Abstract
We report a label-free fluorescent aptamer sensor for adenosine based on the regulation of malachite green (MG) fluorescence, with comparable sensitivity and selectivity to other labeled adenosine aptamer-based sensors. The sensor consists of free MG, an aptamer strand containing an adenosine aptamer next to an MG aptamer, and a bridging strand that partially hybridizes to the aptamer strand. Such a hybridization prevents MG from binding to MG aptamer, resulting in low fluorescence of MG in the absence of adenosine. Addition of adenosine causes the adenosine aptamer to bind adenosine, weakening the hybridization of the aptamer strand with the bridging strand, making it possible for MG to bind to the aptamer strand and exhibits high fluorescence intensity. Since this design is based purely on nucleic acid hybridization, it can be generally applied to other aptamers for the label-free detection of a broad range of analytes.
Aptamers are DNA or RNA molecules that bind a specific target molecule. They have been obtained through a combinatorial selection process called systematic evolution of ligands by exponential enrichment (SELEX).1–10 In such a process, DNA or RNA molecules that are capable of binding a target molecule of interest are selected from a nucleic acid library consisting of 1014–1015 different sequences through iterative steps of selection, amplification and mutation. Mimicking natural evolution, SELEX is a powerful technique in laboratory and has resulted in many aptamers that are specific to a wide range of targets from small organic molecules such as adenosine, to proteins such as thrombin, and even viruses and cells.1–13 The affinity of the aptamers towards their targets can rival that of antibodies, with dissociation constants in as low as the picomolar range.10, 14 Unlike antibodies, however, aptamers can be selected under non-physiological conditions, and for targets that are either too small or too toxic for antibodies to elicit effective immune responses. 3, 11
Because of the wide range of targets that aptamers can bind specifically, an active and emerging area of research is the transformation of aptamers into sensors for in situ and real-time detection and quantification.3, 15–21 Towards this goal, a number of aptamer-based sensors have been designed and optimized.3, 20, 22–24 The design philosophy is to convert the binding event into detectable signal changes, such as fluorescent,25–33 colorimetric,25, 34–43 magnetic,44, 45 electrochemical46–56 and even phase changes.57 Among aptamer-based sensors, fluorescent sensors are particularly interesting, in part due to their high sensitivity and feasibility of quantification. However, most current fluorescent aptamer sensors require fluorophore and quencher labeling either at the ends or at the active site of the aptamer molecules.58 The fluorescence response in a typical fluorescent aptamer sensor is due to a change in the distance between fluorophore and quencher, which in turn is modulated by the presence of the analyte. While these labeled fluorescent sensors have many interesting applications, label-free fluorescent sensors may offer certain advantages, as they cost less due to avoidance of covalent-labeling of fluorophores/quenchers to the nucleic acids. Besides, fluorescent labeling may decrease the performance of the aptamers.58 In addition, the quencher may not quench fluorescence completely, resulting in high background.
Label-free sensors combining fluorescence enhancement of dyes by intercalation with aptamers have been previously developed.26, 59–63 However, they are usually turn-off sensors, which are not preferred in practice, and limited fluorescence decrease at saturation has been observed. Malachite green (MG) is an organic dye with almost no fluorescence when in solution by itself with a quantum yield of 7.9×10−5.64 Grate et al. evolved an RNA aptamer for malachite green65 and it was discovered that the fluorescence of MG increased more than 2000 fold when MG was bound to the aptamer.64 This strong enhancement in fluorescent signal was utilized by Stojanovic and coworkers to design label-free fluorescent sensors.66 To achieve label-free sensing, they used the MG aptamer as a signaling domain and another aptamer, such as the adenosine aptamer, as a target recognition domain. To ensure that the binding of the target to the aptamer results in binding of MG to the MG aptamer, and thus enhancement of fluorescent signals, the authors designed a communication module in which the binding of target triggers change of nucleic acid conformation and binding of MG.66 Approximately 3–5 fold increase of MG fluorescence was indeed observed in the presence of targets and the fluorescence increased with increasing target concentrations. However, the authors pointed out that a major limitation of the method - the design of the communication modules remains a trial and error process. Moreover the communication module may not exist in all aptamers. In addition, it is desirable to increase the fluorescence signal beyond the 3–5 fold increase observed for the adenosine aptamer system noted above. Therefore, to make the label-free method more general and practical, we herein report the regulation of the fluorescence of malachite green by adenosine based merely on hybridization energy differences and its application as a label-free turn-on fluorescent sensor for adenosine, with up to a 12-fold increase in fluorescence signal after optimizations. Moreover, the design of the sensor is general and can be applied to other aptamer-based sensors.
EXPERIMENTAL SECTIONS
Materials
Nucleic acids were purchased from Integrated DNA Technologies, Inc. (Coralville, IA) with standard desalting. No further purification was performed. Malachite green and all other chemicals were purchased from Sigma-Aldrich. Buffers were prepared in Millipore water.
