Traditional chromatographic quantification methods for heparosan produced from the Escherichia coli K5 strain rely on extensive purification requiring laborious sample preparation. These methods are time-consuming, often resulting in sample loss during purification, and thus may not accurately reflect the amount of heparosan in the original mixture. A simple, sensitive 1H-NMR quantification method that directly quantifies heparosan K5 polysaccharide present in E. coli fermentation supernatant is described.
Heparosan is a polysaccharide with a β-1,4-d-glucuronic acid (GlcA) and α-1,4 N-acetyl-d-glucosamine (GlcNAc), [→4)GlcA-β-(1-4) GlcNAc-α (1→]n repeating disaccharide unit. Heparosan is biosynthesized as a bacterial capsule and this polysaccharide is identical to the precursor of the mammalian heparin and heparan sulfate in mammals [1]. Heparin and heparin sulfate participate in many important biological processes, including blood anticoagulation, viral and bacterial infection and entry, angiogenesis, inflammation, cancer and development [2,3,4]. Heparin, extracted from porcine intestines, is one of the oldest drugs and it is currently in widespread use for the prevention of blood clotting [2,5]. In 2008, a new, rapid onset, acute side effect, resulting in hypotension was associated with certain lots of heparin contaminated with oversulfated chondroitin sulfate (OSCS) [6,7]. A bioengineered heparin prepared from heparosan offers a potential alternative for the preparation of a safer heparin [8,9]. Heparosan, of molecular weight > 10,000, is readily obtained from E. coli K5 strain [10] and it can be enzymatically modified to produce an anticoagulant polysaccharide similar to heparin [8,9]. Heparosan itself has also been explored as a biomaterial because of its stability and non-immunogenic characteristics [11].
K5 heparosan is conveniently prepared by E. coli fermentation and recovered directly from the fermentation supernatant [12]. Thus, the heparosan concentration in the fermentation supernatant is a critical parameter for optimizing the fermentation process and calculating the purification efficiency. The carbazole assay has been used in the past to quantify polysaccharides that contain uronic acid [13]. Unfortunately, media components often interfere with this colorimetric assay. Capillary electrophoresis (CE) has been used to quantify purified heparosan, but media components may also interfere with CE analysis [14]. Disaccharide analysis using HPLC and HPLC/MS can also been utilized to quantify heparosan [15], but these methods require a time-consuming enzymatic digestion of the heparosan and the removal of proteins, enzymes and buffer salts prior to sample analysis.
NMR is a powerful technique for elucidating the structure of molecules that can be used in quantitative analysis. 1H-NMR has been used for quantifying carrageenans in blends [16], for monitoring the wine and beer fermentation processes [17,18], for quantifying derivatized Haemophilus influenzae type b polysaccharide intermediate [19], and in many other quantitative applications [20,21]. The major advantages of NMR-based quantification are simple sample preparation and its nondestructive nature.
E. coli K5 (ATCC23506) was cultured in batch on a medium consisting of: 20 g/L glucose, 20 mg/L thiamine, 13.5 g/L KH2PO4, 4.0 g/L (NH4)2HPO4, 1.4 g/L MgSO4·7H2O, 1.7 g/L citric acid to which was added 10.0 mL trace metal solution, consisting of 10.0 g/L FeSO4·7H2O, 2.0 g/L CaCl2, 2.2 g/L ZnSO4·7H2O, 0.5 g/L MnSO4·4H2O, 1.0 g/L CuSO4·5H2O, 0.1 g/L (NH4)6Mo7O24·4H2O, and 0.02 g/L Na2B4O7·10H2O in 5 M hydrochloric acid. The feeding solution during the fed batch cultures consisted of: 250–1000 g/L glucose, 20 g/L MgSO4·7H2O and 0.15 or 0.25 g/L thiamine [22]. The batch growth phase began by inoculating 10 vol % seed culture prepared in a shake flask into 3 L of culture media grown in an Applikon 7 L fermentor. The temperature was maintained at 37°C. The pH was maintained between 6 and 8 by continuously adjusting with 29% ammonia solution. The culture was fed exponentially after the glucose in the medium was depleted. Samples were collected from the fermentor at various time points and centrifuged at 12,000 × g for 30 min to separate supernatant from cells and 1 ml aliquots of supernatant were lyophilized.
