Abstract
Peroxisome proliferator-activated receptor-gamma (PPARγ) regulates the interface between cellular lipid metabolism, redox status and organelle differentiation. Conditional prostatic epithelial knockout of PPARγ in mice resulted in focal hyperplasia which developed into mouse prostatic intraepithelial neoplasia (mPIN). The grade of PIN became more severe with time. Electron microscopy (EM) showed accumulated secondary lysosomes containing cellular organelles and debris suggestive of autophagy. Consistent with this analysis the autophagy marker LC3 was found to be upregulated in areas of PIN in PPARγ KO tissues. We selectively knocked down PPARγ2 isoform in wild-type mouse prostatic epithelial cells and examined the consequences of this in a tissue recombination model. Histopathologically grafted tissues resembled the conditional PPARγ KO mouse prostates. EM studies of PPARγ- and PPARγ2-deficient epithelial cells in vitro were suggestive of autophagy, consistent with the prostatic tissue analysis. This was confirmed by examining expression of beclin-1 and LC3. Gene expression profiling in PPARγ-/γ2-deficient cells indicated a major dysregulation of cell cycle control and metabolic signaling networks related to peroxisomal and lysosomal maturation, lipid oxidation and degradation. The putative autophagic phenotypes of PPARγ-deficient cells could be rescued by re-expression of either γ1 or γ2 isoform. We conclude that disruption of PPARγ signaling results in autophagy and oxidative stress during mPIN pathogenesis.
Keywords: Peroxisome proliferator-activated receptor gamma (γ), PPARγ; Peroxisome proliferator response element, PPRE; Peroxisome; Lysosome; Mitochondria; Autophagy; Fatty acid oxidation; Oxidative Stress; Differentiation; Prostatic Intraepithelial Neoplasia, PIN
Introduction
Peroxisome proliferator-activated receptors (PPARs) regulate gene transcription in response to peroxisome proliferators, fatty acids, and other endogenous ligands (1, 2). There are two isoforms of PPARγ denoted as PPARγ1 and PPARγ2. These represent proteins derived from alternative start sites in the same gene giving rise to two isoforms, with PPARγ2 having an earlier start site with an additional 30 amino acid residues (3). PPARγ regulates differentiation, proliferation and apoptosis in both stromal (4) and parenchymal epithelial cells (5-8), involving in many signaling pathways during the pathogenesis of inflammation and oxidative stress (9, 10). The links between PPARγ target genes and the relevance of alterations of PPARγ functions to human pathology are beginning to be elucidated (11).
Many prostate cancer (Pca) patients with early disease could potentially be treated conservatively. Tissue-based and in vitro studies suggest that alterations in PPARγ activity may be involved in Pca and that PPARγ may be a candidate target for Pca therapy (12-14).
Recent studies have reported autophagy in prostate cancer cell lines (15-17). To gain insights into whether autophagy is involved in the processes of dysregulated lipid metabolism and induced oxidative stress accompanying the prostate proliferative epithelial lesions, we characterized the PPARγ-deficient mouse prostate models. Autophagy accompanied the altered cellular proliferation and differentiation that resulted from PPARγ-deficiency in mouse prostate models. This paper examines the links between PPARγ activity and the subcellular and histopathologic changes taking place in the murine prostate. Details of these changes were examined using parallel in vitro models.
Results
Progressive mPIN in PBCre4 tg/0/PPARγ flox/flox transgenic mice
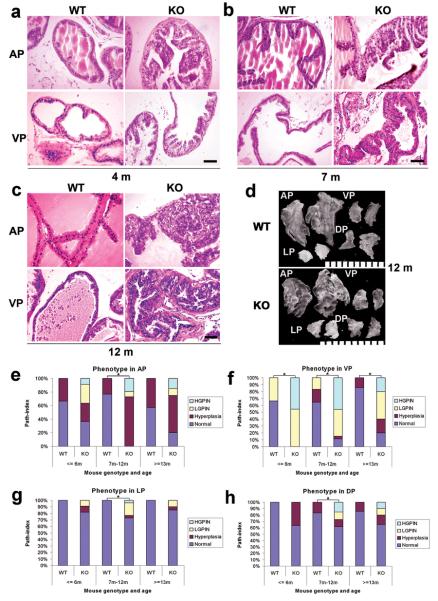
We generated PBCre4 tg/0/PPARγ flox/flox double transgenic mice on a C57/Bl6 background. A PBCre4 tg/0/PPARγ flox/flox mouse line was selected which strongly excised exon 2 of PPARγ (both γ1 and γ2 isoforms) in the anterior (AP) and ventral prostate (VP) with weaker excision in the lateral (LP) and dorsal (DP) prostatic lobes. Loss of prostatic PPARγ protein expression was confirmed using immunohistochemistry (Supplement S1a).
The prostatic phenotypes of mice carrying two flox alleles but no Cre were indistinguishable from their WT counterparts. Beginning at three months of age we observed histologic alterations indicative of mPIN in the KO mice (18, 19). Epithelial hyperplasia with mild cytologic atypia and local inflammatory cell accumulation was observed in the AP and VP (Figure 1a), but not in the DP and LP. The involved lobes showed epithelial stratification, consistent with increased cell proliferation. Nuclei were enlarged, with mild hyperchromasia and focally more prominent nucleoli, compatible with lesions previously categorized as mouse prostatic intraepithelial neoplasia (PIN) I in genetically engineered mouse (GEM) models (19) here designated low grade PIN (LGPIN).
Figure 1. Progressive mouse prostatic intraepithelial neoplasia (mPIN) in conditional PPARγ KO mouse prostates.
(a, b and c) H&E stained sections illustrate the progressive development of mPIN from a low grade at the age of 3 months (a) to a high grade at the age of 7 months (b) and 12 months (c) in the AP and VP of PPARγ KO mice prostatic epithelium, compared to a paired WT control mouse. Scale bar = 100 μm in the panels. (d) Gross appearance of the four mouse prostate lobes, anterior (AP), ventral (VP), lateral (LP) and dorsal prostate (DP), dissected from 12 month old WT and conditional PPARγ KO mice, illustrating similar gross appearance. Scale bar = 1 mm between two small bars. (e, f, g and h) Summary of the incidence of high grade PIN (HGPIN), low grade PIN (LGPIN), Hyperplasia and Normal in the AP, VP, LP and DP of age matched groups of WT and PPARγ KO mouse groups at the ages of ≤ 6 months (3 WT, 11 KO), 7-12 months (48 WT, 26 KO) and ≥ 13 months (7 WT, 20 KO), Total 58 WT mice and 57 KO. *: P < 0.01 by Fisher's Exact Test.
Older KO mice showed progressively more widespread prostatic epithelial hyperplasia with increased nuclear atypia in AP and VP, satisfying key criteria for NCI MMHCC mPIN (18). These progressive changes were similar to lesions designated as PIN II and III (19) (Figures 1b-c) here designated high grade PIN (HGPIN). Cytoplasm was increasingly amphophilic, suggesting decreased secretory differentiation. Nuclear enlargement with chromatin clumping and prominent nucleoli was noted (Figure 1). No foci of invasive carcinoma were identified in PPARγ KO mice up to 15 months of age.
No significant gross differences were seen between prostatic lobe weights from 12 month old KO and WT mice (Supplement S1b). In the AP, decreased branching and associated expansion of some individual AP ducts were noted in KO versus control mice, first discernible in the 7-12 month age group (Figure 1d).
PAS staining illustrated decreased luminal secretions in the conditional PPARγ KO mouse prostatic epithelia (Supplement S1c). AR (Supplement S1c) and cytokeratin 18 (CK-18) (data not shown) were expressed throughout the luminal epithelium of the normal prostate and decreased in the PIN epithelium.
The stroma was thickened around mPIN lesions with strong co-expression of α-smooth muscle (SM)-actin (Supplement S1c) and vimentin (data not shown) consistent with a reactive myofibroblastic phenotype. An inflammatory response composed of mononuclear cells consistent with macrophages and lymphocytes was observed in 25% of the PPARγ KO prostates, whereas it is not commonly noted in WT mouse prostates (18).
