Abstract
The PI3K pathway is a communication hub coordinating critical cell functions including cell survival, cell growth, proliferation, motility and metabolism. Because PI3Kα harbors recurrent somatic mutations resulting in gains of function in human cancers, it has emerged as an important drug target for many types of solid tumors. Various PI3K isoforms are also being evaluated as potential therapeutic targets for inflammation, heart disease, and hematologic malignancies. Structural biology is providing insights into the flexibility of the PI3Ks, and providing basis for understanding the effects of mutations, drug resistance and specificity.
1. Introduction
While each human tumor has its own unique “genetic signature,” certain oncogenes are often found mutated at the same unique positions. These so called “hot-spot” mutations can characterize a specific cancer subtype, confer resistance or sensitivity to individual inhibitors, and in some cases, correlate with cancer prognosis. The ideal “hot-spot” mutant to target with anti-cancer agents would have both an activating effect on the protein and exploitable conformational changes when compared to its wild-type counterpart. These characteristics are embodied by the H1047R mutant of PI3Kα.
PI3Kα, phosphoinositide 3-kinase isoform alpha, is a heterodimeric lipid kinase comprised of p110α, encoded by PIK3CA, and p85α, encoded by PIK3R1. Upon activation by phosphorylated receptor tyrosine kinase (RTK), the enzyme phosphorylates phosphatidylinositol 4,5-bisphosphate, PIP2, at position 3 of the inositol head group to generate phosphatidylinositol 3,4,5-triphosphate, PIP3 [1, 2]. PIP3 recruits proteins that contain a pleckstrin homology domain, such as AKT and PDK-1 to the cell membrane, initiating signaling cascades that result in cellular proliferation, motility, metabolism, and survival [1, 3–6].
Somatic mutations in PI3Kα were identified in a variety of cancer types in 2004 [7]. The most striking feature of the mutation profile was the clustering of the mutations in three hot spots. Changes at three residues, Glu 542, Glu 545, and His 1047, comprised ~80% of the mutations in PIK3CA. All three mutations were found to increase the lipid kinase activity of PI3Kα, with a kcat 2–3 fold higher than that of the wild-type enzyme [7–9]. Subsequent sequencing studies have found PIK3CA to be mutated in 12% of all tumor sequences deposited in the catalog of somatic mutations in cancer database, COSMIC [10]. This high prevalence of mutations in cancer types as diverse as colorectal, breast, gastric and hepatocellular carcinomas makes PIK3CA the most commonly mutated human oncogene.
Histidine 1047 is located in the kinase domain of PI3Kα, and is most often mutated to an Arginine residue. This mutant enzyme is further activated upon binding to phosphorylated receptors, with the activation being independent of Ras-binding but dependent on p85α binding [8, 11]. At least two studies in breast and uterine cancer patients have correlated the H1047R mutation with differential clinical prognoses when compared to patients whose tumors harbor either a wild-type PIK3CA genotype or a different mutation in PIK3CA [12–15]. The crystal structures of wild type and H1047R mutant of PI3Kα provide a critical platform for understanding the mechanism of oncogenic activation and for the structure based design of mutant-specific inhibitors.
2. Structure and activation of Class I Phosphoinositide 3-kinases
Class I phosphoinositide 3-kinases are heterodimeric lipid kinases that catalyze a phosphoryl transfer from ATP to PIP2 to produce PIP3 (for a review of the complete classification see[6, 16]). PI3K enzymes consist of a catalytic subunit p110 (α, β, δ or γ) and a regulatory subunit. Class I is further sub-classified according to the mechanism of enzyme activation, and the regulatory subunit component of the heterodimer. Class 1a enzymes, (PI3Kα, β, δ) are activated by receptor tyrosine kinases or other receptor substrates, and utilize p85α, β, or their splice variants as the regulatory subunit. The class 1b enzyme, PI3kγ, is activated by G-protein coupled receptors (GPCR) and its regulatory domain is p101, or p84/p87, a subunit with no sequence similarity to any of the p85 genes. Vanhaesebroeck and coworkers showed that p110β is also activated by GPCRs, but less is known about this association [17].
