Summary
Bacteria belonging to the genera Photorhabdus and Xenorhabdus participate in a trilateral symbiosis in which they enable their nematode hosts to parasitize insect larvae [1]. The bacteria switch from persisting peacefully in a nematode's digestive tract to a lifestyle in which pathways to produce insecticidal toxins, degrading enzymes to digest the insect for consumption, and antibiotics to ward off bacterial and fungal competitors are activated. This study addresses three questions: 1) What molecular signal triggers antibiotic production in the bacteria? 2) What small molecules are regulated by the signal? and 3) How do the bacteria recognize the signal? Differential metabolomic profiling in Photorhabdus luminescens TT01 and Xenorhabdus nematophila revealed that L-proline in the insect's hemolymph initiates a metabolic shift. Small molecules known to be crucial for virulence and antibiosis in addition to previously unknown metabolites are dramatically upregulated by L-proline, linking the recognition of host environment to bacterial metabolic regulation. To identify the L-proline-induced signaling pathway, proline transporters putP and proU were deleted in P. luminescens TT01. Studies of these strains support a picture where acquisition of L-proline both regulates the metabolic shift and maintains the bacterial proton motive force (PMF) that ultimately regulates the downstream bacterial pathways affecting virulence and antibiotic production.
Results & Discussion
During their infective juvenile stage, insect-killing (entomopathogenic) nematodes sequester symbiotic bacteria in their digestive tracts and hunt insect larvae in the soil [2, 3]. When a nematode successfully invades a larva's circulatory system, it releases the bacteria. In this new food-rich environment, the bacteria switch from quiescent nematode residents to highly effective insect pathogens while the infective juvenile nematodes develop into adults and begin reproducing. Whereas nematode and bacteria have a specific association – Photorhabdus spp. bacteria with Heterorhabditis spp. nematodes and Xenorhabdus spp. bacteria with Steinernema spp. nematodes – a given nematode-bacteria pair can parasitize a wide variety of insect larvae. Identifying the signal(s) driving the switch from one bacterial phenotype to another would provide a more complete description of this trilateral symbiosis and of bacterial virulence regulation. In addition, the signal(s) could provide access to a potentially important suite of antibacterial and antifungal small molecules that have so far eluded characterization (Fig. 1) [4].
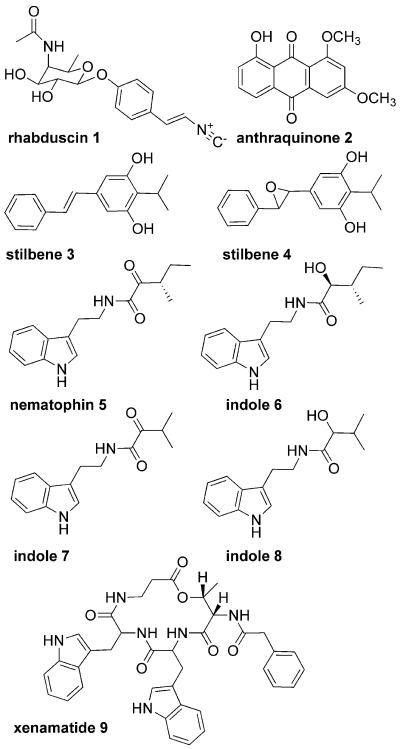
Figure 1.
Selected P. luminescens (1-4) and X. nematophila (1, 5-9) metabolites.
Photorhabdus luminescens TT01, the best studied of these bacterial symbionts, has a sequenced genome containing “a wide array of antibiotic synthesizing genes” with at least 33 genes in 20 loci similar to those used to make nonribosomal peptides (NRPS genes) and polyketides (PKS genes) – two large families of biologically active small molecules [5]. Xenorhabdus nematophila ATCC19061 has recently been sequenced, and it appears to have a similar genome size and comparable number of small molecule biosynthetic genes (www.genoscope.cns.fr/agc/mage/). The majority of these encoded molecules are cryptic, meaning their existence can be inferred from the genome but not yet confirmed in the laboratory, and analyses of sequenced bacterial genomes indicate that cryptic metabolites greatly outnumber known metabolites – arguably by as much as a factor of ten [6]. Most cryptic metabolite biosyntheses are tightly regulated, and discovering the regulatory key for a given metabolite has proved challenging. The Photorhabdus/Xenorhabdus system provides an opportunity to discover the regulatory signal for wholesale production of an entire suite of bioactive small molecules that control a broad swathe of microbial competitors.
Insect hemolymph regulates bacterial secondary metabolism
Crude small molecule extracts of Photorhabdus- and Xenorhabdus-infected insect larvae have enhanced titers of antibacterial, antifungal, and insecticidal activities versus the levels extracted when the bacteria are grown on standard microbiological media [7, 8]. The host factor(s) responsible for turning on production of these activities is likely a shared feature of insect larvae, and we used small molecule production, as judged by chromatographic analysis, to identify it.
