Abstract
Mammalian target of rapamycin (mTOR) is a protein kinase involved in translation control and long-lasting synaptic plasticity. mTOR functions as the central component of two multi-protein signaling complexes, mTORC1 and mTORC2, which can be distinguished from each other based on their unique compositions and substrates. Although majority of evidence linking mTOR function to synaptic plasticity comes from studies utilizing rapamycin, studies in genetically-modified mice also suggest that mTOR couples receptors to the translation machinery for establishing long-lasting synaptic changes that are the basis for higher order brain function, including long-term memory. Finally, perturbation of the mTOR signaling cascade appears to be a common pathophysiological feature of human neurological disorders, including mental retardation syndromes and autism spectrum disorders.
Synaptic plasticity
Memory is ‘stored’ via the carefully regulated interaction of neuronal networks of the nervous system. The synapse is the essential cellular unit of memory and is a site of electrochemical communication between neurons; these connections are ‘plastic’. In other words, the physiological responsiveness, i.e., the ‘strength’ of the synaptic connection is modifiable. A more detailed review of the mechanisms underlying synaptic plasticity can be found elsewhere [1]. Importantly, synaptic plasticity is also defined temporally, with some alterations lasting only seconds while others persist over the lifetime of the organism [2, 3]. In vertebrates, long-term change in synaptic strength is often measured as long-term potentiation (LTP) and long-term depression (LTD) [1, 4, 5]. In invertebrates such as Aplysia, long-term facilitation (LTF) is used as a measure of synaptic plasticity [3, 6].
The more durable forms of synaptic plasticity are conveyed biochemically via the expression of new proteins, both somatically and dendritically [3, 7]. The important role of protein synthesis in synaptic plasticity has been demonstrated in numerous experimental systems using a variety of pharmacological and genetic approaches [8]. Protein synthesis or translation is a highly regulated process that can be separated into three general phases: initiation, elongation, and termination. The majority of known translational regulation occurs at the level of translation initiation. The coordinated activities of numerous initiation factors are required for this process (for reviews see [9] and [5]). Central to the regulation of translation initiation is the activity of a ubiquitously expressed kinase, mammalian target of rapamycin (mTOR). In this review we provide an overview of what is known about mTOR function, its biochemical interactions, and its regulation during protein synthesis-dependent forms of synaptic plasticity and memory. We end the review with a discussion of the potential role of the mTOR signaling cascade in neurological disease and disorders.
mTOR: Central regulator of translational initiation
mTOR function is influenced by the activities of neuronal surface receptors and channels including N-methyl-D-aspartate receptors (NMDA-R), α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors (AMPA-R), brain-dervived neurotropic factor (BDNF), and dopaminergic and metabotropic glutamate receptors (mGluRs), which are vital for the induction and maintenance of LTP and LTD [10–14]. mTOR acts as a node of convergence downstream of these receptors and several signaling pathways that include phosphoinositide dependent kinase-1 (PDK1), phosphatidylinositol 3-kinase (PI3K), Akt, and tuberous scelerous complex proteins 1 and 2 (Tsc1/2) [5, 15–18].
mTOR structure
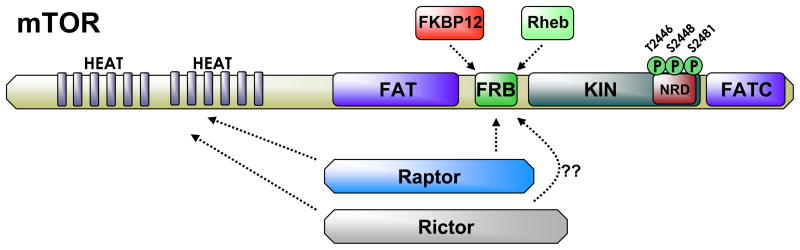
mTOR is a large (2549 amino acid, ~250 kd), ubiquitously expressed multi-effector serine/threonine kinase that is a highly conserved homolog of the yeast protein, target of rapamycin (TOR). Rapamycin (or sirolimus) is a macrolide derived from soil bacterium [19]. The C-terminal end of mTOR contains several important elements, including the kinase catalytic domain (KIN) that is structurally similar to the catalytic site in phosphatidylinositol 3-kinases (PI3Ks), but does not encode lipid kinase activity (Figure 1). The KIN domain also contains a small region that is likely a site of phospho-regulation called the negative regulatory domain (NRD) or “repressor domain” [15, 20]. This domain contains phosphorylation sites conserved in kinases with similar structure. Within this region, phosphorylation at threonine 2446, serine 2448, and serine 2481 are correlated with overall higher levels of mTOR activity. Of particular note is serine 2448, which is a target of Akt as well as p70 S6 kinase (S6K) [21–24]. Some of these residues are autophosphorylated even in the presence of rapamycin, whereas others are substrates of the downstream effectors of mTOR itself, thereby providing multiple mechanisms for feedback regulation [15, 20]. Finally, adjacent to the KIN domain is the FKBP12-rapamycin binding domain (FRB), the site of inhibitory interaction between rapamycin and mTOR. Rapamycin bound to FK506 binding protein 12 (FKBP12, discussed later) disrupts protein-protein interactions that are key to mTOR function.
Figure 1. mTOR structure.
Mammalian target of rapamycin (mTOR) is large multi-domain protein. The N-terminal portion of mTOR contains more than 20 Huntingtin, Elongation Factor 3, A subunit of PP2A, TOR1 (HEAT) repeats. These HEAT repeats form a large helical secondary structure that provides protein interaction activity (dashed lines) with mTOR complex members such as Regulatory-associated protein with TOR (Raptor) and Rapamycin-insensitive companion of TOR (Rictor). The C-terminal portion of mTOR contains several important domains. The first is the FRAP (FKBP12-rapamycin-associated protein)/TOR), ATM, (ataxia-telangiectasia), TRRAP (transactivation/transformation domain-associated protein) (FAT) domain. The FAT domain is a conserved domain among PIKK family members. A second FAT domain (FATC) is located at the distal C-terminal end of mTOR. Both FAT domains are necessary for mTOR catalytic function. Adjacent to the FAT domain is the FKBP12-rapamycin binding (FRB) domain. This domain is bound by the FKBP12-rapamycin complex and is a site of interaction between mTOR and FKBP family members bound to rapamycin. The FRB also is involved in the interaction between mTOR and other mTORC members including (Raptor) and Ras-homolog enriched in the brain (Rheb). The catalytic or kinase (KIN) domain is flanked by the FAT domains and encodes the serine/threonine kinase activity of mTOR. Within the KIN domain is a region that is sometimes referred to as the negative regulatory domain (NRD), which contains serine and threonine residues that are phosphorylated and are involved in the regulation of mTOR activity. Threonine 2246 is targeted by AMPK and S6K, serine 2448 is a target of Akt and S6K, and serine 2481 is autocatalytic target of mTOR.
mTOR complexes: mTORC1 and mTORC2
mTOR controls cell growth and translation via the assembly of multi-protein signaling complexes [25]. Rapamycin does not directly impair mTOR catalytic activity per se, but instead disrupts mTOR-protein complex formation, thereby blocking mTOR signaling [26, 27]. mTOR complexes (mTORCs) consist of numerous proteins that control mTOR signaling, dictate subcellular localization, and regulate substrate specificity. mTORCs share some common constituents such as the G beta-like protein family member, mLST8, but can be largely distinguished by their unique components [20, 27, 28].
mTOR bound to the scaffolding protein, regulatory associated protein of mTOR (Raptor), is called mTOR complex 1 (mTORC1). Raptor acts as a scaffolding companion to mTOR, binding TOR signaling (TOS) motif-containing proteins and shuttling them to the mTOR catalytic domain [15, 27]. mTORC1 is sensitive to rapamycin via competition between Raptor and FKBP12-rapamycin for binding to the FRB domain [27]. The FRB also is a site of positive regulation, albeit through indirect means. Ras-homolog enriched in the brain (Rheb) acts to stimulate mTORC1 function, but does so by blocking the inhibitory binding of FKBP38 to mTORC1 at the FRB. FKBP38 is closely related to FKBP12, the intracellular rapamycin receptor. FKBP38 is formed from an FKBP-C domain (a minimal consensus FKBP12 sequence) and a pair of protein-protein interaction domains. FKBP38 is bound by Rheb-GTP, likely freeing up the mTORC1 FRB for Raptor. Deletion analysis of FKBP38 showed that its FKBP-C domain, a region highly homologous to FKBP12, was sufficient for mTOR binding [29]. Because, FKPB38 is expressed at extremely low levels in the mouse brain, it has been proposed that FKBP12 is the major repressor of Rheb/mTORC1 activity in the brain [30].