Histologically, limited hyperplasia was found in the AP and DP of WT mice from 3 months. More widespread hyperplasia was seen in the AP and VP of PPARγ KO mice from early stages. mPIN was observed in the AP and VP of PPARγ KO mice (Figures 1e-1h). The scores given in Figures 1 e-h represent incidence of disease. When extent of disease was examined it was noted that the individual foci were markedly larger in older versus younger mice. Samples were scored by the presence of the most severe phenotype irrespective of its prevalence. mPIN was not seen in WT mouse prostates. In marked contrast, a 36.4% incidence of PIN (27.3% LGPIN plus 9.1% HGPIN) was observed in the AP of PPARγ KO prostate in ≤6 months, 26.9% PIN (7.7% plus 19.2%) in 7-12 month animals and 25% PIN (10% plus 15%) in ≥13 months old mice. 100% PIN (54.5% plus 45.5%) was found in the VP of PPARγ KO prostates in ≤6 months, 84.6% PIN (38.5% plus 46.1%) in 7-12 months and 60% PIN in (40% plus 20%) in ≥13 months. Histopathologic characterization indicates that PPARγ is a regulator of mouse prostate epithelial cell differentiation and that its loss results in generally progressive mPIN.
Establishment of PPARγ- and PPARγ2-deficient mouse prostate epithelial cell lines
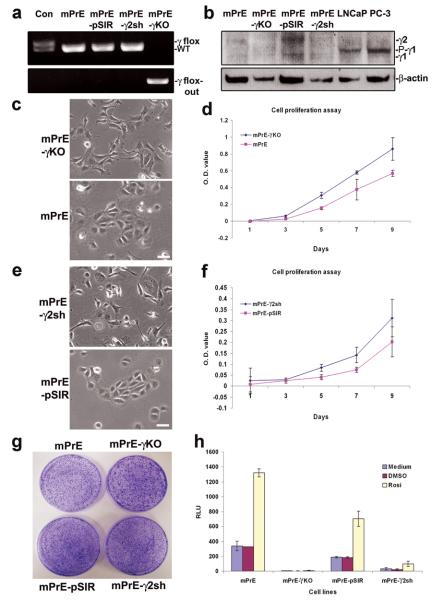
Two PPARγ- and PPARγ2-deficient cell lines were developed. A spontaneously immortalized line was generated from the prostatic epithelium of a KO mouse. These mPrE-PPARγ KO (mPrE-γKO) cells incorporate complete functional deletion of PPARγ, confirmed by PCR analysis (Figure 2a). A human U6-driven mouse PPARγ2 shRNA retroviral construct was introduced into a WT mouse prostatic epithelial cell line (mPrE) (20). These cells are designated mPrE-PPARγ2 shRNA (mPrE-γ2sh). Western blot analysis demonstrated that the mPrE-γKO cells have no PPARγ1 and γ2 protein expression while the PPARγ2 shRNA construct was effective at reducing PPARγ2 protein expression (Figure 2b). The pSIR empty vector was used to make control cells (mPrE-pSIR).
Figure 2. Establishment and characterization of the stable mPrE-PPARγ knockout and mPrE-PPARγ2 knockdown cell lines.
(a) Demonstration of genomic DNA for PPARγ alleles in control mouse tail (Con, PBCre4 0/0/PPARγ flox/WT), mPrE, mPrE-pSIR and mPrE-γ2sh cell lines and its deletion (upper panel) and concurrent presence of a PPARγ flox-out band (lower panel) in the mPrE-KO cell lines by PCR. (b) Western blot showing loss of PPARγ1 (−γ1) and PPARγ2 (−γ2) proteins expression in mPrE-γKO cells and reduced PPARγ2 expression in mPrE-γ2sh cells as compared to mPrE and mPrE-pSIR control cells. A Phosphorylated-PPARγ1 (P-γ1) band showed only in LNCaP and PC-3 cells. (c and d) The effect of loss of PPARγ on cellular morphology and proliferation. Phenotypically the control mPrE cells showed cobblestone morphology while mPrE-γKO cells exhibited a more extended spindle-like phenotype with loss of cell-cell contacts. Scale bar = 50 μm. Loss of PPARγ resulted in increased cellular proliferation as determined by MTT assay. (e and f) The cellular morphology and proliferation of mPrE-γ2sh and mPrE-pSIR cells in culture. Phenotypically these cells showed changes similar to those evoked by the knockout of PPARγ in the mPrE-γKO cells. Suppression of PPARγ2 protein expression resulted in increased cellular proliferation. (g) mPrE-γKO and mPrE-γ2sh cells form larger clones compared to control mPrE and mPrE-pSIR cells when tested using a clonogenicity assay. (h) PPRE activity detected by a luciferase reporter showed a >85% decrease in the mPrE-γ2sh cells as compared to mPrE controls. Signal was undetectable in mPrE-γKO cells. Rosiglitazone has no effect on PPRE activity of mPrE-γKO cells. However it shows mild activation on PPRE activity of mPrE-γ2sh cells, confirming that these cells retain a weakened ability to respond to this PPARγ agonist.
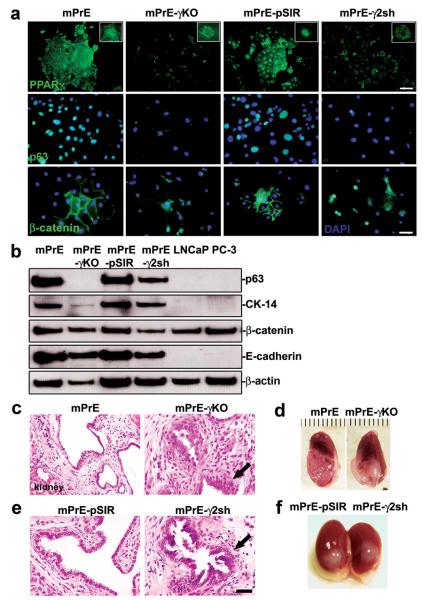
mPrE-γKO and mPrE-γ2sh cells appeared similar, the cells were small and elongated with enlarged nuclei (Figures 2c and 2e). The PPARγ- and PPARγ2-deficient cells grew as discreet individual cells with reduced cell-cell contact and with increased viability and proliferation (Figures 2d and 2f). Cell cycle analysis also showed fewer cells in S phase with mild increase in G0/G1 arrest and essentially no change in G2/M (Supplemental table 1). mPrE-γKO and mPrE-γ2sh cells formed notably larger clones in a clonogenicity assay, than either mPrE or mPrE-pSIR cells (Figure 2g). Suppression of PPARγ activity was confirmed using a luciferase reporter driven by a triple AOX-PPRE (21) (Figure 2h). PPARγ protein was undetectable by immunofluorescence (IF) staining in the nuclei of mPrE-γKO and mPrE-γ2sh cells (Figure 3a). p63 and CK-14 proteins were used to identify basal epithelial cells (18). But, p63 was weak to absent and cytokeratin (CK-14) protein expression was weaker in PPARγ-/γ2-deficient cells in vitro (Figure 3a and Supplement S2a). β-catenin protein expression was lost in the cellular membrane of growing colonies but was detected in the nuclei (Figure 3a). Western blot analysis demonstrated that p63, CK-14, β-catenin and E-cadherin proteins decreased in mPrE-γKO and mPrE-γ2sh cells compared to mPrE and mPrE-pSIR controls (Figure 3b).
Figure 3. Consequences of PPARγ deletion and suppression in prostate epithelial cells.
(a) PPARγ, p63 and β-catenin proteins detected by immunofluorescence in mPrE, mPrE-γKO, mPrE-pSIR and mPrE-γ2Sh cells. Note the loss of nuclear PPARγ staining in mPrE-γKO and mPrE-γ2sh cells as compared to the control cells confirming the efficiency of disruption of PPARγ protein expression (higher magnification in the inset). Loss of PPARγ is associated with a loss of cobblestone morphology, cell-cell contacts, and an associated decrease in nuclear localization of p63 protein and membranous localization of β-catenin as well as nuclear transfer of β-catenin. Scale bar (PPARγ) = 100 μm. Scale bar (p63 and β-catenin) = 50 μm. (b) Western blot analysis demonstrated that p63, CK-14, β-catenin and E-cadherin proteins decreased in mPrE-γKO and mPrE-γ2sh cells compared to mPrE and mPrE-pSIR controls. LNCaP and PC-3 cells were used as the controls. (c and e) Histology of tissue recombinants using rat urogenital sinus mesenchyme (UGM) with mPrE, mPrE-γKO, mPrE-pSIR and mPrE-γ2Sh cells examined at two months post-grafting. The control recombinants resembled prostatic glandular differentiation although with some flattening of epithelial layers consistent with previous descriptions. Tissue recombinants made using mPrE-γKO (c) and mPrE-γ2sh (e) cells grew less readily. Histopathologically these structures exhibited a phenotype consistent with mPIN with epithelial crowding and tufting and out-growths (arrow). Scale bar = 50 μm. (d and f) Gross appearance of tissue recombinants using mPrE and mPrE-pSIR cells were similar, showing glandular differentiation by two months post grafting. In contrast tissue recombinants made using mPrE-γKO (d) cells and mPrE-γ2sh (f) cells grew less readily and exhibited a less obviously glandular phenotype.
