Abstract
Hybrids such as maize (Zea mays) or domestic dog (Canis lupus familiaris) grow bigger and stronger than their parents. This is also true for allopolyploids such as wheat (Triticum spp.) or frog (i.e. Xenopus and Silurana) that contain two or more sets of chromosomes from different species. The phenomenon, known as hybrid vigor or heterosis, was systematically characterized by Charles Darwin (1876). The rediscovery of heterosis in maize a century ago has revolutionized plant and animal breeding and production. Although genetic models for heterosis have been rigorously tested, the molecular bases remain elusive. Recent studies have determined the roles of nonadditive gene expression, small RNAs, and epigenetic regulation, including circadian-mediated metabolic pathways, in hybrid vigor and incompatibility, which could lead to better use and exploitation of the increased biomass and yield in hybrids and allopolyploids for food, feed, and biofuels.
Polyploidy and hybrid vigor – a general view
Hybrids and polyploids (whole genome duplication) are common in plants and animals. Hybrids are formed by hybridizing different strains, varieties, or species. Although heterosis or hybrid vigor is evolutionarily defined as that the heterozygotes have higher fitness in a population than the homozygotes, heterosis generally refers to superior levels of biomass, stature, growth rate, and/or fertility in the hybrid offspring than in the parents. The latter definition is adopted in this review. Polyploidy refers to an organism or cell that contains two or more sets of basic chromosomes. An autopolyploid is formed by duplicating a genome within the same species, such as potato (Solanum tuberosum), alfalfa (Medicago sativa), and sugarcane (Saccharum), whereas an allopolyploid is derived from hybridization between different species followed by chromosome doubling or from fusion of unreduced gametes between species. An allopolyploid is a “doubled interspecific hybrid”, leading to permanent fixation of heterozygosity and hybrid vigor. Many crops, including maize (Zea mays) and rice (Oryza sativa), are grown mainly as hybrids, and many other crops, such as bread wheat (Triticum aestivum), upland cotton (Gossypium hirsutum), and oilseed rape (Brassica napus, also known as canola), are grown as allopolyploids. Despite the evolutionary significance of polyploidy and agricultural importance of hybrid vigor, the mechanisms of polyploidy and hybrid vigor are poorly understood. In this review, I outlined a historical perspective about hybrids, allopolyploids, and hybrid vigor and reevaluated genetic models for heterosis in relation to the recent findings for the roles of nonadditive gene expression, allelic expression variation (see glossary), and small RNAs in hybrid vigor and incompatibility. The molecular mechanisms for single-locus heterosis were highlighted using empirical data on altered epigenetic regulation of master regulators such as circadian clock genes that control physiological and metabolic pathways, leading to increased growth vigor and biomass in hybrids and allopolyploids. A better understanding of the mechanisms for polyploidy and hybrid vigor will help us manipulate gene expression and heterosis in hybrid plants and polyploid crops that are directly relevant to the growing demand of plant materials for food, feed, and fuels.
Hybrids, allopolyploids, and hybrid vigor – a historical perspective
“I raised close together two large beds of self-fertilised and crossed seedlings from the same plant of Linaria vulgaris. To my surprise, the crossed plants when fully grown were plainly taller and more vigorous than the self-fertilised ones.” – Charles Darwin (The Effects of Cross and Self Fertilisation in the Vegetable Kingdom, 1876).
In his book [1], Charles Darwin systematically documented the growth, development, and seed fertility of cross-pollinated plants compared with that of self-fertilized plants for more than 60 different species of plants, including pea (Pisum sativa), tomato (Solnum lycopersicum), and tobacco (Nicotiana tabacum). The overall conclusion was that inbreeding was generally deleterious (later known as inbreeding depression), and cross-fertilization was generally beneficial. Thirty-two years later, George H. Shull published a landmark paper, entitled “The composition of a field of maize” [2], which marked the rediscovery of hybrid vigor or heterosis and the beginning of applying heterosis in plant breeding. Shull indicated that selfing maize (corn; Zea mays) plants led to a reduction of overall growth vigor and yield. The notion was well supported from maize inbreeding experiments by Edward M. East [3]. East predicted that the low seed yield in the inbred lines would impede hybrid production. Shull then demonstrated that the hybrids had uniformly superior growth vigor and yield to the inbreeding parents. The low seed yield in the inbreds was improved by using double-cross (i.e., making the hybrids by crossing two hybrids derived from two pairs of inbred lines). Maize breeders continued to improve seed production in inbred lines until there were sufficient seeds to make the single-cross hybrids with a significant increase in yield [4]. The yield of maize production has steadily increased sixfold since the introduction of hybrids in 1920s [5].
Hybrid rice was first studied in 1964 in China. A rice breeder, Yuan Long Ping, initiated the research on hybrid rice and heterosis in China. The technology of hybrid rice seed production was developed in the 1970s. The most commonly used hybrids are produced between different varieties within a subspecies or between the subspecies Oryza sativa subsp. indica and O. sativa subsp. japonica [6]. Although the grain quality of intraspecific hybrids could be further improved, the yield from hybrid rice is ≥20% greater than that from conventional rice and accounts for 50% of the total rice area in many rice producing countries, including China, India, and Indonesia.
When US scientists produced hybrid maize a century ago, Russian scientists developed a new species named Rhaphanobrassica from the hybrids between two plant genera Raphanus and Brassica [7]. G.D. Karpechenko, a cytologist, hoped to produce plants that would have the roots of radish and the leaves of cabbage. The hybrids were made from artificial crosses between two vegetables, the radish (Raphanus sativus, 2n = 18) and the cabbage (Brassica oleracea, 2n = 18). But instead, the F1 hybrids had the roots of cabbage and the leaves of radish, and were highly sterile, probably because of a failure in chromosome pairing. A few fertile plants were found to be spontaneous allotetraploids that contained 36 chromosomes, and these plants had vegetative growth vigor. Unfortunately, the new species was as short-lived as its creator, who was executed in 1941 for his association with N. Vavilov in an alleged “anti-Soviet group”.
Numerous Nicotiana hybrids and allopolyploids have been produced. Some, such as Nicotiana glutinosa × N. tabacum, were not vigorous but rather dwarf [8], whereas others such as N. glutinosa × Nicotiana tomentosa had great vigor [9].
Triticale (x Triticale Tschermak) is a successful man-made interspecies hybrid or allopolyploid [10, 11]. Triticale is derived from crossing two cereals, hexaploid bread wheat (Triticum aestivum) or tetraploid durum wheat (Triticum turgidum) and rye (Secale cereale). In 1875, A.S. Wilson reported the first hybrid between wheat and rye in Scotland (UK), and a decade later W. Rimpau produced the first doubled-fertile hybrid that showed little heterosis. In Russia during the crop season of 1918, thousands of natural hybrids between wheat and rye appeared in many wheat fields. For the next 16 years, G.K. Meister and his colleagues exploited these vigorous hybrids [11]. In 1935, M. Lindschau and E. Oehler named Triticale after Tschermak, one of the rediscoverers of Mendelian Law. In theory, triticale combines the high yield potential and good grain quality of wheat with the disease and stress tolerance of rye. Triticale has vigor in vegetative growth, biomass, and tolerance to adverse conditions such as limited water and poor soil conditions. It is grown mainly for forage and animal feed because of poor baking quality and seed fertility, which need to be improved. Triticale is primarily grown in Poland, Australia, Germany, France, and China. The Centro Internacional de Mejoramiento de Maíz y Trigo (CIMMYT) has a triticale program that is aimed at improving food production and nutrition in developing countries. Triticale can be considered an energy crop because of its increased levels of biomass heterosis.
Modern view of hybrids, allopolyploids, and hybrid vigor
Humans have simply replicated a few examples of these remarkable natural processes that have produced many hybrid and polyploid plants beyond literature records. Estimates indicate that ∼10% of animal and ∼25% of plant species hybridize with at least one other species[12]. A recent study estimates that 15% of angiosperm and 31% of fern speciation events are accompanied by an increase in ploidy [13]. The proportion of polyploid flowering plants might be over 70% [14], and the majority of them (>75%) are allopolyploids [15, 16]. Many agricultural crops such as wheat, cotton, and oilseed rape are allopolyploids. Allopolyploids are presumably formed spontaneously by crossing related species via unreduced gametes or via spontaneous chromosome doubling of the resulting interspecific hybrids. A large number of hybrids spontaneously form between wheat and rye in wheat fields, suggesting that hybridization between species (and genera) occur frequently if growth and physiological conditions overcome hybridization barriers. Interspecific hybrids and allopolyploids have been formed in Tragopogon [17], Spartina [18], and Senecio [19] in recent centuries. Allopolyploid Spartina townsendii is derived from Spartina alternifolia and Spartina stricta. The allopolyploid is so vigorous that it has replaced the parental forms and spread all over Southern England (UK) and to France [18]. Senecio species are native in France, and the allopolyploids have spread over to England [19]. Tragopogon is native to Euroasia; allopolyploids were formed in the early nineteenth century in North America and become invasive in local environments [17]. Some allopolyploids such as Tragopogon [17] and Brassica [20] are formed through multiple origins and by reciprocal crosses (with different combinations of maternal cytoplasm and paternal nucleus), whereas others such as cotton [21], wheat [22], and Arabidopsis (Arabidopsis thaliana) [23] are formed by a single or a few hybridization events.
Durum wheat (T. turgidum, AABB, 2n = 4x = 28) is an allotetraploid formed by crossing two extant diploid wild grasses, Triticum monococcum or Triticum urartu (AA, 2n = 14) and a wild goatgrass such as Triticum searsii or Triticum speltoides (BB, 2n = 14). The exact donor of B genome is unknown. Approximately 8000–10,000 years ago, hexaploid wheat or bread wheat (T. aestivum, 2n = 6x = 42, AABBDD) was formed in farmers' fields through hybridization between a domesticated tetraploid wheat and a wild diploid grass (Triticum tauschii, DD, 2n = 14). The hexaploid bread wheat has been domesticated and cultivated since the history of human civilization [24].
Cotton belongs to the genus Gossypium, which includes about 45 species split across two ploidy levels, diploid (2n = 2x = 26) and tetraploid (2n = 4x = 52) [21]. A polyploidization event occurred ∼1.5 million years ago (Mya) between AA and DD extant diploid species, and the AADD allotetraploids diverged into five species that are distributed throughout the world [21]. Among them, Upland or American cotton, Gossypium hirsutum, accounts for over 95% of cotton produced worldwide. Pima or Egyptian cotton, Gossypium barbadense, accounts for less than 5% of the cotton produced. The AA progenitor species produce both lint (long) fibers, which are spinnable into yarn, and shorter fibers called fuzz. By contrast, the DD genome progenitor species produce few lint fibers, which are initiated pre-anthesis, but are much shorter in length than the lint fibers of the AA genome progenitor. Interestingly, the allotetraploids produce more abundant and higher quality fibers than the extant descendant species, suggesting strong selection on polyploid cotton for fiber traits.
The genus Brassica offers a textbook example of reciprocal hybrids and allopolyploids formed between three diploid species, which is known as U-triangle [20]. The three diploid species are Brassica nigra (2n = 2x = 16), Brassica oleracea (2n = 2x = 18), and Brassica campestris or rapa (2n = 2x = 20), and each allotetraploid species is formed between two diploid species. For example, B. napus (2n = 4x = 38) is an allotetraploid between Brassica rapa and B. oleracea, Brassica juncea (2n = 4x = 34) is formed between B. nigra and B oleraca, and Brassica carinata (2n = 4x = 36) is formed between B. nigra and B. rapa. B. napus (oilseed rape) has higher oil content and better oil composition than its parents, probably because of natural selection and human domestication for these traits in the interspecific hybrids or allotetraploids.