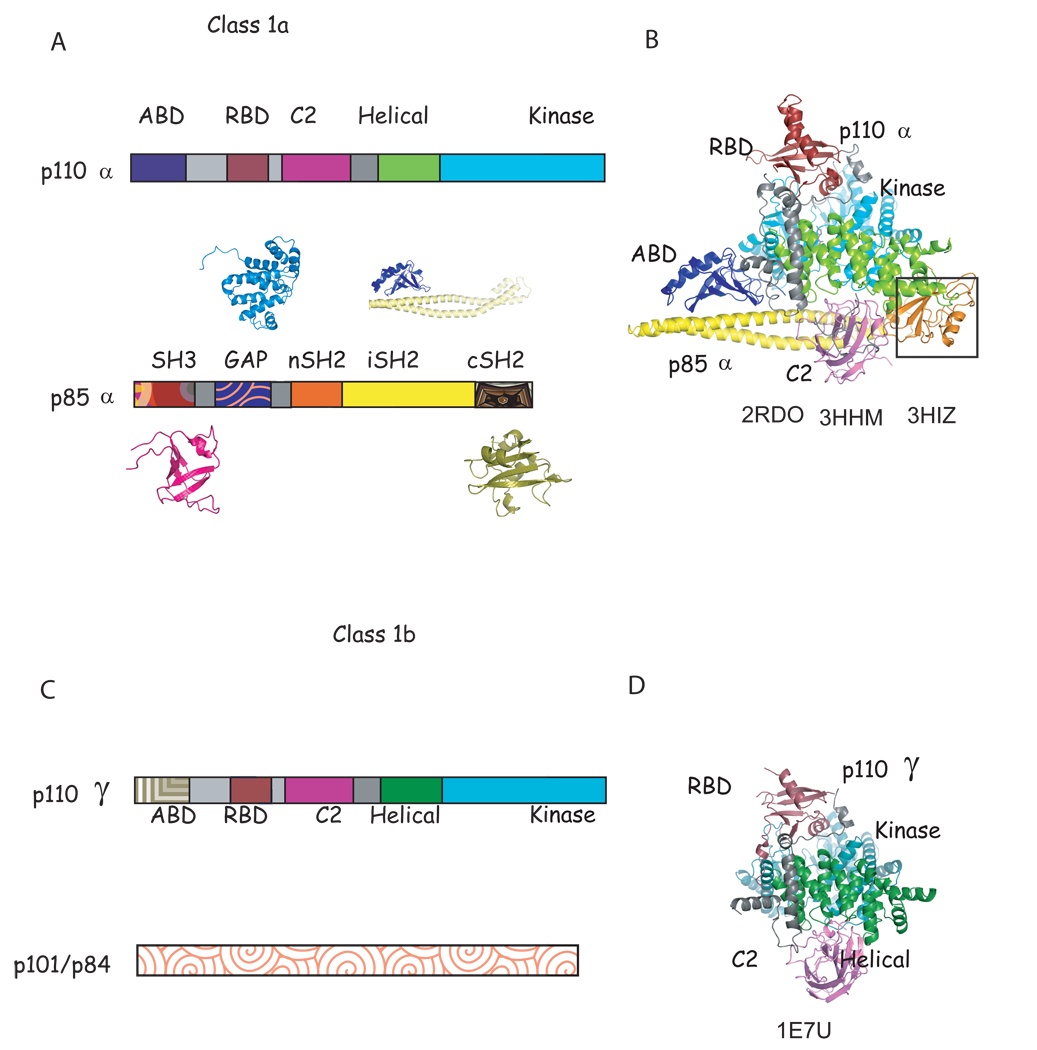
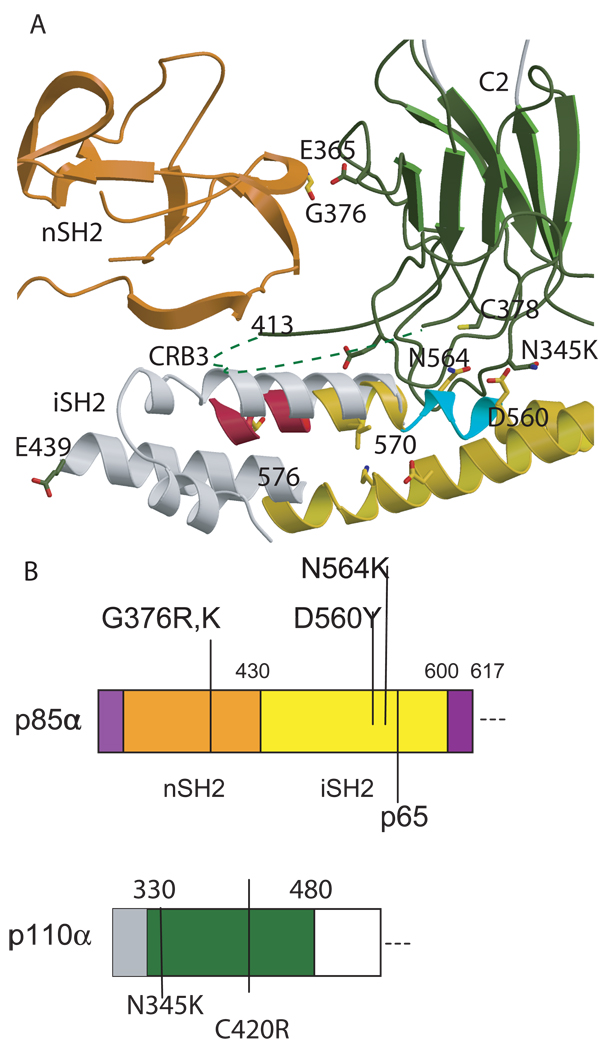
The p110 subunits are comprised of five domains: an adaptor binding domain (ABD), a Ras binding domain (RBD), a C2 domain, a helical domain, and a kinase domain. The last four domains have significant sequence homology between isoforms. The p85 subunits also contain five domains: an Src homology 3 (SH3) domain, a GTPase-activating protein (GAP-like or BH) domain and two SH2 domains separated by an inter-SH2 domain (iSH2, Figure 1).
Figure 1. Characteristic of class 1a and known structures of class 1a and class 1b phosphoinositide 3 kinases.
A. Scheme of the domain structure of the heterodimer of class 1a. The structure of all domains of p85α have been determined individually (shown in patterned colors) (SH3: 1PNJ, 2PNI[43]; GAP: 1PBW[18]; nSH2: 2PNA,2PNB[19, 43], 2IUG, 2IUH, 2IUI[20],1OO4[44]; iSH2: 2V1Y[30]; cSH2: 1H90[21],1QAD[45],1BFI[46], 1PIC[47]); although only nSH2 and iSH2 domain structures(solid colors) have been determined as part of a complex with p110α [23, 24]. B. Ribbon diagram of the known structure of the p110α/niSH2 structure. The black box highlights the nSH2 domain observed in the mutant p110α H1047R/ niSH2 (pdb id 3HMM, 3HIZ[24] but not in native structure (pdb id 2RD0, [23]). C. Scheme of the domain structure of class 1b. D. Ribbon modeled of the known domains of the structure of p110γ. Although more than 20 complex structures of p110γ in complex with inhibitors have been published [22, 27, 40, 41, 48–53], the structures of only four (RBD, C2, helical and kinase) out of the five domains have been determined, and there is no published structural information of the regulatory p101/p84. Homologous domains in p110α and γ are colored in B, C, E and F in the same color in different shade.
The first structural information on the PI3K family was obtained from structures of individual domains of the regulatory domain of p85α (Figure 1,[18, 19]). The structures of the N- and C-terminal SH2 domains bound to phosphopeptides revealed the structural basis for both phosphopeptide specificity and for the preference of a methionine residue at the fourth position of the motif Tyr-Xaa-Xaa-Met [20, 21]. In 1999, the first structure of a catalytic subunit of a PI3K enzyme became available when Williams and coworkers published the structure of four domains of the class 1b p110γ (RBD, C2, Helical and Kinase) [22]. This seminal work was followed by structures of p110γ in complex with the ATP substrate, ATP analogs, and kinase inhibitors (20–22, 37–43). These structures were obtained by soaking the same crystal form in solutions containing the desired small molecules. The proteins that were crystallized did not provide information about the second substrate, PIP2, or about the regulatory subunit p101. In 2007, the structure of the Class 1a PI3Kα, in heterodimeric form was determined. The structure showed all five domains of p110α in complex with the nSH2 and iSH2 domains of the p85α, although no information beyond its general location was available for the nSH2 domain [23]. Recently, the structure of the somatic p110α H1047R/niSH2 mutant alone and in complex with the inhibitor wortmannin was determined. This latter study highlighted the conformational differences between isoforms induced by inhibitor binding as well as the differences between wild type and the H1047R mutant of PI3Kα [24]. As of yet no structural information has been published for p110δ or p110β.