PPARγ- and PPARγ2-suppression in a prostatic tissue recombination model resulted in mPIN pathogenesis
To determine the consequences of PPARγ- and PPARγ2-suppression in mouse epithelial cells in vivo with physiologically relevant epithelial-stromal interactions, we used a tissue recombination model. Recombinants composed of untransfected mPrE cells or control mPrE-pSIR recombined with rat UGM formed glandular structures lined with cuboidal to columnar secretory epithelium surrounded by stroma resembling normal rodent prostate, consistent with previous observations (20) (Figure 3c-f). Tissue recombinants using either mPrE-γKO or mPrE-γ2sh cells exhibited similar mPIN phenotypes within two months of grafting (Figures 3c-f). Mild degrees of epithelial stratification, mild nuclear alterations, and reduced secretory differentiation were detected. These results parallel the mPIN prostate lesions of PBCre4 tg/0/PPARγ flox/flox mice. Reduced luminal secretions were noted in mPrE-γ2sh recombinants versus the controls, further suggesting reduced secretory differentiation (Figure 3). Of note, focal disruption of the basal membrane of reconstructed mPrE-γKO mouse prostate glands was seen on both H&E and Periodic acid Schiff (PAS) stained sections, and in some cases a few epithelial cells had migrated locally from the basal cell layer into surrounding stroma (Figures 3c), reminiscent of microinvasive carcinoma as defined in intact GEM prostates (18).
Autophagocytosis in PPARγ knockout and PPARγ2 knockdown mouse prostate epithelial cells
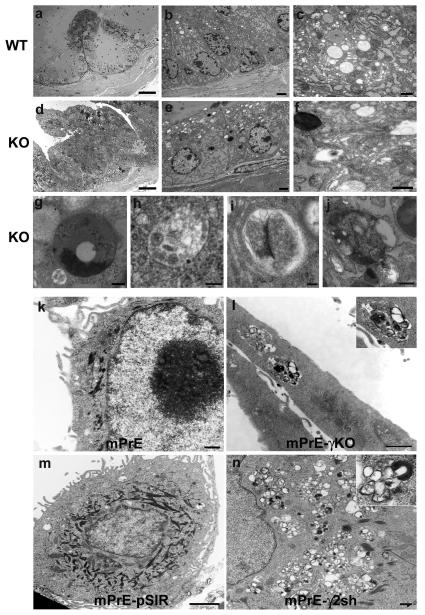
Ultrastructural analysis was performed on AP tissues from 12-month-old conditional PPARγ KO and WT mice. WT AP is illustrated (Figure 4a). Foci of mPIN PPARγ KO mouse were examined by electron microscopy. The secretory luminal cells of WT prostate were cylindrical with supra-nuclear vesicles including some budding from the rough endoplasmic reticulum (RER) (Figures 4b-c). Number, size and distribution of mitochondria, lysosomes and peroxisomes were unremarkable. Secretory vesicles were found budding from the Golgi, and mature and exocytosing secretory vesicles were also observed. In contrast, the PPARγ KO luminal epithelial cells in mPIN regions had a number of abnormal features. There were enlarged nuclei with large nucleoli and few clear storage vesicles were seen in the cytoplasm. Many of the mitochondria appeared to be degenerating and there were 2.4-fold more lysosomes in the HGPIN compared to WT epithelium (Figures 4e-f). The lysosomes varied in size and many showed features suggestive of autophagocytosis, including both early and late autophagosomes (22). The PPARγ KO epithelial cells in mPIN foci contained clusters of variably sized autophagic vacuoles (autophagosomes) containing loose granular to more dense flocculent substance. Features suggesting autophagocytosis included double membranes, internal vesicles, and the presence of organelles such as mitochondria, peroxisomes, and rough endoplasmic reticulum within the lysosome (Figures 4f-j). There were also a number of multivesicular bodies (Figure 4f). Fewer normal-appearing secretory vesicles were present (Figures 4e-f). In some places, the basement membrane was disrupted at the site of basal cell attachment (Figure 4e). These changes were consistent with a reduction in secretory differentiation of prostatic luminal cells and possible increase in autophagy in the proliferating mPIN regions.
Figure 4. Increased autophagic features in the ultrastructure of PPARγ knockout prostate epithelium and mPrE-PPARγ2 shRNA cells.
Light and electron microscopy of prostate tissue indicated significant morphological differences between WT (a-c) and PPARγ KO (d-f) mice at the age of 12 months. Toluidine blue staining of thick plastic sections (a and d) indicated increased cellularity and fewer secretory vesicles in the epithelium from PPARγ KO mice prostate. Scale bar (a and d) = 50 μm. Ultrastructurally, the WT mouse prostate epithelial cells showed a normal distribution of cytoplasmic constituents, organelles and secretory vesicles (b and c). In contrast, the PPARγ KO cells had abnormal appearing mitochondria and increased numbers of lysosomes. The lysosomes had varied morphology. Some exhibited a classical appearance (e and f) or appeared as multivesicular bodies (f) while others had the appearance of autophagosomes (g-j). Scale bars: (b) = 2 μm, (c) = 500 nm, (e) = 2 μm, (f) = 500 nm, (g) = 500 nm, (h) = 100 nm, (i) = 100 nm and (j) = 500 nm. Cytoplasmic organelles in mPrE and mPrE-pSIR cells had a normal distribution and ultrastructural appearance (k and m). In contrast mPrE-γKO cell demonstrated a decrease in mitochondria and increase in secondary lysosomes (l). Inset: increase magnification image lysosome containing numerous membranes and other debris, suggestive of autophagosome. mPrE-γ2sh cells had increased lysosomes, many having the appearance of autophagosomes (n). Inset: autophagosome. Scale bars (k) = 500 nm, (l) = 1 μm, (m) = 2 μm and (n) = 500 nm.
mPrE-pSIR and mPrE cells exhibited normal number, size and distribution of cellular organelles and secretory vesicles (Figures 4k and 4m). In contrast, the mPrE-γKO and mPrE-γ2sh cells had a substantially increased range of lysosomal sizes where the majority were small and distributed as clusters within the cytoplasm. Many of these were complex with lipid whorls and internalized material, including mitochondria, peroxisomes and rough endoplasmic reticulum, in various stages of digestion (Figures 4l and 4n). Thus, while the changes were more dramatic in the mPrE-γ2sh cells, the overall ultrastructural alterations were reminiscent of the cytoplasmic changes noted in the prostatic epithelium from PPARγKO mice and were consistent with increased autophagocytosis (autophagy).
Gene expression profiles in the mPrE-γKO cells vs. mPrE cells or mPrE-γ2sh cells vs. mPrE-pSIR cells demonstrated major changes (Table 1) including alterations in signaling pathways involving genes of the nuclear receptor II subfamily, cell cycle control (dedifferentiation), peroxisomal and mitochondrial lipid transporter and oxidation metabolism (oxidative stress/hypoxia) and ubiquitination/proteosomal degradation. Ingenuity Pathway Analysis (IPA, Ingenuity Systems) was used to evaluate changes based on microarray data. The top canonic pathway and network related to PPARγ-signaling signatures were metabolism pathway/oxidative stress (Supplement S3a-b) and cell cycle control/dedifferentiation (Supplement S4a-b). Effects were generally consistent between PPARγ KO and PPARγ2 shRNA prostate epithelial cells as compared to control cells.
Table 1.