Lyophilized supernatant was dissolved in 400 µl of D2O and lyophilized, then redissolved in 400 µl D2O (99.96 atm. %) again lyophilized and finally in 400 µl of D2O containing 71 µg sodium terephthalate and then transferred to a 5 mm NMR tube. (Water suppression can be used to eliminate the need for lyophilization and D2O exchange steps but results in lower spectral quality.) Standards were prepared by dissolving purified K5 samples (0.2 mg to 1.6 mg) in 400 µl of D2O containing 71 µg sodium terephthalate.
1H-NMR (8 scans) and HMQC NMR were performed on a Bruker 600 MHz NMR spectrometer and acquisition of the spectra was carried out using TOPSPIN 2.0 software. All the spectra were acquired at the temperature of 298 K. The relaxation delay time D1 was set to 20 s to ensure that the protons in the sodium terephthalate and N-acetyl group were adequately relaxed. 1H NMR spectra were processed in MestRe-C software. The phase of the spectra was manually corrected, and the baseline of the spectra was adjusted with the “Baseline Correction-Use Polynomial” function. The integration of the peaks was performed using the “Integration” function, with the peak area selected manually. The 2D NMR spectra were processed and analyzed using the programs Sparky (3.114).
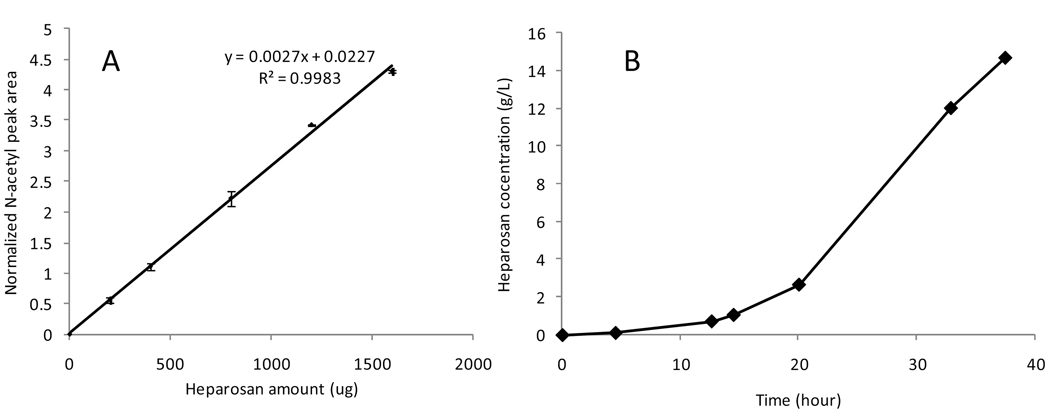
Sodium terephthalate was selected as a water soluble, stable, and non-reactive internal standard for the K5 heparosan quantification because it shows a single peak at 7.91 ppm in the 1H-NMR in a region where there were no interfering peaks from the heparosan and fermentation components (Figure 1). The N-acetyl peaks for heparosan at 2.04 ppm was selected and the peak area was normalized to the sodium terephthalate peak area. A standard curve, prepared from 1H-NMR spectra of triplicate samples at each heparosan concentration, showed good linearity (Figure 2 A).
Figure 1.
