding, we noticed (as revealed by both the kinetic output curve and AFM analysis) the strength of DOP in inhibiting α-syn fibril formation in comparison to the tested ligands, especially tyramine (Figure 3); which was consistent with our modeling (Table 1). Tyramine differs from DOP only by a hydrogen in place of a hydroxyl group. The removal of the polar OH function is associated with a loss of electrostatic interactions with the two negatively charged groups of the NAC region. We thus propose that the dihydroxyphenyl group (or 1,2 benzoquinone as in some dopamine derivatives, like dopaminochrome) may play an important role for binding to α-syn, possibly through long-range electrostatic interactions. Moreover, based on these considerations, we suggest that ligands with dihydroxyphenyl (or 1,2 benzoquinone) groups could be selected for further non-covalent binding assays.
Indoles are compounds that are known to interact with metal ions, which themselves are known to increase in PD brains [42]. Furthermore, metal ions are known to bind to, and facilitate, the aggregation of α-syn [42], [43]. Whilst we cannot exclude the presence of metal ions in our assay, we nevertheless ensured that the highest-grade analytical chemicals with the minimal presence of metal ions were always used.
Interestingly, the ultra-structural analysis of α-syn in the presence of 6-aminoindole not only showed fibrils more isolated and shorter than just α-syn alone (Figure 6A–B ), but at a higher resolution it was possible to clearly identify that the process of ‘fibril’ developing into a mature fibril assembly had been affected by the ligand, when compared to α-syn alone (Figure 6A compared to 6H). Most importantly, the kinetic data for both α-syn with this ligand, and α-syn alone, revealed similar ThT fluorescence with the characteristic growth curve. Therefore, we only used ThT fluorescence as a monitor of amyloid fibril formation. A more detailed quantitative methodology of classification would aid to verify our observations. This is currently under development.
Our conclusions, drawn from the ultra-structural analysis of aggregated α-syn in the presence of 6-aminoindole, are further supported by a recent study by Tashiro et al. [44], which characterized over 76 hours of the fibrillization process of α-syn by electron microscopy and small angle x-ray scattering. The authors observed the progression of the formation of fibrils to a mature form using a discontinuous fibrillization assay. Whilst they clearly saw mature fibrils formed at 60 hours, our images at 100 hours for 6-aminoindole did not show this stage of formation (Fure 6E–H ). Our TEM images for 6-aminoindole (Figure 6E–H and unpublished data) obtained at the end of the assay (i.e.: 100 hour) matched closest (if indeed that) with their images of the aggregation of α-syn at 33 hours, which, they speculated, could have been fibrils forming via a critical nucleus or soluble oligomers. Most importantly, their study clearly supports our implication that 6-aminoindole does indeed affect the nucleation/polymerization mechanism. Whilst the main aim of our study was to use in vitro assays to test our prediction of DOP mimics on α-syn, it is clear that a detailed high resolution study over more time points would lead to a deeper insight into the mechanism of the observed DOP mimics on the aggregation of α-syn.
In conclusion, the combined in silico and in vitro approach employed here is, to our knowledge, the first study where such a detailed approach has been used to predict and evaluate ligands that may affect the aggregation of α-syn by forming non-covalent interactions. Biocomputational methods screened and predicted ligands that could have some inhibitory effect on the aggregation of α-syn protein. Remarkably, in vitro assays in combination with high-resolution microscopy showed similar findings. The consistency between both approaches highlights the importance of a combined in silico and in vitro approach that could be used in predicting and developing new drugs and therapeutic strategies for PD.
Materials and Methods
Biocomputing of α-Syn Structures
A refined set of structures, obtained with an optimized protocol, was kindly provided by M. Vendruscolo (personal communication). A cluster analysis was performed exactly as previously reported [29]. Six representative structures were identified [29].
Biocomputing of Ligands
(i) The structures of dopamine and its oxidation products (Figure 1 in [29]) were taken from a previous publication [23].
(ii) Ligands similar to DOP and to its oxidative derivatives (Figure 1) were identified by virtual screening of the ‘ligand.info’ database [31]. This database contains ≈1,160,000 ligands. Structural similarity was estimated by the Tanimoto's equation [32] using the ROCS algorithm in the OpenEye suite of programs (http://www.openeye.org). The 3D molecular structures were overlapped using atom-centered Gaussians [45]. A bias on the overlay was achieved by adding a positive weight to similar chemical groups. For each template, the 100 best hits were selected (600 molecules overall).