Major gene expression profiling changes identified from microarray in mPrE-γKO cells vs. mPrE cells or mPrE-γ2sh cells vs. mPrE-pSIR cells (> 2 fold and < 2 fold).
| Description | Systematic | Official symbol |
mPrEl -γKO |
mPrE -γ2sh |
|---|---|---|---|---|
| 1. Sub-cellular organelles/Lipid metabolism | ||||
| Catalase | vm059759 | Cat | 0.45 | 0.12 |
| Prostaglandin-endoperoxide synthase2(COX-2) | vm076500 | Ptgs2 | 6.10 | 45.3 |
| Isovaleryl CoA dehydrogenase | vm073968 | Ivd | 0.40 | 0.01 |
| Peroxisomal biogenesis factor 6 | vm081528 | Pex6 | 2.07 | 21.3 |
| Peroxisomal acyl-CoA thioesterase 1 (Pte1) | vm085581 | Acot8 | 2.05 | 13.18 |
| Hydroxy-delta-5-steroid dehydrogenase | vm081035 | Hsd3b7 | 8.02 | 2.42 |
| Solute carrier family 27 (fatty acid transporter) | vm062392 | Slc27a3 | 10.34 | 5.30 |
| ATP-binding cassette, sub-family A, member 3 | vm060761 | Abca3 | 3.82 | 321.9 |
| Sterol carrier protein 2, liver | vm084113 | Scp2 | 2.01 | 16.2 |
| Oxysterol binding protein | vm079937 | Osbp | 71.6 | 18.5 |
| Acetyl-CoA acyltransferase 1 (peroxisomal) | vm083546 | Acaa1 | 0.42 | 0.44 |
| Mitochondrial NADH dehydrogenase 5 | vm083510 | Ndufa5 | 3.20 | 2.16 |
| Acetyl-CoA acyltransferase 2 (mitochondrial) | vm088089 | Acaa2 | 0.45 | 0.15 |
| ATP synthase, H+ transporting, mitochondrial F 1 | vm068155 | Atp5o | 0.44 | 0.38 |
| ATP synthase, H+ transporting, V1 subunit D | vm083645 | Atp6v1d | 0.41 | 0.37 |
| Cytochrome P450, family 26, subfamily b | vm064755 | Cyb26a1 | 0.01 | 0.01 |
| 3-hydroxybutyrate dehydrogenase | vm064957 | Bdh1 | 0.50 | 0.25 |
| Lysosomal mannosidase, beta A | vm071702 | Manba | 2.11 | 16.80 |
| Lysosomal-associated protein transmembrane 4A | vm060108 | Laptm4a | 0.50 | 0.33 |
| Vesicle-associated membrane protein 5 | vm070176 | Vamp5 | 0.20 | 0.13 |
| Vesicle-associated membrane protein 8 | vm068808 | Vamp8 | 0.43 | 0.37 |
|
| ||||
| 2. Oxidative stress (Hypoxia) | ||||
| Hypoxia up-regulated 1 | vm062588 | Hyou1 | 2.17 | 21.2 |
| Hairless | vm086899 | Hr | 2.62 | 76.70 |
| Forkhead box O 6 | vm078799 | Foxo6 | 2.13 | 2.77 |
| Oxidation resistance 1 | vm077473 | Oxr1 | 0.29 | 0.35 |
| Hairy and enhancer of split 1 | vm058417 | Hes1 | 0.5 | 0.28 |
| Microsomal glutathione S-transferase 1 | vm059398 | Mgst1 | 0.01 | 0.09 |
|
| ||||
| 3. Cell cycle control (de-differentiation and EMT) | ||||
| Proliferation cell nuclear antigen | vm072868 | Pcna | 2.03 | 28.5 |
| Ki67 | vm078634 | Mki67 | 2.22 | 13.2 |
| Cell cycle progression 1 | vm085838 | Ccpg1 | 2.18 | 120.2 |
| Cyclin G associated kinase | vm085772 | Gak | 2.45 | 44.8 |
| Cyclin-dependent kinase 2 | vm087446 | Cdk2 | 2.08 | 44.3 |
| Vimentin | vm087332 | Vim | 775.70 | 738.70 |
|
| ||||
| 4. Nuclear receptors/cofactors subfamily | ||||
| Peroxisome proliferative activated receptor, gamma, | vm078414 | Ppargc1b | 0.01 | 0.01 |
| coactivator 1beta | ||||
| Vitamin D receptor | vm076701 | Vdr | 0.01 | 0.01 |
| Cellular retinoic acid binding protein II | vm057044 | Crabp2 | 0.01 | 0.01 |
| cAMP responsive element binding protein 1 | vm086761 | Creb1 | 0.01 | 0.01 |
Global gene expression profiling in mPrE-PPARγ knockout cells vs. mPrE cells or mPrE-PPARγ2 knockdown cells vs. mPrE-pSIR cells. The major signaling pathway changes in mPrE-γKO cells vs. mPrE cells and mPrE-γ2sh cells vs. mPrE-pSIR cells by microarray analysis were involved in metabolic lipid oxidation, subcellular organelles/autophagy, oxidative stress, cell cycle regulation/cellular dedifferentiation and nuclear receptors/PPRE. Expression value of an individual gene was by the comparison of mPrE-γKO cells vs. mPrE cells or mPrE-γ2sh cells vs. mPrE-pSIR control cells.
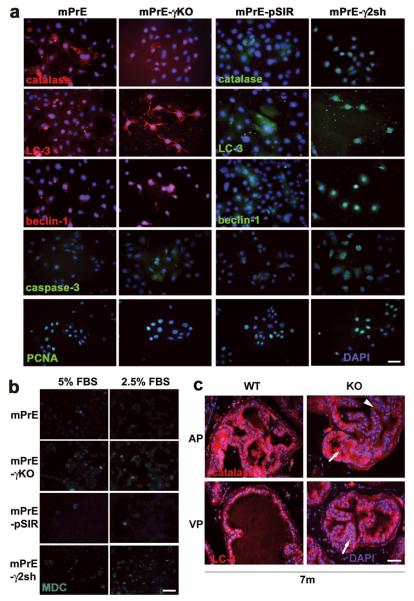
Altered expression patterns of autophagy-associated proteins were detected in mPrE-γKO and mPrE-γ2sh cells compared to the control cells (Figure 5a). LC-3 (Atg8), beclin-1 (Atg6), activated caspase-3, PCNA (Figure 5a), and COX-2 and vimentin (Supplement S2a) were increased, whereas catalase (Figure 5a) and PMP70 (data not shown) were decreased in two PPARγ-/γ2-deficient cell lines.
Figure 5. Alterations in autophagy-associated protein expression in mPrE-PPARγ knockout and mPrE-PPARγ2 shRNA cells.
(a) Catalase, LC-3 (Atg8), beclin-1 (Atg6), caspase-3 and PCNA proteins were detected by immunofluorescence staining in mPrE and mPrE-γKO, mPrE-pSIR and mPrE-γ2sh cells grown on glass slides for three days. Decreased catalase and increased LC-3 and beclin-1 were seen in mPrE-γKO and mPrE-γ2sh cells compared to mPrE and mPrE-pSIR cells. These results suggested active autophagic body formation in the cells. Meanwhile, caspase-3 and PCNA were increased in PPARγ-/γ2-deficient cells. Scale bar = 50μm in the panels. (b) MDC, a marker of autophagy, was visualized in mPrE and mPrE-γKO, mPrE-pSIR and mPrE-γ2sh cells under the 5% FBS regular or 2.5% FBS half-starvation culture conditions. Elevated staining was seen in both mPrE-γKO and mPrE-γ2sh cells as compared to mPrE and mPrE-pSIR cells. The signals were strongly increased in PPARγ-/γ2-deficient cells in the 2.5% FBS culture media. Scale bar = 50 μm in the panels. (c) Immunofluorescence staining of catalase and LC-3 in wild-type (WT) and PPARγ knockout (KO) prostate tissue at ages of 7 months. Low levels of catalase expression and high expression of LC-3 protein were seen in the PIN regions (arrows) of PPARγ KO mouse prostate epithelium compared to WT and more normal-appearing areas (arrowhead). Scale bar = 50 μm in the panels.
Monodansylcadaverine (MDC), a marker of autophagy (22), was visualized in the mPrE and mPrE-γKO, mPrE-pSIR and mPrE-γ2sh cells under 5% FBS (regular growth) or 2.5% FBS (semi-starvation) culture conditions. Elevated MDC signaling was seen in both mPrE-γKO and mPrE-γ2sh as compared to control cells. This was enhanced in 2.5% FBS culture medium (Figure 5b). Lysosomes and mitochondria were tracked, using LysoTracker (DND-99) and MitoTracker (22), respectively, under the 5% or 2.5% FBS culture conditions. The LysoTracker (Supplement S2b) and MitoTracker (data not shown) signaling were strongly increased in PPARγ-/γ2-deficient cell lines in 2.5% FBS culture medium.
In vivo, catalase (Figure 5c) and PMP70 (data not shown) proteins were found to be decreased or lost in mPIN foci of KO mice. In contrast, LC-3 protein (Figure 5c) was increased in the mPIN foci of KO mice compared to the epithelium of WT mice.