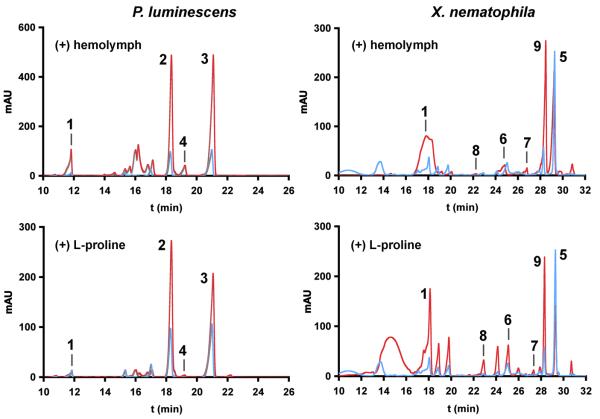
Aqueous and organic extracts from a common insect host, Galleria mellonella (greater waxmoth) larvae, were assessed for their ability to stimulate metabolite production by both P. luminescens TT01 and X. nematophila ATCC19061. Addition of aqueous insect extracts or hemolymph dramatically induced metabolite production in both bacterial strains as judged by high-pressure liquid chromatograms (HPLC) of culture extracts (Fig. 2, top). Individual metabolites were upregulated by as much as an order of magnitude, and some previously cryptic metabolites became evident in the differential profiles. Larval G. mellonella hemolymph was subjected to HPLC-based bioassay-guided fractionation to identify fractions capable of stimulating secondary metabolite production in either strain, and over successive chromatographic steps, identical fractions were shown to induce activity in both. Nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry (MS) of the purified induction factor led to its identification as proline (Fig. S1). Supplementing bacterial cultures with Lproline, but not D-proline, increased secondary metabolite production in both strains (Fig. S2). The differential profiles induced by L-proline were very similar to the hemolymph-induced chromatograms in both Photorhabdus and Xenorhabdus species (Fig. 2, bottom). Since a variety of insect larvae contain high levels of L-proline [9], it could be a generic activation signal.
Figure 2.
Differential metabolomic profiling. Bacterial cultures with or without sterile-filtered insect hemolymph or L-proline were extracted after 72 hours, and organic extracts were assessed by HPLC (280 nm). Hemolymph supplementation (red traces, top) to P. luminescens (left) and X. nematophila (right) alters metabolite production compared to control cultures (blue traces). Proline supplementation (red traces, bottom) similarly regulates metabolite production in both bacteria compared to controls (blue traces). Numbers above peaks refer to metabolites in Fig. 1.
L-proline regulates known virulence factors
Structural characterization by NMR and MS established that known virulence factors and antibiotics were regulated in both bacteria by exogenously supplied L-proline. In P. luminescens, L-proline substantially upregulated an anthraquinone (2) and two stilbenes (3 and 4) previously identified in this bacterium (Figs. 1,2) [10, 11]. In X. nematophila, L-proline significantly altered three indole-containing metabolites: nematophin (5), its reduction product (6), and xenamatide (9) [12, 13]. Production of nematophin (5) decreased with a concomitant increase in its reduction product (6), which illustrates that Lproline (or hemolymph) regulates a metabolic shift rather than a general upregulation.
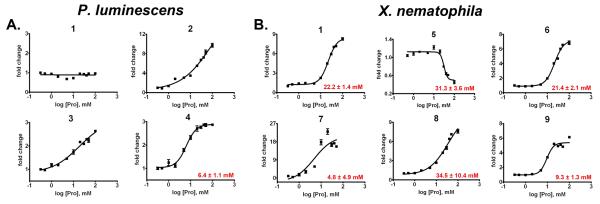
To evaluate whether proline-mediated metabolite regulation occurs at ecologically-relevant Lproline concentrations, dose-response curves were generated for select metabolites in the two model bacterial strains. Marked metabolite regulation was induced by L-proline concentrations greater than 6.4 and 4.8 mM for P. luminescens and X. nematophila, respectively (Fig. 3). Hemolymph, which carries nutrients as insects respire by diffusion, has high free amino acid, carbohydrate, and inorganic salt concentrations [9]. Complete amino acid analysis conducted on hemolymph samples from four representative host insect larvae revealed individual L-proline concentrations of 73, 3, 31, and 59 mM (Table S1). While hemolymph L-proline concentrations have a substantial range (~20 fold), these findings show that the low mM L-proline concentrations needed to regulate metabolite production are physiologically relevant.
Figure 3.