The similarity in electrostatic potential between the selected molecules and their templates in Figure 1 was then calculated using the Tanimoto metric [32]. The EON module of Openeye was used (See Supporting Information S1) [46]. The top 10 hits for each template (60 ligands overall) were docked onto 6 α-syn representative conformations of the protein NMR conformational ensemble; the same procedure as described previously by Herrera et al. [29]. The adducts were ranked based on the number of contacts [29] between the target region 125YEMPS129 [23], [25], [27] and the ligand. Five commercially available ligands forming top ranking adducts were selected for experimental analysis (Figure 2).
MD Simulations of Ligands (i) and (ii) to α-Syn
All of the adducts, with all the five ligands, underwent 6 ns of MD in explicit solvent, as previously reported [29]. Version 2.6 of the NAMD program was used [47]. Overall, 72 different MD simulations were carried out. We identify as bound complexes the adducts in which the distance between Cα of α-syn and ligands center of mass is lower than 8 Å.
Expression, Purification and Characterization of Recombinant Human α-Syn
All chemicals used for the experimental assays were high-grade analytical chemicals >99.9% purity with minimal metal impurities.
Human α-syn nucleotide sequence was cloned into the pET11a expression vector and introduced into E. coli strain BL21 (DE3). Expression of α-syn was obtained by growing cells in 100 µg/mL ampicillin containing Luria-Bertani broth at 37°C until an OD600 of about 0.6, followed by induction with 0.6 mM isopropyl β-thiogalactopyranoside for 5 hours. The protein was purified, based on the method of Huang et al. [34].
Mass Spectrometry
The exact molecular weight of the purified α-syn was analyzed by reverse phase high performance liquid chromatography (HPLC) followed by mass spectrometry. Reverse phase HPLC was carried out using a C4 Phenomenex Jupiter (150×4.6 mm) and eluted with a gradient of acetonitrile and trifluoroacetic acid (TFA) 0.085% versus water and 0.1% TFA: from 5 to 35% over 5 min, from 35 to 55% over 20 min, from 55 to 95% over 2 min, at a flow rate of 0.6 mL/min. The effluent from the column was monitored by recording absorbance at 226 nm wavelength. The eluted peak was collected and analyzed by mass spectrometry using a Mariner System 5220 (Applied Biosystem) spectrometer. Mass measurements were carried out in collaboration with Dr. P. De Laureto (C.R.I.B.I., University of Padua, Italy).
Circular Dichroism (CD) Spectroscopy
CD measurements were carried out on a JASCO J-810 spectrophotometer. Spectra were acquired at room temperature in Tris-HCl 20 mM, pH 8.0 using a HELLMA quartz cell (Cell Bio), with an optical path-length of 0.1 cm. All spectra were recorded in the 196–250 nm wavelength range, using a bandwidth of 1 nm and a time constant of 1 s at a speed of 20 nm/min. The signal-to-noise ratio was improved by accumulating 4 scans.
Preparation of α-Syn Solutions and Dopamine Analogs for the In Vitro Studies
The solutions for the in vitro assay were prepared as follows. All solutions were sterile, filtered through a 0.22 µm filter prior to each assay run in order to reduce the presence of ‘contaminants’. Lyophilized α-syn was dissolved in 20 mM Tris-HCl/150 mM NaCl/pH 7.4 (i.e.: the fibrillization buffer) to achieve a stock concentration of 3 mg/mL. All 5 test ligands (Figure 2) and dopamine were dissolved in DMSO, to achieve a final stock concentration of 10 mM. The ligands and α-syn were then both diluted with the fibrillization buffer, to equimolar concentrations (100 µM) in a final DMSO concentration of 0.1%. Amyloid fibril formation was monitored with the histological dye, ThT. A total of 10 µM ThT was added to each sample. Final working volumes were 200 µL per well. Fibril formation was monitored on either a GEMINI EM plate reader or a Spectramax M5 (Molecular Devices). Each test ligand with α-syn was run in triplicate, in 96 black well plates; each well containing 1 teflon bead. The plate was incubated at 37°C, shaken, and ThT fluorescence readings were recorded every 5 min till 100 hours. For each assay run, the background fluorescence from each ligand (i.e. in the absence of α-syn) was also recorded and run in triplicate.