Active PPARγ1/γ2 signaling suppresses the phenotypes in mPrE-PPARγ KO cells and mPrE-PPARγ2 knockdown tissue recombinants
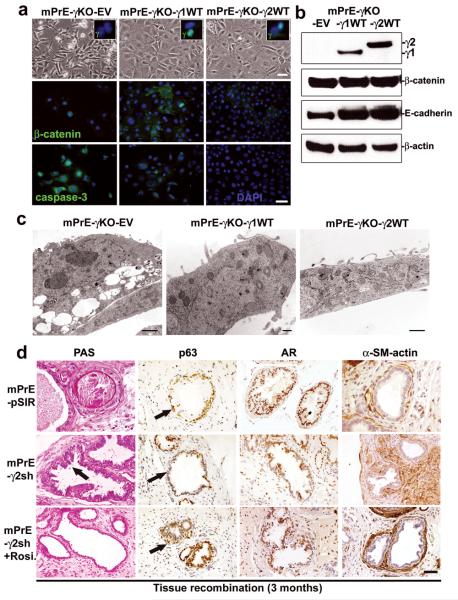
To dissect the biological functions of PPARγ1 and PPARγ2 isoforms we re-expressed PPARγ1 or PPARγ2 cDNA into mPrE-γKO cells viral transduction of wild-type full-length cDNA (23). Three cell lines, mPrE-PPARγ KO-empty vector (mPrE-γKO-EV), mPrE-PPARγ KO-PPARγ1 WT (mPrE-γKO-γ1WT) and mPrE-PPARγ KO-PPARγ2 WT (mPrE-γKO-γ2WT), were established (Figure 6a). Western blotting confirmed re-expression of the PPARγ1 or PPARγ2 isoform (Figure 6b). IF staining showed positive nuclear PPARγ protein in mPrE-γKO-γ1WT and mPrE-γKO-γ2WT cells compared to the control mPrE-γKO-EV cells (Figure 6a). While total β-catenin levels were not greatly altered there was a marked shift in localization from the nuclei in the KO cells to the cytoplasm and membrane in the PPARγ1-/γ2-expressing cells (Figure 6a). Elevated levels of E-cadherin (Figure 6b), but not CK-14 and p63 proteins (data not shown), were seen in mPrE-γKO-γ1WT and mPrE-γKO-γ2WT cells. EM showed decreased lysosome and autophagosome volume and increased numbers of mitochondria in the PPARγ1-/γ2-rescued mPrE-γKO cells compared to the control mPrE-γKO-EV cells (Figure 6c). mPrE-γKO-γ1WT and mPrE-γKO-γ2WT cells showed decreased cell viability and proliferation (Supplement S5a) as well as clonal formation (Supplement S5b). As expected mPrE-γKO-γ1WT and mPrE-γKO-γ2WT cells showed increased PPRE-activity by a luciferase reporter assay (Supplement S5c). IF staining showed increased levels of CK-14 and catalase (Supplement S5d), decreased levels of caspase-3 activation (Figure 6a), LC3 and PCNA (Supplement S5c), and beclin-1 (data not show) in the PPARγ1-/γ2-rescued cells compared to the mPrE-γKO-EV control cells.
Figure 6. Rescue of phenotypes of mouse PPARγ-deficient prostate epithelial cells by re-expression of either PPARγ1 or PPARγ2 isoform or treatment by Rosiglitazone.
(a) Phase-contrast microscopy morphology of control mPrE-γKO-empty vector and mPrE-γKO-PPARγ1 or -PPARγ2 WT cDNA cells, showing a return to a cobblestone morphology following reintroduction of each PPARγ isoform. IF staining confirmed expression of PPARγ protein in the nuclei of mPrE-γKO-γ1WT and mPrE-γKO-γ2WT cells (inset boxes). β-catenin protein was predominantly nuclear in mPrE-γKO whereas in mPrE-γKO-γ1WT and mPrE-γKO-γ2WT cells the protein was found in the cytoplasm and on intercellular membrane interfaces. Caspase-3 was decreased in mPrE-γKO-γ1WT and mPrE-γKO-γ2WT cells compared to mPrE-γKO-EV cells. Scale bar = 50 μm. (b) Western blot analysis demonstrated PPARγ1 protein in mPrE-γKO-γ1WT and PPARγ2 in mPrE-γKO-γ2WT cells. β-catenin and E-cadherin protein levels increased in mPrE-γKO-γ1WT and mPrE-γKO-γ2WT cells compared to mPrE-γKO-EV control cells. (c) mPrE-γKO-EV cells had decreased mitochondria and increased lipid droplets and secondary lysosomes, similar to mPrE-γKO cells. Scale bar = 2 μm. However introduction of the mouse PPARγ1 wild-type cDNA increased mitochondria above wild-type values and decreased the presence of secondary lysosomes and lipid accumulation. Scale bar = 250 nm. Likewise, introduction of the mouse PPARγ2 wild-type cDNA into mPrE-γKO cells restored the level of mitochondria and reduced the instances of secondary lysosomes and lipid droplets. Scale bar = 500 nm. (d) Tissue recombinants made using control (mPrE-pSIR) or mPrE-γ2sh cells with rat UGM. Sections were examined for secretion by PAS. In control recombinants a normal prostatic phenotype with secretion was noted. In contrast in the mPrE-γ2sh containing recombinants a low grade mPIN (arrow) with less secretion into the luminal space and thickened stromal was seen. p63 and AR protein were decreased in the mPrE-γ2sh containing recombinants, but α-SM-actin protein expression was increased compared to tissue recombinants of mPrE-pSIR. Mice carrying tissue recombinants made by mPrE-pSIR and mPrE-γ2sh cells were administered Rosiglitazone chow (0.005% Rosiglitazone) from the time of grafting until sacrifice at three months. Tissue recombinants of mPrE-γ2sh in these mice (designated mPrE-γ2sh+Rosi.) showed secretion, and increased p63 protein in the basal layer and p63 negative-luminal differentiation by IHC staining (arrows). These recombinants showed well-differentiated prostatic glandular structure and a more normal-appearing stroma. Scale bar = 50 μm in the panels.
Tissue recombinants of mPrE-γ2sh cells showed decreased secretion and immunophenotypic features of PIN phenotypes cnsistent with PPARγ KO mouse prostates. These included reduction in p63 positive basal cells and AR (Figure 6d) with lesion progression and phenotypic alterations in stromal cells visualized by α-SM-actin staining (Figure 6d). Basal cells progressively decreased as the mPIN lesion progressed, similar to the pattern recognized in human PIN progression and in progressively severe mPIN in some GEM models (18). To examine the potential for targeted chemoprevention with pharmacologic PPARγ agonists, tissue recombinants composed of mPrE-γ2sh cells plus rat UGM were grown for three months in animals which were continuously fed on chow containing the PPARγ agonist Rosiglitazone. At sacrifice the grafts in these animals were of similar size to mPrE-pSIR controls and on histologic examination, secretory differentiation of luminal epithelial cells and intraluminal secretions were increased compared to mPrE-γ2sh recombinants without drug treatment. mPIN-like changes were reduced compared to grafts of control mice fed on standard animal chow. Rosiglitazone treatment enhanced p63 and AR expression and also modified the stromal compartment in mPrE-γ2sh tissue recombinants (Figure 6d).
These data support the roles of reduced PPARγ function in the development of mPIN and indicate that re-expression of either PPARγ1 or PPARγ2 isoform in PPARγ KO cells is able to rescue the phenotype. Rosiglitazone treatment suppressed mPIN formation in mPrE-γ2sh tissue recombinants, likely by activating transcription through PPARγ1, although PPARγ-independent effects cannot be excluded.
Discussion
Elucidating the molecular pathologic mechanisms underlying prostatic carcinogenesis may allow for more conservative targeted medical treatments for certain low risk patients with precursor lesions or organ confined, small volume, low grade carcinomas. The ability to medically inhibit progression of HGPIN to Pca or the progression of small, lower grade Pcas to larger, higher grade tumors would represent a significant advance for Pca patient management.
We propose that PPARγ is a key regulator in the maintenance of peroxisomal, mitochondrial and lysosomal functions. Genetic disruption of PPARγ or PPARγ2 signaling in mouse prostate epithelial cells resulted in dysregulated expression patterns of peroxisomal and mitochondrial genes whose products are involved in lipid transportation and oxidation pathways. Active autophagosomes and abnormally increased numbers of lysosomes were found in PPARγ- and PPARγ2- deficient prostatic epithelia. In vitro these phenotypes were rescued by re-expression of PPARγ1 and PPARγ2 isoform in mPrE-PPARγ KO cells. In vivo, changes consequent to loss of PPARγ were associated with hyperplasia, PIN formation and progression to malignancy, which in the case of PPARγ2 suppression could be rescued using high levels of the PPARγ agonist Rosiglitazone. The pathophysiological roles of PPARγ in cellular peroxisomal and mitochondrial fatty acid oxidation and autophagy based on results are summarized in Figure 7.