Fold change in metabolite production in P. luminescens (A) and X. nematophila (B) with increasing concentrations of supplementary L-proline. Numbers above curves refer to compounds in Fig. 1, and effective concentration, 50% (EC50) values are shown in red for metabolites with sigmoidal dose-response curves (error = s.d.). Metabolites were extracted at 72 hours, after the bacteria had reached stationary phase (Fig. S2). Rhabduscin (1) production remained constant in P. luminescens cultures upon addition of proline (but were stimulated in the presence of elevated osmolarity, Fig. S5).
The previously described molecules exhibit a range of biological activities, and their regulation by L-proline underscores the connection between recognition of a specific host environment and subsequent bacterial metabolic adjustment. Stilbene 3, for example, possesses antibiotic activity [10], is critical for maintaining symbiosis with nematodes [14], and is a potent inhibitor of phenoloxidase, a component of the insect innate immune system [15]. Nematophin 5 also exhibits antibacterial activity while other X. nematophila compounds (6 and 9) have shown little to no activity in various biological assays to date [12, 13], but their regulation by L-proline implies important biological functions. Differential metabolomic profiling using biologically-relevant triggers provides a systemwide approach to coupling small molecule metabolites to the bacteria's physiological state.
L-proline upregulates cryptic metabolite production
In order to evaluate the ability of Lproline addition to reveal previously cryptic metabolites, we characterized several compounds from X. nematophila that became evident with L-proline addition. After purification of the metabolites, high-resolution MS (Table S2) and a combination of one- and two-dimensional NMR experiments (1H, COSY, HSQC, HMBC, and ROESY) were performed (Tables S3-S6 and Figs. S18-S31), identifying two of them as the indole-containing metabolites 7 and 8 (Fig. 1). Compound 7 is a new natural product, but it has been prepared by synthesis and was shown to be a potent antibiotic against MDR Staphylococcus aureus [12]. Compound 8 is its previously unreported reduction product. The third selected metabolite from X. nematophila, which was upregulated by over an order of magnitude in L-proline-induced cultures, was also characterized. NMR analyses revealed a new isocyanide- and aminoglycosyl-functionalized tyrosine derivative, which we have named rhabduscin (1) (Tables S3-S4).
Biosynthetic genes isnA and isnB were recently discovered from environmental DNA cosmid libraries and found to introduce isocyanide functional groups into their encoded products [16]. The X. nematophila ATCC19061 genome contains a candidate biosynthetic gene cluster for rhabduscin (1): isnA, isnB, and a glycosyltransferase gene (Fig. S3). Heterologous expression of the cluster in Escherichia coli reconstituted rhabduscin (1) production, confirming assignment of the previously cryptic biosynthetic gene cluster (Fig S4). Using these three genes as bioinformatics probes, homologs were also discovered in the genome sequence for P. luminescens TT01. Rhabduscin (1) was, not surprisingly, then identified in P. luminescens culture extracts, and its occurrence in both genera suggests an important role in both systems.
A structurally-related isocyanide metabolite with an altered sugar moiety was previously isolated from an Enterobacter sp., and it was found to possess potent anti-phenoloxidase activity [17]. As noted above, phenoloxidases are critical components of the insect innate immune system. Due to the structural similarity of rhabduscin to the known inhibitor, it is possible that it could enhance the bacteria's immune-evading capabilities.
Genetic analysis of proline transport and response
Because osmolarity is typically much higher in the host than in other bacterial environments, it affects virulence in several bacterial pathogens [18]. To mimic hemolymph's high solute and inorganic salt concentrations [9], we challenged P. luminescens and X. nematophila cultures to osmotic stressors (sorbitol and NaCl) with and without proline. These osmotic challenges contributed to the regulation of virulence factors, and the introduction of L-proline provided an enhanced effect (Fig. S5).
To further probe proline's role and decouple the connection between its transport and metabolite stimulation, we deleted candidate proline transporters in P. luminescens. The gram-negative Gammaproteobacteria E. coli, Photorhabdus spp., and Xenorhabdus spp. share many similarities in physiology and metabolism. E. coli is known to acquire proline through three different proline transport systems: PutP, ProP, and ProU [18]. PutP is a high-affinity uptake system for proline metabolism, while ProP and ProU typically acquire the organic osmolytes proline and betaine for osmoregulation. Two P. luminescens TT01 homologs, putP and proU (proU consists of proV, proX, and proW) were identified and individually excised from the genome by allelic exchange mutagenesis for further interrogation.