3. The Binding Site of PI3Kα
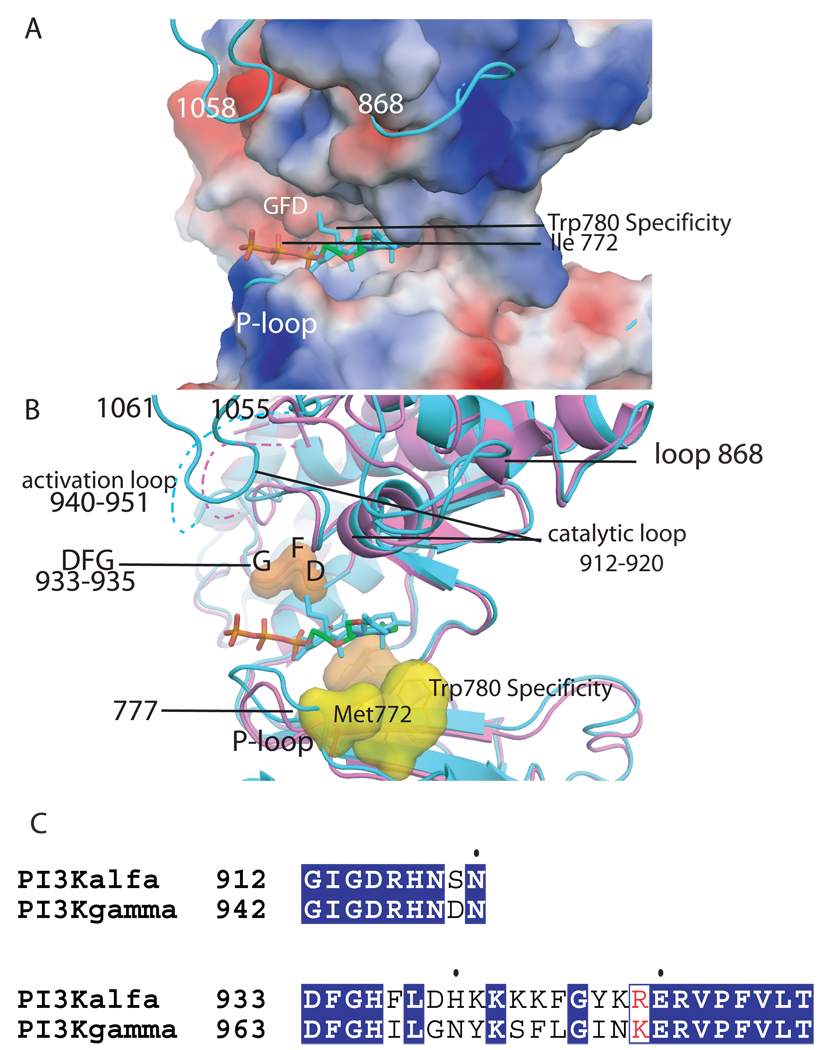
The residues that line the ATP binding pocket are conserved between p110α and p110γ, and in the other two PI3K isoforms (Figure 2). In particular, the residues that form the adenosine binding site, Ile 800, Tyr 836, Phe 930 and Met 922, and the residues that are at hydrogen bonding distance to the adenosine group, Val 851 and Glu 849, are highly conserved. The activation loop, which displays slight sequence diversity between isoforms, is disordered in the crystal structures of PI3Kα (residues 940–950). The catalytic loop (residues 912–920), which has a similar conformation in the p110α and p110γ structures, has one residue that is distinctive: Ser 919 in p110α is an aspartate residue in all other isoforms (Figure 2). Conformational changes of the loop formed by residues 933–935 in p110α (Aspartate, phenylalanine, glycine, DFG-loop), have been exploited in other protein kinases to design inhibitors that target the “DFG-out” or “DFG-in” conformations. However, the DFG loops of the p110α and p110γ PI3K isoforms are identical and do not change conformation upon substrate or inhibitor binding [24]. It remains to be seen whether specificity determinants such as the DFG loop and the gatekeeper residue that were used in the design of protein kinase inhibitors will be equally useful in the design of isozyme-specific PI3K inhibitors [25, 26]. The inhibitors reported to date all target the “active conformation” with the “DFG in”. Residues such as Met 772 which opens a new pocket when the inhibitor PIK39 is bound to p110γ, may emerge as isoform specificity determinants for PI3Ks (Figure 2) [27]. Additionally, exploitation of such conformational changes may not only lead to PI3K isoform specific inhibitors, but also minimize drug’s inhibition of other cellular kinases. The initial successes of isoform selective inhibitors in preclinical studies have led to phase I and phase II trials of compounds such as PX866 and GSK615. Concurrently, efforts to develop dual pan-PI3K/mTOR inhibitors have yielded NVPBEZ235, which is now in clinical trials for advanced solid malignancies [28].
Figure 2. ATP binding site of the kinase domain of p110α.
A. Electrostatic surface of the kinase domain as in the wild type p110α/niSH2 (pdb id 2RD0) structure with the ATP modeled based on the p110γ complex structure (pdb id 1E8X). The complex of wortmannin bound H1047R p110α/niSH2 is shown in turquoise to highlight the conformational changes in loops 772–777 (P-loop), 864–872 and 1047–1062 which protrude from the surface. B. Structural overlap of the wild type p110α/niSH2 (magenta, pdb id 2RD0) structure with the wortmannin bound H1047R p110α/niSH2 (turquoise, 3HMM) overlapping a molecule of ATP. The recognition and catalytically important residues are shown as surfaces. The DFG (residues 933–935) is shown in orange; the protein kinase “gatekeeper residue” in p110γ (Ile 848) is shown in light blue; specificity residues Met 772 and Trp780 are shown in yellow; the activation loop is shown in both structures as a dashed line; the last residues of the p110α H1047R mutant which change conformations in the mutant are also shown in turquoise. C. Sequence alignment of the activation and catalytic loop of p110α and p110γ with identical residues shown in blue background.
Efforts to determine the structure of PI3Ks in the presence of PIP2 or its short chain analogs have thus far been unsuccessful. Interestingly, one of the loops, amino acids 772–776, that shifts conformation between the native and mutant structures of p110α in complex with wortmannin could provide part of the binding site for PIP2 (Figure 2b). Modeling suggests that a PIP2 bound at this position would be at the correct distance to accept the phosphoryl from ATP (unpublished results). However, this model does not explain the result of the enzymatic analysis of loop exchanged hybrids, which demonstrate that basic residues of the catalytic loop are involved in specificity for PIP2 [29].
4. Somatic mutations in p110α
In 2004, it was shown that the PIK3CA gene is somatically mutated in > 25% of the colorectal and breast tumors as well as other tumor types[7]. The distribution of these mutations was striking, with ~80% of these mutations occurring at three “hotspot” residues. This discovery combined with the determination of the structures of PI3Kα mutants will open the door for the design of mutant-specific drugs. Ideally, these targeted drugs would circumvent the side effects associated with inhibiting wild type PI3Kα and other PI3K isoforms [7].