Figure 7. Disruption of PPARγ resulted in mouse prostate carcinogenesis involving oxidative stress and autophagy.
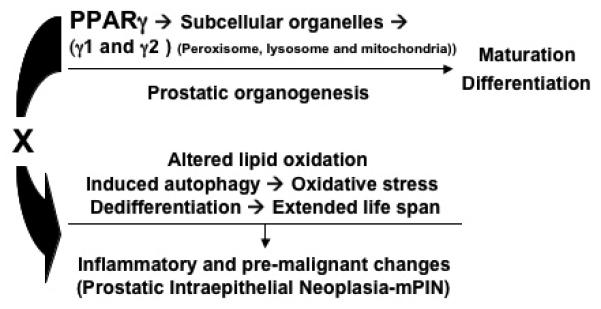
The simplified diagram brings together data from the studies and presents a model illustrating how the PPARγ signaling contributes to prostate carcinogenesis from wild-type to mPIN formation and to set up conditions that would predispose cells to further malignant progression. These data suggested an important role for the PPARγ gene in maintaining the maturation, differentiation and turnover of subcellular organelles (peroxisomes, mitochondria and lysosomes) during mouse prostatic organogenesis and development. In particular this model suggests that a mechanism by which loss of PPARγ could lead to mouse PIN related to the disruption of cellular peroxisomal and mitochondrial lipid metabolism and oxidative stress (hypoxia) and active autophagy for the extended life span and cellular dedifferentiation.
Alterations in lipid metabolism resulting in loss of PPARγ-signaling have been suggested to predispose the prostate to premalignant or malignant changes (24-26). The current study demonstrates, for the first time, that PPARγ loss-of-function in wild-type mouse prostatic epithelium results in progressive epithelial hyperplasia with atypia, indicative of mPIN. Using both intact mice with targeted knockout of PPARγ in prostate epithelium and a tissue recombination model with selective gene modulation in the epithelial compartment reduced PPARγ function was associated with increased activation of oxidative stress, autophagic activity, and activation of pro-inflammatory signaling pathways. This establishes conditions for subsequent malignant transformation which would be expected to occur stochastically, resulting from epithelial genomic damage potentially caused by reactive oxygen species (ROS) (26). The results described here parallel reported changes in gene expression resulting in reduced ligands for PPARγ in the human prostate and provide the first direct evidence that loss of PPARγ expression or function can lead to prostatic neoplasia in vivo.
Reduced activation of PPARγ due to reduced formation of endogenous ligands for PPARγ most likely explains its role early in human Pca development (8, 27, 28). Our studies demonstrate that PPARγ plays a role in maintaining peroxisomal, mitochondrial and lysosomal biogenesis and maturation during prostatic epithelial cellular growth and differentiation. Catalase and PMP70 proteins were decreased in PPARγ- and PPARγ2-deficient prostate epithelial cells. Degenerated mitochondria were found in two PPARγ- and -γ2-deficient prostate epithelial cell lines. Active autophagosomes and abnormally increased numbers of lysosomes were found in PPARγ- and PPARγ2-deficient prostatic epithelia. These changes were associated with mouse PIN formation and progression. Microarray analysis revealed alterations to peroxisomal and mitochondrial fatty acid transporter synthesis and oxidative metabolism in both PPARγ- and -γ2-deficient mPrE cells as compared to the control cells. Disruption of PPARγ-signaling results in altered fatty acid metabolism and induction of oxidative stress and hypoxia, as supported by the increased Hypoxia up-regulated 1 (Hyou-1) and Hairless (Hr), and decreased Hairy and enhancer of split1 (Hes1) gene expression in addition COX-2, GSTs and uPAR which show similar changes in expression patterns in clinical samples of human PIN and in other mouse models (29, 30). A number of these altered genes contain a PPRE domain (31, 32) suggesting that these might be directly regulated. However, regulation of crucial genes could also occur indirectly, as a secondary consequence of reduced PPARγ-signaling. The combination of light microscopic, ultrastructural, immunophenotypic, and gene expression data are consistent with dramatic changes in multiple subcellular organelles including mitochondria, peroxisomes and lysosomes, which likely contribute to the observed neoplastic phenotypes.
Our data suggest that the oxidative stress induced by PPARγ loss-of-function results in lysosomal autophagy which can contribute to malignant progression (33). Staining using lysosome tracker Red DNT-99 and autophagy markers MDC, LC-3 and beclin-1 showed increased signal strength in PPARγ- and -γ2-deficient mPrE cells in vitro and in vivo confirming autophagosome formation induced by active lysosomes, consistent with electron microscopy showing lysosomal changes. Changes included increased lysosomal number with variable sized including many small structures, often accumulating in clusters in PPARγ-deficient mPrE cells. This appears to be an abnormal autophagic response which does not result in cell death, possibly allowing individual cells to avoid an increase in apoptotic activity (indicated by increased caspase-3 activation) induced by loss of PPARγ activity.
The role of autophagy in cancer is complex (34). Autophagy may be pro-tumorigenic, promoting tumor cell survival and restricting necrosis (35, 36). Alternatively autophagy could represent either a barrier, or an adaptive response, to cancer. Here, using both deletion (PPARγ KO) and suppression (PPARγ2 shRNA) of PPARγ in mouse prostate epithelial cells, we show increased autophagic activity. We suggest that PIN may indirectly result from the deregulation of pro-inflammatory pathways following PPARγ inactivation combined with extension of life span via increased autophagic capacity. This outcome may be related to disruption of peroxisomal lipid oxidation/metabolism signaling pathways.
Early prostate cancer has been linked to a loss of enzymes including 15-lipoxygenase-2 (15-LOX-2) which is involved in the generation of 15(S)-hydroxyeicosatetraenoic acid (15-HETE) (8, 27). Such a scenario justifies the consideration of PPARγ agonists as chemopreventive agents to inhibit the genesis of early stage prostate cancer. Based upon the data presented here we would suggest that modulation of PPARγ signaling by glitazone drugs be considered as an addition to anti-oxidant diets to inhibit progression of HGPIN to prostate cancer.
Materials and methods
Animal experiments
Floxed PPARγ (37) and PBCre4 (38) transgenic mouse lines have been described previously. PBCre4 mice were backcrossed in C57/Bl6 for more than 10 generations. Double transgenic PBCre4 tg/0/PPARγ flox/flox mice were generated by breeding the PBCre4 and floxed PPARγ transgenic lines and were maintained in a C57/Bl6 background. Wild-type littermates were used as the control groups. PCR primers for PPARγ genotyping and for detecting DNA recombination and excision have been previously reported (37). Adult male severe combined immunodeficient (SCID) mice [C.B.17/IcrHsd-scid] were purchased (Harlan, Indianapolis, IN). All work involving animals was performed under protocols reviewed and approved by the Vanderbilt IACUC.
H&E, immunofluorescence (IF) and immunohistochemical (IHC) staining
Mouse prostate lobes and tissue recombinants were dissected and fixed in 10% phosphate-buffered formalin overnight, transferred to 50% ethanol, then embedded in paraffin. Samples were sectioned for 8 successive layers at 5 μm intervals and stained with hematoxylin and eosin (H&E). IF and IHC were performed as previously described (27). The observation was under a fluorescence microscope (ZEISS, Axio imager M1) equipped with an appropriate filter system.
Electron microscopy (EM)
Prostate tissue and fresh cell pellets were fixed in 2.5% glutaraldehyde in cacodylate buffer (0.1M, pH 7.2) overnight at 4°C and washed in the same buffer for 30 minutes followed by post-fixation for 2 hours at 4°C in 1% osmium tetroxide (OsO4) in cacodylate buffer. After fixation, the material was dehydrated through a graded series of alcohols and embedded in Spurr Resin. For light microscopic analysis, semi-thin sections (2 μm) were stained with toluidine blue. Ultrathin sections (70 nm) were cut and ultrastructural analysis was performed on a Phillips CM-12 Transmission Electron Microscope operated at 80 KeV. Images were acquired using an AMT digital camera system. For quantification of lysosome number per area of cell cytoplasm examined, four separate thin sections were analyzed. The number of cross sections through lysosomes was counted and the area of cell cytoplasm represented in the four thin sections was determined. Given that most lysosomes are roughly spherical in shape, a lysosome would not be sampled twice in this procedure. Although, the exact number of lysosomes per cell cannot be obtained using this procedure, differences in the relative occurrence of lysosomes between wild type and PPARγ-deficient cells can be estimated with reasonable accuracy.