In addition to Raptor, mTORC1 contains other proteins such as proline-rich Akt/PKB substrate 40 kD (PRAS40). PRAS40 regulates mTOR-raptor interactions and negatively regulates mTOR signaling by blocking mTORC1 access to its substrates [31]. However, in another study, PRAS40 was required for substrate activation, suggesting that in some contexts PRAS40 facilitates mTORC1 function. The interaction between mTOR and PRAS40 is disrupted by rapamycin, indicating that FKBP12 may mediate its nascent inhibitory activity [15, 31–33].
A second distinct mTOR complex termed mTORC2 contains the rapamycin-insensitive companion of mTOR (Rictor) [28] and as the name implies, is resistant to rapamycin. Although insensitive to acute rapamycin treatment, mTORC2 is disrupted by prolonged rapamycin exposure [34]. In addition, newly synthesized Rictor is susceptible to rapamycin, suggesting that only preformed mTORC2 is resistant to rapamycin, perhaps through steric occlusion, blocking access to the FRB [20, 34]. mTORC2 contains its own unique protein components, such as SAPK interacting protein 1 (Sin1). Sin1 encodes an essential function because the deletion of Sin1 is embryonically lethal [35]. The role of Sin1 in the brain is not well understood, but it is required for proper mTORC2 formation and activity in cell culture [35, 36]. There are at least five alternatively spliced isoforms of Sin1 in mammals, suggesting the possibility of brain-specific and/or neuronal subtype-specific mTORC2 isoforms [37]. Another recently identified mTORC2 component is protein observed with rictor (Protor). Protor binds Rictor independent of mTOR and does not appear to be required for mTORC2-mediated Akt activation [35, 38–40]. Despite the presence of its mRNA, little is known about the function of Protor in the brain [41]. Although there is nothing known about the role of mTORC2 signaling in synaptic plasticity, given the fact that mTORC2 is known to regulate Akt and PKC in other cell types, it is likely only a matter of time before a role for mTORC2 in plasticity is elucidated.
mTORC substrates
The best-characterized function of mTORC1 is the regulation of translation where it regulates two critical core components of the translation initiation machinery: p70 ribosomal S6 kinase 1 and 2 (S6K1/2) and the eIF4E-binding proteins (4E-BPs) [1]. mTORC1 also regulates the activity of phosphatases such as protein phosphatase 2A (PP2A), and these phosphatases can regulate mTOR substrates, thereby generating mTOR-dependent feedback loops that control initiation rates.
Increased mTORC1 formation is involved in the activation of S6K(1/2) and repression of the 4E-BPs [1, 25] (Figure 2). S6K controls several aspects of protein synthesis. S6K can phosphorylate and regulate the biosynthesis of the ribosomal subunit protein S6 (for which S6K was originally named). S6K also phosphorylates eIF4B, a non-catalytic cofactor of the RNA helicase eIF4A. Increased eIF4A activity is critical for the translation of mRNA substrates with complex 5′ UTR secondary structure. In addition, S6K is involved in regulating translation elongation via phosphorylation of the eukaryotic elongation factor 2 kinase (eEF2) kinase (Figure 2). S6K inhibits eEF2 kinase activity reducing inhibitory phosphorylation of eEF2, thereby increasing elongation rates [42]. Finally, S6K can phosphorylate mTOR itself at Ser 2448, but the function of this phosphorylation is unknown.
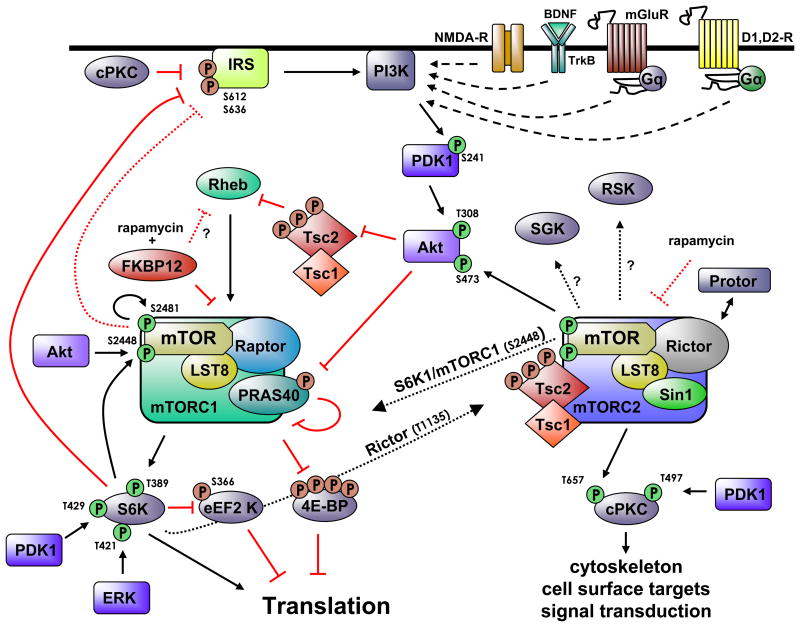
Figure 2. Signaling upstream and downstream of mTORC1 and mTORC2.
Neuronal receptors and channels (NMDAR, Trk-B, mGluR D1R, and D2R) activate downstream signaling pathways leading to mTORC1 activation. The upstream signaling regulating mTORC2 activity in neurons is currently unknown. mTORC1 activity regulates several downstream effectors of translation (S6K, 4E-BP, eEF2K) both somatically and dendritically in neurons. mTORC2 may modulate the activity of mTORC1 either directly (S2448) or indirectly (Akt, cPKC, S6K). mTORC1 substrates (S6K) can phosphorylate Rictor enabling crosstalk between the two TORC complexes. mTORCs regulate several critical neuronal metabolisms including translation, cytoskeletal structure, protein stability and signal transduction. Abbreviations used: Akt/protein kinase B (Akt/PKB), brain-derived neurotrophic factor (BDNF), conventional protein kinase C (cPKC), Dopamine receptor type 1 (D1R), dopamine receptor type 2, (D2R) extracellular signal-regulated protein kinase (ERK), eukaryotic initiation factor 4E-binding protein (4E-BP), eukaryotic elongation factor 2 Kinase (eEF2 K), FK506-binding protein 12 (FKBP12), Gq-protein (Gq), insulin response element (IRS), mammalian lethal with sec 13 (mLST8), metabotropic glutamate receptor (mGluR), mammalian target of rapamycin (mTOR), N-methyl-D-aspartate receptor (NMDAR), phosphoinositide-3 kinase (PI3K), mTOR complex 1 (mTORC1), mTOR complex 2 (mTORC2), phosphoinositide-dependent kinase 1 (PDK1), proline-rich Akt/PKB substrate 40 kD (PRAS40), protein observed with rictor (Protor), Regulatory-associated protein with TOR (Raptor), Ras homolog enriched in brain (Rheb), Rapamycin- insensitive companion of mTOR (Rictor), p90 ribosomal S6K kinase 2 (Rsk2), SAPK (Sin1), p70 S6 Kinase (S6K), serum- and glucorticoid inducible kinase (SGK), tyrosine receptor kinase-B (TrkB) tuberous sclerosis complex 1 and 2 (TSC1/2). T=threonine, S=serine (number denotes residue phosphorylated), Red (P) denotes inhibitory regulation; Green (P) denotes stimulatory regulation.