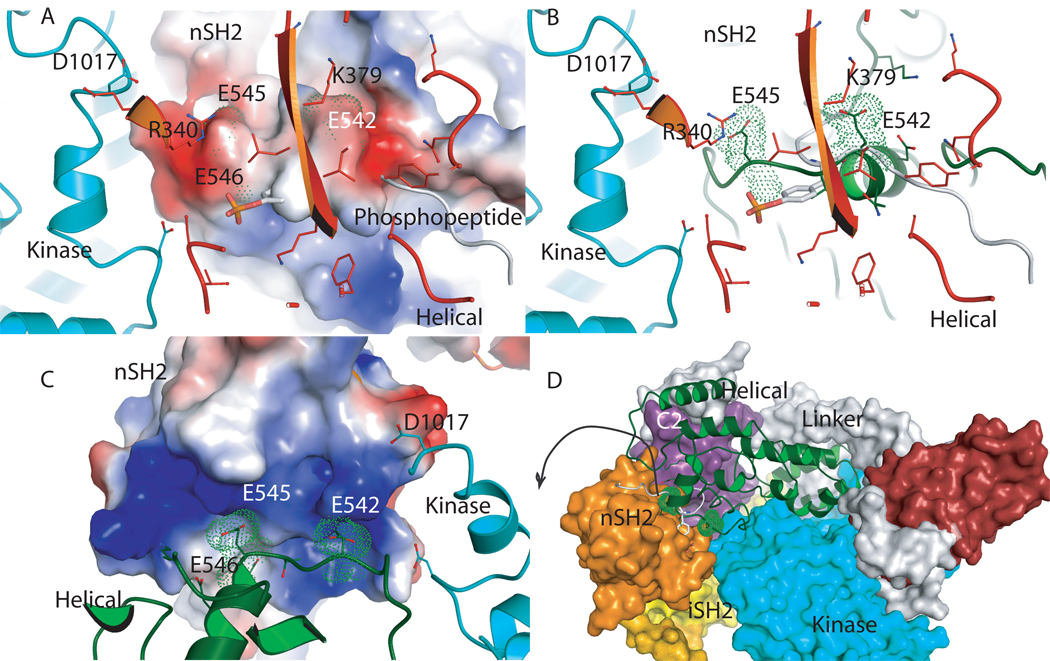
Analysis of the location of somatic PI3K mutations in the structure of p110α/niSH2 shows that most of the observed mutations localize to domain interfaces. Moreover, the hotspot mutants of the helical domain of p110α are at the interface with the nSH2 domain (Figure 4). Although no structure of the helical domain mutants has yet been determined, analysis of the wild type p110α/niSH2 and the H1047R mutant p110α/niSH2 suggests a possible mechanism of PI3K activation by these mutations. The helical domain Hot spot mutations, Glu 542, Glu 545, Gln 546 are in the loop region of the helical domain that contacts the nSH2 domain. Biochemical and mutational studies showed that these residues interact with Lys 379 and Arg 340 of nSH2 [30]. In the structure of PI3Kα Glu 542 and Glu 545 are localized in a groove formed by positively charged residues in nSH2 that includes K379 and R340 (Figure 4). This domain “locking” suggests that in order to bind the phosphorylated activation loop of the effector protein, the nSH2 domain must move outward. The observed helical domain mutations have a similar effect: they weaken the interaction between the helical and nSH2 domains and allow the nSH2 to move outward, resulting in PI3Kα activation.
Figure 4. nSH2 p85α and helical p110α interaction.
A. Electrostatic surface of the helical p110α domain with the location of the hotspots mutations 542, 545, 546 marked with green dots. Residues Arg 340 and Lys 379 of the nSH2 domain are shown as orange sticks. The helix of the kinase domain is shown for orientation purposes (turquoise). B. Same orientation as in A with the helical domain shown in green; the PDGFR phosphopeptide in grey overlapping the loop of the helical domain, and nSH2 residues in orange. C. Electrostatic surface of the nSH2 domain of p85α with the residues of the helical domain shown in green, and the kinase in turquoise. The Van der Waals surface of the hot spot mutations is shown with green dots showing their position in the nSH2. D. Interface between the helical domain and nSH2 showing how nSH2 could move to allow binding of the activating phosphopeptides of receptor tyrosine kinases and other activating proteins. The structure suggests that the nSH2 moves outwards from the complex since the helical domain is locked by nSH2, the linker and the kinase domains.
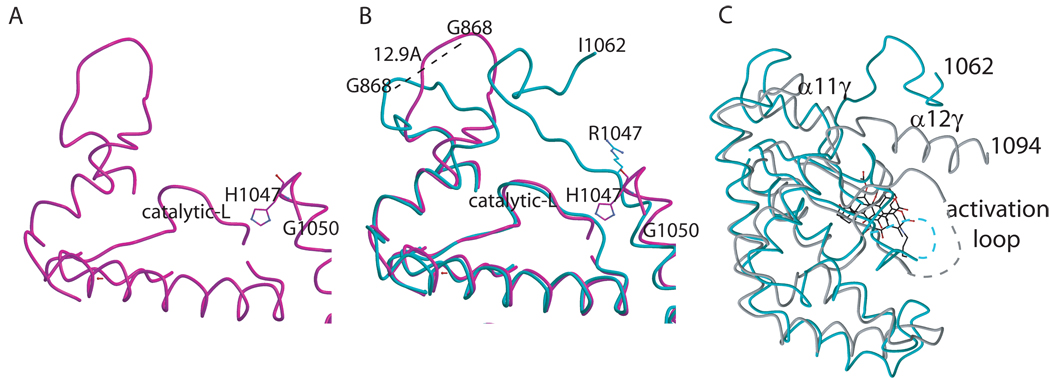
The H1047R mutation elicits conformational changes that are not at a domain interface, but at the interface between p110α and the membrane, most likely altering membrane-protein interactions ([24], Figure 5). The structure of the H1047R mutant of p110α in complex with the niSH2 domains of p85α demonstrates three conformational changes caused by the H1047R mutation. First, the orientation of the 1047 residue is changed. The wild-type His1047 points towards the activation loop of p110α, while the mutant Arg1047 points at a 90° angle from the wild-type orientation, upward into a cavity in the p110α kinase domain, and towards the cell membrane. Second, the last ordered C-terminal residues of p110α curl away from the cell membrane in the wild-type structure (Figure 5A). In contrast, in the H1047R p110α structure the last 12 residues (1050–1062) loop up making contact to the cell membrane. Third, loop 864–874, which also contacts the cell membrane, shifts orientation by 12.9Å in the H1047R mutant when compared to the wild type. These changes in the kinase domain conformation caused by the H1047R mutation result in altered protein-membrane contacts. Mutant-specific inhibitors could possibly target these differential protein-lipid interactions. Molecular dynamic simulations of the possible effects of the PI3Kα H1047R mutation were carried out with a model based on the PI3Kγ structure [31, 32]. These simulations failed to predict the observed conformational changes in the p110α H1047R mutant [31, 32]. In contrast, the changes predicted by the simulations were a rotation of the residues of the DFG loop and rearrangements of the catalytic with respect to the activation loop, both comprising movements not observed between the wild type and the mutant experimental structures [23, 24].
Figure 5. Conformational changes due to the H1047R mutation.
A. Ribbon diagram showing the loop 864–874 in the wild type structure. B. Ribbon diagram showing the overlap of the loops (residues 864–874 and 1050–1062) in the mutant (cyan) and the wild type (magenta). C. Structural overlap of the C-lobe PI3Kα (turquoise) and PI3Kγ (grey) kinase domains showing the differences after helix α11γ and α12α where in PI3Kα is a loop and in PI3Kγ is another helix (α12γ).
5. Somatic mutations in p85α
Mutations in the p85α regulatory subunit have been reported less frequently than those in p110α [33, 34]. A survey of all somatic mutations identified in p85α shows that these mutations predominantly occur in the nSH2 and iSH2 domains. Contrary to the distribution of p110α mutations, somatic mutations in iSH2 include in-frame deletions (Figure 3). In glioblastomas, mutations cluster around the interface of the p85α iSH2 domain and the p110α C2 domain [33, 34]. This interface can communicate structural changes to the inhibitory nSH2 domain. Interestingly, the Asn 564 and Asp 560 mutations in iSH2 are at hydrogen bonding distance of residue Asn 345 of the Calcium Binding Region 1 (CBR1) within the C2 domain, a residue found somatically mutated in p110α [23]. Another frequent C2 mutation is C420R positioned in the CBR3. Although the residues 414–420 are disordered in all PI3Kα structures, the packing of the interface suggests that Cys 420 and CRB3 would interact with iSH2[23, 24]. The six amino acid deletion (560–565) of iSH2 found in glioblastomas is likely to cause an unraveling of the helix in this coiled-coiled domain. As a consequence of this deletion, the contacts between the nSH2 and p110α might be disrupted. Also, Gly 376 of nSH2 is at hydrogen bonding distance of Glu 365 of C2. The G376R mutation most likely affects the interface with C2 by pushing away the domain or by forming a salt bridge [24].