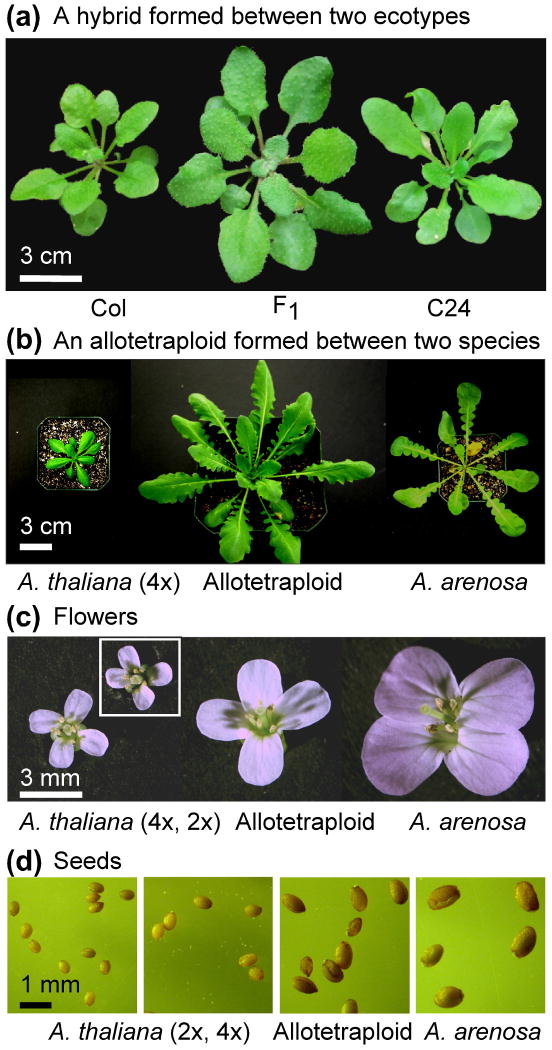
Hybrids and allopolyploids also occur in Arabidopsis, a member of the Brassicaeae family. Many hybrids formed between different ecotypes do not have obvious growth vigor. Only a handful of hybrid combinations give rise to vigor in growth [25] and other traits such as cold tolerance [26] (Figure 1a). The available genetic resources such as recombinant inbred lines have been used to dissect and study quantitative trait loci (QTL) that are associated with growth-related and life history traits [25, 27, 28].
Figure 1.
Arabidopsis hybrids and allotetraploids. (a) Seedlings of the F1 hybrid produced by crossing Arabidopsis thaliana Columbia × A. thaliana C24. (b) A stable allotetraploid (in F8 generation) was maintained by self-pollination. (a) and (b) were reproduced from [101] with permission. The F1 interspecific hybrid or allotetraploid was produced by pollinating A. thaliana Ler autotetraploid with pollen from the outcrossing A. arenosa tetraploid [48, 132]. (c) Typical flowers of the allotetraploid and its progenitors, A. thaliana tetraploid (inset, diploid) and A. aresona. (d) Seeds of the allotetraploid and its progenitors, A. thaliana Ler tetraploid and Arabidopsis arenosa. Seeds of A. thaliana Ler diploid are also shown.
Arabidopsis suecica (2n = 4x = 26) is a natural allotetraploid formed between extant A. thaliana and Arabidopsis arenosa 12,000–300,000 years ago [29]. New allotetraploids can be readily resynthesized by crossing these two species A. thaliana (2n = 4x = 20) and A. arenosa (2n = 4x = 32) (Figure 1b). During vegetative growth, the allotetraploids are 3–5 times larger than A. thaliana and at least onefold larger than A. arenosa. Under long-day conditions (light:dark of 16:8 hours), the allotetraploids flower slightly later than the late-flowering parent A. arenosa, and produce 18–25 rosette leaves, whereas A. arenosa has 10–12 leaves at flowering. The flowers of allotetraploids are intermediate between those of the two parents. The seeds are roughly twice the size of A. thaliana and slightly smaller than that of A. arenosa, a natural outcrossing autotetraploid. The seed germination rates are much higher in the stable allotetraploids (after 7–8 generations of selfing) than in A. arenosa. A large portion of A. arenosa seeds are not fully developed, probably resulting from failure of embryo and endosperm development as a consequence of being an autotetraploid [30, 31].
By definition, most heterozygous animals, including humans, are hybrids that carry different alleles from female and male parents. Mating among siblings leads to accumulation of deleterious mutations and recessive alleles, a phenomenon known as inbreeding depression [32]. Although interspecific hybrids and polyploids are rarer in animals than in plants [33, 34], interspecific hybrids do occur in mammals (e.g., a mule is a hybrid between a horse and a donkey). Mammalian interspecific hybrids are sterile, probably because of incompatibility and/or imbalance in imprinting and sex chromosome dosage, as proposed by H. Muller [33]. The number of homoploid hybrid species in animals is growing rapidly [35]. They include a recent invasive sculpin, a hybrid fish (Cottus gobio) derived from Cottus perifretum and Cottus rhenanum, a cyrinid fish Gila seminuda that is formed between Gila robusta and Gila elgans, Rhagoletis fruitflies, and Heliconius butterflies [36, 37]. Like plant hybrids, animal hybrids behave generally better than their parents. For example, mules are generally tougher than horses, and they endure heat better than the horses. They have denser muscling from their donkey parents than the horses and have fewer leg problems than horses, but they do not run as fast as the horses, which is probably a trait inherited from their donkey parents.
Many interspecific hybrids have reduced viability and fertility. The Bateson-Dobzhansky-Muller model suggests that the hybrid incompatibilities are caused by interactions between the genes that have functionally diverged in the respective hybridizing species [38, 39]. These incompatibilities appear concurrently with speciation or consequently after species divergence. The incompatibility genes include hybrid lethality genes found in Drosophila [40, 41], Caenorhabditis elegans [42], and Arabidopsis [43, 44]. In Drosophila, the lethality in F1 hybrid males is caused by the interaction between Lethal hybrid rescue (Lhr), which has functionally diverged in Drososphila simulans and Hybrid male rescue (Hmr), which has functionally diverged in Drosophila melanogaster [40]. In another study, hybrid lethality is caused by the nucleoporin 160 kDa (Nup160) gene of D. simulans, which is incompatible with one or more factors from the D. melanogaster X chromosome [41]. In C. elegans, the interactions between two tightly linked but diverged alleles zeel-1 and peel-1 causes widespread genetic incompatibility [42]. Recent work in Arabidopsis supports functional divergence between duplicate genes that lead to hybrid incompatibilities between ecotypes [44] or hybrid necrosis in intraspecific hybrids [43]. In mammals, hybrid incompatibilities are related to abnormal expression patterns of imprinting genes in interspecific hybrids in Peromyscus [45] or epigenetic activation of retrolelements in marsupial hybrids [46]. In plants, some imprinted genes were abnormally silenced in Arabidopsis interspecific hybrids [31, 47], and many protein-coding genes are epigenetically regulated in allotetraploids [48, 49].
For the genetically viable hybrids, the degree of heterosis is proportional to the genic differences in two parental strains [50]. In other words, the levels of heterosis increase as the genetic distances between the parents increase. After evaluating the phenotypic data from 37 genera, including Zea, Solanum, and Nicotiana, E.M. East (1936) noted that interspecific hybrids generally show more heterosis than intraspecific hybrids, if the genetic difference between the species or genera does not prevent them from forming compatible hybrids. The hybrids formed between different subgenera show more heterosis than the hybrids formed between species within the same subgenera. If the hybrids are incompatible, they are dwarf and stunted, probably because dramatic differences in growth and reproductive development inherited from the divergent parents fail to be reconciled. Indeed, the hybrids formed between subgenera often have more heterosis as well as more dwarfs. For example, most intergenic or interspecific hybrids are abnormal, and yet the greatest amount of heterosis is found in the hybrids derived from Raphanus and Brassica [7]. In rice, the hybrids between two subspecies show more heterosis than the hybrids between varieties within a subspecies. However, the notion may not be generalized across all hybrids. In maize (Z. mays) and tobacco, although the varieties (inbred lines) are genetically similar, the hybrids formed between different combinations of varieties show dramatic levels of heterosis. This suggests that the interaction between a few genes or the combination of a few genes in a genetic cross plays an important role in heterosis, as observed in tomato [51]. Alternatively, large-scale recombination suppression accompanied by a high level of residual heterozygosity could be associated with inbreeding depression and heterosis in maize [52, 53]. Notably, genetic mechanisms responsible for heterosis may be different between the species that are naturally self-pollinating and out-crossing. Heterosis is more predominant in outcrossing than inbreeding species, and the inbreeding populations do not have obvious heterosis of fitness.
Notably, heterosis present in the interspecific hybrids is permanently fixed in the respective allopolyploids in which the chromosomes are doubled. This is facilitated by many allopolyploids that become self-pollinating irrespective of pollinating patterns in the parents. Thus, the heterosis is heritable and selected in the allopolyploid progeny. Although heterosis in interspecific hybrids and allopolyploids is generally high, the heterosis in autopolyploids is not obvious [50, 54]. In Arabidopsis, diploids and autotetraploids often have similar morphology, leaf sizes, and plant stature. The autotetraploids have slightly larger flowers and seeds (Figure 1c and d), and flower later than the diploids, depending on the combination of genotypes. For example, the difference in flowering time between a diploid and an autotetraploid is greater in Columbia ecotype than in Landsberg erecta ecotype.
The degree of heterosis may shift during different stages of growth and development [51]. If growth vigor is shown in the early stages, it is often exhibited not only in seedlings, vegetative tissues and organs such as rosettes, and overall biomass, but also in the late stages of reproduction such as in the flowers and fruits. In some plants, heterosis in vegetative growth is different from that in reproductive development because they are controlled by different sets of genes and regulatory pathways. It is notable that biomass heterosis in plants is largely dependent on flowering time. For example, late flowering and indefinite inflorescent plants often have greater biomass than the early flowering and definite inflorescent plants. The flowering time is controlled by a few loci in inbreeding Arabidopsis and rice [55-58]. The single-locus heterosis in tomato could be controlled by a FT-like locus that regulates the transition from definite to indefinite inflorescence [51]. In outcrossing maize, the flowering time is mediated by additive effects of numerous (two-dozen or more) quantitative loci (QTLs), each with only a small effect on the trait [59]. Interestingly, in Brassica napus late flowering is heterotic, whereas in maize hybrids early flowering is, suggesting different effects of gene actions (repression or activation) on heterosis.
Genetic models for hybrid vigor
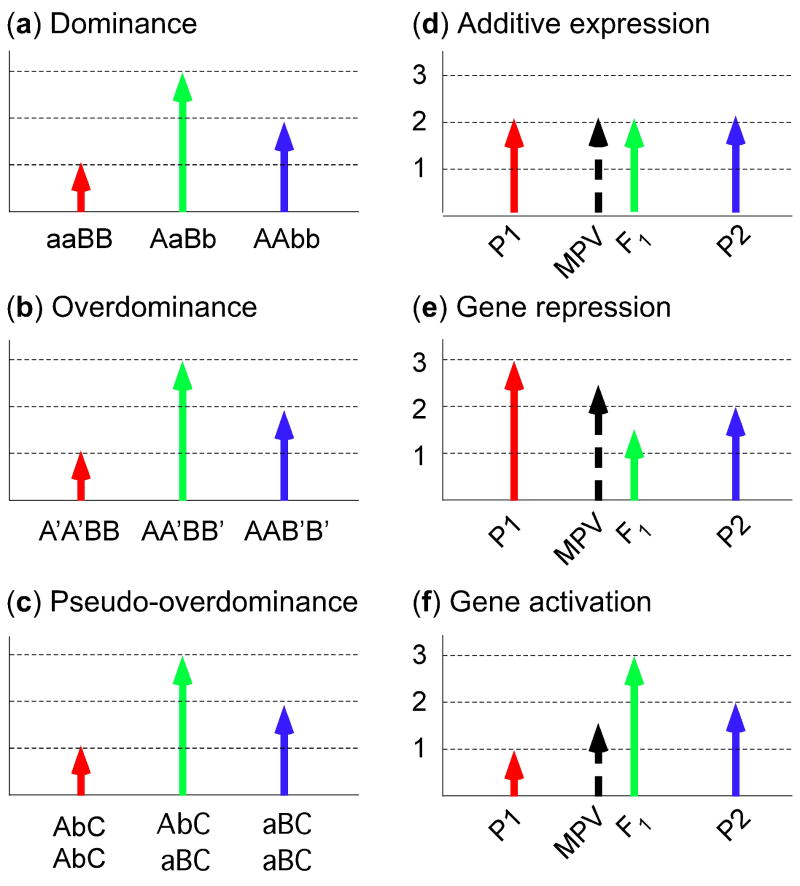
The genetic basis for hybrid vigor or heterosis has been debated for over a century, but little consensus has been reached. Several hypotheses including dominance, overdominance, and pesudo-overdominance are available to explain the phenomenon of hybrid vigor. According to the dominance model [60, 61], inbred parents contain inferior or deleterious alleles in several loci that inhibit overall good performance, whereas in the hybrids these inferior alleles in one parent are complemented by the superior or dominant alleles from the other parent (Figure 2a). As a result, the hybrids have an overall better performance than the parents. The model is based on the dominance (wild type) and recessive (mutant) aspect of trait performance, and genetic complementation is likely to occur in the combination of alleles from respective parents. Moreover, one can apply statistical models to dissect additive and dominant components of genetic variation. In theory, the parent that contains homozygous superior or dominant alleles for all possible loci would perform better than the hybrids, but hybrid maize breeding practice has indicated otherwise. In spite of dramatic improvement of inbred parents by eliminating deleterious alleles, the heterotic (or allelomorphic) responses in the hybrids often exceed those in the parents [50]. Maize is naturally outcrossing and requires a certain amount of combinational dominant and recessive alleles in some genetic loci to avoid inferior performance or lethality from being completely inbred. In other words, the parent with recessive alleles in all genetic loci would be deleterious, as would the parent with dominant alleles in all genetic loci.