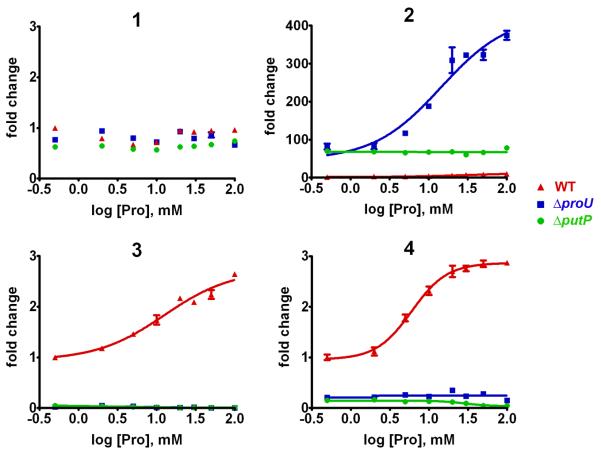
Stilbene production (3 and 4) was abolished in P. luminescens ΔproU compared to wild-type cultures, linking L-proline transport or accumulation to production of secondary metabolites (Fig. 4). Conversely, anthraquinone 2 was dramatically upregulated in the ΔproU mutant, implying altered downstream regulatory control of the two natural product classes (Fig. 4). P. luminescens ΔputP cultures showed similar trends, with virtually no production of stilbenes 3 and 4 and upregulated production of anthraquinone 2 compared to wild-type. To validate the specificity of PutP for L-proline, P. luminescens putP driven by the lactose operon was introduced into the E. coli proline auxotroph WG389 [19], and fully restored growth under conditions lacking osmotic challenges compared to controls (Fig. S5). Furthermore, the observed preference for transporting and metabolizing L-proline led us to determine that both P. luminescens and X. nematophila could grow on a defined MOPS minimal medium with L-proline substituted as the sole carbon source (Fig. S6). Taken together, the results argue that osmolarity alone is insufficient to account for the regulation of virulence factors contributing to pathogenesis and support the view that L-proline acquisition and its subsequent metabolism serve as indicators of environmental conditions to direct bacterial gene expression.
Figure 4.
Metabolic shift of wild-type (red triangles) versus ΔproU (blue squares) and ΔputP (green circles) P. luminescens with increasing proline. Stilbene production (3 and 4) was abolished in both mutants, but anthraquinone production (2) was upregulated in the mutants compared to WT (error bars = s.d.). While elevation of anthraquinone production by the ΔproU mutant was even more dramatic with addition of L-proline to the medium, levels were high, but unchanged over the proline concentration window for the ΔputP mutant, indicating relative insensitivity of the ProU transporter to high levels of proline in a non-osmotically challenged medium.
PMF manipulation alters metabolite regulation
For reasons not understood, Photorhabdus and Xenorhabdus species undergo phenotypic variation in stationary phase denoted as primary and secondary forms [1, 20], which differ in proline consumption, respiratory enzyme activity, and PMF depending on their growth conditions [21] (see “Bacterial lifestyle and proline utilization” in the SI). The primary form maintains an ability to produce antibiotics in rich medium, whereas the secondary form does not. Because PMF is implicated in this phenotypic switch, and because it contains osmotic and electrical components, we reasoned that it could play a role in insect host recognition and metabolite stimulation as described above. Indeed, proline oxidation to pyrroline-5-carboxylate by the tri-functional proline dehydrogenase PutA provides an electron source to generate a PMF in E. coli [22], and PMF has been implicated in many membrane-dependent roles such as sensory transduction and solute accumulation.
To evaluate the link between membrane energy levels and proline utilization, we first evaluated growth on a solid rich medium with increasing osmotic stress (NaCl or sorbitol) with and without proline and/or the PMF diffusion agent carbonyl cyanide-m-chlorophenylhydrazone (CCCP) (Figs. S8-S16). P. luminescens was incapable of forming colonies on a rich medium containing 10 M CCCP. Proline rescued growth of P. luminescens with or without osmotic stress, presumably by providing an electron source to out-compete CCCP-dependent membrane energy diffusion.
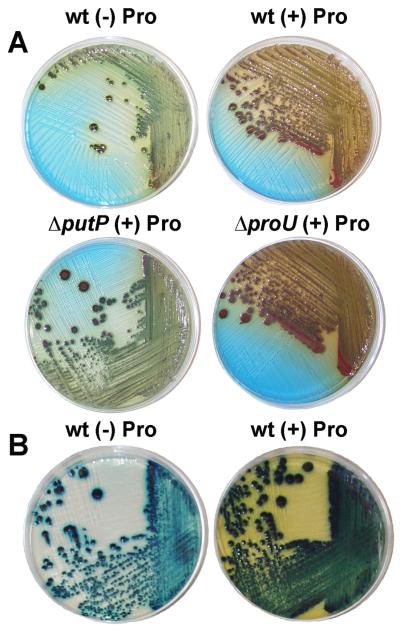
Another characteristic difference between primary and secondary forms is the differential staining patterns on agar plates containing the dye bromothymol blue and the redox indicator 2,3,5-triphenyl tetrazolium chloride (TTC) [20]. Both forms reduce TTC giving a red color phenotype, but only the antibiotic-producing primary form takes up bromothymol blue. Proline addition to both P. luminescens and X. nematophila cultures showed a darkening of the colonies indicating increased dye uptake (Fig. 5). Additionally, intense reddening of the colonies was observed with proline, showing markedly increased reduction of TTC when proline was present, which further supports proline's role as an electron source.