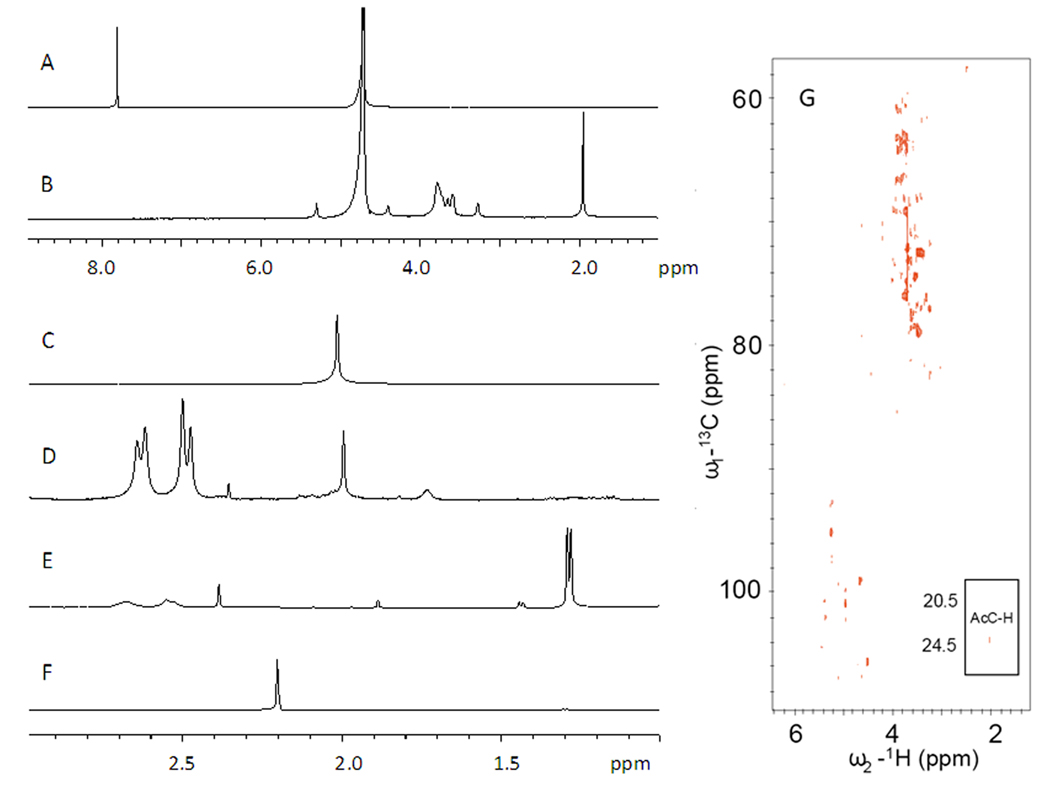
1H NMR spectra of (A) sodium terephthalate and (B) K5 heparosan. 1H NMR spectra expanded between 1 and 3 ppm of (C) K5 heparosan, (D) E. coli K5 heparosan fermentation supernatant, (E) E. coli BL21 culture supernatant, (F) E. coli BL21 lysate, and (G) HMQC NMR of purified K5 heparosan showing the correlation of 1H and 13C signals confirming assignments.
Figure 2.
(A) Standard curve for the quantification of K5 heparosan. x-axis is the amounts of heparosan in mg in the NMR samples. The y-axis is the normalized N-acetyl peak area from the 1H NMR spectra. The equation and R2 value are displayed. Data was acquired from triplicate experiments. (B) Time course of the K5 heparosan concentration during the fermentation process.
E. coli K5 fermentation supernatant is a complicated mixture, containing complex medium, proteins, metabolic products, and K5 polysaccharide (Figure 1D). Most of the peaks in the 1HNMR spectrum overlap with media components and cannot be used to quantify K5 polysaccharide. However, the peak at 2.04 ppm, corresponding to the methyl protons in N-acetyl groups of heparosan, was well resolved and diagnostic of heparosan in the fermentation supernatant. Heteronuclear HMQC NMR confirmed the assignment of the peak at 2.04 ppm in the 1H spectrum through its correlation to the 13C signal 23.9 ppm (Figure 1G) [9]. In a control experiment, E. coli BL21, a strain not producing K5 polysaccharide, was grown for 16 h in the same medium. The supernatant and cell pellet (after solubilization by sonication and centrifuged at 7000 × g for 30 min) were examined by 1H-NMR and showed no peaks at or around 2.04 ppm eliminating the possibility that cell wall or cell lysis components from E. coli might interfere with the NMR quantification.