Figure 2.
Genetic models and nonadditive gene expression for heterosis. (a) The dominance model. The F1 with both dominant alleles (AaBb) of two loci is superior to the parents that contain only one pair of dominant alleles (aaBB and AAbb) because the superior or dominant allele complements the inferior or recessive allele. (b) The overdominance model. The interactions between heterozygous alleles in F1 (AA′BB′) causes superior phenotypes compared with the combinations of homozygous alleles in the parents (A′A′BB and AAB′B′). (c) The pseudo-overdominance model. The combination of dominant alleles (AaBb) in repulsion (AbC/aBC) in the F1 acts as overdominance compared with homozygous parents (AAbbCC and aaBBCC). The presence of dominant alleles in F1 complements the recessive alleles, leading to a better phenotype. (d) Additive expression. The expression level of a gene, genotype or phenotype is additive. Abbreviations: MPV, mid-parent value (1/2P1 + 1/2P2); P1, parent 1; P2, parent 2. P1, P2, MPV, and F1 represent the values of gene expression, genotype or phenotype. (e, f) Nonadditive expression. (e) Gene repression. The expression of a gene, genotype or phenotype is lower than the MPV. (f) Gene activation. The expression of a gene, genotype or phenotype is higher than the MPV, which includes dominance, overdominance, and pseudo-overdominance models. Gene repression and activation also explain epistatic interactions. Relative expression levels (1, 2 and 3) are shown on y-axis.
The overdominance model [2, 50, 62] suggests novel allelic interactions within each of many genetic loci lead to superior function over the homozygous states in the inbred parents (Figure 2b). This model is favored because hybrids always outperform the parents that have been excessively inbred and selected and contain many superior or dominant genetic loci [50]. Moreover, it is the allelic combination in the hybrids that determines the levels of heterosis. The genetic composition of inbred parents does not necessarily predict the levels of hybrid vigor. A challenge for this model is to identify the best combination of a single genetic locus or a few loci that contribute to the overall heterosis, which seems to contradict the hybrid performance of many agronomic traits that are controlled by multiple genetic loci.
A recent study [63] has suggested an alternative model, pseudo-overdominance (Figure 2c). The overdominance is associated with the complementation of two or more linked dominant and recessive alleles in repulsion, in which the dominant and recessive alleles are located on opposite homologs of the two genes, acting as overdominance. The heterosis associated with pseudo-overdominance can dissipate in the selfing progeny because genetic recombination leads to the dissociation of the alleles from the repulsion state, which is exactly what is observed in a study with tomato hybrids [63]. This pseudo-overdominance can also arise from numerous alleles in recombination suppression regions where good and bad allele combinations are in repulsion [52, 53].
These genetic models have limitations. For example, heterosis in rice has been found to be associated with three different models, namely, dominance [64], overdominance [65], and epistasis [66]. These different conclusions are probably related to the complexity of genetic bases and trait variability for heterosis. First, heterosis can result from epistatic interactions among the alleles in different loci, which cannot be easily explained by statistical models. Epistasis is involved in many QTLs associated with inbreeding depression and heterosis in maize [67] and rice[66, 68]. Second, heterosis is affected by genetic backgrounds. For example, one of the two QTLs controlling differences in morphology and inflorescence architecture between maize and its ancestor (teosinte, Zea mays ssp. parviglumis) has strong phenotypic effects in the teosinte background but reduced effects in the maize genetic background [69]. When the two QTLs are combined into one genotype, both morphology and inflorescence architecture are altered. In an extensive analysis of heterosis for dry biomass in 63 Arabidopsis accessions that are crossed with three reference lines (Col-0, C24, and Nd), the authors of Ref. [25] found that 29 out of 169 crosses showed significant heterosis for shoot biomass, and the biomass heterosis is higher in some hybrids (e.g., Col × C24) than in others. This is consistent with the higher levels of growth vigor in interspecific hybrids than in the ecotype hybrids (Figure 1). Third, altered levels of heterosis observed in different genetic backgrounds also suggest a role for maternal and paternal effects of genetic loci in hybrid performance [70], although allelic expression variation is not commonly observed in reciprocal crosses [71, 72]. However, a recent study suggested otherwise, and nearly 50% genes show paternal dominant expression patterns in the seedlings of maize reciprocal hybrids [73], which is different from similar phenotypes observed in reciprocal hybrids [54, 71, 72]. It is likely that some of these changes in gene expression may dissipate over time. Fourth, heterosis is affected by many genetic loci. Statistical and genetic models cannot accurately estimate the relative contribution of individual loci to a particular pathway or trait. Some transcription factors and chromatin proteins may control the expression of many other genes in various biological pathways. Finally, these genetic models do not explain well the heterosis in polyploid plants because allelic and genomic dosage may play a more important role than the allelic complementation or interactions. Changes in dosage-dependent gene expression may be more profound than alteration in allelic interactions. In maize, the increased number of gene and genome dosage appears to have a negative effect on growth vigor and increased levels of inbreeding depression [54].
Nonadditive gene expression in the hybrids and allotetraploids
At gene expression levels, the dominance model suggests that the expression of genes in the hybrids is a result of combined or additive expression of two alleles in the parents (e.g., 1 + 1 = 2) (Figure 2d), whereas the overdominance model indicates that allelic interactions in the hybrids lead to nonadditive expression of the alleles derived from the parents (1 + 1 ≠ 2) (Figure 2e and f). If the interactions lead to positive effects or gene activation, the outcome is expected to be 1 + 1 >2. If the interactions result in negative effects or gene repression, the expected outcome would be 1 + 1 <2. The expression of some genes falls in the range between additive and nonadditive expression. Nonadditive expression explains positive as well as negative epistatic interactions.
Nonadditive expression of 30 selected genes were studied in maize diploid and triploid hybrids using RNA blots and normalized expression values with internal controls [74]. The expression values of 19–20 genes in reciprocal hybrids are different from the mid-parent values (MPV). The transcript levels in the diploid hybrids correlate negatively with the levels in diploid inbreds. Moreover, genome dosage affects transcript levels in diploid and triploid hybrids. The transcript levels are higher in triploids than those of diploids. The transcript levels for nearly half of the genes tested are different in reciprocal triploid crosses, suggesting strong maternal effects of gene expression in triploid hybrids.
In a study using cDNA microarrays, the authors [75] found that ∼10% of ESTs are expressed differently between the two inbred parents. Among them, 78% (1062 of 1367) of ESTs were additively expressed in the hybrids relative to the MPV, and 22% are nonadditively expressed. The expression patterns include all possible modes of nonadditive expression: high- and low-parent dominance, underdominance, and overdominance. The data suggest that multiple molecular mechanisms, including overdominance, contribute to heterosis. In a similar study using microarrays, the authors [72] found that ∼80% of the genes that are expressed differently between the two parents are additively expressed in the hybrids. However, among 20% of nonadditively expressed genes, many are expressed at the levels within the parental range. Few genes showed expression levels higher than the high parent or lower than the low parent. Further analysis of allele-specific expression patterns in the hybrid indicates that gene expression variation is largely associated with cis-regulatory variation. The data suggest that cis-regulatory variation between the alleles maintains allelic expression levels in the F1 hybrid, leading to additive expression. Another study [76] suggested that hybrid yield and heterosis are associated positively with the proportion of additively expressed genes, negatively with the proportion of paternally expressed genes, and not correlated with over- or under-expression of some specific genes. These different conclusions related to the relative contribution of additive and nonadditive expression to the hybrid performance in similar studies using the same pair of inbred parents might be caused by developmental variation among different tissues examined, various normalization methods and and/or different statistical tools used in microarray and RNA blot analyses. Moreover, it is not surprising to identify positive effects of additive expression on heterosis because the proportion of additively expressed genes is generally high (∼80%).
Allelic expression variation varies from unequal expression of both alleles (biallelic) to expression of a single allele (monoallelic) in the hybrids, which is reminiscent of developmental reactivation of silenced rRNA genes in Brassica allotetraploids [77] and organ-specific reciprocal silencing in cotton allotetraploids [78], although they involve two homoeologous loci. In maize hybrids, the allelic expression variation can respond to planting density and drought stress [71]. For example, biallelic expression for 7 of 15 genes examined is found in a genetically improved modern hybrid, whereas mono-allelic expression is observed in a less improved old hybrid. The two alleles of stress responsive genes in the hybrid are differentially expressed in response to density and drought stresses. Although maternal or paternal effects on allelic expression are not commonly observed in vegetative tissues and seedlings, expression of many genes (∼8%) is deviated from a 1:1 ratio, the expected ratio in the hybrids of reciprocal crosses, and 2:1, the expected ratio in three stages of endosperm development in the hybrids of reciprocal crosses [70]. These genes resemble maternally or paternally expressed genes, which is probably associated with genomic imprinting. The gene encoding a no-apical-meristem (NAM) related protein 1 (nrp1) is expressed only in the endosperm, in which the maternally transmitted alleles are expressed, whereas the paternally transmitted alleles are silenced throughout the three developmental stages.
Genome-wide nonadditive expression of homoeologous loci has been extensively studied in many interspecific hybrids and allopolyploids, including Arabidopsis, Brassica, cotton, Drosophila, Senecio, and wheat (see review and ref. [79]). Although the levels of gene expression detected vary from one experimental species to another, the trends are similar. The levels of differentially expressed genes between the related species are higher than those within species. Over 15–50% of genes are differentially expressed between the related species in plants or animals. The number of nonadditively expressed genes ranges from 5–38% in Arabidopsis allotetraploids to ∼30% in cotton allotetraploids [80]. In Senecio, the number of differentially expressed genes between the natural and synthetic allopolyploids can be as high as ∼60% [81], although some of which could be related to genotypic differences between synthetic and natural allopolyploids. In Arabidopsis allotetraploids, >65% of the nonadditively expressed genes are repressed, and >94% of the repressed genes in the allotetraploids are expressed at higher levels in A. thaliana than in A. arenosa, consistent with the silencing of A. thaliana rRNA genes subjected to nucleolar dominance [77] and with overall suppression of the A. thaliana phenotype in the synthetic allotetraploids and natural A. suecica [82]. The data suggest transcriptome and phenotypic dominance of A. arenosa over A. thaliana in the allotetraploids. In cotton, the A-genome ESTs are selectively enriched in the allotetraploid [83], a result consistent with the production of long lint fibers in A-genome species. However, in another study, the expression of homoeologous loci is shifted toward the D-genome species [80], which does not produce spinnable fibers. Moreover, ∼20% of the genes show locus-specific expression patterns in different stages of fiber development. The data support the role of developmental regulation in the expression rRNA genes and protein-coding genes found in Arabidopsis and Brassica allotetraploids [77, 82].