AFM Analysis
The assay end product was imaged with a NanoWizard-II BioAFM (JPK Instruments AG) operating in dynamic mode. Scans were made using an ARROW™ silicon probe with Al coating at the detector side, a tip radius <10 nm, a nominal spring constant of 42 N/m and a nominal resonance frequency of 285 kHz (NanoWorld). Fibrils were deposited onto a freshly cleaved piece of mica at a concentration of 15 µM and left to adhere for 60 min. Samples were then washed with distilled water and blow-dried under a flow of nitrogen. Optically clear regions were chosen for the scanning analysis. The images were collected at a line scan rate of 0.5 - 2 Hz in ambient conditions. The AFM free oscillation amplitudes were ranging from 25 nm to 40 nm, with characteristic set points ranging from 75% to 90% of these free oscillation amplitudes.
TEM Analysis
The assay end product was analyzed on a Jeol 2010F UHR TEM/STEM microscope operated at an accelerating voltage of 200 kV. Samples were absorbed to 300 µm holey formvar/carbon coated grids for ≈1 minute before a brief rinse in water, and negatively stained for 1 minute with 1% phosphotungstic acid. Typical magnifications ranged from 20000-180000.
Image Analysis of AFM Scans and TEM Micrographs
AFM data were analyzed with Gwyddion (gwyddion.net) and SPIP™, (www.imagemet.com). TEM images were analyzed, using the NIH Image processing program, Image J (rsbweb.nih.gov/ij/).
Supporting Information
Supporting information manuscript including references.
(0.04 MB DOC)
Molecules selected from the ligand.info database. Ten molecules have been selected for each of the six compounds reported in Figure 1. These are the molecules which feature the largest shape and electrostatic similarity with dopamine, as calculated using the Tanimoto's definition. The compounds for each set are listed in the order of the priority score. Five commercially available ligands among these 60 molecules have undergone the in vitro assay reported in this study. They are highlighted in bold.
(0.06 MB DOC)
Hydrogen bonds and hydrophobic contacts between the ligands (as depicted in Figure 2) or the dopamine oxidation products (Figure 1) and the target region of α-syn conformations. Column titles from left to right: (i) number of the representative conformation of α-syn, (ii) ligand name, (iii) hydrogen-bonds and distances, (iv) hydrophobic contacts and distances. The contacts are listed for the adducts where the ligands are bound to the target region for more than 50% of their time. Highlighted in grey are the compounds used in the experiments.
(0.10 MB DOC)
Binding regions of ligands other than the target region. Column titles from left to right: (i) The representative conformation of α-syn, (ii) ligand name, (iii) binding region. The contacts are listed for those adducts in which the ligands bind to regions other than the target region for more than 50% of their time. Highlighted in grey are the compounds used in the experiments.
(0.05 MB DOC)
The average lag phase time of α-syn aggregation in different concentrations of DMSO. Notice that above 2.5% DMSO concentration, the lag phase time becomes extremely variable.
(0.45 MB TIF)
AFM images of the aggregation of α-syn. AFM height images were acquired from assay end products after 100 hours of incubation which had been deposited onto freshly cleaved mica surfaces. The height images (A, B, D) are displayed using the same color scale as shown on the right hand side. (A) Height image of long straight fibrils with occasional ‘putative’ looking oligomers. (B) Height image and (C) phase image (range 0–13 deg) of a cluster of fibrils. (D) Height image showing a high degree of clustering of fibrils.
(3.15 MB TIF)
AFM images of the aggregation of α-syn in 0.1% DMSO. AFM height images were acquired from assay end products after 100 hours of incubation which had been deposited onto freshly cleaved mica surfaces. The height images (A, C, D) are displayed using the same color scale as shown on the right hand side. (A) Height image and (B) phase image (range: 0–40 deg) of long straight fibrils with branching. (C–D) Height images of long, straight and circular fibrils, fragments, and both clustered and scattered ‘putative’ oligomers.