Tissue recombinants and sub-renal capsule xenografting
Single cell suspensions of rat UGM were prepared from E18.5 embryonic fetuses as previously described (20).Viable cells were counted using a hemacytometer. To prepare tissue recombinants, rat UGM was mixed with genetically modified mouse prostate epithelial cells at a ratio of 250,000 to 400,000. The cell mixture was pelleted and resuspended in 50 μL of rat-tail collagen (pretitrated to pH 7.4). After polymerization, the collagen was overlaid with growth medium. After incubation at 37°C overnight, the tissue recombinants were grafted under the renal capsule of intact CB17Icr/Hsd-SCID mouse. Hosts were sacrificed at 4 weeks, 8 weeks and 12 weeks post-grafting. The kidneys with the grafts were removed and imaged before processing for histology.
Administration of Rosiglitazone to mice
Host SCID mice were fed by prepared BRL-49653 (Avandia or Rosiglitazone) chow (0.005 % Rosiglitazone) RTD C056 (TestDiet, Richmond, IN) (39) for seven days before xenografting surgery. And then subsequently for three months until sacrifice. The control mice were fed matched regular rodent chow.
Statistical analysis
A Statistical Analysis System (version 9.1, SAS Institute, Cary, NC) was used. Pathological index of wild-type and PPARγ knockout mice were divided into four groups of Normal, Hyperplasia, LGPIN and HGPIN. Percent of each index in each lobe, (AP, VP, LP or DP), at different ages; ≤6 months, 7-12 months and ≥13 months was analyzed by Fisher's Exact Test. Differences were considered statistically significant at P < 0.05.
Accession number
All microarray data have been deposited at the GEO database, accession number GSE13867 and a link for review: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=dbgfxmkkaieiuny&acc=GSE13867.
Supplementary Material
Acknowledgements
The authors appreciate EM core lab members for help with E.M., Vanderbilt MCN III Animal facility for animal care, Vanderbilt Microarray Shared Resource (VMSR) facility for microarray work. The pCMX-mouse PPARγ1 and PPARγ2 wild-type full-length cDNA expression plasmids and PPARγ-Luciferase reporter plasmid (PPRE)3-tk-luciferase were gifts from Dr. V.K.K. Chatterjee, University of Cambridge, Cambridge, UK. The pQCXIP-mouse PPARγ1 and PPARγ2 wild-type full-length cDNA vectors were gifts from Drs. Y. Eugene Chen and Jifeng Zhang, University of Michigan Medical Center. The CK-14 (LL001) antibody was a gift from Dr. EB Lane, Universty of Dundee, Scotland and the α-LC-3 antiserum from Dr. Tamotsu Yoshimori, National Institute of Genetics (NIG), Mishima, Japan. The work was supported by NIH R01 DK67049 and DOD-PCRP, W81XWH-07-1-0479 grants to S.W.H. NIH R01 CA113392 and NIH R01 CA59705 grants to P.R-B, NIH R01 DK055748 to R.J.M. The VUMC Research EM Resource is supported by NIH grants DK20539, CA68485 and DK58404. The Vanderbilt Microarray Shared Resource is supported by the Vanderbilt Ingram Cancer Center (P30 CA68485), the Vanderbilt Digestive Disease Center (P30 DK58404) and the Vanderbilt Vision Center (P30 EY08126). We also thank the Frances Williams Preston Laboratories of the TJ Martell Foundation for support. The content of this paper is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations
- Pca
prostate cancer or prostate carcinoma
- PIN
prostatic intraepithelial neoplasia
- PPRE
peroxisome proliferator response element
- mPrE
a non-malignant mouse prostate epithelial cell line spontaneously immortalized from an adult wild-type C57Bl6 mouse
- mPrE-PPARγ KO (mPrE-γKO)
a PPARγ knockout mouse prostate epithelial cell line spontaneously immortalized from an adult PBCre4 tg/0/PPARγ flox/flox mouse prostate
- mPrE-pSIR-PPARγ2 shRNA (mPrE-γ2sh)
a PPARγ2 knockdown mouse prostate epithelial cell line transfected by U6-mouse PPARγ2 shRNA in pSIR retroviral vector into mPrE cells
- mPrE-pSIR-empty vector (mPrE-pSIR)
a control pSIR vector-transfected mouse prostate epithelial cell line
- mPrE-PPARγ KO-PPARγ1 wild-type (WT) (mPrE-γKO-γ1WT)
mPrE-γKO overexpressing mouse PPAR γ1 WT full-length cDNA. mPrE-PPARγ KO-PPARγ2 wild-type (WT) (mPrE-γKO-γ2WT), mPrE-γKO overexpressing mouse PPAR γ2 WT full-length cDNA. mPrE-PPARγ KO-empty vector (EV) (mPrE-γKO-EV) transduction control, mPrE-γKO retrovirally transduced using a control empty vector
- UGS
urogenital sinus
- UGM
urogenital mesenchymal cell
- KO
knockout
- KD
knockdown
References
- 1.Yu K, Bayona W, Kallen CB, Harding HP, Ravera CP, McMahon G, et al. Differential activation of peroxisome proliferator-activated receptors by eicosanoids. J Biol Chem. 1995 Oct 13;270(41):23975–23983. doi: 10.1074/jbc.270.41.23975. [DOI] [PubMed] [Google Scholar]
- 2.Hihi AK, Michalik L, Wahli W. PPARs: transcriptional effectors of fatty acids and their derivatives. Cell Mol Life Sci. 2002 May;59(5):790–798. doi: 10.1007/s00018-002-8467-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tontonoz P, Hu E, Graves RA, Budavari AI, Spiegelman BM. mPPAR gamma 2: tissue-specific regulator of an adipocyte enhancer. Genes & development. 1994 May 15;8(10):1224–1234. doi: 10.1101/gad.8.10.1224. [DOI] [PubMed] [Google Scholar]
- 4.Michalik L, Desvergne B, Dreyer C, Gavillet M, Laurini RN, Wahli W. PPAR expression and function during vertebrate development. The International journal of developmental biology. 2002 Jan;46(1):105–114. [PubMed] [Google Scholar]
- 5.Mao-Qiang M, Fowler AJ, Schmuth M, Lau P, Chang S, Brown BE, et al. Peroxisome-proliferator-activated receptor (PPAR)-gamma activation stimulates keratinocyte differentiation. J Invest Dermatol. 2004 Aug;123(2):305–312. doi: 10.1111/j.0022-202X.2004.23235.x. [DOI] [PubMed] [Google Scholar]
- 6.Sarraf P, Mueller E, Jones D, King FJ, DeAngelo DJ, Partridge JB, et al. Differentiation and reversal of malignant changes in colon cancer through PPARgamma. Nature medicine. 1998 Sep;4(9):1046–1052. doi: 10.1038/2030. [DOI] [PubMed] [Google Scholar]
- 7.Saez E, Tontonoz P, Nelson MC, Alvarez JG, Ming UT, Baird SM, et al. Activators of the nuclear receptor PPARgamma enhance colon polyp formation. Nature medicine. 1998 Sep;4(9):1058–1061. doi: 10.1038/2042. [DOI] [PubMed] [Google Scholar]
- 8.Shappell SB, Gupta RA, Manning S, Whitehead R, Boeglin WE, Schneider C, et al. 15S-Hydroxyeicosatetraenoic acid activates peroxisome proliferator-activated receptor gamma and inhibits proliferation in PC3 prostate carcinoma cells. Cancer research. 2001 Jan 15;61(2):497–503. [PubMed] [Google Scholar]
- 9.Hsi LC, Wilson LC, Eling TE. Opposing effects of 15-lipoxygenase-1 and -2 metabolites on MAPK signaling in prostate. Alteration in peroxisome proliferator-activated receptor gamma. J Biol Chem. 2002 Oct 25;277(43):40549–40556. doi: 10.1074/jbc.M203522200. [DOI] [PubMed] [Google Scholar]
- 10.Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 11.Michalik L, Desvergne B, Wahli W. Peroxisome-proliferator-activated receptors and cancers: complex stories. Nature reviews. 2004 Jan;4(1):61–70. doi: 10.1038/nrc1254. [DOI] [PubMed] [Google Scholar]