Sensor preparation
The sensor solution was prepared by dissolving the nucleic acid strands and malachite green in buffer. The solution was then heated to 80 °C and cooled down to room temperature in about one hour to ensure the system to fold into the structure described in Figure 1.
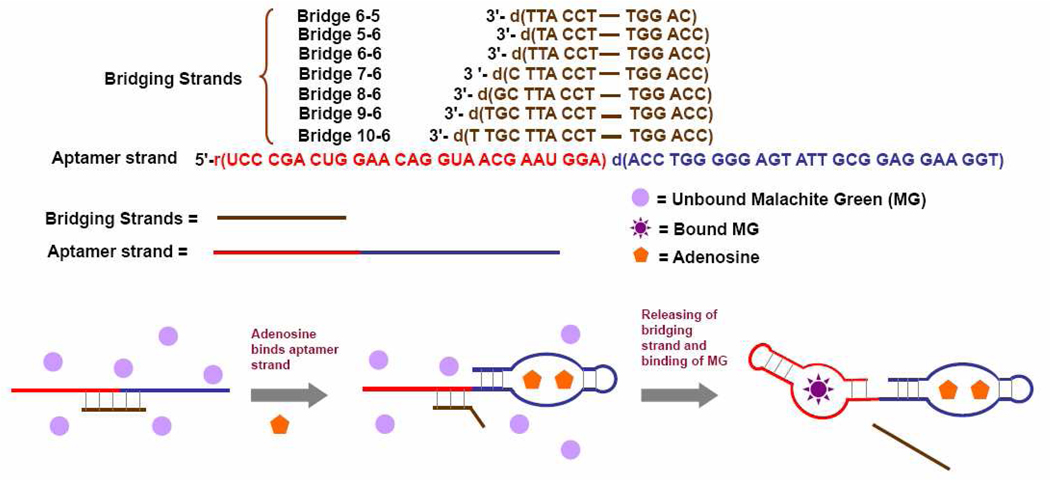
Figure 1.
Scheme showing the regulation of the fluorescence of malachite green by adenosine. Without adenosine, the affinity of the aptamer strand (in red-blue) is inhibited by the bridging strand (in brown). With adenosine, the bridging strand separates from the aptamer strand, which then binds malachite green, leading to an enhancement of fluorescence.
Because of the poor solubility of adenosine, the highest concentration of the stock solution we can prepare is 50 mM. Thus, during the sensor preparation step, the dilution effect by the addition of adenosine was considered. In a typical procedure at the optimized conditions, 1.11 µM aptamer strand, 1.56 µM Bridge 9-6 and 0.67 µM MG were dissolved in 22 mM Tris buffer containing 5.56 mM NaCl, 155.6 mM KCl and 5.56 MgCl2. After heating and cooling the solution as described above, 0.45 mL of the prepared solution was mixed with 0.05 mL 10-times adenosine stock solution. After 30 min, the fluorescence of the sample was measured and recorded.
Fluorescent measurements and data analysis
Fluorescence experiments were carried out on a Fluromax-2 fluorimeter (HORIBA Jobin Yvon inc., Edison, NJ). Emission acquisition mode was used. The fluorescence was excited by 615 nm excitation. The fluorescence at 650 nm was recorded. F/F0 was plotted as sensor signal. F is the fluorescence of MG after the addition of adenosine. During optimization procedure, F0 is the fluorescence of MG before addition of adenosine stock solution. For the sensor calibration curve acquisition, F0 is the fluorescence of the sensor solution with addition of 0 mM adenosine (water). All the measurements were performed for 3 times and the standard deviation was plotted as the error bar.
Nucleic acids sequences
The sequences of the nucleic acids used in the paper are presented in Figure 1.65,67 Bridging strands are named as bridge M-N, in which M is the number of the complimentary bases to the MG aptamer and N is the number of the complimentary bases to the adenosine aptamer.
Hybridization energy calculation
The energy was calculated using IDT SciTools, provided free of charge on the web by Integrated DNA Technologies, Inc. (Coralville, IA) to assist in the design of the system.
RESULTS AND DISCUSSIONS
Sensor Design
The design of the label-free aptamer fluorescent sensor is shown in Figure 1. The sensor contains two nucleic acid strands. One is a chimeric conjugate of the adenosine DNA aptamer sequence (in blue) and MG RNA aptamer sequence (in red), called the “aptamer strand” in this work. The other is a DNA strand, called the “bridging strands” (in brown), that contain sequences complimentary to both the adenosine and MG aptamers. The sequence of the bridging strand is designed so that the aptamer and bridging strands form a stable complex in buffer at room temperature to prevent the MG aptamer strand from binding MG in the solution if there is no adenosine present. Under this condition, the MG remains free in solution and almost non-fluorescent. In the presence of adenosine, however, the aptamer strand binds adenosine, leaving much less number of complimentary base pairs between the aptamer strand and bridging strand, which is less stable at room temperature, resulting in release of the bridging strand from the aptamer strand. As a consequence, the fluorescence of the MG is recovered.