Integration of N-acetyl peak at 2.04 ppm against the peak at 7.91 ppm for the internal standard afforded an accurate determination of heparosan concentration in the fermentation supernatant (Figure 2B). The concentrations determined by 1H-NMR were in excellent agreement with concentrations determined by carbazole assay after heparosan recovery and purification. Heparosan concentration in the supernatant increased over the fermentation time, as expected, correlating to the increase in cell density.
In conclusion, 1H NMR affords a simple and reliable method to quantify K5 heparosan from the fermentation. Other polysaccharides containing N-acetylhexosamine residues such as chondroitin (from E. coli K4) or hyaluronan should also be quantifiable using this method.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lindahl U, Kusche-Gullberg M, Kjellén L. Regulated diversity of heparan sulfate. J. Biol. Chem. 1998;273:24979–24982. doi: 10.1074/jbc.273.39.24979. [DOI] [PubMed] [Google Scholar]
- 2.Linhardt RJ. Heparin: an important drug enters its seventh decade. Chem. Ind. 1991;2:45–50. [Google Scholar]
- 3.Chuang YJ, Swanson R, Raja SM, Olson ST. Heparin enhances the specificity of antithrombin for thrombin and factor Xa independent of the reactive center loop sequence. J. Biol. Chem. 2001;276:14961–14971. doi: 10.1074/jbc.M011550200. [DOI] [PubMed] [Google Scholar]
- 4.Linhardt RJ, Toida T. Heparin oligosaccharides: new analogues-development and applications. In: Witczak ZJ, Nieforth KA, editors. Carbohydrates in Drug Design. New York: Marcel Dekker; 1997. pp. 277–341. [Google Scholar]
- 5.Coyne E. Heparin - past, present and future. In: Lundblad RL, Brown WV, Mann KG, Roberts HR, editors. Chemistry and Biology of Heparin. Amsterdam, Holland: Elsevier North Holland; 1981. pp. 9–17. [Google Scholar]
- 6.Kishimoto TK, Viswanathan K, Ganguly T, Elankumaran S, Smith S, Pelzer K, Lansing JC, Sriranganathan N, Zhao G, Galcheva-Gargova Z, Al-Hakim A, Bailey GS, Fraser B, Roy S, Rogers-Cotrone T, Buhse L, Whary M, Fox J, Nasr M, Dal Pan GJ, Shriver Z, Langer RS, Venkataraman G, Austen KF, Woodcock J, Sasisekharan R. Contaminated heparin associated with adverse clinical events and activation of the contact system. N. Engl. J. Med. 2008;358:2457–2467. doi: 10.1056/NEJMoa0803200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guerrini M, Beccati D, Shriver Z, Naggi A, Viswanathan K, Bisio A, Capila I, Lansing JC, Guglieri S, Fraser B, Al-Hakim A, Gunay NS, Zhang Z, Robinson L, Buhse L, Nasr M, Woodcock J, Langer R, Venkataraman G, Linhardt RJ, Casu B, Torri G, Sasisekharan R. Oversulfated chondroitin sulfate is a contaminant in heparin associated with adverse clinical events. Nat. Biotechnol. 2008;26:669–675. doi: 10.1038/nbt1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindahl U, Li JP, Kusche-Gullberg M, Salmivirta M, Alaranta S, Veromaa T, Emeis J, Roberts I, Taylor C, Oreste P, Zoppetti G, Naggi A, Torri G, Casu B. Generation of "neoheparin" from E. coli K5 capsular polysaccharide. J. Med. Chem. 2005;48:349–352. doi: 10.1021/jm049812m. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Z, McCallum SA, Xie J, Nieto L, Corzana F, Jiménez-Barbero J, Chen M, Liu J, Linhardt RJ. Solution structures of chemoenzymatically synthesized heparin and its precursors. J. Am. Chem. Soc. 2008;130:12998–13007. doi: 10.1021/ja8026345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vann WF, Schmidt MA, Jann B, Jann K. The structure of the capsular polysaccharide (K5 antigen) of urinary-tract-infective Escherichia coli 010:K5:H4. Eur. J. Biochem. 1981;116:359–364. doi: 10.1111/j.1432-1033.1981.tb05343.x. [DOI] [PubMed] [Google Scholar]
- 11.Deangelis PL. Heparosan - based biomaterials and coatings and methods of production and use. PCT Int. Application. 2009 Pub. No.: WO 2009014559.