(3.18 MB TIF)
Acknowledgments
We thank Maurizio Polano, Alessandra Chesi, Joanna Narkiewicz and Razvan Stoian for their comments and assistance in the development of the in vitro assays. We thank Vincenzo Grillo for technical assistance in TEM analyses. We thank Polly Beidas for her comments and critical reading of the manuscript. The authors wish to thank Gabriella Furlan for editing and proofreading the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by grants from the Italian Institute of Technology to PC and GL and by FP6, Bio-Nano European Infrastructure (BINASP) - SSA 011936 grant to AB and ML. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Moore DJ, West AB, Dawson VL, Dawson TM. Molecular pathophysiology of Parkinson's disease. Annu Rev Neurosci. 2005;28:57–87. doi: 10.1146/annurev.neuro.28.061604.135718. [DOI] [PubMed] [Google Scholar]
- 2.de Rijk MC, Tzourio C, Breteler MM, Dartigues JF, Amaducci L, et al. Prevalence of parkinsonism and Parkinson's disease in Europe: the EUROPARKINSON Collaborative Study. European Community Concerted Action on the Epidemiology of Parkinson's disease. J Neurol Neurosurg Psychiatry. 1997;62:10–15. doi: 10.1136/jnnp.62.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lewy FH. Berlin: Springer; 1912. Handbuch der Neurologie. [Google Scholar]
- 4.Fearnley JM, Lees AJ. Ageing and Parkinson's disease: substantia nigra regional selectivity. Brain. 1991;114 (Pt 5):2283–2301. doi: 10.1093/brain/114.5.2283. [DOI] [PubMed] [Google Scholar]
- 5.Forno LS. Neuropathology of Parkinson's disease. J Neuropathol Exp Neurol. 1996;55:259–272. doi: 10.1097/00005072-199603000-00001. [DOI] [PubMed] [Google Scholar]
- 6.Baba M, Nakajo S, Tu PH, Tomita T, Nakaya K, et al. Aggregation of alpha-synuclein in Lewy bodies of sporadic Parkinson's disease and dementia with Lewy bodies. Am J Pathol. 1998;152:879–884. [PMC free article] [PubMed] [Google Scholar]
- 7.Conway KA, Lee SJ, Rochet JC, Ding TT, Harper JD, et al. Accelerated oligomerization by Parkinson's disease linked alpha-synuclein mutants. Ann N Y Acad Sci. 2000;920:42–45. doi: 10.1111/j.1749-6632.2000.tb06903.x. [DOI] [PubMed] [Google Scholar]
- 8.Goedert M. Parkinson's disease and other alpha-synucleinopathies. Clin Chem Lab Med. 2001;39:308–312. doi: 10.1515/CCLM.2001.047. [DOI] [PubMed] [Google Scholar]
- 9.Dawson TM, Dawson VL. Rare genetic mutations shed light on the pathogenesis of Parkinson disease. J Clin Invest. 2003;111:145–151. doi: 10.1172/JCI17575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, et al. Alpha-synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 11.Jenco JM, Rawlingson A, Daniels B, Morris AJ. Regulation of phospholipase D2: selective inhibition of mammalian phospholipase D isoenzymes by alpha- and beta-synucleins. Biochemistry. 1998;37:4901–4909. doi: 10.1021/bi972776r. [DOI] [PubMed] [Google Scholar]
- 12.Lee FJ, Liu F, Pristupa ZB, Niznik HB. Direct binding and functional coupling of alpha-synuclein to the dopamine transporters accelerate dopamine-induced apoptosis. Faseb J. 2001;15:916–926. doi: 10.1096/fj.00-0334com. [DOI] [PubMed] [Google Scholar]
- 13.Paxinou E, Chen Q, Weisse M, Giasson BI, Norris EH, et al. Induction of alpha-synuclein aggregation by intracellular nitrative insult. J Neurosci. 2001;21:8053–8061. doi: 10.1523/JNEUROSCI.21-20-08053.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharon R, Goldberg MS, Bar-Josef I, Betensky RA, Shen J, et al. alpha-Synuclein occurs in lipid-rich high molecular weight complexes, binds fatty acids, and shows homology to the fatty acid-binding proteins. Proc Natl Acad Sci U S A. 2001;98:9110–9115. doi: 10.1073/pnas.171300598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lotharius J, Brundin P. Pathogenesis of Parkinson's disease: dopamine, vesicles and alpha-synuclein. Nat Rev Neurosci. 2002;3:932–942. doi: 10.1038/nrn983. [DOI] [PubMed] [Google Scholar]