The other major substrate trafficked to the mTORC1 complex by Raptor is eIF4E-binding protein, (4E-BP). 4E-BPs sequester the 5′ methylated-GTP cap-binding protein eIF4E. mTOR phosphorylation of 4E-BP disassociates it from eIF4E, thereby derepressing its cap-binding activity. eIF4E is a member of the multimeric eIF4F (composed of eIF4E, eIF4G, eIF4A, eIF4B). This complex is vital for translation initiation; it mediates circularization of mRNA (by eIF4G association with poly-A binding proteins) and promotes the unwinding of proximal 5′ UTR mRNA secondary structure that may impair ribosomal access to the start AUG codon. It is believed that eIF4E greatly facilitates this process through binding of the m7-GTP cap of the substrate mRNA. Finally, eIF4G, the primary scaffolding component of eIF4F, is phosphorylated at serine 1108 by mTORC1. The exact effect of this phosphorylation is not clear, but is believed to enhance eIF4F formation [5].
As might be expected given its unique composition, mTORC2 acts on a different set of targets than mTORC1. There is little information about how mTORC2 is regulated during synaptic plasticity and memory. However, it is possible to speculate about mTORC2 targets in the brain from what is known about mTORC2 signaling in response to stress and growth factor stimulation [35, 43, 44]. mTORC2 was recently identified as the elusive “PDK2” (see [45] for description), acting to promote Akt signaling [35]. This is intriguing because Akt lies upstream of mTORC1, raising the possibility that mTORCs may be involved in signaling crosstalk with each other. Direct evidence of mTORC crosstalk has recently been demonstrated, as S6K phosphorylates Rictor at threonine 1135 in a growth factor-dependent and rapamycin-sensitive manner [46]. Another target of mTORC2 is the conventional calcium-dependent protein kinase Cα (PKCα) [25, 28, 35, 43, 47]. PKC is critical for several forms of synaptic plasticity [48]. mTORC2 phosphorylates serine 657 in the hydrophobic motif (HM) domain of PKC. Phosphorylation of this residue is involved in regulating the activity, stability and localization of PKC [44, 49]. Moreover, LTP was shown to be associated with enhanced phosphorylation of PKC at this residue [50]. Given the conservation between AGC kinase family members, it is also plausible that mTORC2 is involved in the regulation of related kinases such as cAMP-dependent protein kinase (PKA), p90 Rsk, SGK, and PDK1 [44].
mTORC substrate specificity is thought to be conveyed by the binding activities of the mTOR scaffolding partner found in the TORC (Raptor or Rictor). Raptor accomplishes this with its Raptor N-terminal conserved (RNC), and WD40 protein-protein interaction domains [15, 51]. Unlike Raptor, the domains of Rictor are not well characterized, so the domains involved in substrate binding are unknown. Another layer of substrate specificity is likely defined by other mTORC binding partners (mLST8, SIN, Protor) whose activities probably control subcellular localization and transport, and thus, access to local substrates [20]. Future plasticity studies using Raptor and Rictor mutant mice with altered substrate binding activities should help to identify signaling components and mRNA substrates with different roles in memory formation.
Although the majority of studies of mTOR have focused on translation, an emerging body of evidence demonstrates that mTOR signaling also is involved in other cellular processes such as transcription, protein degradation, autophagy, and cytoskeletal assembly [15, 42, 52]. Thus, in addition to translation, mTOR also is likely to regulate other processes involved in long-lasting synaptic change.
mTOR and synaptic plasticity
Evidence linking mTOR signaling to synaptic plasticity has largely been derived from studies using rapamycin, which was first used in LTF studies in Aplysia and crayfish [53–56]. Rapamycin also blocked eEF2 phosphorylation during LTF-mediated elongation in Apylsia [57]. These findings highlight that TOR signaling is crucial to multiple phases of long-lasting plastic change in invertebrates.
Rapamycin was first used to conclusively demonstrate the role of mTOR in late phase NMDA-receptor dependent hippocampal LTP (L-LTP) by Tang et al. [16]. These authors also showed that mTOR, translation initiation components (eIF4E), and mTOR substrates (4E-BP), co-localized with post-synaptic markers, strongly suggesting that mTOR inhibition with rapamycin targeted synaptic translation machinery. These findings were extended by Cammalleri et al. by determining the temporal window required for mTORC1 activation to induce long-lasting LTP. PI3K-dependent dendritic activation of mTOR, as determined by S6K phosphorylation at threonine 389, also was demonstrated following several pharmacological stimulation protocols [58]. It was also shown that L-LTP-inducing stimulation in hippocampal slices promoted mTOR signaling sufficient to result in the translation of the elongation factor, eEF1A [59]. This study is of particular interest because it demonstrated that a bona fide 5 terminal oligopyrimidine tract (TOP) containing mRNA, eEF1A, was dendritically induced during L-LTP, suggesting that the translation machinery itself is rate limiting. This finding suggests that in the process of establishing protein synthesis-dependent plasticity, mTOR regulates the availability of factors involved in the translational apparatus.
mTOR is also important to the establishment of another form of protein synthesis-dependent synaptic plasticity in vertebrates, mGluR-dependent-LTD [11], which is altered in mice that model fragile X syndrome (FXS) [60]. mTORC1 signaling is activated following 3,5 dihydroxyphenylglycine (DHPG), a mGluR1/5 agonist that induces mGluR-LTD [13, 61]. Furthermore, pharmacological blockade of PI3K, which is upstream of mTORC1, attenuated DHPG-induced LTD [62]. mGluR1/5 stimulation also results in the phosphorylation of the S6K target, S6 and the synthesis of eEF1A [61]. Moreover, rapamycin also blocks mGluR-dependent phosphorylation of eIF4E and 4E-BP [63]. Finally, activation of mGluR1/5 can also cause hippocampal depotentiation, which also is mTOR-dependent [64].
An interesting question is how the activation of NMDA-Rs and mGluRs can both promote mTOR signaling, but generate LTP and LTD, respectively, which are completely different physiological outcomes. This specificity must be transduced via the activation of specific pools of mTOR localized to highly organized receptor-signaling regions at the plasma membrane. The synaptic response to stimulation could also be determined via the specific trafficking of mRNA substrates to the different neuronal compartments, thereby shaping the response of mTOR-dependent protein apparatus to available transcripts.
Genetic evidence for mTOR signaling in plasticity and memory
The importance of mTOR signaling in memory was first revealed with rapacmycin studies, but more specific of targeting of the mTOR signaling cascade has been achieved with recent addition of genetic reagents [54, 65, 66]. Unfortunately, no genetic knockout for mTOR, in either invertebrate or vertebrate systems, has been generated and used for either synaptic plasticity or memory studies. In fact, deletion of several mTORC elements including mTOR, Raptor, Rictor and mLST8 is developmentally lethal [67]. Perhaps this is not surprising given the overarching importance of mTOR in developmental and post-developmental cellular processes. Although an mTOR mutant would be ideal to confirm the role mTOR in synaptic plasticity, numerous mutant and transgenic lines for gene products both upstream and downstream of mTOR have been developed and utilized in studies that highlight the importance of mTOR in the formation of long-lasting synaptic change.