The DNA/RNA chimeric aptamer described herein is expected to be less stable than all-DNA aptamer, but more stable than all-RNA aptamers. The stability can be further improved if the RNA aptamer can be replaced by a DNA aptamer that can be obtained through in vitro selection.
Sensor Optimization
Since there are two aptamers in the design, each with its own optimal buffer condition for performance, a common condition suitable for both aptamer functions should be found first. Although adenosine aptamer was in vitro selected in a 20 mM Tris-acetate buffer with pH 7.6,67 it has been reported that adenosine aptamer-based biosensors are effective in a range of pH between 7.4 and 8.3.33, 57, 68, 69 In addition, the Na+ and Mg2+ concentrations can be varied from 100 mM to 300 mM and from 0 to 5 mM, respectively.33, 57, 68, 69 Encouraged by these facts, we used the system developed by Li and co-workers30 to test the performance of adenosine aptamer in different buffers, and found that the sensing performance is similar at pH between 7.4 and 8.4 and concentrations of NaCl between 150 mM and 300 mM and MgCl2 between 2 mM and 5 mM (data not shown).
After confirming the workable range of buffer conditions for adenosine aptamer, we next explored a common buffer condition that is also effective for MG aptamer. Since MG aptamer was selected in 10 mM Na-HEPES buffer at pH 7.4, together with 100 mM KCl and 5 mM NaCl, we decided to use 20 mM Tris (pH 7.4), 145 mM NaCl and 5 mM MgCl2 as a compromise between buffers for adenosine and MG aptamers in our first trial to find out if this buffer is effective for MG aptamer. We observed 3 orders of magnitudes of fluorescence increase in the presence of the aptamer strand (sequence displayed in Figure 1), similar to that reported previously (Figure S1A).64 The result shows that the aptamer strand in this work and the common buffer condition result in comparable performance as previously reported MG aptamer and buffer conditions.
Since the MG aptamer was selected in buffer containing potassium, we wondered whether replacing NaCl with KCl in the buffer may help to increase the performance of the sensor. We therefore tested the sensor performance using Bridge 6-5 (Figure 1) in buffers containing 20 mM Tris (pH 7.4), 5 mM MgCl2 and 145 mM in total monovalent metal ions, but with different combination of Na+ and K+. Our results showed that variation of potassium concentration did not chang the signal significantly (by around 10%, Figure S1B). Monovalent metal ions are necessary for the MG aptamer affinity because the DNA strand has to fold, during which the negative charge on the DNA backbone has to be neutralized. As K+ and Na+ has the similar ability to help DNA fold, variation of K+ concentration did not affect the sensor performance significantly. We decided to use 20 mM Tris (pH 7.4), 5 mM MgCl2, 140 mM KCl and 5 mM NaCl as the optimized buffer to make it as close as possible to the buffer in which MG aptamer was selected. It is worthwhile to point that since two aptamers are used in this label-free system, each with its own optimized condition, the range of optimal conditions is much narrower than that of the labeled systems.