Figure 5.
P. luminescens (A) and X. nematophila (B) L-proline growth effects. Bacteria were grown on rich solid media (medium used above: 2 g tryptone and 5 g yeast-extract per L) containing the dye bromothymol blue and the redox indicator TTC for 3-5 days, with and without 100 mM L-proline. Phenotypic growth effects on various media are illustrated in Figs. S8-S16. Staining patterns reveal a reddening of wild type colonies and the ΔproU mutant upon proline supplementation, indicating increased reduction of TTC. A darkening of colonies is also observed with proline, indicating increased dye uptake. Given our differential metabolic observations, it is logical that the P. luminescens ΔputP culture with proline resembled the wild-type strain grown in the absence of proline. P. luminescens ΔproU was visually similar to wild-type in these dye uptake studies in keeping with proline's proposed primary role.
To more directly test the link between membrane energization and metabolite stimulation, we added the PMF antagonists CCCP and valinomycin to proline-supplemented mid-exponential suspension cultures of P. luminescens. The cultures continued to grow until stationary phase before the metabolites were extracted. P. luminescens stilbene virulence factors 3 and 4 were substantially upregulated (2.8 0.3 to 9.8 ± 1.0 fold change) in the presence of both CCCP and valinomycin compared to cultures supplemented with proline alone, indicating that PMF control is linked to metabolite production (Fig. S17).
The connection between PMF and metabolite production may be applicable to other bacterial systems. For example, sub-inhibitory levels of a panel of translation inhibiting antibiotics greatly alter global transcription patterns, and changes to as many as 10% of the gene transcripts by factors as large as 100-fold have been observed [23, 24]. The same families of protein translational inhibitors have also been shown to significantly affect the PMF in Clostridium sporogenes [25], and the link between antibiotic regulated metabolite production could involve PMF regulation. Motility, for example, which is another characteristic of the antibiotic-producing form, is directly controlled by the PMF since the electrical potential component of the PMF affords energy for flagellar motion [26]. Additionally, pathogenic S. aureus mutants defective in PutP showed significantly reduced mammalian infectivity [18], hinting that PMF control could be a general phenomenon for host recognition in bacterial pathogenesis.
Downstream regulation
Investigations of Photorhabdus temperata (a close relative of P. luminescens) and X. nematophila have revealed that these species employ dissimilar downstream regulatory mechanisms for controlling general antibiotic activity as judged by zone of inhibition [27, 28]. P. temperata employs the LysR-type transcriptional regulator HexA to repress antibiotic activity, as evidenced by the ΔhexA mutant exhibiting a derepressed, larger zone of inhibition [27]. Deletion of the X. nematophila hexA homolog (lrhA), however, displayed no observed differences in general antibiotic activity [29]. Instead, X. nematophila utilizes a leucine-responsive regulatory protein (Lrp) homolog [28], which governs many pathogenic-associated activities, including positively influencing flagellar motion and antibiotic production (Δlrp exhibits no zone of inhibition) [3]. In E. coli, Lrp is known as a feast or famine transcriptional regulator, and nutrient sources such as amino acid ligands bind at the dimer interface altering its activity [30]. Similarly, small molecule co-inducers modulate LysR-type transcriptional regulatory activity [31]. Given the drastic proline response observed, proline could either directly act as a transcriptional protein ligand or regulate the production of the cognate ligand. Although it is not yet clear how the initial signal translates to the disparate downstream regulatory machinery in Photorhabdus and Xenorhabdus, both genera evolved to recognize proline as a nutrient signal to induce phenotypic change, linking host physiology to bacterial pathogenesis.