Although mutant mouse lines for PDK1, PI3K, and Akt are available, synaptic plasticity studies utilizing these mice have not yet been conducted extensively [68–72]. Tsc1/2 is the farthest upstream effector of mTOR that has been genetically modified in mice and used in plasticity studies. Tsc1 (+/−) mice demonstrate a host of hippocampus-dependent learning and memory deficits, as well as impairments in social behavior [73]. Tsc2 (+/−) rats display several synaptic phenotypes, including enhanced paired pulse facilitation, which suggests a role for mTOR in short-lasting plasticity independent of translation [74]. Tsc2 (+/−) mice display L-LTP following a single train of HFS that would normally only induce early phase LTP (E-LTP), a transient form of LTP [75]. Moreover, rapamycin treatment was capable of rescuing both behavioral and electrophysiological phenotypes displayed by Tsc2 (+/−) mice, consistent with the notion that Tsc removal results in either the general upregulation of mTORC1 signaling or a reduced threshold for mTORC1 activation. Interestingly, this type of L-LTP threshold reduction has been observed in mutations of other molecules that should increase rates of protein synthesis [12, 76]. Recently, it was reported that TSC1/2 binds mTORC2, and unlike mTORC1, promotes mTORC2 activity [77]. This raises the interesting prospect of mTORC2 and mTORC1 having opposing functions with respect to the regulation of downstream effectors and possibly synaptic plasticity.
The importance of rapamycin for mTOR-related studies is beyond question, but in many ways its use does not provide for a complete understanding of its binding partner, FKBP12. Does this molecule simply act as a receptor for rapamycin, or might it serve to regulate mTORC1 either on its own or with an unknown endogenous inhibitory partner? Indeed, rapamycin is capable of binding mTOR on its own albeit with reduced efficiency [78, 79]. Furthermore, it was demonstrated that FKBP38 was capable of inhibiting mTORC1 in the absence of rapamycin [29]. This makes the idea that FKBP12 has a role in mTORC1 regulation, independent of rapamycin, particularly intriguing. A recent study addressed this question by examining the role of FKBP12 in regulating mTORC1 in the brain. Like mTOR, complete FKBP12 removal, is developmentally lethal [80], so FKBP12 conditional knockout (cKO) mice were generated [30, 81]. These mice displayed enhanced perseverative behaviors and enhanced L-LTP [30]. Moreover, removal of neuronal FKBP12 resulted in enhanced mTORC1 formation and increased phosphorylation of some mTORC1 targets (S6K1) but left others unchanged (4E-BP) [30]. Thus, FKBP12 appears to repress mTORC1 activity (similar to TSC) either as a direct inhibitor or by acting as the intracellular adaptor/receptor for a yet unknown endogenous inhibitor(s) that regulates mTOR signaling. In contrast to Tsc2 (+/−) mice [75], the threshold for induction of L-LTP was not changed in FKBP12 cKO mice; instead the potentiation was exaggerated. This could be because FKBP12 regulation of mTORC1 occurs more acutely than TSC. Alternatively, TSC may have regulatory roles affecting mTORC2 function [20]. The recent availability of non-FKBP12 mediated inhibitors of mTORC1 activity will allow further elucidation of mTORC1 regulation following disruption of FKBP12 [82].
Mouse mutants also are available for the two most prominent mTOR substrates, S6K and 4E-BP [12, 83, 84] and have been used to study both synaptic plasticity and behavior. The S6K1 KO mice display memory impairments and impaired E-LTP but, surprisingly, display normal L-LTP [85]. Like S6K1 KO mice, S6K2 mutants displayed memory impairment but exhibited normal expression of both forms of LTP [85]. S6K1 KO mice have normal mGluR-dependent LTD, whereas S6K2 KO mice have enhanced LTD. The enhanced LTD observed in S6K2 KO mice is resistant to protein synthesis inhibition [61]. Given the important role of S6K at several points in translation initiation (Figure 2), a rational explanation of the synaptic plasticity findings is challenging. This may be a result of either a functional overlap between the S6Ks or that other related kinases can compensate for the loss S6K activity. Regardless, because S6K KO mice display memory impairments, S6K function is required for proper memory formation, a process undoubtedly related to synaptic plasticity.
Another key target of mTOR signaling is 4E-BP. There are three 4E-BP genes, 4E-BP1, 4E-BP2 and 4E-BP3, with 4E-BP2 being the most highly expressed in the hippocampus [12]. Similar to S6K KO mice, 4E-BP2-deficient mice display multiple behavioral abnormalities, but unlike S6K KO mice, the abnormalities displayed by the 4E-BP2 KO mice are more pronounced and include severe spatial learning and memory impairments [12, 86]. 4E-BP2 KO mice display a reduced threshold for the induction of protein synthesis-dependent L-LTP, similar to what has been observed in Tsc2 (+/−) mice. Surprisingly, 4E-BP2-deficient mice fail to exhibit L-LTP [12]. Although direct measurements of mTOR signaling and protein synthesis rates have not been examined, supporting biochemical data suggest that protein synthesis rates, which enhance responses to weak stimuli, are increased in the 4E-BP2 KO mice. Finally, 4E-BP2 KO mice exhibit enhanced mGluR-dependent LTD. Although this enhancement was sensitive to ERK inhibition, it was insensitive to rapamycin, suggesting that 4E-BP2 removal supercedes mTORC1 blockade [63].
mTOR related diseases and disorders
Although most of the experimental results described thus far approach the essential question of mTOR signaling from the perspective of protein synthesis inhibition (either direct inhibition of mTORC1 with rapamycin or the genetic ablation of signaling that promotes mTOR activity), strategies have been employed to disrupt signaling mechanisms that act normally to inhibit mTOR signaling or removed repressors inactivated by mTOR signaling (Tsc, FKBP12, 4E-BP2 mutant mice). In most cases, excessive protein synthesis results in significant plasticity and behavioral deficits, rather than ‘improved’ plasticity. This suggests that mTOR does not simply act as a translational ‘on/off’ switch promoting generalized beneficial protein synthesis, but more likely acts as a valve carefully modulating translational rates during a distinct temporal window. Failure to properly curtail mTOR signaling once activated may be just as detrimental as blocking it. Indeed excessive mTOR signaling has long been known to play a role in human cancers [87]. This extensive body of work combined with what is known about excessive mTOR signaling in the brain highlight a potential link between mTOR, abnormal translation, and human neurological disorders.
Neurofibromatosis type 1 (NF1) is a familial cancer syndrome characterized by the formation of neurofibromas and other nerve tumors. Additionally, NF1 patients are often afflicted with cognitive deficits. In NF1, a mutation disrupts neurofibromin, a regulator of Ras signaling. Enhanced Ras can upregulate the PI3K signaling pathway activating mTOR (Figure 2) and is in fact a feature of the disease pathology [87]. Tuberous sclerosis complex (TSC) is defined clinically by the appearance and growth of benign hamartomas throughout the body and brain. Though the severity of is variable, a large number of TSC patients suffer from epilepsy and mental retardation [88]. Other neurological diseases involving benign tumor growth such as Lhermitte-Duclos disease, Cowden syndrome, and von Hippel-Lindau disease may also involve aberrant mTOR signaling [87, 89].