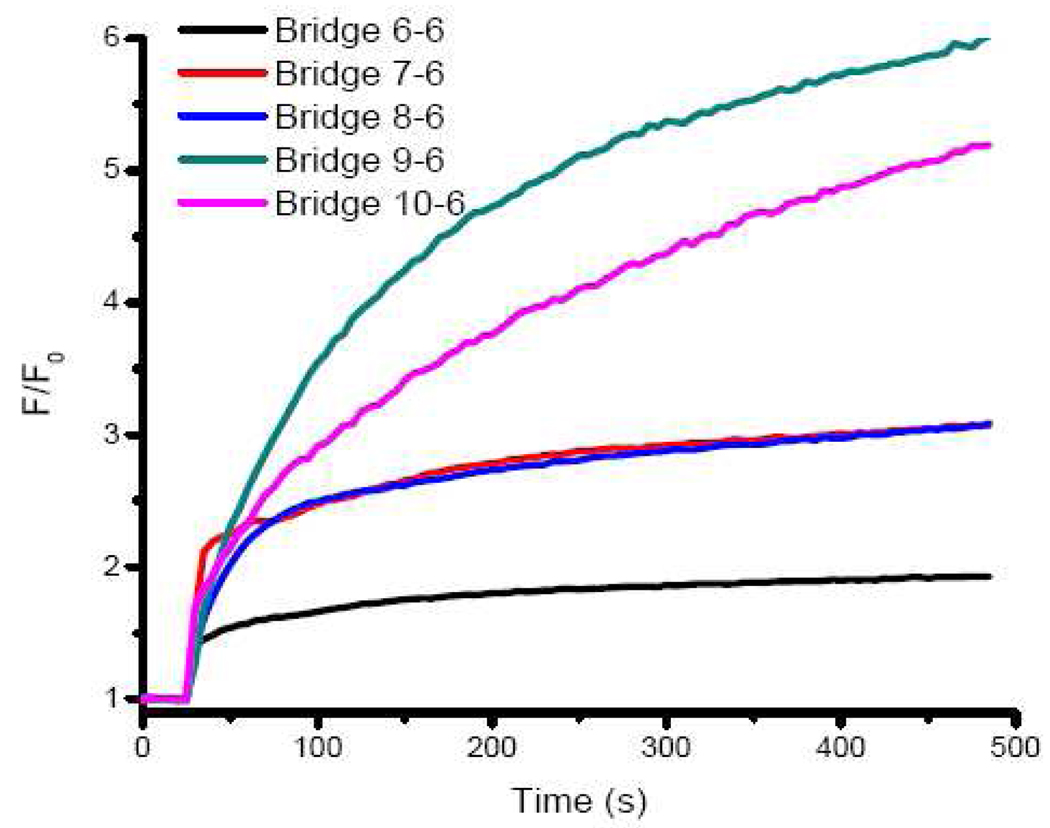
Having arrived at the buffer conditions, we then investigated the optimal sequence for the bridging DNA strands. The design shown in Figure 1 depends on thermodynamics of DNA hybridization and adenosine and MG bindings to their respective aptamers, which include the binding of adenosine to its aptamer, the binding MG to its aptamer, and the interaction between the aptamer strand and the bridging strand. Although design can be generally applicable to any aptamers, optimization for the sequence of the bridging DNA is still necessary. For this design to work, several conditions must be met. First, the base parings between the aptamer strand and the bridging strand have to prevent the MG aptamer from binding MG in the absence of adenosine. At the same time, however, the base parings between the aptamer strand and the bridging strand should not inhibit the binding of adenosine if adenosine is present. Finally, once adenosine binds to adenosine aptamer and starts structural switching, the hybridization between the MG aptamer part and the bridging DNA should not be very stable at room temperature to ensure the release of it and binding of the aptamer strand to MG. We first investigated the bridging strand that contains 6 bases that are complementary to adenosine aptamer70 and various numbers of bases that are complementary to MG aptamer (Bridge 6-6, 7-6, 8-6, 9-6 and 10-6. See Figure 1 for the sequences). During the optimization procedure, we tried to maximize the fluorescence fold increase upon the addition of the adenosine which was chosen as the signal output of the sensor. Meanwhile kinetics is our consideration. We ensured that the equilibrium can be reached in less than 30 min. The sensor was prepared by dissolving 1 µM aptamer strand, bridging strand and MG in the optimized buffer. Upon addition of 5 mM adenosine to the sensor solution, fluorescence (F/F0, Ex: 615 nm, Em: 650 nm) increases when the number of bridging strand base complementary to MG aptamer increases from 6 to 9, and then deceases when the number is 10 (Figure 2). Based on these results we conclude Bridge 9-6 is the optimal for the sensing application. These aptamer and bridging strands form a stable complex in buffer at room temperature, with a hybridization energy (ΔG) of −29.52 kcal/mol, calculated based on IDT SciTools. In the absence of adenosine, such a stable complex prevent the MG aptamer strand from binding MG. However, the 6 base pairs of the bridging strand to the adenosine aptamer (with a ΔG of −11.03 kcal/mol) are not strong enough to inhibit the binding of the aptamer strand towards adenosine. Therefore, in the presence of adenosine, the aptamer strand binds adenosine, leaving only 9 base pairs between the aptamer strand and bridging strand. Such a 9 base pair duplex has a ΔG of only 16.55 kcal/mol, and is not very stable at room temperature. Thus, some bridging strands are released from the aptamer strands and the MG aptamers are recovered. As a result, part of the free MG molecules bind the aptamer strand, giving an increase in the fluorescent signal.
Figure 2.