- 12.Mueller E, Smith M, Sarraf P, Kroll T, Aiyer A, Kaufman DS, et al. Effects of ligand activation of peroxisome proliferator-activated receptor gamma in human prostate cancer. Proceedings of the National Academy of Sciences of the United States of America. 2000 Sep 26;97(20):10990–10995. doi: 10.1073/pnas.180329197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hisatake JI, Ikezoe T, Carey M, Holden S, Tomoyasu S, Koeffler HP. Down-Regulation of prostate-specific antigen expression by ligands for peroxisome proliferator-activated receptor gamma in human prostate cancer. Cancer research. 2000 Oct 1;60(19):5494–5498. [PubMed] [Google Scholar]
- 14.Smith MR, Kantoff PW. Peroxisome proliferator-activated receptor gamma (PPargamma) as a novel target for prostate cancer. Invest New Drugs. 2002 May;20(2):195–200. doi: 10.1023/a:1015670126203. [DOI] [PubMed] [Google Scholar]
- 15.Li M, Jiang X, Liu D, Na Y, Gao GF, Xi Z. Autophagy protects LNCaP cells under androgen deprivation conditions. Autophagy. 2008 Jan 1;4(1):54–60. doi: 10.4161/auto.5209. [DOI] [PubMed] [Google Scholar]
- 16.Chang CL, Liao JJ, Huang WP, Lee H. Lysophosphatidic acid inhibits serum deprivation-induced autophagy in human prostate cancer PC-3 cells. Autophagy. 2007 May-Jun;3(3):268–270. doi: 10.4161/auto.3909. [DOI] [PubMed] [Google Scholar]
- 17.Cao C, Subhawong T, Albert JM, Kim KW, Geng L, Sekhar KR, et al. Inhibition of mammalian target of rapamycin or apoptotic pathway induces autophagy and radiosensitizes PTEN null prostate cancer cells. Cancer research. 2006 Oct 15;66(20):10040–10047. doi: 10.1158/0008-5472.CAN-06-0802. [DOI] [PubMed] [Google Scholar]
- 18.Shappell SB, Thomas GV, Roberts RL, Herbert R, Ittmann MM, Rubin MA, et al. Prostate pathology of genetically engineered mice: definitions and classification. Cancer research. 2004 Mar 15;64(6):2270–2305. doi: 10.1158/0008-5472.can-03-0946. The consensus report from the Bar Harbor meeting of the Mouse Models of Human Cancer Consortium Prostate Pathology Committee. [DOI] [PubMed] [Google Scholar]
- 19.Park JH, Walls JE, Galvez JJ, Kim M, Abate-Shen C, Shen MM, et al. Prostatic intraepithelial neoplasia in genetically engineered mice. The American journal of pathology. 2002 Aug;161(2):727–735. doi: 10.1016/S0002-9440(10)64228-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Day KC, McCabe MT, Zhao X, Wang Y, Davis JN, Phillips J, et al. Rescue of embryonic epithelium reveals that the homozygous deletion of the retinoblastoma gene confers growth factor independence and immortality but does not influence epithelial differentiation or tissue morphogenesis. J Biol Chem. 2002 Nov 15;277(46):44475–44484. doi: 10.1074/jbc.M205361200. [DOI] [PubMed] [Google Scholar]
- 21.Brockman JA, Gupta RA, Dubois RN. Activation of PPARgamma leads to inhibition of anchorage-independent growth of human colorectal cancer cells. Gastroenterology. 1998 Nov;115(5):1049–1055. doi: 10.1016/s0016-5085(98)70072-1. [DOI] [PubMed] [Google Scholar]
- 22.Munafo DB, Colombo MI. A novel assay to study autophagy: regulation of autophagosome vacuole size by amino acid deprivation. Journal of cell science. 2001 Oct;114(Pt 20):3619–3629. doi: 10.1242/jcs.114.20.3619. [DOI] [PubMed] [Google Scholar]
- 23.Zhang J, Fu M, Cui T, Xiong C, Xu K, Zhong W, et al. Selective disruption of PPARgamma 2 impairs the development of adipose tissue and insulin sensitivity. Proceedings of the National Academy of Sciences of the United States of America. 2004 Jul 20;101(29):10703–10708. doi: 10.1073/pnas.0403652101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang MS, Ji KA, Jeon SB, Jin BK, Kim SU, Jou I, et al. Interleukin-13 enhances cyclooxygenase-2 expression in activated rat brain microglia: implications for death of activated microglia. J Immunol. 2006 Jul 15;177(2):1323–1329. doi: 10.4049/jimmunol.177.2.1323. [DOI] [PubMed] [Google Scholar]
- 25.Jiang M, Shappell SB, Hayward SW. Approaches to understanding the importance and clinical implications of peroxisome proliferator-activated receptor gamma (PPARg) signaling in prostate cancer. J Cell Biochem. 2004;91:513–527. doi: 10.1002/jcb.10770. [DOI] [PubMed] [Google Scholar]
- 26.Pathak SK, Sharma RA, Steward WP, Mellon JK, Griffiths TR, Gescher AJ. Oxidative stress and cyclooxygenase activity in prostate carcinogenesis: targets for chemopreventive strategies. Eur J Cancer. 2005 Jan;41(1):61–70. doi: 10.1016/j.ejca.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 27.Shappell SB, Boeglin WE, Olson SJ, Kasper S, Brash AR. 15-lipoxygenase-2 (15-LOX-2) is expressed in benign prostatic epithelium and reduced in prostate adenocarcinoma. The American journal of pathology. 1999 Jul;155(1):235–245. doi: 10.1016/S0002-9440(10)65117-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shappell SB, Keeney DS, Zhang J, Page R, Olson SJ, Brash AR. 15-Lipoxygenase-2 expression in benign and neoplastic sebaceous glands and other cutaneous adnexa. J Invest Dermatol. 2001 Jul;117(1):36–43. doi: 10.1046/j.1523-1747.2001.01378.x. [DOI] [PubMed] [Google Scholar]
- 29.Kirschenbaum A, Liotta DR, Yao S, Liu XH, Klausner AP, Unger P, et al. Immunohistochemical localization of cyclooxygenase-1 and cyclooxygenase-2 in the human fetal and adult male reproductive tracts. The Journal of clinical endocrinology and metabolism. 2000 Sep;85(9):3436–3441. doi: 10.1210/jcem.85.9.6780. [DOI] [PubMed] [Google Scholar]
- 30.Bostwick DG, Meiers I, Shanks JH. Glutathione S-transferase: differential expression of alpha, mu, and pi isoenzymes in benign prostate, prostatic intraepithelial neoplasia, and prostatic adenocarcinoma. Human pathology. 2007 Sep;38(9):1394–1401. doi: 10.1016/j.humpath.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 31.Keen HL, Ryan MJ, Beyer A, Mathur S, Scheetz TE, Gackle BD, et al. Gene expression profiling of potential PPARgamma target genes in mouse aorta. Physiological genomics. 2004 Jun 17;18(1):33–42. doi: 10.1152/physiolgenomics.00027.2004. [DOI] [PubMed] [Google Scholar]
- 32.Perera RJ, Marcusson EG, Koo S, Kang X, Kim Y, White N, et al. Identification of novel PPARgamma target genes in primary human adipocytes. Gene. 2006 Mar 15;369:90–99. doi: 10.1016/j.gene.2005.10.021. [DOI] [PubMed] [Google Scholar]
- 33.Hippert MM, O'Toole PS, Thorburn A. Autophagy in cancer: good, bad, or both? Cancer research. 2006 Oct 1;66(19):9349–9351. doi: 10.1158/0008-5472.CAN-06-1597. [DOI] [PubMed] [Google Scholar]
- 34.Bergmann A. Autophagy and cell death: no longer at odds. Cell. 2007 Dec 14;131(6):1032–1034. doi: 10.1016/j.cell.2007.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer cell. 2006 Jul;10(1):51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007 Sep;8(9):741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 37.Jones JR, Shelton KD, Guan Y, Breyer MD, Magnuson MA. Generation and functional confirmation of a conditional null PPARgamma allele in mice. Genesis. 2002 Feb;32(2):134–137. doi: 10.1002/gene.10042. [DOI] [PubMed] [Google Scholar]
- 38.Wu X, Wu J, Huang J, Powell WC, Zhang J, Matusik RJ, et al. Generation of a prostate epithelial cell-specific Cre transgenic mouse model for tissue-specific gene ablation. Mechanisms of development. 2001 Mar;101(1-2):61–69. doi: 10.1016/s0925-4773(00)00551-7. [DOI] [PubMed] [Google Scholar]
- 39.Duan SZ, Ivashchenko CY, Russell MW, Milstone DS, Mortensen RM. Cardiomyocyte-specific knockout and agonist of peroxisome proliferator-activated receptor-gamma both induce cardiac hypertrophy in mice. Circulation research. 2005 Aug 19;97(4):372–379. doi: 10.1161/01.RES.0000179226.34112.6d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.