- 16.Perez RG, Waymire JC, Lin E, Liu JJ, Guo F, et al. A role for alpha-synuclein in the regulation of dopamine biosynthesis. J Neurosci. 2002;22:3090–3099. doi: 10.1523/JNEUROSCI.22-08-03090.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu J, Kao SY, Lee FJ, Song W, Jin LW, et al. Dopamine-dependent neurotoxicity of alpha-synuclein: a mechanism for selective neurodegeneration in Parkinson disease. Nat Med. 2002;8:600–606. doi: 10.1038/nm0602-600. [DOI] [PubMed] [Google Scholar]
- 18.Maries E, Dass B, Collier TJ, Kordower JH, Steece-Collier K. The role of alpha-synuclein in Parkinson's disease: insights from animal models. Nat Rev Neurosci. 2003;4:727–738. doi: 10.1038/nrn1199. [DOI] [PubMed] [Google Scholar]
- 19.Fountaine TM, Wade-Martins R. RNA interference-mediated knockdown of alpha-synuclein protects human dopaminergic neuroblastoma cells from MPP(+) toxicity and reduces dopamine transport. J Neurosci Res. 2007;85:351–363. doi: 10.1002/jnr.21125. [DOI] [PubMed] [Google Scholar]
- 20.Burke WJ, Kumar VB, Pandey N, Panneton WM, Gan Q, et al. Aggregation of alpha-synuclein by DOPAL, the monoamine oxidase metabolite of dopamine. Acta Neuropathol. 2008;115:193–203. doi: 10.1007/s00401-007-0303-9. [DOI] [PubMed] [Google Scholar]
- 21.Conway KA, Rochet JC, Bieganski RM, Lansbury PT., Jr Kinetic stabilization of the alpha-synuclein protofibril by a dopamine-alpha-synuclein adduct. Science. 2001;294:1346–1349. doi: 10.1126/science.1063522. [DOI] [PubMed] [Google Scholar]
- 22.Cappai R, Leck SL, Tew DJ, Williamson NA, Smith DP, et al. Dopamine promotes alpha-synuclein aggregation into SDS-resistant soluble oligomers via a distinct folding pathway. Faseb J. 2005;19:1377–1379. doi: 10.1096/fj.04-3437fje. [DOI] [PubMed] [Google Scholar]
- 23.Mazzulli JR, Mishizen AJ, Giasson BI, Lynch DR, Thomas SA, et al. Cytosolic catechols inhibit alpha-synuclein aggregation and facilitate the formation of intracellular soluble oligomeric intermediates. J Neurosci. 2006;26:10068–10078. doi: 10.1523/JNEUROSCI.0896-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mazzulli JR, Armakola M, Dumoulin M, Parastatidis I, Ischiropoulos H. Cellular oligomerization of alpha-synuclein is determined by the interaction of oxidized catechols with a C-terminal sequence. J Biol Chem. 2007;282:31621–31630. doi: 10.1074/jbc.M704737200. [DOI] [PubMed] [Google Scholar]
- 25.Li HT, Lin DH, Luo XY, Zhang F, Ji LN, et al. Inhibition of alpha-synuclein fibrillization by dopamine analogs via reaction with the amino groups of alpha-synuclein. Implication for dopaminergic neurodegeneration. Febs J. 2005;272:3661–3672. doi: 10.1111/j.1742-4658.2005.04792.x. [DOI] [PubMed] [Google Scholar]
- 26.Norris EH, Giasson BI, Hodara R, Xu S, Trojanowski JQ, et al. Reversible inhibition of alpha-synuclein fibrillization by dopaminochrome-mediated conformational alterations. J Biol Chem. 2005;280:21212–21219. doi: 10.1074/jbc.M412621200. [DOI] [PubMed] [Google Scholar]
- 27.Bisaglia M, Mammi S, Bubacco L. Kinetic and structural analysis of the early oxidation products of dopamine: analysis of the interactions with alpha-synuclein. J Biol Chem. 2007;282:15597–15605. doi: 10.1074/jbc.M610893200. [DOI] [PubMed] [Google Scholar]
- 28.Dedmon MM, Lindorff-Larsen K, Christodoulou J, Vendruscolo M, Dobson CM. Mapping long-range interactions in alpha-synuclein using spin-label NMR and ensemble molecular dynamics simulations. J Am Chem Soc. 2005;127:476–477. doi: 10.1021/ja044834j. [DOI] [PubMed] [Google Scholar]