Autism spectrum disorders (ASDs) are a collection of clinically related human neurological disorders categorized by varying degrees of cognitive impairment that can be divided into three general categories: impaired social interaction, impaired communication and language ability, and the exhibition of perseverative and ritualistic, repetitive behaviors [90–92]. The evidence for ASDs being largely a family of inherited genetic disorders is strong [93]. Unfortunately, identifying the genetic causes of ASDs has proven to be elusive, as they are believed to be polygenic (>3 genes sources) [91, 94]. However, some monogenetic sources of ASD have been identified [93] and although these single gene sources account for only 8% to 15% of all ASDs, more than half are involved in direct regulation of either mTOR signaling or translation control [95]. Phosphatase tensin homolog on chromosome ten (PTEN) is a phosphatase involved in the inhibition of the PI3K-Akt-mTOR signal cascade, and disruptions in PTEN are found at rates of 1–2% in ASD patients [96, 97]. Fully 25–50% of the aforementioned TSC patients with cognitive deficits satisfy the clinical diagnosis for ASD [98]. Finally, fragile X syndrome (FXS), caused by the transcriptional silencing of the FMR1 gene, is the leading genetic cause of autism (2%–8%) [99, 100]. Fragile X mental retardation protein (FMRP) is a RNA-binding protein capable of binding several hundred different mRNAs [101]. FMRP likely acts to sequester mRNA, preventing or limiting the translation of messages bound to it. The most commonly used FXS model mouse, the Fmr1 KO, displays several behavioral deficits similar to those observed in human patients [102]. Importantly, these mice exhibit enhanced mGluR-dependent LTD [11]. The link between FMRP, mTOR and translation is unclear, but it has been shown that the mTORC1 substrate S6K can phosphorylate and regulate the mRNA-binding activity of FMRP [103]. Moreover, exciting new evidence indicates that mTORC1 function is enhanced in Fmr1 KO mice [104]. This finding may explain why the enhanced mGluR-LTD in Fmr1 KO mice is resistant to protein synthesis inhibitors [14, 105]. Combined this information suggests that mTOR signaling dysregulation may be a common biochemical feature of many ASDs.
Alzheimer’s disease (AD) is a progressive neurodegenerative disorder characterized by gradual and severe loss of memory, reasoning, and ultimately basic neurological function. Pathologically, the disease is characterized by the accumulation of beta-amyloid containing plaques, neurofibrillary tangles, and the loss of cortical neurons. mTOR signaling deregulation is a feature of AD, as PTEN, Akt, S6K and mTOR have been all been shown to be dysregulated in post-mortem samples obtained from the brains of AD patients [87, 106, 107]. Finally, a growing body of evidence suggests mTOR plays a critical role in autophagy following ischemic insult and in neurodegenerative diseases such as AD and Huntington’s disease (HD). Indeed, rapamycin was demonstrated to promote clearance of huntingtin aggregates in mouse models of HD [108, 109].
Conclusions and future directions
Although there is abundant evidence linking mTOR signaling to synaptic change, memory and neurological disease, significant gaps in our knowledge remain. Perhaps the most important role of mTOR is as a signal integrator, shaping the neuronal response from the myriad of activation/inhibition signals generated by synaptic activity. Most work investigating mTOR signaling in synaptic plasticity have been performed in either hippocampal slices or cell cultures. There is much less known about mTOR function in other brain regions and how mTOR is regulated in different classes of neurons, such as inhibitory interneurons, and in glia. Importantly, neuronal mTOR function has largely been studied only in the context of translation.
There is a paucity of knowledge with respect to mTORC2 in synaptic plasticity. All key components of mTORC2 are present in the brain. Recent data suggest that mTORC2 might interact with mTORC1 and that it may be regulated in a fashion opposite to mTORC1 [77]. What is the nature of the mTORC1 and mTORC2 relationship with respect to plasticity? Is mTORC2 involved in a separate class of synaptic plasticity-induced change involving either cytoskeletal rearrangement or in the modulation of other signaling pathways? Moreover, it was only very recently that several mTORC complex members were identified. Are there other components of the complex yet to be identified? Clearly, the composition of mTORCs is dynamic, which suggests that complex formation influences subcellular localization, target specificity, and duration of signaling activity. Although some mTORC substrates have been well studied, it is clear that this signaling complex targets numerous molecules in the neuron, so better knowledge of these targets will improve our understanding of synaptic plasticity and memory.
Nearly all of the genetic analyses of mTOR signaling in synaptic plasticity and behavior have been performed in global knockout mice. The availability of sophisticated genetic driver tools such as dox on/off, improved brain region-specific drivers, and optico-genetic switches should permit much better spatio-temporal resolution of mTOR signaling. Pharmacological manipulations have relied almost entirely on the use of rapamycin to block mTORC1. Although rapamycin is fairly specific and potent [110, 111], no drug is perfect. Continued mTOR studies would benefit from the use of drugs such as CCI-779 [82] or drugs that directly block mTOR catalytic activity rather than disrupt mTORC1 [112, 113]. This would allow for both mTORC1 and mTORC2 to be blocked simultaneously to more completely assess the role of mTOR in synaptic plasticity.
Finally, what is the identity of the mRNAs translated in response to mTORC1 activation? Does mTORC1 activation induce the expression of a specific subset of ‘plasticity’ molecules via translation initiation and/or translation elongation? These are all important issues that remain to be addressed. Given the requirement for protein synthesis in long-term memory formation, and the increasingly important role of misregulated protein synthesis in human autism and mental retardation disorders (ASD, FXS), expanding our knowledge of how mTOR functions in the brain is key to understanding human learning and cognition, as well as for developing new therapies and treatments related to neurological disorders with mTOR dysfunction.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Klann E, Dever TE. Biochemical mechanisms for translational regulation in synaptic plasticity. Nat Rev Neurosci. 2004;5(12):931–42. doi: 10.1038/nrn1557. [DOI] [PubMed] [Google Scholar]
- 2.Dudai Y. The neurobiology of consolidations, or, how stable is the engram? Annu Rev Psychol. 2004;55:51–86. doi: 10.1146/annurev.psych.55.090902.142050. [DOI] [PubMed] [Google Scholar]
- 3.Kandel ER. The molecular biology of memory storage: a dialog between genes and synapses. Biosci Rep. 2001;21(5):565–611. doi: 10.1023/a:1014775008533. [DOI] [PubMed] [Google Scholar]
- 4.Bliss TV, Collingridge GL. A synaptic model of memory: long-term potentiation in the hippocampus. Nature. 1993;361(6407):31–9. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- 5.Costa-Mattioli M, et al. Translational control of long-lasting synaptic plasticity and memory. Neuron. 2009;61(1):10–26. doi: 10.1016/j.neuron.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malenka RC, Bear MF. LTP and LTD: an embarrassment of riches. Neuron. 2004;44(1):5–21. doi: 10.1016/j.neuron.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 7.Lynch MA. Long-term potentiation and memory. Physiol Rev. 2004;84(1):87–136. doi: 10.1152/physrev.00014.2003. [DOI] [PubMed] [Google Scholar]