Sensor performance with different sequences of the bridging strands. 1 µM aptamer strand, bridging strand and MG were used as the sensor. 20 mM Tris (pH 7.4), 140 mM KCl, 5 mM NaCl and 5 mM MgCl2 were used as buffer. 5 mM adenosine was added at t = 25 s and fluorescence versus time was recorded and plotted
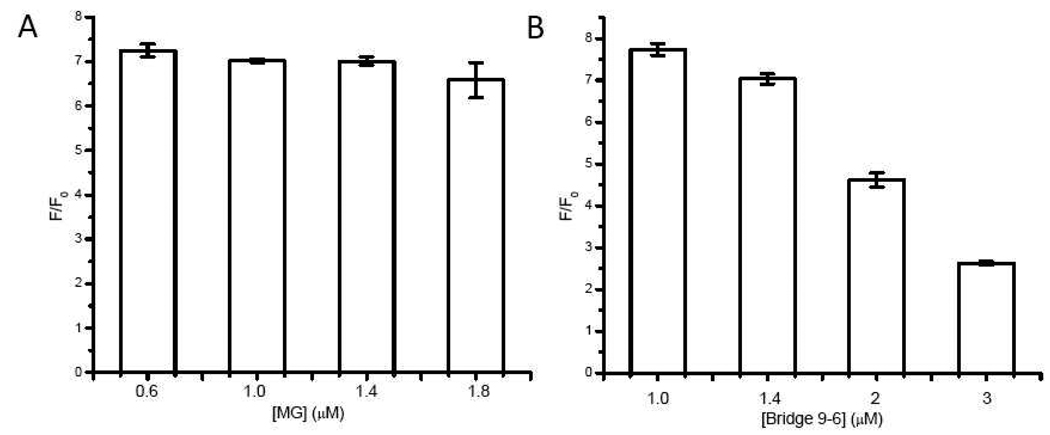
Since there are three DNA species (the aptamer and bridging strands and MG) in the system, it offers a chance to optimize the ratio of the three species separately to increase the performance of the sensor. First, both DNA strands were kept 1 µM and MG concentration was varied. As presented in Figure 3A, MG = 0.6 µM has the best performance. We then kept the aptamer strand and MG concentrations to be 1 µM and 0.6 µM respectively and varied the concentration of the bridging strand (bridge 9-6). Based on the data in Figure 4B, 1.0 µM and 1.4 µM of the Bridge 9-6 have similar fluorescence fold increase in the presence of 5 mM adenosine (around 10% difference). We decided to choose 1.4 µM of the Bridge 9-6 to ensure the complete hybridization of the aptamer strand to the bridge strand. Therefore the optimal sensor design is for 1.4 µM Bridge 9-6 to be complementary to 1 µM aptamer strand while in the presence of 0.6 µM MG.
Figure 3.
Optimization of the ratio of the aptamer and bridge 9-6 strands and MG. The nucleic strands were kept 1 µM and MG concentration varied (A); the aptamer strands and MG were kept 1 µM and 0.6 µM respectively and the concentration of bridge 9-6 varied.
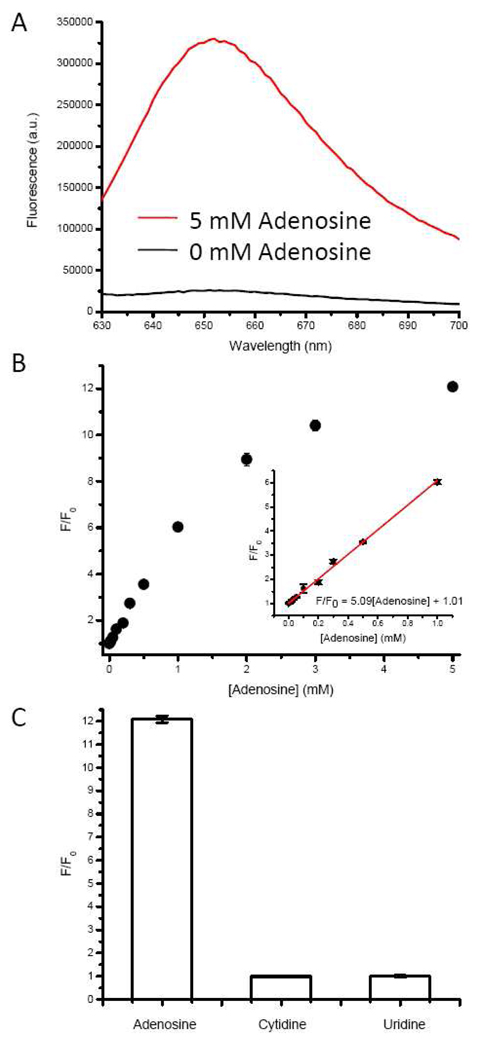
Figure 4.
(A) The fluorescence enhancement of the optimized sensor upon the addition of adenosine; (B) the saturated fluorescence of malachite green with various concentrations of adenosine. The inset shows the fluorescence response at low concentrations of adenosine and the red line shows a linear fitting of the data. (C) selectivity of the sensor towards other nucleosides. Cytidine and uridine did not increase the fluorescence of MG.
Performance of the Sensor
Under the optimal sensor design and buffer conditions, minimal fluorescence was observed in the absence of adenosine (black trace in Figure 4A). Upon addition of 5 mM adenosine, however, a 12 fold increase in fluorescence was observed (red trace in Figure 4A), indicating that the binding of adenosine by the adenosine aptamer caused the release of the bridging strand, recovering the affinity of the MG aptamer, which then binds MG, resulting in a fluorescence increase. To quantify concentrations of adenosine based on the increase in MG fluorescence, we measured the saturated fluorescence upon addition of different concentrations of adenosine. The sensor solution concentration was diluted to 20% of what was described to save the material. The results shown in Figure 4B indicate that the fluorescence of MG increases with an increasing concentration of adenosine, reaching ~12 fold fluorescence increase around 5 mM adenosine. The inset of Figure 5B shows the region between 0 and 1 mM adenosine where a linear relationship was observed between adenosine concentration and MG fluorescence. A detection limit of 20 µM at 90% confidence level was calculated, which is comparable with other adenosine aptamer-based sensors. The sensitivity of the sensor is dependent on the binding affinity of the aptamer. Usually a good aptamer-based sensor has a detection limit that is comparable to the Kd value of the aptamer. We are optimistic that a higher sensitivity for the sensor will be obtained if an aptamer with higher binding affinity is used. It is worthwhile to point out that the binding affinities of the aptamer strand towards adenosine and MG are decreased because of the steric hindrance. However, the detection limit of our sensor is comparable to other adenosine aptamer-based sensors because the affinity of both aptamers is not significantly affected and the label-free design helps to lower the background.