- 29.Herrera FE, Chesi A, Paleologou KE, Schmid A, Munoz A, et al. Inhibition of alpha-synuclein fibrillization by dopamine is mediated by interactions with five C-terminal residues and with E83 in the NAC region. PLoS ONE. 2008;3:e3394. doi: 10.1371/journal.pone.0003394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bostrom J, Hogner A, Schmitt S. Do structurally similar ligands bind in a similar fashion? J Med Chem. 2006;49:6716–6725. doi: 10.1021/jm060167o. [DOI] [PubMed] [Google Scholar]
- 31.von Grotthuss M, Koczyk G, Pas J, Wyrwicz LS, Rychlewski L. Ligand.Info small-molecule Meta-Database. Comb Chem High Throughput Screen. 2004;7:757–761. doi: 10.2174/1386207043328265. [DOI] [PubMed] [Google Scholar]
- 32.Tanimoto T. IBM Internal Report. 1957.
- 33.Slutzki M, Jaitin DA, Yehezkel TB, Schreiber G. Variations in the unstructured C-terminal tail of interferons contribute to differential receptor binding and biological activity. J Mol Biol. 2006;360:1019–1030. doi: 10.1016/j.jmb.2006.05.069. [DOI] [PubMed] [Google Scholar]
- 34.Huang C, Ren G, Zhou H, Wang CC. A new method for purification of recombinant human alpha-synuclein in Escherichia coli. Protein Expr Purif. 2005;42:173–177. doi: 10.1016/j.pep.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 35.Conway KA, Harper JD, Lansbury PT., Jr Fibrils formed in vitro from alpha-synuclein and two mutant forms linked to Parkinson's disease are typical amyloid. Biochemistry. 2000;39:2552–2563. doi: 10.1021/bi991447r. [DOI] [PubMed] [Google Scholar]
- 36.Narhi L, Wood SJ, Steavenson S, Jiang Y, Wu GM, et al. Both familial Parkinson's disease mutations accelerate alpha-synuclein aggregation. J Biol Chem. 1999;274:9843–9846. doi: 10.1074/jbc.274.14.9843. [DOI] [PubMed] [Google Scholar]
- 37.Wood SJ, Wypych J, Steavenson S, Louis JC, Citron M, et al. alpha-synuclein fibrillogenesis is nucleation-dependent. Implications for the pathogenesis of Parkinson's disease. J Biol Chem. 1999;274:19509–19512. doi: 10.1074/jbc.274.28.19509. [DOI] [PubMed] [Google Scholar]
- 38.Jarrett JT, Lansbury PT., Jr Amyloid fibril formation requires a chemically discriminating nucleation event: studies of an amyloidogenic sequence from the bacterial protein OsmB. Biochemistry. 1992;31:12345–12352. doi: 10.1021/bi00164a008. [DOI] [PubMed] [Google Scholar]
- 39.Kaylor J, Bodner N, Edridge S, Yamin G, Hong DP, et al. Characterization of oligomeric intermediates in alpha-synuclein fibrillation: FRET studies of Y125W/Y133F/Y136F alpha-synuclein. J Mol Biol. 2005;353:357–372. doi: 10.1016/j.jmb.2005.08.046. [DOI] [PubMed] [Google Scholar]
- 40.van Raaij ME, van Gestel J, Segers-Nolten IM, de Leeuw SW, Subramaniam V. Concentration dependence of alpha-synuclein fibril length assessed by quantitative atomic force microscopy and statistical-mechanical theory. Biophys J. 2008;95:4871–4878. doi: 10.1529/biophysj.107.127464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Raaij ME, Segers-Nolten IM, Subramaniam V. Quantitative morphological analysis reveals ultrastructural diversity of amyloid fibrils from alpha-synuclein mutants. Biophys J. 2006;91:L96–98. doi: 10.1529/biophysj.106.090449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wright JA, Brown DR. Alpha-synuclein and its role in metal binding: relevance to Parkinson's disease. J Neurosci Res. 2008;86:496–503. doi: 10.1002/jnr.21461. [DOI] [PubMed] [Google Scholar]
- 43.Uversky VN, Li J, Fink AL. Metal-triggered structural transformations, aggregation, and fibrillation of human alpha-synuclein. A possible molecular NK between Parkinson's disease and heavy metal exposure. J Biol Chem. 2001;276:44284–44296. doi: 10.1074/jbc.M105343200. [DOI] [PubMed] [Google Scholar]