- 8.Neves G, Cooke SF, Bliss TV. Synaptic plasticity, memory and the hippocampus: a neural network approach to causality. Nat Rev Neurosci. 2008;9(1):65–75. doi: 10.1038/nrn2303. [DOI] [PubMed] [Google Scholar]
- 9.Richter JD, Klann E. Making synaptic plasticity and memory last: mechanisms of translational regulation. Genes Dev. 2009;23(1):1–11. doi: 10.1101/gad.1735809. [DOI] [PubMed] [Google Scholar]
- 10.Zheng F, Gallagher JP. Metabotropic glutamate receptors are required for the induction of long-term potentiation. Neuron. 1992;9(1):163–72. doi: 10.1016/0896-6273(92)90231-2. [DOI] [PubMed] [Google Scholar]
- 11.Huber KM, Roder JC, Bear MF. Chemical induction of mGluR5- and protein synthesis--dependent long-term depression in hippocampal area CA1. J Neurophysiol. 2001;86(1):321–5. doi: 10.1152/jn.2001.86.1.321. [DOI] [PubMed] [Google Scholar]
- 12.Banko JL, et al. The translation repressor 4E-BP2 is critical for eIF4F complex formation, synaptic plasticity, and memory in the hippocampus. J Neurosci. 2005;25(42):9581–90. doi: 10.1523/JNEUROSCI.2423-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hou L, Klann E. Activation of the phosphoinositide 3-kinase-Akt-mammalian target of rapamycin signaling pathway is required for metabotropic glutamate receptor-dependent long-term depression. J Neurosci. 2004;24(28):6352–61. doi: 10.1523/JNEUROSCI.0995-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hou L, et al. Dynamic translational and proteasomal regulation of fragile X mental retardation protein controls mGluR-dependent long-term depression. Neuron. 2006;51(4):441–54. doi: 10.1016/j.neuron.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 15.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18(16):1926–45. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 16.Tang SJ, Schuman EM. Protein synthesis in the dendrite. Philos Trans R Soc Lond B Biol Sci. 2002;357(1420):521–9. doi: 10.1098/rstb.2001.0887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Slipczuk L, et al. BDNF activates mTOR to regulate GluR1 expression required for memory formation. PLoS One. 2009;4(6):e6007. doi: 10.1371/journal.pone.0006007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schicknick H, et al. Dopaminergic modulation of auditory cortex-dependent memory consolidation through mTOR. Cereb Cortex. 2008;18(11):2646–58. doi: 10.1093/cercor/bhn026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kunz J, Hall MN. Cyclosporin A, FK506 and rapamycin: more than just immunosuppression. Trends Biochem Sci. 1993;18(9):334–8. doi: 10.1016/0968-0004(93)90069-y. [DOI] [PubMed] [Google Scholar]
- 20.Jacinto E. What controls TOR? IUBMB Life. 2008;60(8):483–96. doi: 10.1002/iub.56. [DOI] [PubMed] [Google Scholar]
- 21.Scott PH, et al. Evidence of insulin-stimulated phosphorylation and activation of the mammalian target of rapamycin mediated by a protein kinase B signaling pathway. Proc Natl Acad Sci U S A. 1998;95(13):7772–7. doi: 10.1073/pnas.95.13.7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reynolds THt, Bodine SC, Lawrence JC., Jr Control of Ser2448 phosphorylation in the mammalian target of rapamycin by insulin and skeletal muscle load. J Biol Chem. 2002;277(20):17657–62. doi: 10.1074/jbc.M201142200. [DOI] [PubMed] [Google Scholar]
- 23.Holz MK, Blenis J. Identification of S6 kinase 1 as a novel mammalian target of rapamycin (mTOR)-phosphorylating kinase. J Biol Chem. 2005;280(28):26089–93. doi: 10.1074/jbc.M504045200. [DOI] [PubMed] [Google Scholar]
- 24.Chiang GG, Abraham RT. Phosphorylation of mammalian target of rapamycin (mTOR) at Ser-2448 is mediated by p70S6 kinase. J Biol Chem. 2005;280(27):25485–90. doi: 10.1074/jbc.M501707200. [DOI] [PubMed] [Google Scholar]
- 25.Guertin DA, Sabatini DM. An expanding role for mTOR in cancer. Trends Mol Med. 2005;11(8):353–61. doi: 10.1016/j.molmed.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 26.Beretta L, et al. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. Embo J. 1996;15(3):658–64. [PMC free article] [PubMed] [Google Scholar]
- 27.Kim DH, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110(2):163–75. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 28.Sarbassov DD, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14(14):1296–302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 29.Bai X, et al. Rheb activates mTOR by antagonizing its endogenous inhibitor, FKBP38. Science. 2007;318(5852):977–80. doi: 10.1126/science.1147379. [DOI] [PubMed] [Google Scholar]
- 30.Hoeffer CA, et al. Removal of FKBP12 enhances mTOR-Raptor interactions, LTP, memory, and perseverative/repetitive behavior. Neuron. 2008;60(5):832–45. doi: 10.1016/j.neuron.2008.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L, et al. PRAS40 regulates mTORC1 kinase activity by functioning as a direct inhibitor of substrate binding. J Biol Chem. 2007;282(27):20036–44. doi: 10.1074/jbc.M702376200. [DOI] [PubMed] [Google Scholar]
- 32.Gingras AC, Raught B, Sonenberg N. mTOR signaling to translation. Curr Top Microbiol Immunol. 2004;279:169–97. doi: 10.1007/978-3-642-18930-2_11. [DOI] [PubMed] [Google Scholar]
- 33.Vander Haar E, et al. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol. 2007;9(3):316–23. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- 34.Sarbassov DD, et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22(2):159–68. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 35.Jacinto E, et al. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 2006;127(1):125–37. doi: 10.1016/j.cell.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 36.Yang Q, et al. Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev. 2006;20(20):2820–32. doi: 10.1101/gad.1461206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frias MA, et al. mSin1 is necessary for Akt/PKB phosphorylation, and its isoforms define three distinct mTORC2s. Curr Biol. 2006;16(18):1865–70. doi: 10.1016/j.cub.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 38.Polak P, Hall MN. mTORC2 Caught in a SINful Akt. Dev Cell. 2006;11(4):433–4. doi: 10.1016/j.devcel.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 39.Pearce LR, et al. Identification of Protor as a novel Rictor-binding component of mTOR complex-2. Biochem J. 2007;405(3):513–22. doi: 10.1042/BJ20070540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akcakanat A, et al. Rapamycin regulates the phosphorylation of rictor. Biochem Biophys Res Commun. 2007;362(2):330–3. doi: 10.1016/j.bbrc.2007.07.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnstone CN, et al. PRR5 encodes a conserved proline-rich protein predominant in kidney: analysis of genomic organization, expression, and mutation status in breast and colorectal carcinomas. Genomics. 2005;85(3):338–51. doi: 10.1016/j.ygeno.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 42.Proud CG. mTORC1 signalling and mRNA translation. Biochem Soc Trans. 2009;37(Pt 1):227–31. doi: 10.1042/BST0370227. [DOI] [PubMed] [Google Scholar]
- 43.Shiota C, et al. Multiallelic disruption of the rictor gene in mice reveals that mTOR complex 2 is essential for fetal growth and viability. Dev Cell. 2006;11(4):583–9. doi: 10.1016/j.devcel.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 44.Jacinto E, Lorberg A. TOR regulation of AGC kinases in yeast and mammals. Biochem J. 2008;410(1):19–37. doi: 10.1042/BJ20071518. [DOI] [PubMed] [Google Scholar]