Figure 3. p85α somatic mutations at the interfase of the C2 domain the nSH2 and iSH2 domains.
A. Ribbon model of the nSH2 (orange) and the C2 domain (dark green) as observed in the structure of p110α/niSH2; the iSH2 is shown in yellow as in H1047R (pdb id 3HMM) and in grey as in 2VYI. As the connection of the coiled coil is not observed in the heterodimeric structures of p85α (pdb id 2RD0, 3HMM and 3HMI), the figure of this region in 2VY1 represents a model. The iSH2 deletions observed in glioblastoma by TCARN [33] are shown in turquoise and red ribbons. B. Scheme of the p85αniSH2 and the p110αC2 sequence with the mutations discussed in the text marked.
6. Inhibitors of PI3K
The initial validation of PI3Ks as drug target was established with wortmannin, a fungal metabolite with nanomolar IC50 that binds covalently to the enzyme[35]. Viridin, and other furanosteroids, were investigated as clinical agents, but this family of compounds shares the properties of low solubility, instability, and in vivo toxicity, and was subsequently dismissed as a viable drug candidate[36]. LY294002 followed as the first synthetic and reversible PI3K inhibitor with micromolar IC50, but also showed unacceptable toxicity. However, two second generation derivatives of wortmannin and LY294002, PX-866 and SF1126, have more favorable pharmacokinetic properties[37–39]}. Both of these pan-PI3K inhibitors are currently in phase I trials for advanced malignancies. Since wortmannin binding to p110α elicits different conformational changes in p110α compared to p110γ, perhaps future generations of PI3K inhibitors can exploit these differences for isofrm specificity.
The third generation of PI3K inhibitors encompasses a variety of chemotypes including arylthiazolidinones and pyridinylfuranopyrimidines. Some thiazolidnedione compounds (AS252424 and AS605240 by Merck-Serono) show PI3Kγ specificity, and may be therapeutic for chronic inflammatory disorders [40]. The structure of PI3Kα H1047R in complex with wortmannin made it clear that these thialidinedione compounds would be too short to reach to the loops of the PI3Kα mutant H1047 that display conformational changes. Likewise, comparing the effect of the PI3Kδ specific quinazolinone derivative, PIK-39 on the active site of p110γ to our structure of p110α suggests that the compound will require a conformational change of loop 770–776 to fit in the p110α ATP binding site[27]. In particular, Met 772 would clash in with the edge of the quinazolinenone in its current position as we also noted in the wild type structure [40]. Compounds that target the metal binding site such as the organorutheniums may be isoform-specific, as they are at hydrogen bonding distance of residues in the helix 890–897 of p110γ, which is displaced in PI3Kα when compared to PI3Kγ [51]. Notably, the equivalent helix in p110α is the secondary structure element prior to one of the loops that changes conformation in the Pi3KαH1047R mutant and more sequence divergent within isoforms.
Both pan-PI3K inhibitors such as BEZ235, and SF1126, and specific PI3K inhibitors, such as the PI3Kδ specific CAL-101 show promising results in early clinical trials for malignancies[28]. Because PI3K signaling is involved in several critical physiological functions, the toxicity of PI3K inhibition is a main concern. Thus far, most clinical trials have shown a tolerable side effect profile for PI3K inhibitors. The discovery that inhibitor binding differentially changes the active site of p110γ and p110α should facilitate the development of isoform-specific inhibitors. A deeper insight into isoform-selectivity requires studying the p110δ and p110β isoforms to minimize the off-target effects of the next generation of PI3K inhibitors.
7. Concluding remarks: Implications for isoform-selective inhibitors
The four isoforms of PI3K, α, β, δ, γ have distinct biological functions. Among others, PI3Kα is involved in tumorigenesis and insulin signaling, PI3Kβ plays a role in platelet aggregation, PI3Kγ is expressed in leukocytes and is a component of the inflammatory response, and PI3Kδ is implicated in allergic responses and hematological cancers. Therefore, a pan-PI3K inhibitor used as an anti-cancer agent may induce undesirable phenotypes due to the inhibition of all other isoforms.
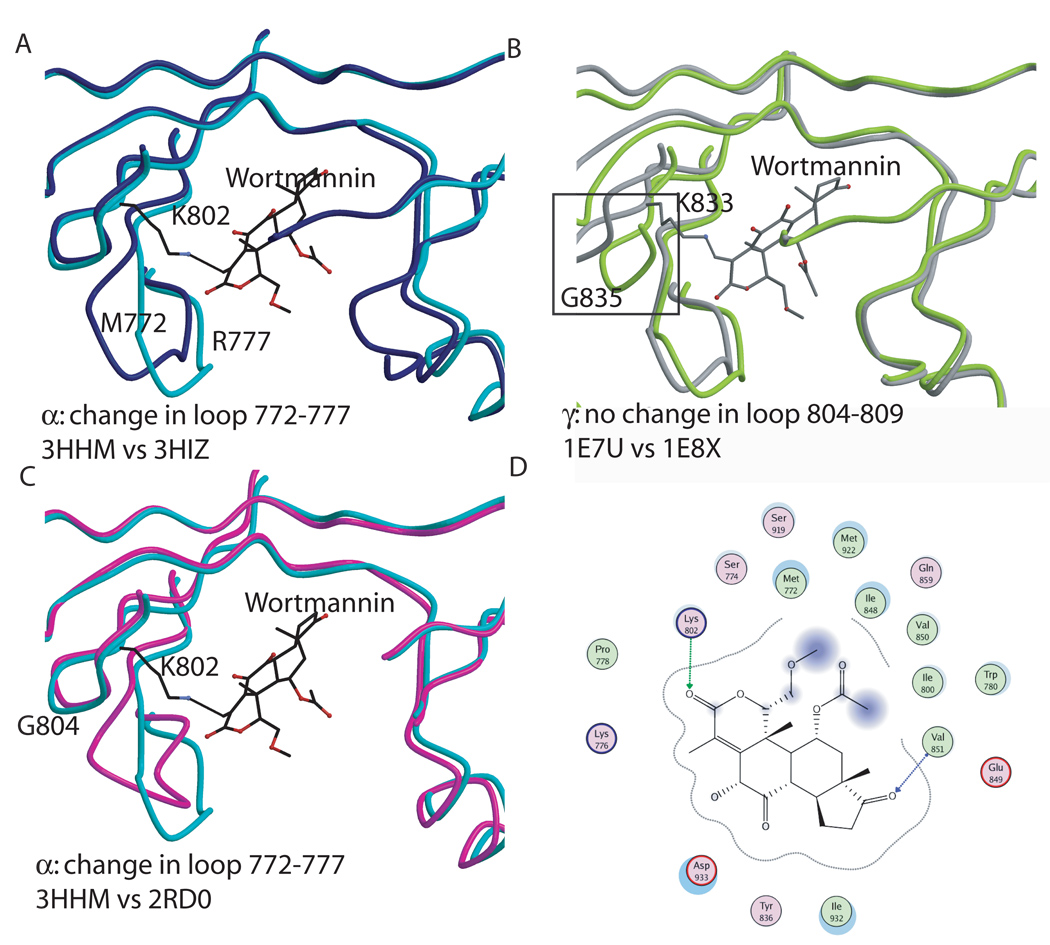
The structure of PI3Kγ in complex with wortmannin has been previously described [41]. Now that the crystal structure of the H1047R mutant of p110α in complex with the nSH2 and the iSH2 domains of p85α has been determined alone and in complex with the kinase inhibitor wortmannin, the conformational changes elicited by wortmannin binding to these different PI3K isoforms can be compared (Figure 6a [24, 41]). In the H1047R mutant of p110α in complex with p85α niSH2, a 3Å shift in the loop spanning residues 772–779 of the kinase domain was observed upon wortmannin binding. The corresponding loop in PI3Kγ (residues 804–809) displays no conformational change in response to wortmannin. Therefore, this loop shift in p110α represents an isoform-specific conformational change in the ATP binding site in response to an inhibitor. Three loops in p110γ experience structural changes upon wortmannin binding (residues 748–750, 832–838 and 871–876,) while the equivalent residues in p110α, 718–720, 801–807 and 840–845, show no conformational changes that can be attributed to wortmannin binding (Figure 6b).