Genome-wide gene expression data collectively support the genetic models of dominance and overdominance at the levels of individual genes but do not provide mechanistic insights into the molecular basis for heterosis.
A molecular clock model for growth vigor in hybrids and allopolyploids
At the molecular levels, both dominance and overdominance models suggest nonadditive expression of alleles in the hybrids relative to the parents. The dominant mode of gene expression represents one extreme: monoallelic expression in the hybrids, whereas overdominant mode of gene expression indicates another: biallelic expression in the hybrids at the levels either higher than the high-parent value or lower than the low-parent value. Neither the dominance nor the overdominance model can explain the epistatic interactions among different genes and gene products that are involved in the same or different regulatory and/or biological pathways, leading to an altered trait or phenotype. Moreover, heterosis changes over time or during growth and development of plants and animals. For example, heterosis in biomass such as vigorous growth in seedlings, roots, and other vegetative tissues may not be directly translated into large fruits or seeds because different sets of genes in the biological pathways control vegetative growth and reproductive development, although some pathways are intricately related. Therefore, a molecular model for heterosis should define individual genes in specific regulatory pathways. One model is that epigenetic regulation induces nonadditive expression of one or more key regulator genes in the hybrids, which in turn mediates the expression of many other genes in the same regulatory networks associated with changes in developmental and physiological pathways, leading to heterosis in specific stages of growth and development. As a result, nonadditive expression of many genes collectively in various biological pathways gives rise to an overall vigor of vegetative growth and yield.
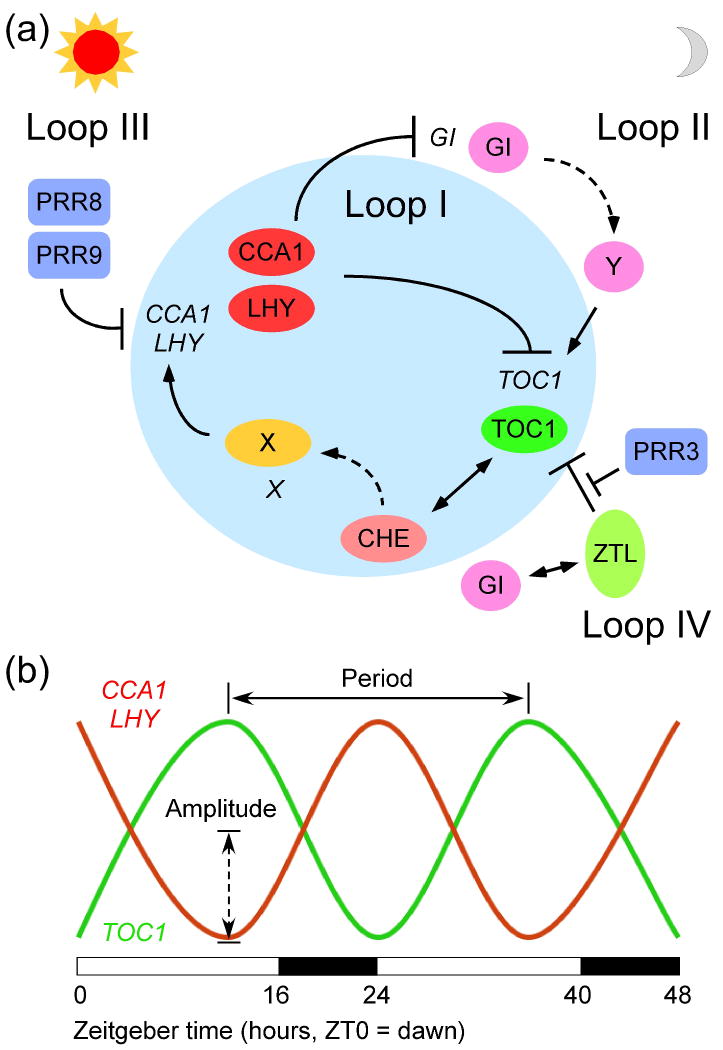
Circadian clocks affect many physiological and developmental processes, including various metabolic pathways and fitness traits in animals and plants, and photosynthesis and starch metabolism in plants (see box 1) [84-87]. In Arabidopsis, the central oscillators of circadian clock consist of negative regulators CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) [88, 89] and reciprocal positive regulators TIMING OF CAB EXPRESSION 1 (TOC1), CCA1 Hiking Expedition (CHE) [90], and GIGANTEA (GI) [89, 91, 92]. CHE, a transcription factor belonging to the TCP class, represses CCA1 expression [90]. CCA1 and LHY are partially redundant MYB-domain transcription factors with incompletely overlapping functions [88, 89]. CCA1 and LHY negatively regulate TOC1 and GI expression, whereas TOC1 binds to the CCA1 promoter and interacts with CHE, positively regulating CCA1 and LHY expression [89-91, 93]. This circular feedback regulation affects the rhythms, amplitude and/or period of circadian clock as well as its input and output pathways in Arabidopsis [94, 95]. At least ∼15% of genes, including those involved in photosynthesis and starch metabolism [96, 97], and up to 90% of transcriptome [98] are affected by the circadian clock regulators. Moreover, day-length and circadian effects on transitory starch degradation and maltose metabolism correlate with the diurnal expression patterns of these metabolic genes [99]. Consequently, maintaining circadian regulation increases CO2 fixation and growth, whereas disrupting circadian rhythms reduces fitness [87, 100].
Box 1. Central role of the circadian clock in plant growth and development.
Every organism under the sun lives by day and night with a constant cycle of ∼24 hours. Plants, in particular, during the day, convert sunlight, water, and carbon dioxide into carbohydrates and eventually biomass, and emit oxygen as a byproduct of photosynthesis. At night, plants store, transport, and use the carbohydrates, and release energy, carbon dioxide, and water as a byproduct of respiration. Moreover, the temperature and growth conditions change during day and night. These rhythmic cycles are known as the circadian clock, which is derived from the Latin words “circa” (about) and “dies” (day) [136]. The scientific literature on circadian rhythms began with the daily leaf movements of heliotrope plants even in continuous darkness [137], suggesting an internal circadian rhythm. Figure I (a) Internal time keepers or circadian clock regulators include CCA1, LHY and TOC1 in a major negative feedback loop (Loop I) of the circadian oscillator in Arabidopsis, which produces a self-sustaining and constant periodicity of 24 hours, even when plants are grown under constant light and temperature. CCA1 Hiking Expedition (CHE) has recently been shown to be a negative regulator of CCA1 [90]. In addition to CCA1, LHY, and TOC1, other regulatory loops include one (Loop III) consisting of PSEUDO-RESPONSE REGULATOR (PRR) 7 and 9, another (Loop II) of GI and unknown protein, and another (Loop IV) of ZEITLUPE (ZTL), GI, and PRR3. Figure I (b) Diagram of CCA1 and LHY (red line) and TOC1 (green line) expression rhythms in a 24-hour clock with 16 hours of light (open bar) and 8 hours of darkness (filled bar). Zeitgeber (ZT) is German for time giver, and dawn is defined as ZT0. Period is the time for completing one cycle of rhythms and is shown from one peak to another (or form one trough to another). The expression amplitude of rhythm is defined as one-half the distance between the peak and trough. Many aspects of plant physiology, metabolism and development are under circadian control, and a large proportion of transcriptome (from 15% up to ∼90%) shows circadian regulation [96, 98]. For further information, see the many excellent reviews in the field, including historical perspectives of circadian rhythms [94], how plants tell time [138], regulation of output from the circadian clock [139], and the most recent reviews of circadian systems in higher plants [95, 140].
Figure I.
Analyzing genome-wide nonadditively expressed genes in Arabidopsis allotetraploids [82], the authors [101] found that among ∼130 genes upregulated in the allotetraploids, two thirds of them in their upstream regions contain at least one (CCA1)-binding site (CBS; AAAAATCT) or evening element (AAAATATCT) [96]. One subset of the genes encodes protochlorophyllide (pchlide) oxidoreductases a and b, PORA and PORB, that mediate the light-requiring step in chlorophyll biosynthesis in higher plants [102]. Both PORA and PORB are upregulated in the allotetraploids. In A. thaliana, PORA and PORB are expressed at high levels in seedlings and young leaves, and overexpression of PORA and PORB increases chlorophyll a and b content [103]. The other subset of genes encodes all major enzymes in starch metabolism and sugar transport [104, 105], many of which contain EE/CBS and are upregulated in the allotetraploids. As a result, the allotetraploids accumulate ∼60% more starch than the low parent and ∼30% more than the high parent, and ∼70% more chlorophyll than the low parent. The starch amount in the allotetraploids is twofold to fourfold more than the low parent and 70% more than the high parent, and the sugar content is 50–100% more in the allotetraploids than in the parents.
The study further established a direct connection between epigenetic repression of CCA1 and LHY and upregulation of the genes involved in the light-requiring processes of photosynthesis, starch metabolism, and sugar biosynthesis in the hybrids and allopolyploids [101]. This daytime-specific repression of clock genes has an epigenetic cause because it correlates with loss of histone modifications (e.g., H3k9 acetylation and H3K4 dimethylation) that are normally associated with active transcription from the CCA1 and LHY genes. By contrast, upregulation of TOC1 and GI correlates with increased levels of H3K9 acetylation and H3K4 dimethylation. Interestingly, similar repression of CCA1 and LHY and upregulation of TOC1 are also found in the F1 hybrids made by crossing C24 and Colombia strains of A. thaliana without ploidy changes. However, the levels of changes in gene expression, chlorophyll and starch content in the hybrids are lower than in the allotetraploids. This observation is consistent with a positive correlation between the levels of heterosis and genetic distances among the parents used in the hybrids. Similar expression changes of a CCA1-like gene were observed in maize hybrids as those observed in Arabidopsis hybrids [101] (Z.J. Chen, unpublished).
Altering the clock amplitude but maintaining the rhythmic phase increases growth vigor in the hybrids and allotetraploids (Figure 3). Expressing TOC1::CCA1 and TOC1::cca1(RNAi) in the diploid transgenic plants mimics alteration in the CCA1 expression amplitude. Repressing or over-expressing CCA1 under the TOC1 promoter might also slightly affect rhythmic phase and have pleiotropic (but mild) effects on flowering time and plant growth [101], but these effects may be minimal. Completely knocking out clock genes affects other aspects of plant growth and development, and the plants may lose their fitness and growth vigor. Although the results obtained in cca1 and lhy mutants also show increased growth vigor in the early stages [101], over time the constant loss of rhythmic phase in the mutants induces many other changes, including flowering time and physiological syndromes, leading to low fitness and small plants in the late stages of development [84]. The mutant plants are likely to develop indirect effects independent of original cca1 lhy double mutations such as flowering time defects [106]. Moreover, the genetic interaction between CCA1 or LHY and TOC1 is complex. TOC1 mediates the floral transition in a CCA1 or LHY-dependent manner, whereas CCA1/LHY function upstream of TOC1 in regulating a photomorphogenic process [107]. In Arabidopsis C24 × Columbia F1 hybrids, heterosis for biomass (leaf size and dry shoot mass) is 2–3 times higher at high light intensity than at low and intermediate light intensities [25]. The relative growth rates of the hybrids are high in the early developmental stages under low and intermediate light intensities and constantly high over the entire vegetative phase under high light intensity. The above data suggest other factors such as light intensities and light signaling pathways affect the degree and early onset of heterosis for biomass.
Figure 3.