- 45.Franke TF. Intracellular signaling by Akt: bound to be specific. Sci Signal. 2008;1(24):e29. doi: 10.1126/scisignal.124pe29. [DOI] [PubMed] [Google Scholar]
- 46.Dibble CC, Asara JM, Manning BD. Characterization of Rictor phosphorylation sites reveals direct regulation of mTOR complex 2 by S6K1. Mol Cell Biol. 2009 doi: 10.1128/MCB.00735-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sarbassov DD, et al. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307(5712):1098–101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 48.Sossin WS. Isoform specificity of protein kinase Cs in synaptic plasticity. Learn Mem. 2007;14(4):236–46. doi: 10.1101/lm.469707. [DOI] [PubMed] [Google Scholar]
- 49.Facchinetti V, et al. The mammalian target of rapamycin complex 2 controls folding and stability of Akt and protein kinase C. Embo J. 2008;27(14):1932–43. doi: 10.1038/emboj.2008.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sweatt JD, et al. Protected-site phosphorylation of protein kinase C in hippocampal long-term potentiation. J Neurochem. 1998;71(3):1075–85. doi: 10.1046/j.1471-4159.1998.71031075.x. [DOI] [PubMed] [Google Scholar]
- 51.Dunlop EA, et al. Mammalian target of rapamycin complex 1-mediated phosphorylation of eukaryotic initiation factor 4E-binding protein 1 requires multiple protein-protein interactions for substrate recognition. Cell Signal. 2009;21(7):1073–84. doi: 10.1016/j.cellsig.2009.02.024. [DOI] [PubMed] [Google Scholar]
- 52.Salminen A, Kaarniranta K. Regulation of the aging process by autophagy. Trends Mol Med. 2009;15(5):217–24. doi: 10.1016/j.molmed.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 53.Yanow SK, et al. Biochemical pathways by which serotonin regulates translation in the nervous system of Aplysia. J Neurochem. 1998;70(2):572–83. doi: 10.1046/j.1471-4159.1998.70020572.x. [DOI] [PubMed] [Google Scholar]
- 54.Casadio A, et al. A transient, neuron-wide form of CREB-mediated long-term facilitation can be stabilized at specific synapses by local protein synthesis. Cell. 1999;99(2):221–37. doi: 10.1016/s0092-8674(00)81653-0. [DOI] [PubMed] [Google Scholar]
- 55.Khan A, Pepio AM, Sossin WS. Serotonin activates S6 kinase in a rapamycin-sensitive manner in Aplysia synaptosomes. J Neurosci. 2001;21(2):382–91. doi: 10.1523/JNEUROSCI.21-02-00382.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beaumont V, et al. Phosphorylation and local presynaptic protein synthesis in calcium- and calcineurin-dependent induction of crayfish long-term facilitation. Neuron. 2001;32(3):489–501. doi: 10.1016/s0896-6273(01)00483-4. [DOI] [PubMed] [Google Scholar]
- 57.Carroll M, et al. 5-HT stimulates eEF2 dephosphorylation in a rapamycin-sensitive manner in Aplysia neurites. J Neurochem. 2004;90(6):1464–76. doi: 10.1111/j.1471-4159.2004.02634.x. [DOI] [PubMed] [Google Scholar]
- 58.Cammalleri M, et al. Time-restricted role for dendritic activation of the mTOR-p70S6K pathway in the induction of late-phase long-term potentiation in the CA1. Proc Natl Acad Sci U S A. 2003;100(24):14368–73. doi: 10.1073/pnas.2336098100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsokas P, et al. Local protein synthesis mediates a rapid increase in dendritic elongation factor 1A after induction of late long-term potentiation. J Neurosci. 2005;25(24):5833–43. doi: 10.1523/JNEUROSCI.0599-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huber KM, et al. Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proc Natl Acad Sci U S A. 2002;99(11):7746–50. doi: 10.1073/pnas.122205699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Antion MD, et al. mGluR-dependent long-term depression is associated with increased phosphorylation of S6 and synthesis of elongation factor 1A but remains expressed in S6K-deficient mice. Mol Cell Biol. 2008;28(9):2996–3007. doi: 10.1128/MCB.00201-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Banko JL, Hou L, Klann E. NMDA receptor activation results in PKA- and ERK-dependent Mnk1 activation and increased eIF4E phosphorylation in hippocampal area CA1. J Neurochem. 2004;91(2):462–70. doi: 10.1111/j.1471-4159.2004.02734.x. [DOI] [PubMed] [Google Scholar]
- 63.Banko JL, et al. Regulation of eukaryotic initiation factor 4E by converging signaling pathways during metabotropic glutamate receptor-dependent long-term depression. J Neurosci. 2006;26(8):2167–73. doi: 10.1523/JNEUROSCI.5196-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zho WM, et al. The group I metabotropic glutamate receptor agonist (S)-3,5-dihydroxyphenylglycine induces a novel form of depotentiation in the CA1 region of the hippocampus. J Neurosci. 2002;22(20):8838–49. doi: 10.1523/JNEUROSCI.22-20-08838.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tischmeyer W, et al. Rapamycin-sensitive signalling in long-term consolidation of auditory cortex-dependent memory. Eur J Neurosci. 2003;18(4):942–50. doi: 10.1046/j.1460-9568.2003.02820.x. [DOI] [PubMed] [Google Scholar]
- 66.Parsons RG, Gafford GM, Helmstetter FJ. Translational control via the mammalian target of rapamycin pathway is critical for the formation and stability of long-term fear memory in amygdala neurons. J Neurosci. 2006;26(50):12977–83. doi: 10.1523/JNEUROSCI.4209-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guertin DA, et al. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell. 2006;11(6):859–71. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 68.Cho H, et al. Insulin resistance and a diabetes mellitus-like syndrome in mice lacking the protein kinase Akt2 (PKB beta) Science. 2001;292(5522):1728–31. doi: 10.1126/science.292.5522.1728. [DOI] [PubMed] [Google Scholar]
- 69.Cho H, et al. Akt1/PKBalpha is required for normal growth but dispensable for maintenance of glucose homeostasis in mice. J Biol Chem. 2001;276(42):38349–52. doi: 10.1074/jbc.C100462200. [DOI] [PubMed] [Google Scholar]
- 70.Chen J, et al. Impaired platelet responses to thrombin and collagen in AKT-1-deficient mice. Blood. 2004;104(6):1703–10. doi: 10.1182/blood-2003-10-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barber DF, et al. PTEN regulation, a novel function for the p85 subunit of phosphoinositide 3-kinase. Sci STKE. 2006;2006(362):e49. doi: 10.1126/stke.3622006pe49. [DOI] [PubMed] [Google Scholar]
- 72.Bayascas JR, et al. Hypomorphic mutation of PDK1 suppresses tumorigenesis in PTEN(+/−) mice. Curr Biol. 2005;15(20):1839–46. doi: 10.1016/j.cub.2005.08.066. [DOI] [PubMed] [Google Scholar]
- 73.Goorden SM, et al. Cognitive deficits in Tsc1+/− mice in the absence of cerebral lesions and seizures. Ann Neurol. 2007;62(6):648–55. doi: 10.1002/ana.21317. [DOI] [PubMed] [Google Scholar]
- 74.von der Brelie C, et al. Impaired synaptic plasticity in a rat model of tuberous sclerosis. Eur J Neurosci. 2006;23(3):686–92. doi: 10.1111/j.1460-9568.2006.04594.x. [DOI] [PubMed] [Google Scholar]
- 75.Ehninger D, et al. Reversal of learning deficits in a Tsc2+/− mouse model of tuberous sclerosis. Nat Med. 2008;14(8):843–8. doi: 10.1038/nm1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Costa-Mattioli M, et al. Translational control of hippocampal synaptic plasticity and memory by the eIF2alpha kinase GCN2. Nature. 2005;436(7054):1166–73. doi: 10.1038/nature03897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang J, Manning BD. A complex interplay between Akt, TSC2 and the two mTOR complexes. Biochem Soc Trans. 2009;37(Pt 1):217–22. doi: 10.1042/BST0370217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Banaszynski LA, Liu CW, Wandless TJ. Characterization of the FKBP.rapamycin.FRB ternary complex. J Am Chem Soc. 2005;127(13):4715–21. doi: 10.1021/ja043277y. [DOI] [PubMed] [Google Scholar]