Figure 6. Wortmannin binding to the p110α and γ kinase domains.
A. Conformational changes elicited by wortmannin binding to p110α (H1047R p110α + wortmannin in turquoise; H1047Rp110α in blue). B. Conformational changes observed in p110γ in response to wortmannin binding. The black box highlights one of the observed differences in residues 748–750, 832–838, 871–876 which moved away from the wortmannin. C. Comparison between the changes due to wortmanninn binding in p110α (turquoise) and p110γ (grey). D. Interaction between wortmannin and the p110α structure.
Overall, wortmannin binds similarly to p110α and p110γ as the residues in the inhibitor binding pocket are conserved between the two isoforms, and the covalent bond occurs between equivalent lysines (802 is p110α and 833 in p110γ, Figure 6d.) However, the conformational shifts induced by wortmannin binding in the ATP binding pockets of the two isoforms differ significantly. These differences could potentially be exploited in the design of a p110α specific inhibitor targeting the ATP binding site of the enzyme.
Finally, the last twenty residues of p110α and p110γ exhibit large conformational differences [24, 42]. The residues following helix αK11γ form a helix in p110γ (residues 1077–1092, αK12γ in pdb id 1E8X) while in the p110α they are either disordered in the wild type structure or fold as a loop in the H1047R mutant (residues 1047–1062, Figure 5C). These differences might affect enzymatic activity by modifying membrane affinity and provide additional targets for isoform-specific inhibitors.
Acknowledgments
Support was provided by the Virginia and D.K. Ludwig Fund for Center Research and National Institutes of Health Grants CA43460, GM07309, and GM07184.
References
- 1.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 2.Vanhaesebroeck B, Higashi K, Raven C, Welham M, Anderson S, Brennan P, Ward SG, Waterfield MD. Autophosphorylation of p110delta phosphoinositide 3-kinase: a new paradigm for the regulation of lipid kinases in vitro and in vivo. Embo J. 1999;18:1292–1302. doi: 10.1093/emboj/18.5.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marone R, Cmiljanovic V, Giese B, Wymann MP. Targeting phosphoinositide 3-kinase-Moving towards therapy. Biochim Biophys Acta. 2008;1784:159–185. doi: 10.1016/j.bbapap.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 4.Vanhaesebroeck B, Alessi DR. The PI3K-PDK1 connection: more than just a road to PKB. Biochem J. 2000;346(Pt 3):561–576. [PMC free article] [PubMed] [Google Scholar]
- 5.Vanhaesebroeck B, Waterfield MD. Signaling by distinct classes of phosphoinositide 3-kinases. Exp Cell Res. 1999;253:239–254. doi: 10.1006/excr.1999.4701. [DOI] [PubMed] [Google Scholar]
- 6.Katso R, Okkenhaug K, Ahmadi K, White S, Timms J, Waterfield MD. Cellular function of phosphoinositide 3-kinases: implications for development, homeostasis, and cancer. Annu Rev Cell Dev Biol. 2001;17:615–675. doi: 10.1146/annurev.cellbio.17.1.615. [DOI] [PubMed] [Google Scholar]
- 7.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, Willson JK, Markowitz S, Kinzler KW, Vogelstein B, Velculescu VE. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 8.Carson JD, Van Aller G, Lehr R, Sinnamon RH, Kirkpatrick RB, Auger KR, Dhanak D, Copeland RA, Gontarek RR, Tummino PJ, Luo L. Effects of oncogenic p110alpha subunit mutations on the lipid kinase activity of phosphoinositide 3-kinase. Biochem J. 2008;409:519–524. doi: 10.1042/BJ20070681. [DOI] [PubMed] [Google Scholar]
- 9.Gymnopoulos M, Elsliger MA, Vogt PK. Rare cancer-specific mutations in PIK3CA show gain of function. Proc Natl Acad Sci U S A. 2007;104:5569–5574. doi: 10.1073/pnas.0701005104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. http://www.sanger.ac.uk/perl/genetics/CGP/cosmic?action=bygene&ln=PIK3CA&start=1&end=1069&coords=AA:AA.