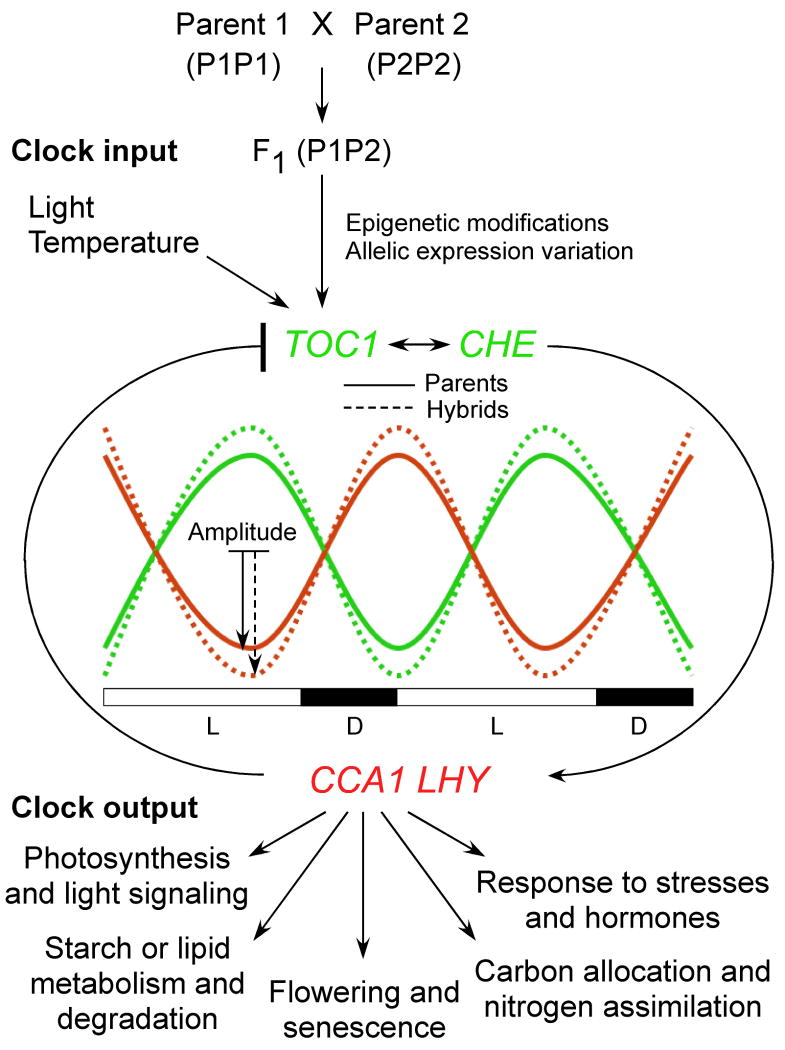
Growing around the clock: a molecular mechanism for hybrid vigor. A molecular clock model explains the basis of heterosis. The internal clocks of plants are controlled by multiple feedback loops, including a major loop that consists of two transcription repressors CCA1 and LHY with redundant but incompletely overlapping functions and feedback regulators TOC1 and CHE (see Box 1). The clock receives input signals such as lights and temperature and controls output traits and pathways, including photosynthesis and light signaling, flowering, starch biosynthesis and metabolism, responses to stresses and hormones, and carbon allocation and nitrogen assimilation, through the expression of evening element (EE) or CCA1 binding site (CBS)-associated genes. The expression amplitude and periodicity of circadian clock regulators can be changed or fine-tuned in response to input (external) signals such as light and temperature, as well as internal mechanisms such as allelic expression variation. L and D indicate the length of light (L) and darkness (D) in a circadian cycle. In the hybrids, the allelic interactions between parent 1 (P1) and parent 2 (P2) induce epigenetic repression of CCA1 and LHY expression amplitudes (red dashed line) and upregulation of TOC1 expression amplitudes (green dashed line) relative to the expression values in the parents (solid red and green lines, respectively), whereas the periodicity of the clock remains the same [101] because maintaining clock periodicity and rhythm is important for plant growth and fitness [84]. The reduced amount of CCA1 repressors in the hybrids during the day induces the expression of circadian-clock-associated genes (CCGs) in various output pathways, including chlorophyll biosynthesis and starch metabolism and degradation. As a result, the hybrids produce more chlorophyll and starch than the parents, which promotes vegetative growth and morphological vigor. The CCA1 expression amplitude is regulated by chromatin modifications, where the levels of active histone marks are reduced during the day and increased at night. The hybrid-induced changes in the CCA1 expression amplitude are reminiscent of expression alterations in response to changes in input signals such as light (intensities) and temperature. The clock modulates auxin signaling and responses [141]. In addition, the output pathways also produce feedback regulation for the internal clocks. For example, circadian oscillator regulation requires organic nitrogen signals [142] and free cytosolic Ca2+ [143]. Allelic interactions in the hybrids induce superior performance of physiological pathways for chlorophyll biosynthesis and starch metabolism. The overdominant performance is caused by epigenetic repression (nonadditive expression) of a key regulator in the feedback loop of the clock oscillator, which mediates the downstream genes in chlorophyll biosynthesis and starch metabolism. Clock-mediated heterosis is probably universal because internal clocks mediate physiological and metabolic pathways in plants and animals. Moreover, this model can be extrapolated to explain superior traits of many other biological pathways.
Do the changes in circadian clock genes affect other traits in hybrids? Many life history traits, including plant height and leaf length and number, were coincidently mapped in the locations of CCA1 (bottom of chromosome 2) and LHY (top of chromosome 1) in the recombinant inbred lines (RILs) derived from Ler and Cvi [27] (Z.J. Chen, unpublished). Another locus CRY2 in the vicinity of LHY was also a candidate for fruit length and ovule number but not for other traits [28], suggesting a role of epistatic interactions among CCA1, LHY, and CRY2 in life history traits. As noted above, heterosis is manifested in many different forms during growth and development. Other key regulators and/or environmental factors such as light intensities, photoperiod, nutrients, and the conditions for optimal growth can also affect many other pathways and traits such as plant stature, flower size, seed fertility, and yield.
How is the allelic or locus-specific expression of CCA1 and other clock regulators established in the hybrids and allotetraploids? Although allelic expression variation of clock genes has not been studied in the Arabidopsis hybrids, the locus-specific expression was observed in two allotetraploid lines examined [101]. In respective parents, A. thaliana and A. arenosa loci were equally expressed. In the allotetraploids A. thaliana CCA1 (AtCCA1) was repressed 2-3-fold more than the A. arenosa CCA1 (AaCCA1) whose expression was slightly reduced. Similarly, the repression of AaLHY was 1.5-fold lower than the AtLHY in the allotetraploids. Conversely, AtTOC1 and AtGI loci were upregulated more than AaTOC1 and AaGI in the allotetraploids. The data collectively indicate that A. thaliana genes are more sensitive to expression changes (repression or activation) than the homoeologous A. arenosa genes through epigenetic modifications in the allotetraploids [82, 101, 108]. Moreover, both A. thaliana C24 and Columbia alleles in the hybrids or both A. thaliana and A. arenosa loci in the allotetraploids are expressed but either upregulated or repressed relative to the MPV, suggesting a role for expression overdominance or repression in hybrid vigor.
Altering expression of a few genes in the circadian clock regulation to promote growth vigor is reminiscent of single locus heterosis, which has been documented for the erecta and augustifolia loci in A. thaliana [109]. These loci also show an overdominant mode of expression and encode regulatory proteins, namely, a receptor-like kinase [110] and a transcription factor [111], respectively. This offers a solution to clone QTLs that have been extensively studied in the hybrids of Arabidopsis, tomato, maize, and rice. For example, the genetic basis of heterosis in an elite rice hybrid is controlled by single-locus heterotic effects and dominance-by-dominance interactions [112].
A good example is the domestication of maize (Zea mays spp. mays), which involves a transition of apical dominance (a collection of stem cells for the development of main stem and axillary branches) from its probable wild ancestor, teosinte (Zea mays ssp. parviglumis). The apical dominance is controlled by a major genetic locus named teosinte branched 1 (tb1). tb1 encodes a protein with homology to the cycloidea in snapdragon. tb1 represses the growth of axillary organs and promotes the formation of female inflorescences. The maize allele of tb1 is expressed at twice the level of the teosinte allele, suggesting that gene regulatory changes underlie the evolutionary divergence of maize from teosinte [113]. Another example is the domestication of tomato (Solanaceae). The wild type produces few-flowered inflorescences, but the mutants compound inflorescence (s) and anantha (an) are highly branched, and s produces hundreds of flowers [51]. The S and AN encode a homeobox transcription factor and an F-box protein, respectively. Apical dominance and branch formation are controlled by a few regulatory genes, suggesting a molecular basis for single-locus heterosis. However, the connection between the gene function and morphological variation in these studies has yet to be established, and also it is debatable whether the control of inflorescence architecture (e.g., from definite and indefinite) by promoting progression of an inflorescence meristem to floral organs is part of heterosis or developmental variation.
Allelic activation and repression through cis- and trans-acting effects in hybrids or allopolyploids is reminiscent of paramutation [114, 115], X-inactivation [116, 117], and repeat-associated gene silencing [118]. However, in hybrids and allopolyploids allelic- and locus-specific expression occurs in a genome-wide scale, which occurs on any chromosomes but does not occur at every locus in a specific chromosome or even in a small chromosomal segment [49]. In some cases, epigenetic regulation is stochastic and takes several generations to establish [48]. In contrast to random inactivation of paternal and maternal X-chromosomes in somatic cells, there is a dominance hierarchy for locus-specific gene expression in allopolyploids. The expression of homoeologous genes, including rDNA loci, is dominant from one parent over the other in the interspecific hybrids or allopolyploids. The dominance phenomenon is similar to paramutagenic and paramutable alleles in paramutation, but the expression of two alleles and loci in the hybrids and allopolyploids is additive, whereas the paramutagenic allele exerts transgenerational effects on the expression of the paramutable allele. Compared with epigenetic silencing of endogenous repeat gene loci, the alleles or homoeologous loci examined in the hybrids and allopolyploids do not have obvious internal repeats. If epigenetic mechanisms are responsible for allelic- and locus-specific gene expression in hybrids and allopolyploids, they probably operate through cis- and trans-acting effects [119, 120], chromatin modifications, and/or small RNAs that discriminate between homoeologous loci [108, 121].
Roles for small RNAs in hybrid vigor and incompatibility in allotetraploids
The above models suggest that epigenetic and transcriptional regulation of key regulatory genes leads to heterosis. Nonadditive gene expression is also controlled by post-transcriptional mechanisms via RNA-mediated pathways [108, 122]. Small RNAs, including microRNAs (miRNAs) [123], small interfering RNAs (siRNAs) [124], and trans-acting siRNAs (tasiRNAs) [125, 126], mediate post-transcriptional regulation, RNA-directed DNA methylation, and chromatin remodeling. miRNAs are produced from genetic loci independent of their targets and serve as negative regulators of gene expression by targeting RNA degradation or translational repression [123]. tasiRNAs arise in plants from specific TAS loci that are transcribed into precursors, which are cleaved by miRNA-guided mechanisms. The resulting 21-nt tasiRNAs direct the degradation of target mRNAs [125, 126]. miRNAs and tasiRNAs control the expression of genes that encode transcription factors and proteins that are important for growth and development. It is conceivable that different ecotypes and species might have developed specific growth and developmental patterns, which are partly mediated by miRNAs and tasiRNAs. Combination of miRNAs and their targets of different parental origins in the hybrids or new allopolyploid species may reprogram expression of miRNAs and tasiRNAs and their targets [127]. Indeed, many miRNA targets are nonadditively expressed in the allotetraploids [82], suggesting a role for miRNAs in buffering genetic clashes between species [127]. In a recent study using massive parallel sequencing of small RNAs and microarray analysis of miRNAs in resynthesized and natural Arabidopsis allotetraploids and their progenitors, the authors found that miRNAs and tasiRNAs but not the siRNAs are associated with nonadditive expression of target genes in the allotetraploids [122]. Although the sequences of many miRNAs are conserved, miRNA accumulation levels are nonadditive in the leaves or flowers of interspecific hybrids and allotetraploids relative to the parents. Nonadditive accumulation levels of miRNAs are associated positively with the expression levels of miRNA biogenesis genes such as AGO1 and DCL1 but negatively with many miRNA targets. The data suggest that expression variation of miRNAs and their targets in the hybrids and allotetraploids are controlled by epigenetic mechanisms at transcriptional and post-transcriptional levels. The genome merger in the allotetraploids induces epigenetic modifications [108], leading to nonadditive expression of some miRNA targets, miRNA primary transcripts, and miRNA biogenesis genes. At the post-transcriptional level, nonadditive expression of miRNA biogenesis genes can affect the processing efficiency of miRNA precursors, resulting in nonadditive accumulation of miRNAs. Moreover, differential expression of A. thaliana and A. arenosa miRNAs and their targets in the allotetraploids leads to biased target degradation, probably because the efficiency of target mRNA degradation is dependent on a threshold of miRNA concentration [128]. In addition, although the target loci of different parental origins are conserved, their secondary structures might have diverged, which affects the efficiency of miRNA-triggered degradation [129].