Conclusion
Systematic examination of the insect host's small molecule repertoire with differential metabolomic profiling provides a straightforward approach to bacterial metabolite discovery with an ecological rationale. The hemolymph nutrient availability and high osmolarity common in the wide spectrum of insect prey restricted bacterial evolution to only a handful of carbon/nitrogen sources – primarily free amino acids (the 20 proteinogenic amino acids varied among insect species from undetectable to 100 mM, Table S1). The chemical and microbiological analyses reported here support a model in which both Photorhabdus and Xenorhabdus species evolved to utilize L-proline in the hemolymph as a preferred nutrient source, which can also protect against osmotic stress, to regulate the production of metabolites linked to pathogenesis through its ability to enhance the PMF. Because proline also functions as a preferred energy source for insects [32], its consumption by the bacteria depletes the insect's resources in the conflict. These results define a link between metabolic status (PMF) and virulence factor regulation in Photorhabdus and Xenorhabdus species and an approach to the wholesale discovery of cryptic metabolites with biomedical potential involved in this trilateral symbiosis.
Experimental Procedures
Metabolite stimulation
P. luminescens and X. nematophila were grown on Luria Bertani (LB) agar for two days at 30 °C. Single colonies were selected and grown two additional days in LB broth. For metabolite stimulation assays, 50 μL was used to inoculate 5 mL of a rich tryptone-yeast extract based medium (2 g tryptone, 5 g yeast extract, and 10 g NaCl per L) with and without indicated supplements. Triplicate cultures were extracted at 72 h with ethyl acetate, dried, and resuspended in methanol for HPLC analysis.
Induction factor identification
Hemolymph was collected from 500 G. mellonella larvae. Fresh hemolymph stored on ice was diluted with 100 mM potassium phosphate buffer (pH = 7.4) and filtered through a 3,000 molecular weight cutoff membrane (Amicon Ultra, Millipore); the activity resided in the flow through. The flow through was concentrated and separated using reversed-phase HPLC. The activity eluted within the first isocratic step (2:98, acetonitrile:water). The polar fraction was iteratively collected, pooled, and concentrated for further purification using a HYPERCARB (Thermo Scientific) column and again the compound eluted in the initial isocratic step (100% water). NMR experiments in D2O showed chemical shifts (1H) matching that of a proline standard (Fig. S1). LC/MS analysis and two-dimensional (gCOSY, gHSQC, and gHMBC) NMR correlations were also consistent with proline.
NMR analysis
NMR experiments (1H, gCOSY, gHSQC, gHMBC, and ROESY) were performed in deuterated methanol, water, chloroform, or dimethyl sulfoxide with a symmetrical NMR microtube susceptibility-matched with the appropriate solvent (Shigemi, Inc.) on a Varian INOVA 600 MHz NMR.
Allelic-exchange mutagenesis
Deletion constructs were generated in pDS132 [33] and introduced by conjugation from the dap auxotroph E. coli WM6026 lambda pir [34] using standard molecular and microbiological procedures. The P. luminescens ΔputP and ΔproU deletions were validated by PCR and sequence verified.
Phenotypic growth effects
P. luminescens and X. nematophila single colonies were selected and grown for two days in LB at 30 °C. 30 μL of the suspensions were streaked onto agar plates (salt experiments: 2 g tryptone, 5 g yeast-extract, and 0, 10, or 35 g NaCl per L agar; sorbitol experiment: 2 g tryptone and 5 g yeast-extract per L agar, with 750 mM sorbitol) containing bromothymol blue and TTC and grown for 3-5 days [20]. These experiments were conducted with and without 100 mM L-proline and/or 10 uM CCCP.
PMF and metabolite stimulation
Bacteria were grown as described in the metabolite stimulation experiments. Proline-supplemented (50 mM) cultures were grown to mid-log phase and then antagonized by 10 uM CCCP; 10 uM valinomycin and 150 mM KCl; or 20 uM valinomycin and 150 mM KCl. The cultures continued to grow for a total time of 72 hours before extraction.
Supplementary Material
Acknowledgements
This work was supported by NIH grant CA24487 to JC. JMC is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-2002-09). We are grateful to Drs. Roberto Kolter and Christopher T. Walsh (Harvard Medical School) for helpful suggestions and critical reading of a preliminary manuscript. We thank Drs. Janet Wood (Univ. of Guelph) and William Metcalf (Univ. of Illinois) for providing E. coli strains WG389 and WM6026, respectively.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Data
Supplemental Data include 31 figures, 6 tables, a statement regarding “Bacterial lifestyle and proline utilization,” and additional experimental details and can be found with this article online at http://www.cell.com/current-biology/supplemental/xxx.