- 11.Zhao L, Vogt PK. Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc Natl Acad Sci U S A. 2008;105:2652–2657. doi: 10.1073/pnas.0712169105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Catasus L, Gallardo A, Cuatrecasas M, Prat J. PIK3CA mutations in the kinase domain (exon 20) of uterine endometrial adenocarcinomas are associated with adverse prognostic parameters. Mod Pathol. 2008;21:131–139. doi: 10.1038/modpathol.3800992. [DOI] [PubMed] [Google Scholar]
- 13.Kalinsky K, Jacks LM, Heguy A, Patil S, Drobnjak M, Bhanot UK, Hedvat CV, Traina TA, Solit D, Gerald W, Moynahan ME. PIK3CA mutation associates with improved outcome in breast cancer. Clin Cancer Res. 2009;15:5049–5059. doi: 10.1158/1078-0432.CCR-09-0632. [DOI] [PubMed] [Google Scholar]
- 14.Lai Y, Mau B, Cheng W, Chen H, Chiu H, Tzen C. PIK3CA exon 20 mutation is independently associated with a poor prognosis in breast cancer patients. Ann Surg Oncol. 2008;15:1064–1069. doi: 10.1245/s10434-007-9751-7. [DOI] [PubMed] [Google Scholar]
- 15.Lerma E, Catasus L, Gallardo A, Peiro G, Alonso C, Aranda I, Barnadas A, Prat J. Exon 20 PIK3CA mutations decreases survival in aggressive (HER-2 positive) breast carcinomas. Virchows Arch. 2008;453:133–139. doi: 10.1007/s00428-008-0643-4. [DOI] [PubMed] [Google Scholar]
- 16.Marone R, Erhart D, Mert AC, Bohnacker T, Schnell C, Cmiljanovic V, Stauffer F, Garcia-Echeverria C, Giese B, Maira SM, Wymann MP. Targeting melanoma with dual phosphoinositide 3-kinase/mammalian target of rapamycin inhibitors. Mol Cancer Res. 2009;7:601–613. doi: 10.1158/1541-7786.MCR-08-0366. [DOI] [PubMed] [Google Scholar]
- 17.Guillermet-Guibert J, Bjorklof K, Salpekar A, Gonella C, Ramadani F, Bilancio A, Meek S, Smith AJ, Okkenhaug K, Vanhaesebroeck B. The p110beta isoform of phosphoinositide 3-kinase signals downstream of G protein-coupled receptors and is functionally redundant with p110gamma. Proc Natl Acad Sci U S A. 2008;105:8292–8297. doi: 10.1073/pnas.0707761105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Musacchio A, Cantley LC, Harriso SC. Crystal structure of the breakpoint cluster region-homology domain from phosphoinositide 3-kinase p85 alpha subunit. Proc Natl Acad Sci U S A. 1996;93:14373–14378. doi: 10.1073/pnas.93.25.14373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Booker GW, Breeze AL, Downing AK, Panayotou G, Gout I, Waterfield MD, Campbell ID. Structure of an SH2 domain of the p85 alpha subunit of phosphatidylinositol-3-OH kinase. Nature. 1992;358:684–687. doi: 10.1038/358684a0. [DOI] [PubMed] [Google Scholar]
- 20.Nolte R, Eck M, Schlessinger J, Shoelson S, Harrison S. Crystal structure of the PI 3-kinase p85 amino-terminal SH2 domain and its phosphopeptide complexes. Nat Struct Biol. 1996;3:364–374. doi: 10.1038/nsb0496-364. [DOI] [PubMed] [Google Scholar]
- 21.Pauptit RA, Dennis CA, Derbyshire DJ, Breeze AL, Weston SA, Rowsell S, Murshudov GN. NMR trial models: experiences with the colicin immunity protein Im7 and the p85alpha C-terminal SH2-peptide complex. Acta Crystallogr D Biol Crystallogr. 2001;57:1397–1404. doi: 10.1107/s0907444901012434. [DOI] [PubMed] [Google Scholar]
- 22.Walker EH, Perisic O, Ried C, Stephens L, Williams RL. Structural insights into phosphoinositide 3-kinase catalysis and signalling. Nature. 1999;402:313–320. doi: 10.1038/46319. [DOI] [PubMed] [Google Scholar]
- 23.Huang CH, Mandelker D, Schmidt-Kittler O, Samuels Y, Velculescu VE, Kinzler KW, Vogelstein B, Gabelli SB, Amzel LM. The structure of a human p110alpha/p85alpha complex elucidates the effects of oncogenic PI3Kalpha mutations. Science. 2007;318:1744–1748. doi: 10.1126/science.1150799. [DOI] [PubMed] [Google Scholar]
- 24.Mandelker D, Gabelli S, Schmidt-Kittler O, Zhu J, Huang C, Kinsler K, Vogelstein B, Amzel L. A frequent kinase domain mutation that changes the interaction between PI3Ka and the membrane. PNAS X. 2009 doi: 10.1073/pnas.0908444106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schindler T, Bornmann W, Pellicena P, Miller WT, Clarkson B, Kuriyan J. Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science. 2000;289:1938–1942. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- 26.Schindler T, Sicheri F, Pico A, Gazit A, Levitzki A, Kuriyan J. Crystal structure of Hck in complex with a Src family-selective tyrosine kinase inhibitor. Mol Cell. 1999;3:639–648. doi: 10.1016/s1097-2765(00)80357-3. [DOI] [PubMed] [Google Scholar]
- 27.Knight ZA, Gonzalez B, Feldman ME, Zunder ER, Goldenberg DD, Williams O, Loewith R, Stokoe D, Balla A, Toth B, Balla T, Weiss WA, Williams RL, Shokat KM. A pharmacological map of the PI3-K family defines a role for p110alpha in insulin signaling. Cell. 2006;125:733–747. doi: 10.1016/j.cell.2006.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maira SM, Stauffer F, Brueggen J, Furet P, Schnell C, Fritsch C, Brachmann S, Chene P, De Pover A, Schoemaker K, Fabbro D, Gabriel D, Simonen M, Murphy L, Finan P, Sellers W, Garcia-Echeverria C. Identification and characterization of NVP-BEZ235, a new orally available dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor with potent in vivo antitumor activity. Mol Cancer Ther. 2008;7:1851–1863. doi: 10.1158/1535-7163.MCT-08-0017. [DOI] [PubMed] [Google Scholar]
- 29.Pirola L, Zvelebil MJ, Bulgarelli-Leva G, Van Obberghen E, Waterfield MD, Wymann MP. Activation loop sequences confer substrate specificity to phosphoinositide 3-kinase alpha (PI3Kalpha). Functions of lipid kinase-deficient PI3Kalpha in signaling. J Biol Chem. 2001;276:21544–21554. doi: 10.1074/jbc.M011330200. [DOI] [PubMed] [Google Scholar]
- 30.Miled N, Yan Y, Hon WC, Perisic O, Zvelebil M, Inbar Y, Schneidman-Duhovny D, Wolfson HJ, Backer JM, Williams RL. Mechanism of two classes of cancer mutations in the phosphoinositide 3-kinase catalytic subunit. Science. 2007;317:239–242. doi: 10.1126/science.1135394. [DOI] [PubMed] [Google Scholar]
- 31.Mankoo PK, Sukumar S, Karchin R. PIK3CA somatic mutations in breast cancer: Mechanistic insights from Langevin dynamics simulations. Proteins. 2009;75:499–508. doi: 10.1002/prot.22265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams R, Berndt A, Miller S, Hon WC, Zhang X. Form and flexibility in phosphoinositide 3-kinases. Biochem Soc Trans. 2009;37:615–626. doi: 10.1042/BST0370615. [DOI] [PubMed] [Google Scholar]