- 44.Tashiro M, Kojima M, Kihara H, Kasai K, Kamiyoshihara T, et al. Characterization of fibrillation process of alpha-synuclein at the initial stage. Biochem Biophys Res Commun. 2008;369:910–914. doi: 10.1016/j.bbrc.2008.02.127. [DOI] [PubMed] [Google Scholar]
- 45.Frisch MJ. Pittsburgh: Gaussian Inc; 2001. Gaussian98. revision A.1x ed. [Google Scholar]
- 46.Muchmore SW, Souers AJ, Akritopoulou-Zanze I. The use of three-dimensional shape and electrostatic similarity searching in the identification of a melanin-concentrating hormone receptor 1 antagonist. Chem Biol Drug Des. 2006;67:174–176. doi: 10.1111/j.1747-0285.2006.00341.x. [DOI] [PubMed] [Google Scholar]
- 47.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information manuscript including references.
(0.04 MB DOC)
Molecules selected from the ligand.info database. Ten molecules have been selected for each of the six compounds reported in Figure 1. These are the molecules which feature the largest shape and electrostatic similarity with dopamine, as calculated using the Tanimoto's definition. The compounds for each set are listed in the order of the priority score. Five commercially available ligands among these 60 molecules have undergone the in vitro assay reported in this study. They are highlighted in bold.
(0.06 MB DOC)
Hydrogen bonds and hydrophobic contacts between the ligands (as depicted in Figure 2) or the dopamine oxidation products (Figure 1) and the target region of α-syn conformations. Column titles from left to right: (i) number of the representative conformation of α-syn, (ii) ligand name, (iii) hydrogen-bonds and distances, (iv) hydrophobic contacts and distances. The contacts are listed for the adducts where the ligands are bound to the target region for more than 50% of their time. Highlighted in grey are the compounds used in the experiments.
(0.10 MB DOC)
Binding regions of ligands other than the target region. Column titles from left to right: (i) The representative conformation of α-syn, (ii) ligand name, (iii) binding region. The contacts are listed for those adducts in which the ligands bind to regions other than the target region for more than 50% of their time. Highlighted in grey are the compounds used in the experiments.
(0.05 MB DOC)
The average lag phase time of α-syn aggregation in different concentrations of DMSO. Notice that above 2.5% DMSO concentration, the lag phase time becomes extremely variable.
(0.45 MB TIF)
AFM images of the aggregation of α-syn. AFM height images were acquired from assay end products after 100 hours of incubation which had been deposited onto freshly cleaved mica surfaces. The height images (A, B, D) are displayed using the same color scale as shown on the right hand side. (A) Height image of long straight fibrils with occasional ‘putative’ looking oligomers. (B) Height image and (C) phase image (range 0–13 deg) of a cluster of fibrils. (D) Height image showing a high degree of clustering of fibrils.
(3.15 MB TIF)
AFM images of the aggregation of α-syn in 0.1% DMSO. AFM height images were acquired from assay end products after 100 hours of incubation which had been deposited onto freshly cleaved mica surfaces. The height images (A, C, D) are displayed using the same color scale as shown on the right hand side. (A) Height image and (B) phase image (range: 0–40 deg) of long straight fibrils with branching. (C–D) Height images of long, straight and circular fibrils, fragments, and both clustered and scattered ‘putative’ oligomers.
(3.18 MB TIF)