Repeat-associated siRNAs (rasiRNAs) are predominately derived from transposons and repeats and highly enriched in centromeres and heterochomatic regions [130], and diverge rapidly among closely related species. The rasiRNA population is relatively low in F1, and many rasiRNAs absent in F1 are restored in late and natural allotetraploids, indicating that it takes several generations to establish stable expression patterns of siRNAs of protein-coding genes [48]. Although the proportion of rasiRNAs is lower in F1 than in A. thaliana, the number of miRNA reads is higher in F1 than in A. thaliana, indicating rapid and dynamic changes of siRNAs and miRNAs in early stages of allopolyploid formation. A few transposons generated new siRNAs in F1, F7 allotetraploids, and/or A. suecica. This might be related to sequencing depth or activation of these elements in allopolyploids. Reduction of siRNAs in F1 may activate some transposable elements in response to “genomic shock” [131] in marsupial interspecific hybrids [46] and induce genome instability and infertility in Arabidopsis allotetraploids [48, 132]. siRNA-directed DNA methylation and chromatin modifications are required for the establishment and maintenance of heterochromatin and centromeres [124, 130], leading to genome stability. Consistent with the notion, siRNA accumulation is related to DNA hypermethylation of A. thaliana homoeologous centromeres in natural allotetraploid A. suecica [121]. During F1 and early stages of allotetraploid formation, genomic shock causes meiotic disorders and genome instability [131], probably resulting from a temporary loss of siRNAs. Over time, genome stability is restored through regeneration of rasiRNAs in genetically stable allotetraploids.
Some rasiRNAs are associated with gene repression in diploids but weakly with gene expression changes between the related species or in allotetraploids. The correlation between siRNA-generating genes and the genes that are nonadditively expressed in the allotetraploids is insignificant, which is consistent with a few genes that are affected by DNA hypomethylation in A. suecica [121]. This is because siRNAs are tightly regulated for the maintenance of heterochromatin and genome stability. It is also likely that the majority of nonadditively expressed genes encode proteins, and siRNA-containing transposons and repeats are underrepresented in microarrays [82].
A probable model is that siRNAs are inherited maternally to silence transposons that are reactivated during gametogenesis. The repression of A. thaliana homoeologous loci [82] and accumulation of A. thaliana centromeric siRNAs [121] are similar to the repression of transposons through maternal transmission of endogenous siRNAs in Drosophila [133]. Indeed, interspecific hybrids and allotetraploids can only be produced using A. thaliana as the maternal parent [48, 132], suggesting an important role of maternal inheritance in overcoming hybrid incompatibility. A recent study has shown that the expression of PolIV-dependent siRNAs (p4siRNAs) is initiated in the female gametophyte and persists during seed development [134], suggesting a role for maternally inherited siRNAs in maintaining genomic stability of the new hybrids and offspring. Unlike conventional imprinting genes, the inheritance of maternal p4siRNAs is independent of DNA methylation. It is proposed that activating factors related to the maternal expression of RNAi genes such as NRPD1A, RDR2 and DCL3 are responsible for maternal p4siRNA production. Alternatively, repressive factors in the paternal genome can also be involved. The loss of p4siRNAs in the sperm cells is consistent with expression loss of chromatin remodeling factor DDM1, suggesting transcriptional repression of paternal p4siRNAs during male gamete formation, which persists after fertilization [135].
The rasiRNAs may be directly related to suppression of transposons and indirectly related to genomic stability and growth vigor in the hybrids. The maternal inheritance of p4siRNAs and paternal suppression of rasiRNAs occur only in the hybrids, which may lead to morphological and developmental changes in the hybrids but not in the parents. As a result, both increase in growth vigor and post-zygotic failures are frequently observed in hybrid plants, depending on the presence or absence of rasiRNAs that are required to maintain genome stability and fertility.
Future perspectives
Heterosis or hybrid vigor results from genome-wide changes and interactions between paternal and maternal alleles. Heterozygosity is a prerequisite to changes in gene expression and phenotypic variation in hybrids and allopolyploids. The heterotic effects on gene expression changes in the hybrids can be augmented in polyploids (e.g., diploid versus tetraploid hybrids). Expression alteration of the genes that encode transcription factors and chromatin proteins is expected to cause cascade effects on the expression of downstream genes and their biological processes. In that sense, heterosis can be explained by a single gene or a few genes in the biological pathways. Epigenetic regulation of circadian-mediated changes in chlorophyll biosynthesis and starch metabolism offers one of the direct links to growth vigor in plant hybrids and allopolyploids. Maternal inheritance and paternal suppression of rasiRNAs affect post-zygotic failures and seed fertility and development, whereas reprogramming of miRNAs and tasiRNAs in the hybrids leads to nonadditive phenotypes and growth vigor. Several questions remain to be answered. First, what causes the allelic expression variation in the hybrids and allopolyploids? For example, how and why does the genomic mixture turn down the expression amplitude of circadian clock genes without affecting the duration of internal clocks? Why are the rasiRNAs maternally inherited? How are allelic expression variation and genetic divergence established and maintained? Is heterosis caused by genome-wide chromatin modifications or modifications of a few regulatory genes? Second, how can heterosis be permanently fixed? Apomixis (seed production without paternal genetic material) has been extensively pursued as a means for fixation of hybrid vigor. Doubling chromosomes in hybrids, particularly in the intraspecific or interspecific hybrids, offers an alternative solution to the permanent fixation of hybrid vigor. Finally, many hybrids, particularly intraspecific and interspecific hybrids, cannot survive, probably because of speciation or lethality genes that existed before speciation or diverged after speciation, which cause hybrid incompatibilities. Piwi-piRNA and transposons are associated with germline defects in Drosophila, a phenomenon known as hybrid dysgenesis. Hybrid vigor and hybrid incompatibility are two-edges of a magic sword that is hidden in the parents but revealed in the hybrids and allopolyploids. A better understanding of the genes and regulatory mechanisms for polyploidy and hybrid vigor will help us effectively select the best combinations of parents for producing best-performing hybrids and polyploids, as well as genetically manipulate the expression of key regulatory genes in the hybrid and polyploid plants for the increased production of seeds, fruits, biomass, and metabolites, such as carbohydrates, celluloses, sugars, lipids, and oils, for the growing demand of these materials to produce food, feed, and biofuels.
Acknowledgments
I thank many former and current members, including but not limited to Hyeon-Se Lee, Jianlin Wang, Lu Tian, Zhongfu Ni, Meng Chen, Erika Lackey, Misook Ha, Eun-Deok Kim, Danny Ng, Changqing Zhang, Gyoungju Nah, Jie Lu, Marisa Miller, and Dae Kawn Ko, for their invaluable contributions to the research program. I am grateful to Edward Buckler and an anonymous reviewer for their insightful and constructive suggestions to improve the manuscript. I apologize for not citing additional relevant references owing to space limitations. The work was supported by the grants from the National Institutes of Health (GM067015) and the National Science Foundation (DBI0733857 and DBI0624077).
Glossary
- Allelic expression variation
the expression pattern or level of the alleles in the hybrids is different from that in the parents. This can also refer to the expression of homoeologous loci in interspecific hybrids
- Allopolyploid
an organism or individual that contains two or more sets of genetically distinct chromosomes, usually by hybridization between different species
- Amphidiploid
synonymous to allopolyploid. Contains a diploid set of chromosomes derived from each parent. Strictly speaking, only bivalents are formed in an amphidiploid, whereas multivalents are formed in an allopolyploid
- Aneuploid
an individual in which the chromosome number is not an exact multiple of the typical haploid set for that species
- Apomixis
only one parent (usually female) contributes genes to the offspring
- Autopolyploid
a polyploid created by the multiplication of one basic set of chromosomes (in one species)
- Epigenetics
non-Mendelian inheritance, heritable changes in gene expression without changes in primarily DNA sequences
- Gametic imprinting
the expression of a gene is dependent on its parental origin in the offspring
- Genomic shock
the release of genome-wide chromatin constraints of gene expression, including activation of transposons in response to environmental changes and genomic hybridization
- Heterosis
the greater vigor of growth, survival, and fertility in hybrids than in the parents
- Homoeologous
chromosomes or genes in related species that are derived from the same ancestor and coexist in an allopolyploid
- Homologous
genes or structures that share a common evolutionary ancestor
- Homoploid hybrids
hybrids formed between different species, in some cases, resulting in a derivative hybrid species without a change in chromosome number
- Imprinting or genomic imprinting
unequal expression of maternal and paternal alleles in the offspring
- Nonadditive gene expression
the expression level of a gene in an allotetraploid is not equal to the sum of two parental loci (1 + 1 ≠ 2), leading to activation (>2), repression (<2), dominance, or overdominance
- Orthologous
chromosomes or genes in different species that have evolved from the same ancestor
- Paralogous
two or more genes in the same species that share a single ancestral origin
- Paramutation
heritable changes in gene expression induced by allelic interactions
- Ploidy
the number of basic chromosome sets
- Polyploid
an individual or cell that has two or more basic sets of chromosomes
- X-chromosome inactivation
during mammalian development, the repression of one of the two X chromosomes in the somatic cells of females as a method of dosage compensation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Darwin CR. The Effects of Cross- and Self-fertilization in the Vegetable Kingdom. John Murry; 1876. [Google Scholar]
- 2.Shull GH. The composition of a field of maize. Amer Breeders Assoc Rep. 1908;4:296–301. [Google Scholar]
- 3.East EM. Reports of the Connecticut Agricultural Experiment Station for Years 1907-1908. Connecticut Agricultural Experiment Station; 1908. Inbreeding in corn; pp. 419–428. [Google Scholar]
- 4.Duvick DN. Biotechnology in the 1930s: the development of hybrid maize. Nat Rev Genet. 2001;2:69–74. doi: 10.1038/35047587. [DOI] [PubMed] [Google Scholar]
- 5.Crow JF. 90 years ago: the beginning of hybrid maize. Genetics. 1998;148:923–928. doi: 10.1093/genetics/148.3.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng SH, et al. Progress in research and development on hybrid rice: a super-domesticate in China. Ann Bot (Lond) 2007;100:959–966. doi: 10.1093/aob/mcm121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karpechenko GD. Polyploid hybrids of Raphanus sativus L. X Brassica oleracea. L Bull Appl Bot. 1927;17:305–410. [Google Scholar]
- 8.Clausen RE, Goodspeed TH. Interspecific Hybridization in Nicotiana. II. a Tetraploid GLUTINOSA-TABACUM Hybrid, an Experimental Verification of Winge's Hypothesis. Genetics. 1925;10:278–284. doi: 10.1093/genetics/10.3.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodspeed TH. Chromosome Number and Morphology in Nicotiana VI: Chromosome Numbers of Forty Species. Proc Natl Acad Sci U S A. 1933;19:649–653. doi: 10.1073/pnas.19.7.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Mara JG. Cytogenetics of Triticale. Botanical Review. 1953;19:587–605. [Google Scholar]
- 11.Guedes-Pinto H, et al. Triticale: Today and Tomorrow. Springer; 1996. [Google Scholar]