To test the selectivity of the sensor, 5 mM cytidine or uridine, the highest concentration tested for adenosine, was added to the sensor solution and no fluorescence increase was observed (see Figure 4C). Our results suggest that the sensor is specific to adenosine. Guanidine was not used due to poor solubility.
CONCLUSIONS
We have demonstrated coupling of adenosine aptamer strand with MG aptamer strand through a bridging strand in which the fluorescence of malachite green can be regulated by the presence of adenosine. This system has sensitivity and selectivity comparable to other adenosine sensor systems, with the benefit of no need for labeling of the DNA with fluorophores. More importantly, the design is based purely on DNA hybridizations and therefore can be more generally applied to other aptamers for the sensing of a broad range of analytes.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the US Department of Energy (DE-FG02-08ER64568), National Institute of Health (Grant No. ES016865), and National Science Foundation (Grant No. CTS-0120978 and DMI-0328162). We thank Nandini Nagraj for proofreading the manuscript.
Footnotes
SUPPORTING INFORMATION AVAILABLE
Additional information as noted in the texts.
References
- 1.Ellington AD, Szostak JW. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 2.Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 3.Liu J, Cao Z, Lu Y. Chem. Rev. 2009;109:1948–1998. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shamah SM, Healy JM, Cload ST. Acc. Chem. Res. 2008;41:130–138. doi: 10.1021/ar700142z. [DOI] [PubMed] [Google Scholar]
- 5.Famulok M, Hartig JS, Mayer G. Chem. Rev. 2007;107:3715–3743. doi: 10.1021/cr0306743. [DOI] [PubMed] [Google Scholar]
- 6.Manimala JC, Rajendran M, Ellington AD. Recent Dev. Nucleic Acids Res. 2004;1:207–231. [Google Scholar]
- 7.Famulok M, Mayer G, Blind M. Acc. Chem. Res. 2000;33:591–599. doi: 10.1021/ar960167q. [DOI] [PubMed] [Google Scholar]
- 8.Hesselberth J, Robertson MP, Jhaveri S, Ellington AD. Rev. Mol. Biotech. 2000;74:15–25. doi: 10.1016/s1389-0352(99)00005-7. [DOI] [PubMed] [Google Scholar]
- 9.Wilson DS, Szostak JW. Annu. Rev. Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 10.Morris KN, Jensen KB, Julin CM, Weil M, Gold L. Proc. Natl. Acad. Sci. U. S. A. 1998;95:2902–2907. doi: 10.1073/pnas.95.6.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J, Lu Y. J. Fluoresc. 2004;14:343–354. doi: 10.1023/b:jofl.0000031816.06134.d3. [DOI] [PubMed] [Google Scholar]
- 12.Famulok M. Curr. Opin. Struct. Biol. 1999;9:324–329. doi: 10.1016/S0959-440X(99)80043-8. [DOI] [PubMed] [Google Scholar]
- 13.Jayasena SD. Clin. Chem. 1999;45:1628–1650. [PubMed] [Google Scholar]
- 14.Green LS, Jellinek D, Jenison R, Oestman A, Heldin C-H, Janjic N. Biochemistry. 1996;35:14413–14424. doi: 10.1021/bi961544+. [DOI] [PubMed] [Google Scholar]
- 15.Li Y, Lu Y, editors. Functional Nucleic Acids for Sensing and Other Analytical Applications. New York, NY: Springer; 2009. [Google Scholar]
- 16.Fowler CC, Li Y. Chem. Biol. 2007;14:736–738. doi: 10.1016/j.chembiol.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 17.Lu Y, Liu J. Curr. Opin. Biotechnol. 2006;17:580–588. doi: 10.1016/j.copbio.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 18.Navani NK, Li Y. Curr. Opin. Chem. Biol. 2006;10:272–281. doi: 10.1016/j.cbpa.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Potyrailo RA, Conrad RC, Ellington AD, Hieftje GM. Anal.Chem. 1998;70:3419–3425. doi: 10.1021/ac9802325. [DOI] [PubMed] [Google Scholar]