References
- 1.Goodrich-Blair H, Clarke DJ. Mutualism and pathogenesis in Xenorhabdus and Photorhabdus: two roads to the same destination. Mol. Microbiol. 2007;64:260–268. doi: 10.1111/j.1365-2958.2007.05671.x. [DOI] [PubMed] [Google Scholar]
- 2.Waterfield NR, Ciche T, Clarke DJ. Photorhabdus and a host of hosts. Annu. Rev. Microbiol. 2009;63:557–574. doi: 10.1146/annurev.micro.091208.073507. [DOI] [PubMed] [Google Scholar]
- 3.Richards GR, Goodrich-Blair H. Masters of conquest and pillage: Xenorhabdus nematophila global regulators control transitions from virulence to nutrient acquisition. Cell. Microbiol. 2009;11:1025–1033. doi: 10.1111/j.1462-5822.2009.01322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bode HB. Entomopathogenic bacteria as a source of secondary metabolites. Curr. Opin. Chem. Biol. 2009;13:224–230. doi: 10.1016/j.cbpa.2009.02.037. [DOI] [PubMed] [Google Scholar]
- 5.Duchaud E, Rusniok C, Frangeul L, Buchrieser C, Givaudan A, Taourit S, Bocs S, Boursaux-Eude C, Chandler M, Charles JF, et al. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nature Biotechnol. 2003;21:1307–1313. doi: 10.1038/nbt886. [DOI] [PubMed] [Google Scholar]
- 6.Scherlach K, Hertweck C. Triggering cryptic natural product biosynthesis in microorganisms. Org. Biomol. Chem. 2009;7:1753–1760. doi: 10.1039/b821578b. [DOI] [PubMed] [Google Scholar]
- 7.Maxwell PW, Chen G, Webster JM, Dunphy GB. Stability and activities of antibiotics produced during infection of the insect Galleria mellonella by two isolates of Xenorhabdus nematophilus. Appl. Environ. Microbiol. 1994;60:715–721. doi: 10.1128/aem.60.2.715-721.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu K, Webster JM. Antibiotic production in relation to bacterial growth and nematode development in Photorhabdus-Heterorhabditis infected Galleria mellonella larvae. FEMS Microbiol. Lett. 2000;189:219–223. doi: 10.1111/j.1574-6968.2000.tb09234.x. [DOI] [PubMed] [Google Scholar]
- 9.Wyatt GR. The biochemistry of insect hemolymph. Annu. Rev. Entomol. 1961;6:75–102. [Google Scholar]
- 10.Li J, Chen G, Wu H, Webster JM. Identification of two pigments and a hydroxystilbene antibiotic from Photorhabdus luminescens. Appl. Environ. Microbiol. 1995;61:4329–4333. doi: 10.1128/aem.61.12.4329-4333.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu K, Li J, Li B, Webster JM, Chen G. A novel antimicrobial epoxide isolated from larval Galleria mellonella infected by the nematode symbiont, Photorhabdus luminescens (Enterobacteriaceae) Bioorg. Med. Chem. 2006;14:4677–4681. doi: 10.1016/j.bmc.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 12.Li J, Chen G, Webster JM. Synthesis and antistaphylococcal activity of nematophin and its analogs. Bioorg. Med. Chem. Lett. 1997;7:1349–1352. [Google Scholar]
- 13.Lang G, Kalvelage T, Peters A, Wiese J, Imhoff JF. Linear and cyclic peptides from the entomopathogenic bacterium Xenorhabdus nematophilus. J. Nat. Prod. 2008;71:1074–1077. doi: 10.1021/np800053n. [DOI] [PubMed] [Google Scholar]
- 14.Joyce SA, Brachmann AO, Glazer I, Lango L, Schwar G, Clarke DJ, Bode HB. Bacterial biosynthesis of a multipotent stilbene. Angew. Chem. Int. Ed. 2008;47:1942–1945. doi: 10.1002/anie.200705148. [DOI] [PubMed] [Google Scholar]
- 15.Eleftherianos I, Boundy S, Joyce SA, Aslam S, Marshall JW, Cox RJ, Simpson TJ, Clarke DJ, ffrench-Constant RH, Reynolds SE. An antibiotic produced by an insect-pathogenic bacterium suppresses host defenses through phenoloxidase inhibition. Proc. Natl. Acad. Sci. U.S.A. 2007;104:2419–2424. doi: 10.1073/pnas.0610525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brady SF, Clardy J. Cloning and heterologous expression of isocyanide biosynthetic genes from environmental DNA. Angew. Chem. Int. Ed. 2005;44:7063–7065. doi: 10.1002/anie.200501941. [DOI] [PubMed] [Google Scholar]
- 17.Nakagawa A, Fuchu-shi Y, Takahashi S, Miyazaki T, Osanai Y, Kosaka K, Nagai K, Arao N, Tanaka K. US Patent US 7,115,721 B2 Microorganism, its product and utilization thereof. 2006
- 18.Sleator RD, Hill C. Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol. Rev. 2002;26:49–71. doi: 10.1111/j.1574-6976.2002.tb00598.x. [DOI] [PubMed] [Google Scholar]