- 12.Mallet J. Hybridization as an invasion of the genome. Trends Ecol Evol. 2004;20:229–237. doi: 10.1016/j.tree.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 13.Wood TE, et al. The frequency of polyploid speciation in vascular plants. Proc Natl Acad Sci U S A. 2009;106:13875–13879. doi: 10.1073/pnas.0811575106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masterson J. Stomatal size in fossil plants: evidence for polyploidy in majority of angiosperms. Science. 1994;264:421–424. doi: 10.1126/science.264.5157.421. [DOI] [PubMed] [Google Scholar]
- 15.Grant V. Plant Speciation. Columbia University Press; 1981. [Google Scholar]
- 16.Brochmann C, et al. Polyploidy in arctic plants. Biological Journal of the Linnean Society. 2004;82:521–536. [Google Scholar]
- 17.Soltis DE, Soltis PS. Polyploidy: recurrent formation and genome evolution. Trends Ecol Evolu. 1999;14:348–352. doi: 10.1016/s0169-5347(99)01638-9. [DOI] [PubMed] [Google Scholar]
- 18.Baumel A, et al. Molecular investigations in populations of Spartina anglica C.E. Hubbard (Poaceae) invading coastal Brittany (France) Mol Ecol. 2001;10:1689–1701. doi: 10.1046/j.1365-294x.2001.01299.x. [DOI] [PubMed] [Google Scholar]
- 19.Abbott RJ, Lowe AJ. Origins, establishment and evolution of new polyploid species: Senecio cambrensis and S-eboracensis in the British Isles. Biological Journal of the Linnean Society. 2004;82:467–474. [Google Scholar]
- 20.U, N. Genome analysis in Brassica with special references to the experimental formation of B. napus and peculiar mode of fertilization. Jpn J Genet. 1935;7:389–452. [Google Scholar]
- 21.Wendel JF, Cronn RC. Polyploidy and the evolutionary history of cotton. Advances in Agronomy. 2003;78:139–186. [Google Scholar]
- 22.Salamini F, et al. Genetics and geography of wild cereal domestication in the near east. Nat Rev Genet. 2002;3:429–441. doi: 10.1038/nrg817. [DOI] [PubMed] [Google Scholar]
- 23.Sall T, et al. Chloroplast DNA indicates a single origin of the allotetraploid Arabidopsis suecica. J Evol Biol. 2003;16:1019–1029. doi: 10.1046/j.1420-9101.2003.00554.x. [DOI] [PubMed] [Google Scholar]
- 24.Dubcovsky J, Dvorak J. Genome plasticity a key factor in the success of polyploid wheat under domestication. Science. 2007;316:1862–1866. doi: 10.1126/science.1143986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyer RC, et al. Heterosis of biomass production in Arabidopsis. Establishment during early development. Plant Physiol. 2004;134:1813–1823. doi: 10.1104/pp.103.033001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rohde P, et al. Heterosis in the freezing tolerance of crosses between two Arabidopsis thaliana accessions (Columbia-0 and C24) that show differences in non-acclimated and acclimated freezing tolerance. Plant J. 2004;38:790–799. doi: 10.1111/j.1365-313X.2004.02080.x. [DOI] [PubMed] [Google Scholar]
- 27.Alonso-Blanco C, et al. Natural allelic variation at seed size loci in relation to other life history traits of Arabidopsis thaliana. Proc Natl Acad Sci U S A. 1999;96:4710–4717. doi: 10.1073/pnas.96.8.4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.el-Assal SE, et al. Pleiotropic effects of the Arabidopsis cryptochrome 2 allelic variation underlie fruit trait-related QTL. Plant Biol (Stuttg) 2004;6:370–374. doi: 10.1055/s-2004-820890. [DOI] [PubMed] [Google Scholar]
- 29.Jakobsson M, et al. A unique recent origin of the allotetraploid species Arabidopsis suecica: Evidence from nuclear DNA markers. Mol Biol Evol. 2006;23:1217–1231. doi: 10.1093/molbev/msk006. [DOI] [PubMed] [Google Scholar]
- 30.Comai L, et al. FISH analysis of meiosis in Arabidopsis allopolyploids. Chromosome Res. 2003;11:217–226. doi: 10.1023/a:1022883709060. [DOI] [PubMed] [Google Scholar]
- 31.Bushell C, et al. The basis of natural and artificial postzygotic hybridization barriers in Arabidopsis species. Plant Cell. 2003;15:1430–1442. doi: 10.1105/tpc.010496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Charlesworth B, Charlesworth D. The genetic basis of inbreeding depression. Genet Res. 1999;74:329–340. doi: 10.1017/s0016672399004152. [DOI] [PubMed] [Google Scholar]
- 33.Muller HJ. Why polyploidy is rarer in animals than in plants. Amer Nat. 1925;59:346–353. [Google Scholar]
- 34.Mable BK. ‘Why polyploidy is rarer in animals than in plants’: myths and mechanisms. Biological Journal of the Linnean Society. 2004;82:453–466. [Google Scholar]
- 35.Dowling TE, Secor CL. The role of hybridization and introgression in the diversification of animals. Annual Review of Ecology and Systematics. 1997;28:593–619. [Google Scholar]
- 36.Mavarez J, et al. Speciation by hybridization in Heliconius butterflies. Nature. 2006;441:868–871. doi: 10.1038/nature04738. [DOI] [PubMed] [Google Scholar]
- 37.Mallet J. Hybrid speciation. Nature. 2007;446:279–283. doi: 10.1038/nature05706. [DOI] [PubMed] [Google Scholar]
- 38.Muller HJ. Isolating mechanisms, evolution and temperature. Biol Symp. 1942;6:71–125. [Google Scholar]
- 39.Dobzhansky T. Studies on Hybrid Sterility. II. Localization of Sterility Factors in Drosophila Pseudoobscura Hybrids. Genetics. 1936;21:113–135. doi: 10.1093/genetics/21.2.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brideau NJ, et al. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science. 2006;314:1292–1295. doi: 10.1126/science.1133953. [DOI] [PubMed] [Google Scholar]
- 41.Tang S, Presgraves DC. Evolution of the Drosophila nuclear pore complex results in multiple hybrid incompatibilities. Science. 2009;323:779–782. doi: 10.1126/science.1169123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seidel HS, et al. Widespread genetic incompatibility in C. elegans maintained by balancing selection. Science. 2008;319:589–594. doi: 10.1126/science.1151107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bomblies K, et al. Autoimmune Response as a Mechanism for a Dobzhansky-Muller-Type Incompatibility Syndrome in Plants. PLoS Biol. 2007;5:e236. doi: 10.1371/journal.pbio.0050236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bikard D, et al. Divergent evolution of duplicate genes leads to genetic incompatibilities within A. thaliana. Science. 2009;323:623–626. doi: 10.1126/science.1165917. [DOI] [PubMed] [Google Scholar]
- 45.Vrana PB, et al. Genetic and epigenetic incompatibilities underlie hybrid dysgenesis in Peromyscus. Nat Genet. 2000;25:120–124. doi: 10.1038/75518. [DOI] [PubMed] [Google Scholar]
- 46.O'Neill RJ, et al. Undermethylation associated with retroelement activation and chromosome remodelling in an interspecific mammalian hybrid. Nature. 1998;393:68–72. doi: 10.1038/29985. [DOI] [PubMed] [Google Scholar]
- 47.Josefsson C, et al. Parent-Dependent Loss of Gene Silencing during Interspecies Hybridization. Curr Biol. 2006;16:1322–1328. doi: 10.1016/j.cub.2006.05.045. [DOI] [PubMed] [Google Scholar]
- 48.Wang J, et al. Stochastic and epigenetic changes of gene expression in Arabidopsis polyploids. Genetics. 2004;167:1961–1973. doi: 10.1534/genetics.104.027896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee HS, Chen ZJ. Protein-coding genes are epigenetically regulated in Arabidopsis polyploids. Proc Natl Acad Sci U S A. 2001;98:6753–6758. doi: 10.1073/pnas.121064698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.East EM. Heterosis. Genetics. 1936;21:375–397. doi: 10.1093/genetics/21.4.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lippman ZB, et al. The making of a compound inflorescence in tomato and related nightshades. PLoS Biol. 2008;6:e288. doi: 10.1371/journal.pbio.0060288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gore MA, et al. A first-generation haplotype map of maize. Science. 2009;326:1115–1117. doi: 10.1126/science.1177837. [DOI] [PubMed] [Google Scholar]
- 53.McMullen MD, et al. Genetic properties of the maize nested association mapping population. Science. 2009;325:737–740. doi: 10.1126/science.1174320. [DOI] [PubMed] [Google Scholar]
- 54.Birchler JA, et al. In search of the molecular basis of heterosis. Plant Cell. 2003;15:2236–2239. doi: 10.1105/tpc.151030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Michaels SD, Amasino RM. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. 1999;11:949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Corbesier L, et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316:1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- 57.Valverde F, et al. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science. 2004;303:1003–1006. doi: 10.1126/science.1091761. [DOI] [PubMed] [Google Scholar]
- 58.Sasaki A, et al. Green revolution: a mutant gibberellin-synthesis gene in rice. Nature. 2002;416:701–702. doi: 10.1038/416701a. [DOI] [PubMed] [Google Scholar]
- 59.Buckler ES, et al. The genetic architecture of maize flowering time. Science. 2009;325:714–718. doi: 10.1126/science.1174276. [DOI] [PubMed] [Google Scholar]
- 60.Jones DF. Dominance of linked factors as a means of accounting for heterosis. Genetics. 1917;2:466–479. doi: 10.1093/genetics/2.5.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bruce AB. The Mendelian theory of heredity and the augmentation of vigor. Science. 1910;32:627–628. doi: 10.1126/science.32.827.627-a. [DOI] [PubMed] [Google Scholar]
- 62.Crow JF. Alternative hypothesis of hybrid vigor. Genetics. 1948;33:477–487. doi: 10.1093/genetics/33.5.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Semel Y, et al. Overdominant quantitative trait loci for yield and fitness in tomato. Proc Natl Acad Sci U S A. 2006;103:12981–12986. doi: 10.1073/pnas.0604635103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xiao J, et al. Dominance is the major genetic basis of heterosis in rice as revealed by QTL analysis using molecular markers. Genetics. 1995;140:745–754. doi: 10.1093/genetics/140.2.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li ZK, et al. Overdominant epistatic loci are the primary genetic basis of inbreeding depression and heterosis in rice. I. Biomass and grain yield. Genetics. 2001;158:1737–1753. doi: 10.1093/genetics/158.4.1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu SB, et al. Importance of epistasis as the genetic basis of heterosis in an elite rice hybrid. Proc Natl Acad Sci U S A. 1997;94:9226–9231. doi: 10.1073/pnas.94.17.9226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stuber CW, et al. Identification of genetic factors contributing to heterosis in a hybrid from two elite maize inbred lines using molecular markers. Genetics. 1992;132:823–839. doi: 10.1093/genetics/132.3.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Luo LJ, et al. Overdominant epistatic loci are the primary genetic basis of inbreeding depression and heterosis in rice. II. Grain yield components. Genetics. 2001;158:1755–1771. doi: 10.1093/genetics/158.4.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Doebley J, et al. teosinte branched1 and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics. 1995;141:333–346. doi: 10.1093/genetics/141.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guo M, et al. Genome-wide mRNA profiling reveals heterochronic allelic variation and a new imprinted gene in hybrid maize endosperm. Plant J. 2003;36:30–44. doi: 10.1046/j.1365-313x.2003.01852.x. [DOI] [PubMed] [Google Scholar]
- 71.Guo M, et al. Allelic variation of gene expression in maize hybrids. Plant Cell. 2004;16:1707–1716. doi: 10.1105/tpc.022087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stupar RM, Springer NM. Cis-transcriptional variation in maize inbred lines B73 and Mo17 leads to additive expression patterns in the F1 hybrid. Genetics. 2006;173:2199–2210. doi: 10.1534/genetics.106.060699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Swanson-Wagner RA, et al. Paternal Dominance of Trans-eQTL Influences Gene Expression Patterns in Maize Hybrids. Science. 2009;326:1118–1120. doi: 10.1126/science.1178294. [DOI] [PubMed] [Google Scholar]
- 74.Auger DL, et al. A test for a metastable epigenetic component of heterosis using haploid induction in maize. Theor Appl Genet. 2004;108:1017–1023. doi: 10.1007/s00122-003-1521-8. [DOI] [PubMed] [Google Scholar]
- 75.Swanson-Wagner RA, et al. All possible modes of gene action are observed in a global comparison of gene expression in a maize F1 hybrid and its inbred parents. Proc Natl Acad Sci U S A. 2006;103:6805–6810. doi: 10.1073/pnas.0510430103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guo M, et al. Genome-wide transcript analysis of maize hybrids: allelic additive gene expression and yield heterosis. Theor Appl Genet. 2006;113:831–845. doi: 10.1007/s00122-006-0335-x. [DOI] [PubMed] [Google Scholar]