- 79.Leone M, et al. The FRB domain of mTOR: NMR solution structure and inhibitor design. Biochemistry. 2006;45(34):10294–302. doi: 10.1021/bi060976+. [DOI] [PubMed] [Google Scholar]
- 80.Shou W, et al. Cardiac defects and altered ryanodine receptor function in mice lacking FKBP12. Nature. 1998;391(6666):489–92. doi: 10.1038/35146. [DOI] [PubMed] [Google Scholar]
- 81.Tang W, et al. Altered excitation-contraction coupling with skeletal muscle specific FKBP12 deficiency. Faseb J. 2004;18(13):1597–9. doi: 10.1096/fj.04-1587fje. [DOI] [PubMed] [Google Scholar]
- 82.Shor B, et al. A new pharmacologic action of CCI-779 involves FKBP12-independent inhibition of mTOR kinase activity and profound repression of global protein synthesis. Cancer Res. 2008;68(8):2934–43. doi: 10.1158/0008-5472.CAN-07-6487. [DOI] [PubMed] [Google Scholar]
- 83.Shima H, et al. Disruption of the p70(s6k)/p85(s6k) gene reveals a small mouse phenotype and a new functional S6 kinase. Embo J. 1998;17(22):6649–59. doi: 10.1093/emboj/17.22.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pende M, et al. S6K1(−/−)/S6K2(−/−) mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol Cell Biol. 2004;24(8):3112–24. doi: 10.1128/MCB.24.8.3112-3124.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Antion MD, et al. Removal of S6K1 and S6K2 leads to divergent alterations in learning, memory, and synaptic plasticity. Learn Mem. 2008;15(1):29–38. doi: 10.1101/lm.661908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Banko JL, et al. Behavioral alterations in mice lacking the translation repressor 4E-BP2. Neurobiol Learn Mem. 2007;87(2):248–56. doi: 10.1016/j.nlm.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 87.Rosner M, et al. The mTOR pathway and its role in human genetic diseases. Mutat Res. 2008;659(3):284–92. doi: 10.1016/j.mrrev.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 88.Kwiatkowski DJ. Tuberous sclerosis: from tubers to mTOR. Ann Hum Genet. 2003;67(Pt 1):87–96. doi: 10.1046/j.1469-1809.2003.00012.x. [DOI] [PubMed] [Google Scholar]
- 89.Krab LC, Goorden SM, Elgersma Y. Oncogenes on my mind: ERK and MTOR signaling in cognitive diseases. Trends Genet. 2008;24(10):498–510. doi: 10.1016/j.tig.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 90.Kanner L, Eisenberg L. Early infantile autism, 1943–1955. Psychiatr Res Rep Am Psychiatr Assoc. 1957;(7):55–65. doi: 10.4159/harvard.9780674367012.c2. [DOI] [PubMed] [Google Scholar]
- 91.DiCicco-Bloom E, et al. The developmental neurobiology of autism spectrum disorder. J Neurosci. 2006;26(26):6897–906. doi: 10.1523/JNEUROSCI.1712-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Persico AM, Bourgeron T. Searching for ways out of the autism maze: genetic, epigenetic and environmental clues. Trends Neurosci. 2006;29(7):349–58. doi: 10.1016/j.tins.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 93.Moldin SO, Rubenstein JL, Hyman SE. Can autism speak to neuroscience? J Neurosci. 2006;26(26):6893–6. doi: 10.1523/JNEUROSCI.1944-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Risch N, et al. A genomic screen of autism: evidence for a multilocus etiology. Am J Hum Genet. 1999;65(2):493–507. doi: 10.1086/302497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kelleher RJ, 3rd, Bear MF. The autistic neuron: troubled translation? Cell. 2008;135(3):401–6. doi: 10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 96.Butler MG, et al. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. J Med Genet. 2005;42(4):318–21. doi: 10.1136/jmg.2004.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Herman GE, et al. Increasing knowledge of PTEN germline mutations: Two additional patients with autism and macrocephaly. Am J Med Genet A. 2007;143(6):589–93. doi: 10.1002/ajmg.a.31619. [DOI] [PubMed] [Google Scholar]
- 98.Wiznitzer M. Autism and tuberous sclerosis. J Child Neurol. 2004;19(9):675–9. doi: 10.1177/08830738040190090701. [DOI] [PubMed] [Google Scholar]
- 99.Jacquemont S, et al. Fragile-X syndrome and fragile X-associated tremor/ataxia syndrome: two faces of FMR1. Lancet Neurol. 2007;6(1):45–55. doi: 10.1016/S1474-4422(06)70676-7. [DOI] [PubMed] [Google Scholar]
- 100.Penagarikano O, Mulle JG, Warren ST. The pathophysiology of fragile x syndrome. Annu Rev Genomics Hum Genet. 2007;8:109–29. doi: 10.1146/annurev.genom.8.080706.092249. [DOI] [PubMed] [Google Scholar]
- 101.Brown V, et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107(4):477–87. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- 102.Bagni C, Greenough WT. From mRNP trafficking to spine dysmorphogenesis: the roots of fragile X syndrome. Nat Rev Neurosci. 2005;6(5):376–87. doi: 10.1038/nrn1667. [DOI] [PubMed] [Google Scholar]
- 103.Narayanan U, et al. S6K1 phosphorylates and regulates fragile X mental retardation protein (FMRP) with the neuronal protein synthesis-dependent mammalian target of rapamycin (mTOR) signaling cascade. J Biol Chem. 2008;283(27):18478–82. doi: 10.1074/jbc.C800055200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sharma A, et al. Dysregulation of mTOR signaling in fragile X syndrome. Journal of Neuroscience. doi: 10.1523/JNEUROSCI.3696-09.2010. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nosyreva ED, Huber KM. Metabotropic receptor-dependent long-term depression persists in the absence of protein synthesis in the mouse model of fragile X syndrome. J Neurophysiol. 2006;95(5):3291–5. doi: 10.1152/jn.01316.2005. [DOI] [PubMed] [Google Scholar]
- 106.Li X, et al. Levels of mTOR and its downstream targets 4E-BP1, eEF2, and eEF2 kinase in relationships with tau in Alzheimer’s disease brain. Febs J. 2005;272(16):4211–20. doi: 10.1111/j.1742-4658.2005.04833.x. [DOI] [PubMed] [Google Scholar]
- 107.Chano T, Okabe H, Hulette CM. RB1CC1 insufficiency causes neuronal atrophy through mTOR signaling alteration and involved in the pathology of Alzheimer’s diseases. Brain Res. 2007;1168:97–105. doi: 10.1016/j.brainres.2007.06.075. [DOI] [PubMed] [Google Scholar]
- 108.Ravikumar B, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36(6):585–95. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 109.Rami A. Autophagy in neurodegeneration: fire-fighter and/or incendiarist? Neuropathol Appl Neurobiol. 2009 doi: 10.1111/j.1365-2990.2009.01034.x. [DOI] [PubMed] [Google Scholar]
- 110.Yu DY, et al. Effects of cyclosporin A, FK506 and rapamycin on calcineurin phosphatase activity in mouse brain. IUBMB Life. 2006;58(7):429–33. doi: 10.1080/15216540600791555. [DOI] [PubMed] [Google Scholar]
- 111.Weiwad M, et al. Comparative analysis of calcineurin inhibition by complexes of immunosuppressive drugs with human FK506 binding proteins. Biochemistry. 2006;45(51):15776–84. doi: 10.1021/bi061616p. [DOI] [PubMed] [Google Scholar]
- 112.Xue Q, et al. Palomid 529, a novel small-molecule drug, is a TORC1/TORC2 inhibitor that reduces tumor growth, tumor angiogenesis, and vascular permeability. Cancer Res. 2008;68(22):9551–7. doi: 10.1158/0008-5472.CAN-08-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Garcia-Martinez JM, et al. Ku-0063794 is a specific inhibitor of the mammalian target of rapamycin (mTOR) Biochem J. 2009;421(1):29–42. doi: 10.1042/BJ20090489. [DOI] [PMC free article] [PubMed] [Google Scholar]