- 20.Mok W, Li Y. Sensors. 2008;8:7050–7084. doi: 10.3390/s8117050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan L, Li Y, Drake TJ, Moroz L, Wang K, Li J, Munteanu A, Yang CJ, Martinez K, Tan W. Analyst. 2005;130:1002–1005. doi: 10.1039/b500308n. [DOI] [PubMed] [Google Scholar]
- 22.Barrick JE, Breaker RR. Sci. Am. 2006;296:50–57. doi: 10.1038/scientificamerican0107-50. [DOI] [PubMed] [Google Scholar]
- 23.O'Sullivan CK. Anal. Bioanal. Chem. 2002;372:44–48. doi: 10.1007/s00216-001-1189-3. [DOI] [PubMed] [Google Scholar]
- 24.Jhaveri S, Rajendran M, Ellington AD. Nat. Biotechnol. 2000;18:1293–1297. doi: 10.1038/82414. [DOI] [PubMed] [Google Scholar]
- 25.Pei R, Rothman J, Xie Y, Stojanovic MN. Nucleic Acids Res. 2009;37:e59/51–e59/56. doi: 10.1093/nar/gkp154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang C-C, Chang H-T. Chem. Commun. 2008:1461–1463. doi: 10.1039/b718752a. [DOI] [PubMed] [Google Scholar]
- 27.Tang Z, Mallikaratchy P, Yang R, Kim Y, Zhu Z, Wang H, Tan W. J. Am. Chem. Soc. 2008;130:11268–11269. doi: 10.1021/ja804119s. [DOI] [PubMed] [Google Scholar]
- 28.Shlyahovsky B, Li D, Weizmann Y, Nowarski R, Kotler M, Willner I. J. Am. Chem. Soc. 2007;129:3814–3815. doi: 10.1021/ja069291n. [DOI] [PubMed] [Google Scholar]
- 29.Cao Z, Suljak SW, Tan W. Curr. Proteomics. 2005;2:31–40. [Google Scholar]
- 30.Nutiu R, Li Y. Angew. Chem., Int. Ed. 2005;44:5464–5467. doi: 10.1002/anie.200501214. [DOI] [PubMed] [Google Scholar]
- 31.Nutiu R, Li Y. Methods. 2005;37:16–25. doi: 10.1016/j.ymeth.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 32.Merino EJ, Weeks KM. J. Am. Chem. Soc. 2003;125:12370–12371. doi: 10.1021/ja035299a. [DOI] [PubMed] [Google Scholar]
- 33.Nutiu R, Li Y. J. Am. Chem. Soc. 2003;125:4771–4778. doi: 10.1021/ja028962o. [DOI] [PubMed] [Google Scholar]
- 34.Elbaz J, Moshe M, Shlyahovsky B, Willner I. Chem. Eur. J. 2009;15:3411–3418. doi: 10.1002/chem.200802004. [DOI] [PubMed] [Google Scholar]
- 35.Li F, Zhang J, Cao X, Wang L, Li D, Song S, Ye B, Fan C. Analyst. 2009;134:1355–1360. doi: 10.1039/b900900k. [DOI] [PubMed] [Google Scholar]
- 36.Zhang J, Wang L, Pan D, Song S, Boey FYC, Zhang H, Fan C. Small. 2008;4:1196–1200. doi: 10.1002/smll.200800057. [DOI] [PubMed] [Google Scholar]
- 37.Zhao W, Chiuman W, Lam JCF, McManus SA, Chen W, Cui Y, Pelton R, Brook MA, Li Y. J. Am. Chem. Soc. 2008;130:3610–3618. doi: 10.1021/ja710241b. [DOI] [PubMed] [Google Scholar]
- 38.Zhao W, Brook MA, Li Y. Chembiochem. 2008;9:2363–2371. doi: 10.1002/cbic.200800282. [DOI] [PubMed] [Google Scholar]
- 39.Wang J, Wang L, Liu X, Liang Z, Song S, Li W, Li G, Fan C. Adv. Mater. 2007;19:3943–3946. [Google Scholar]
- 40.Zhao W, Chiuman W, Brook MA, Li Y. Chembiochem. 2007;8:727–731. doi: 10.1002/cbic.200700014. [DOI] [PubMed] [Google Scholar]
- 41.Liu J, Lu Y. Angew. Chem., Int. Ed. 2006;45:90–94. [Google Scholar]
- 42.Liu J, Lu Y. Nat. Protoc. 2006;1:246–252. doi: 10.1038/nprot.2006.38. [DOI] [PubMed] [Google Scholar]
- 43.Liu J, Lu Y. Anal. Chem. 2004;76:1627–1632. doi: 10.1021/ac0351769. [DOI] [PubMed] [Google Scholar]
- 44.Yigit MV, Mazumdar D, Lu Y. Bioconjugate Chem. 2008;19:412–417. doi: 10.1021/bc7003928. [DOI] [PubMed] [Google Scholar]