- 77.Chen ZJ, Pikaard CS. Transcriptional analysis of nucleolar dominance in polyploid plants: biased expression/silencing of progenitor rRNA genes is developmentally regulated in Brassica. Proc Natl Acad Sci U S A. 1997;94:3442–3447. doi: 10.1073/pnas.94.7.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Adams KL, et al. Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc Natl Acad Sci U S A. 2003;100:4649–4654. doi: 10.1073/pnas.0630618100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jackson SA, Chen ZJ. Genomic and expression plasicity of polyploidy. Curr Opin Plant Biol. 2009 doi: 10.1016/j.pbi.2009.11.004. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hovav R, et al. Partitioned expression of duplicated genes during development and evolution of a single cell in a polyploid plant. Proc Natl Acad Sci U S A. 2008;105:6191–6195. doi: 10.1073/pnas.0711569105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hegarty MJ, et al. Transcriptome shock after interspecific hybridization in senecio is ameliorated by genome duplication. Curr Biol. 2006;16:1652–1659. doi: 10.1016/j.cub.2006.06.071. [DOI] [PubMed] [Google Scholar]
- 82.Wang J, et al. Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics. 2006;172:507–517. doi: 10.1534/genetics.105.047894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang SS, et al. Accumulation of genome-specific transcripts, transcription factors and phytohormonal regulators during early stages of fiber cell development in allotetraploid cotton. Plant J. 2006;47:761–775. doi: 10.1111/j.1365-313X.2006.02829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dodd AN, et al. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309:630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- 85.Wijnen H, Young MW. Interplay of circadian clocks and metabolic rhythms. Annu Rev Genet. 2006;40:409–448. doi: 10.1146/annurev.genet.40.110405.090603. [DOI] [PubMed] [Google Scholar]
- 86.Panda S, et al. Circadian rhythms from flies to human. Nature. 2002;417:329–335. doi: 10.1038/417329a. [DOI] [PubMed] [Google Scholar]
- 87.Michael TP, et al. Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science. 2003;302:1049–1053. doi: 10.1126/science.1082971. [DOI] [PubMed] [Google Scholar]
- 88.Mizoguchi T, et al. LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev Cell. 2002;2:629–641. doi: 10.1016/s1534-5807(02)00170-3. [DOI] [PubMed] [Google Scholar]
- 89.Alabadi D, et al. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science. 2001;293:880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- 90.Pruneda-Paz JL, et al. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science. 2009;323:1481–1485. doi: 10.1126/science.1167206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Strayer C, et al. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science. 2000;289:768–771. doi: 10.1126/science.289.5480.768. [DOI] [PubMed] [Google Scholar]
- 92.Park DH, et al. Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science. 1999;285:1579–1582. doi: 10.1126/science.285.5433.1579. [DOI] [PubMed] [Google Scholar]
- 93.Wang ZY, Tobin EM. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell. 1998;93:1207–1217. doi: 10.1016/s0092-8674(00)81464-6. [DOI] [PubMed] [Google Scholar]
- 94.McClung CR. Plant circadian rhythms. Plant Cell. 2006;18:792–803. doi: 10.1105/tpc.106.040980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Harmer SL. The Circadian System in Higher Plants. Annu Rev Plant Biol. 2009;60:357–377. doi: 10.1146/annurev.arplant.043008.092054. [DOI] [PubMed] [Google Scholar]
- 96.Harmer SL, et al. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- 97.Smith SM, et al. Diurnal changes in the transcriptome encoding enzymes of starch metabolism provide evidence for both transcriptional and posttranscriptional regulation of starch metabolism in Arabidopsis leaves. Plant Physiol. 2004;136:2687–2699. doi: 10.1104/pp.104.044347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Covington MF, et al. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 2008;9:R130. doi: 10.1186/gb-2008-9-8-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lu Y, et al. Daylength and circadian effects on starch degradation and maltose metabolism. Plant Physiol. 2005;138:2280–2291. doi: 10.1104/pp.105.061903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dodd AN, et al. The plant clock shows its metal: circadian regulation of cytosolic free Ca(2+) Trends Plant Sci. 2005;10:15–21. doi: 10.1016/j.tplants.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 101.Ni Z, et al. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature. 2009;457:327–331. doi: 10.1038/nature07523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Reinbothe S, et al. PORA and PORB, Two Light-Dependent Protochlorophyllide-Reducing Enzymes of Angiosperm Chlorophyll Biosynthesis. Plant Cell. 1996;8:763–769. doi: 10.1105/tpc.8.5.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sperling U, et al. Overexpression of light-dependent PORA or PORB in plants depleted of endogenous POR by far-red light enhances seedling survival in white light and protects against photooxidative damage. Plant J. 1997;12:649–658. doi: 10.1046/j.1365-313x.1997.00649.x. [DOI] [PubMed] [Google Scholar]
- 104.Lloyd JR, et al. Leaf starch degradation comes out of the shadows. Trends Plant Sci. 2005;10:130–137. doi: 10.1016/j.tplants.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 105.Smith AM, et al. Starch degradation. Annu Rev Plant Biol. 2005;56:73–98. doi: 10.1146/annurev.arplant.56.032604.144257. [DOI] [PubMed] [Google Scholar]
- 106.Fujiwara S, et al. Circadian clock proteins LHY and CCA1 regulate SVP protein accumulation to control flowering in Arabidopsis. Plant Cell. 2008;20:2960–2971. doi: 10.1105/tpc.108.061531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ding Z, et al. A complex genetic interaction between Arabidopsis thaliana TOC1 and CCA1/LHY in driving the circadian clock and in output regulation. Genetics. 2007;176:1501–1510. doi: 10.1534/genetics.107.072769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen ZJ. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu Rev Plant Biol. 2007;58:377–406. doi: 10.1146/annurev.arplant.58.032806.103835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Redei GP. Single locus heterosis. Mol Gen Genet. 1962;93:164–170. [Google Scholar]
- 110.Shpak ED, et al. Synergistic interaction of three ERECTA-family receptor-like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation. Development. 2004;131:1491–1501. doi: 10.1242/dev.01028. [DOI] [PubMed] [Google Scholar]
- 111.Kim GT, et al. The ANGUSTIFOLIA gene of Arabidopsis, a plant CtBP gene, regulates leaf-cell expansion, the arrangement of cortical microtubules in leaf cells and expression of a gene involved in cell-wall formation. Embo J. 2002;21:1267–1279. doi: 10.1093/emboj/21.6.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hua J, et al. Single-locus heterotic effects and dominance by dominance interactions can adequately explain the genetic basis of heterosis in an elite rice hybrid. Proc Natl Acad Sci U S A. 2003;100:2574–2579. doi: 10.1073/pnas.0437907100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Doebley J, et al. The evolution of apical dominance in maize. Nature. 1997;386:485–488. doi: 10.1038/386485a0. [DOI] [PubMed] [Google Scholar]
- 114.Chandler VL, Stam M. Chromatin conversations: mechanisms and implications of paramutation. Nat Rev Genet. 2004;5:532–544. doi: 10.1038/nrg1378. [DOI] [PubMed] [Google Scholar]
- 115.Mittelsten Scheid O, et al. Formation of stable epialleles and their paramutation-like interaction in tetraploid Arabidopsis thaliana. Nat Genet. 2003;34:450–454. doi: 10.1038/ng1210. [DOI] [PubMed] [Google Scholar]
- 116.Lee JT, Jaenisch R. The (epi)genetic control of mammalian X-chromosome inactivation. Curr Opin Genet Dev. 1997;7:274–280. doi: 10.1016/s0959-437x(97)80138-4. [DOI] [PubMed] [Google Scholar]
- 117.Lee JT. Regulation of X-chromosome counting by Tsix and Xite sequences. Science. 2005;309:768–771. doi: 10.1126/science.1113673. [DOI] [PubMed] [Google Scholar]
- 118.Bender J, Fink GR. Epigenetic control of an endogenous gene family is revealed by a novel blue fluorescent mutant of Arabidopsis. Cell. 1995;83:725–734. doi: 10.1016/0092-8674(95)90185-x. [DOI] [PubMed] [Google Scholar]
- 119.Wittkopp PJ, et al. Evolutionary changes in cis and trans gene regulation. Nature. 2004;430:85–88. doi: 10.1038/nature02698. [DOI] [PubMed] [Google Scholar]
- 120.Wang J, et al. Nonadditive Regulation of FRI and FLC Loci Mediates Flowering-Time Variation in Arabidopsis Allopolyploids. Genetics. 2006;173:965–974. doi: 10.1534/genetics.106.056580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen M, et al. RNAi of met1 reduces DNA methylation and induces genome-specific changes in gene expression and centromeric small RNA accumulation in Arabidopsis allopolyploids. Genetics. 2008;178:1845–1858. doi: 10.1534/genetics.107.086272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ha M, et al. Small RNAs serve as a genetic buffer against genomic shock in Arabidopsis interspecific hybrids and allopolyploids. Proc Natl Acad Sci U S A. 2009;106:17835–17840. doi: 10.1073/pnas.0907003106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 124.Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- 125.Vazquez F, et al. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell. 2004;16:69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 126.Peragine A, et al. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ha M, et al. Interspecies regulation of microRNAs and their targets. Biochim Biophys Acta. 2008;1779:735–742. doi: 10.1016/j.bbagrm.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Brown BD, et al. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat Biotechnol. 2007;25:1457–1467. doi: 10.1038/nbt1372. [DOI] [PubMed] [Google Scholar]
- 129.Long D, et al. Potent effect of target structure on microRNA function. Nat Struct Mol Biol. 2007;14:287–294. doi: 10.1038/nsmb1226. [DOI] [PubMed] [Google Scholar]
- 130.Lippman Z, Martienssen R. The role of RNA interference in heterochromatic silencing. Nature. 2004;431:364–370. doi: 10.1038/nature02875. [DOI] [PubMed] [Google Scholar]
- 131.McClintock B. The significance of responses of the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- 132.Comai L, et al. Phenotypic instability and rapid gene silencing in newly formed Arabidopsis allotetraploids. Plant Cell. 2000;12:1551–1568. doi: 10.1105/tpc.12.9.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Brennecke J, et al. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322:1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mosher RA, et al. Uniparental expression of PolIV-dependent siRNAs in developing endosperm of Arabidopsis. Nature. 2009;460:283–286. doi: 10.1038/nature08084. [DOI] [PubMed] [Google Scholar]
- 135.Slotkin RK, et al. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell. 2009;136:461–472. doi: 10.1016/j.cell.2008.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Halberg F, et al. Phase relations of 24-hour periodicities in blood corticosterone, mitoses in cortical adrenal parenchyma, and total body activity. Endocrinology. 1959;64:222–230. doi: 10.1210/endo-64-2-222. [DOI] [PubMed] [Google Scholar]
- 137.de Mairan J. Observation botanique. Hist 1729 [Google Scholar]
- 138.Gardner MJ, et al. How plants tell the time. Biochem J. 2006;397:15–24. doi: 10.1042/BJ20060484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yakir E, et al. Regulation of output from the plant circadian clock. Febs J. 2007;274:335–345. doi: 10.1111/j.1742-4658.2006.05616.x. [DOI] [PubMed] [Google Scholar]
- 140.McClung CR. Comes a time. Curr Opin Plant Biol. 2008;11:514–520. doi: 10.1016/j.pbi.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 141.Covington MF, Harmer SL. The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol. 2007;5:e222. doi: 10.1371/journal.pbio.0050222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gutierrez RA, et al. Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proc Natl Acad Sci U S A. 2008;105:4939–4944. doi: 10.1073/pnas.0800211105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Dodd AN, et al. The Arabidopsis circadian clock incorporates a cADPR-based feedback loop. Science. 2007;318:1789–1792. doi: 10.1126/science.1146757. [DOI] [PubMed] [Google Scholar]