- 19.Culham DE, Lasby B, Marangoni AG, Milner JL, Steer BA, van Nues RW, Wood JM. Isolation and sequencing of Escherichia coli gene proP reveals unusual structural features of the osmoregulatory proline/betaine transporter, ProP. J. Mol. Biol. 1993;229:268–276. doi: 10.1006/jmbi.1993.1030. [DOI] [PubMed] [Google Scholar]
- 20.Akhurst RJ. Morphological and functional dimorphism in Xenorhabdus spp., bacteria symbiotically associated with the insect pathogenic nematodes Neoaplectana and Heterorhabditis. J. Gen. Micro. 1980;121:303–309. [Google Scholar]
- 21.Smigielski AJ, Akhurst RJ, Boemare NE. Phase variation in Xenorhabdus nematophilus and Photorhabdus luminescens: differences in respiratory activity and membrane energization. Appl. Environ. Microbiol. 1994;60:120–125. doi: 10.1128/aem.60.1.120-125.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abrahamson JL, Baker LG, Stephenson JT, Wood JM. Proline dehydrogenase from Escherichia coli K12. Properties of the membrane-associated enzyme. Eur. J. Biochem. FEBS. 1983;134:77–82. doi: 10.1111/j.1432-1033.1983.tb07533.x. [DOI] [PubMed] [Google Scholar]
- 23.Goh EB, Yim G, Tsui W, McClure J, Surette MG, Davies J. Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. Proc. Natl. Acad. Sci. U.S.A. 2002;99:17025–17030. doi: 10.1073/pnas.252607699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsui WH, Yim G, Wang HH, McClure JE, Surette MG, Davies J. Dual effects of MLS antibiotics: transcriptional modulation and interactions on the ribosome. Chem. Biol. 2004;11:1307–1316. doi: 10.1016/j.chembiol.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 25.Flythe MD, Russell JB. The ability of acidic pH, growth inhibitors, and glucose to increase the proton motive force and energy spilling of amino acid-fermenting Clostridium sporogenes MD1 cultures. Arch. Microbiol. 2005;183:236–242. doi: 10.1007/s00203-005-0765-x. [DOI] [PubMed] [Google Scholar]
- 26.Paul K, Erhardt M, Hirano T, Blair DF, Hughes KT. Energy source of flagellar type III secretion. Nature. 2008;451:489–492. doi: 10.1038/nature06497. [DOI] [PubMed] [Google Scholar]
- 27.Joyce SA, Clarke DJ. A hexA homologue from Photorhabdus regulates pathogenicity, symbiosis and phenotypic variation. Mol. Microbiol. 2003;47:1445–1457. doi: 10.1046/j.1365-2958.2003.03389.x. [DOI] [PubMed] [Google Scholar]
- 28.Cowles KN, Cowles CE, Richards GR, Martens EC, Goodrich-Blair H. The global regulator Lrp contributes to mutualism, pathogenesis and phenotypic variation in the bacterium Xenorhabdus nematophila. Cell. Microbiol. 2007;9:1311–1323. doi: 10.1111/j.1462-5822.2006.00873.x. [DOI] [PubMed] [Google Scholar]
- 29.Richards GR, Herbert EE, Park Y, Goodrich-Blair H. Xenorhabdus nematophila lrhA is necessary for motility, lipase activity, toxin expression, and virulence toward Manduca sexta insects. J. Bacteriol. 2008;190:4870–4879. doi: 10.1128/JB.00358-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yokoyama K, Ishijima SA, Clowney L, Koike H, Aramaki H, Tanaka C, Makino K, Suzuki M. Feast/famine regulatory proteins (FFRPs): Escherichia coli Lrp, AsnC and related archaeal transcription factors. FEMS. Microbiol. Rev. 2006;30:89–108. doi: 10.1111/j.1574-6976.2005.00005.x. [DOI] [PubMed] [Google Scholar]
- 31.Maddocks SE, Oyston PC. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiol. 2008;154:3609–3623. doi: 10.1099/mic.0.2008/022772-0. [DOI] [PubMed] [Google Scholar]
- 32.Bursell E. The role of proline in energy metabolism. In: Downer RGH, editor. Energy Metabolism of Insects. Plenum Press; New York, London: 1981. pp. 135–154. [Google Scholar]
- 33.Philippe N, Alcaraz JP, Coursange E, Geiselmann J, Schneider D. Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid. 2004;51:246–255. doi: 10.1016/j.plasmid.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 34.Blodgett JA, Thomas PM, Li G, Velasquez JE, van der Donk WA, Kelleher NL, Metcalf WW. Unusual transformations in the biosynthesis of the antibiotic phosphinothricin tripeptide. Nature Chem. Biol. 2007;3:480–485. doi: 10.1038/nchembio.2007.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.