- 45.Yigit MV, Mazumdar D, Kim HK, Lee JH, Odintsov B, Lu Y. Chembiochem. 2007;8:1675–1678. doi: 10.1002/cbic.200700323. [DOI] [PubMed] [Google Scholar]
- 46.Du Y, Li B, Wang F, Dong S. Biosens. Bioelectron. 2009;24:1979–1983. doi: 10.1016/j.bios.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 47.Zuo X, Xiao Y, Plaxco KW. J. Am. Chem. Soc. 2009;131:6944–6945. doi: 10.1021/ja901315w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu G, Wan Y, Gau V, Zhang J, Wang L, Song S, Fan C. J. Am. Chem. Soc. 2008;130:6820–6825. doi: 10.1021/ja800554t. [DOI] [PubMed] [Google Scholar]
- 49.Li B, Du Y, Wei H, Dong S. Chem. Commun. 2007:3780–3782. doi: 10.1039/b707057h. [DOI] [PubMed] [Google Scholar]
- 50.Willner I, Zayats M. Angew. Chem., Int. Ed. 2007;46:6408–6418. doi: 10.1002/anie.200604524. [DOI] [PubMed] [Google Scholar]
- 51.Xiao Y, Lai RY, Plaxco KW. Nat. Protoc. 2007;2:2875–2880. doi: 10.1038/nprot.2007.413. [DOI] [PubMed] [Google Scholar]
- 52.Zuo X, Song S, Zhang J, Pan D, Wang L, Fan C. J. Am. Chem. Soc. 2007;129:1042–1043. doi: 10.1021/ja067024b. [DOI] [PubMed] [Google Scholar]
- 53.Baker BR, Lai RY, Wood MS, Doctor EH, Heeger AJ, Plaxco KW. J. Am. Chem. Soc. 2006;128:3138–3139. doi: 10.1021/ja056957p. [DOI] [PubMed] [Google Scholar]
- 54.Zayats M, Huang Y, Gill R, Ma C-a, Willner I. J. Am. Chem. Soc. 2006;128:13666–13667. doi: 10.1021/ja0651456. [DOI] [PubMed] [Google Scholar]
- 55.Xiao Y, Lubin AA, Heeger AJ, Plaxco KW. Angew. Chem., Int. Ed. 2005;44:5456–5459. doi: 10.1002/anie.200500989. [DOI] [PubMed] [Google Scholar]
- 56.Xiao Y, Piorek BD, Plaxco KW, Heeger AJ. J. Am. Chem. Soc. 2005;127:17990–17991. doi: 10.1021/ja056555h. [DOI] [PubMed] [Google Scholar]
- 57.Yang H, Liu H, Kang H, Tan W. J. Am. Chem. Soc. 2008;130:6320–6321. doi: 10.1021/ja801339w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jhaveri SD, Kirby R, Conrad R, Maglott EJ, Bowser M, Kennedy RT, Glick G, Ellington AD. J. Am. Chem. Soc. 2000;122:2469–2473. [Google Scholar]
- 59.Li B, Qin C, Li T, Wang L, Dong S. Anal. Chem. 2009;81:3544–3550. doi: 10.1021/ac900110a. [DOI] [PubMed] [Google Scholar]
- 60.Wang Y, Liu B. Analyst. 2008;133:1593–1598. doi: 10.1039/b806908e. [DOI] [PubMed] [Google Scholar]
- 61.Wang M, Zhang D, Zhang G, Tang Y, Wang S, Zhu D. Anal. Chem. 2008;80:6443–6448. doi: 10.1021/ac801020v. [DOI] [PubMed] [Google Scholar]
- 62.Wang J, Liu B. Chem. Commun. 2008:4759–4761. doi: 10.1039/b806885b. [DOI] [PubMed] [Google Scholar]
- 63.Li B, Wei H, Dong S. Chem. Commun. 2007:73–75. doi: 10.1039/b612080f. [DOI] [PubMed] [Google Scholar]
- 64.Babendure JR, Adams SR, Tsien RY. J. Am. Chem. Soc. 2003;125:14716–14717. doi: 10.1021/ja037994o. [DOI] [PubMed] [Google Scholar]
- 65.Grate D, Wilson C. Proc. Natl. Acad. Sci. U. S. A. 1999;96:6131–6136. doi: 10.1073/pnas.96.11.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stojanovic MN, Kolpashchikov DM. J. Am. Chem. Soc. 2004;126:9266–9270. doi: 10.1021/ja032013t. [DOI] [PubMed] [Google Scholar]
- 67.Huizenga DE, Szostak JW. Biochemistry. 1995;34:656–665. doi: 10.1021/bi00002a033. [DOI] [PubMed] [Google Scholar]
- 68.Lu N, Shao C, Deng Z. Chem. Commun. 2008:6161–6163. doi: 10.1039/b810812a. [DOI] [PubMed] [Google Scholar]
- 69.Liu J, Lu Y. J. Am. Chem. Soc. 2007;129:8634–8643. doi: 10.1021/ja072075+. [DOI] [PubMed] [Google Scholar]
- 70.Xiang Y, Tong A, Lu Y. J. Am. Chem. Soc. 2009;131:15352–15357. doi: 10.1021/ja905854a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.