- 33.Cancer GAR. Network, Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA, Hartigan J, Jr, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Norman BH, Shih C, Toth JE, Ray JE, Dodge JA, Johnson DW, Rutherford PG, Schultz RM, Worzalla JF, Vlahos CJ. Studies on the mechanism of phosphatidylinositol 3-kinase inhibition by wortmannin and related analogs. J Med Chem. 1996;39:1106–1111. doi: 10.1021/jm950619p. [DOI] [PubMed] [Google Scholar]
- 36.Wipf P, Minion DJ, Halter RJ, Berggren MI, Ho CB, Chiang GG, Kirkpatrick L, Abraham R, Powis G. Synthesis and biological evaluation of synthetic viridins derived from C(20)-heteroalkylation of the steroidal PI-3-kinase inhibitor wortmannin. Org Biomol Chem. 2004;2:1911–1920. doi: 10.1039/b405431h. [DOI] [PubMed] [Google Scholar]
- 37.Vlahos CJ, Matter WF, Hui KY, Brown RF. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002) J Biol Chem. 1994;269:5241–5248. [PubMed] [Google Scholar]
- 38.Ihle NT, Williams R, Chow S, Chew W, Berggren MI, Paine-Murrieta G, Minion DJ, Halter RJ, Wipf P, Abraham R, Kirkpatrick L, Powis G. Molecular pharmacology and antitumor activity of PX-866, a novel inhibitor of phosphoinositide-3-kinase signaling. Mol Cancer Ther. 2004;3:763–772. [PubMed] [Google Scholar]
- 39.Garlich JR, De P, Dey N, Su JD, Peng X, Miller A, Murali R, Lu Y, Mills GB, Kundra V, Shu HK, Peng Q, Durden DL. A vascular targeted pan phosphoinositide 3-kinase inhibitor prodrug, SF1126, with antitumor and antiangiogenic activity. Cancer Res. 2008;68:206–215. doi: 10.1158/0008-5472.CAN-07-0669. [DOI] [PubMed] [Google Scholar]
- 40.Camps M, Ruckle T, Ji H, Ardissone V, Rintelen F, Shaw J, Ferrandi C, Chabert C, Gillieron C, Francon B, Martin T, Gretener D, Perrin D, Leroy D, Vitte PA, Hirsch E, Wymann MP, Cirillo R, Schwarz MK, Rommel C. Blockade of PI3Kgamma suppresses joint inflammation and damage in mouse models of rheumatoid arthritis. Nat Med. 2005;11:936–943. doi: 10.1038/nm1284. [DOI] [PubMed] [Google Scholar]
- 41.Walker EH, Pacold ME, Perisic O, Stephens L, Hawkins PT, Wymann MP, Williams RL. Structural determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin, and staurosporine. Mol Cell. 2000;6:909–919. doi: 10.1016/s1097-2765(05)00089-4. [DOI] [PubMed] [Google Scholar]
- 42.Amzel LM, Huang CH, Mandelker D, Lengauer C, Gabelli SB, Vogelstein B. Structural comparisons of class I phosphoinositide 3-kinases. Nat Rev Cancer. 2008;8:665–669. doi: 10.1038/nrc2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Booker GW, Gout I, Downing AK, Driscoll PC, Boyd J, Waterfield MD, Campbell ID. Solution structure and ligand-binding site of the SH3 domain of the p85 alpha subunit of phosphatidylinositol 3-kinase. Cell. 1993;73:813–822. doi: 10.1016/0092-8674(93)90259-s. [DOI] [PubMed] [Google Scholar]
- 44.Gunther UL, Weyrauch B, Zhang X, Schaffhausen B. Nuclear magnetic resonance structure of the P395S mutant of the N-SH2 domain of the p85 subunit of PI3 kinase: an SH2 domain with altered specificity. Biochemistry. 2003;42:11120–11127. doi: 10.1021/bi034353x. [DOI] [PubMed] [Google Scholar]
- 45.Hoedemaeker FJ, Siegal G, Roe SM, Driscoll PC, Abrahams JP. Crystal structure of the C-terminal SH2 domain of the p85alpha regulatory subunit of phosphoinositide 3-kinase: an SH2 domain mimicking its own substrate. J Mol Biol. 1999;292:763–770. doi: 10.1006/jmbi.1999.3111. [DOI] [PubMed] [Google Scholar]
- 46.Siegal G, Davis B, Kristensen SM, Sankar A, Linacre J, Stein RC, Panayotou G, Waterfield MD, Driscoll PC. Solution structure of the C-terminal SH2 domain of the p85 alpha regulatory subunit of phosphoinositide 3-kinase. J Mol Biol. 1998;276:461–478. doi: 10.1006/jmbi.1997.1562. [DOI] [PubMed] [Google Scholar]
- 47.Breeze AL, Kara BV, Barratt DG, Anderson M, Smith JC, Luke RW, Best JR, Cartlidge SA. Structure of a specific peptide complex of the carboxy-terminal SH2 domain from the p85 alpha subunit of phosphatidylinositol 3-kinase. EMBO J. 1996;15:3579–3589. [PMC free article] [PubMed] [Google Scholar]
- 48.Apsel B, Blair JA, Gonzalez B, Nazif TM, Feldman ME, Aizenstezn B, Hoffman R, Williams RL, Shokat KM, Knight ZA. Targeted polypharmacology: discovery of dual inhibitors of tyrosine and phosphoinositide kinases. Nat Chem Biol. 2008;4:691–699. doi: 10.1038/nchembio.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Folkes AJ, Ahmadi K, Alderton WK, Alix S, Baker SJ, Box G, Chuckowree IS, Clarke PA, Depledge P, Eccles SA, Friedman LS, Hayes A, Hancox TC, Kugendradas A, Lensun L, Moore P, Olivero AG, Pang J, Patel S, Pergl-Wilson GH, Raynaud FI, Robson A, Saghir N, Salphati L, Sohal S, Ultsch MH, Valenti M, Wallweber HJ, Wan NC, Wiesmann C, Workman P, Zhyvoloup A, Zvelebil MJ, Shuttleworth SJ. The identification of 2-(1H-indazol-4-yl)-6-(4-methanesulfonyl-piperazin-1-ylmethyl)-4-morpholin -4-yl-thieno[3,2-d]pyrimidine (GDC-0941) as a potent, selective, orally bioavailable inhibitor of class I PI3 kinase for the treatment of cancer. J Med Chem. 2008;51:5522–5532. doi: 10.1021/jm800295d. [DOI] [PubMed] [Google Scholar]
- 50.Pacold ME, Suire S, Perisic O, Lara-Gonzalez S, Davis CT, Walker EH, Hawkins PT, Stephens L, Eccleston JF, Williams RL. Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase gamma. Cell. 2000;103:931–943. doi: 10.1016/s0092-8674(00)00196-3. [DOI] [PubMed] [Google Scholar]
- 51.Perry B, Alexander R, Bennett G, Buckley G, Ceska T, Crabbe T, Dale V, Gowers L, Horsley H, James L, Jenkins K, Crepy K, Kulisa C, Lightfoot H, Lock C, Mack S, Morgan T, Nicolas AL, Pitt W, Sabin V, Wright S. Achieving multi-isoform PI3K inhibition in a series of substituted 3,4-dihydro-2H-benzo[1,4]oxazines. Bioorg Med Chem Lett. 2008;18:4700–4704. doi: 10.1016/j.bmcl.2008.06.104. [DOI] [PubMed] [Google Scholar]
- 52.Xie P, Williams DS, Atilla-Gokcumen GE, Milk L, Xiao M, Smalley KS, Herlyn M, Meggers E, Marmorstein R. Structure-based design of an organoruthenium phosphatidyl-inositol-3-kinase inhibitor reveals a switch governing lipid kinase potency and selectivity. ACS Chem Biol. 2008;3:305–316. doi: 10.1021/cb800039y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zask A, Verheijen JC, Curran K, Kaplan J, Richard DJ, Nowak P, Malwitz DJ, Brooijmans N, Bard J, Svenson K, Lucas J, Toral-Barza L, Zhang WG, Hollander I, Gibbons JJ, Abraham RT, Ayral-Kaloustian S, Mansour TS, Yu K. ATP-Competitive Inhibitors of the Mammalian Target of Rapamycin: Design and Synthesis of Highly Potent and Selective Pyrazolopyrimidines. J Med Chem. 2009 doi: 10.1021/jm900851f. [DOI] [PubMed] [Google Scholar]