- 12.Manzoni M, Bergomi S, Cavazzoni V. Extracellular K5 polysaccharide of Escherichia coli: production and characterization. Journal of Bioactive and Compatible Polymers. 1993;8:251–257. [Google Scholar]
- 13.Bitter T, Muir HM. A modified uronic acid carbazole reaction. Anal. Biochem. 1962;4:330–334. doi: 10.1016/0003-2697(62)90095-7. [DOI] [PubMed] [Google Scholar]
- 14.Volpi Nicola. Purification of the Escherichia coli K5 capsular polysaccharide and use of high-performance capillary electrophoresis to qualitative and quantitative monitor the process. Electrophoresis. 2004;25:3307–3312. doi: 10.1002/elps.200305856. [DOI] [PubMed] [Google Scholar]
- 15.Viskov C, Lux F, Gervier R, Colas G. Method for producing K5 polysaccharide. U.S. Patent Application. 2008 Pub. No.: US 2008/0032349 A1.
- 16.Tojo E, Prado J. A simple 1H NMR method for the quantification of carrageenans in blends. Carbohydrate Polymers. 2003;53:325–329. [Google Scholar]
- 17.López-Rituerto E, Cabredo S, López M, Avenoza A, Busto JH, Peregrina JM. A thorough study on the use of quantitative 1H NMR in Rioja red wine fermentation processes. J. Agric. Food Chem. 2009;57:2112–2118. doi: 10.1021/jf803245r. [DOI] [PubMed] [Google Scholar]
- 18.Nord LI, Vaag P, Duus JØ. Quantification of organic and amino acids in beer by 1H NMR spectroscopy. Anal. Chem. 2004;76:4790–4798. doi: 10.1021/ac0496852. [DOI] [PubMed] [Google Scholar]
- 19.Xu Q, Klees J, Teyral J, Capen R, Huang M, Sturgess AW, Hennessey JP, Jr, Washabaugh M, Sitrin R, Abeygunawardana C. Quantitative nuclear magnetic resonance analysis and characterization of the derivatized Haemophilus influenzae type b polysaccharide intermediate for PedvaxHIB. Anal. Biochem. 2005;337:235–245. doi: 10.1016/j.ab.2004.11.019. [DOI] [PubMed] [Google Scholar]
- 20.Pauli GF, Jaki BU, Lankin DC. Quantitative 1H NMR: development and potential of a method for natural products analysis. J. Nat. Prod. 2005;68:133–149. doi: 10.1021/np0497301. [DOI] [PubMed] [Google Scholar]
- 21.Pauli GF, Jaki BU, Lankin DC. A routine experimental protocol for qHNMR illustrated with Taxol. J. Nat. Prod. 2007;70:589–595. doi: 10.1021/np060535r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang F, Lee SY. High cell density culture of metabolically engineered Escherichia coli for the production of poly(3-hydroxybutyrate) in a defined medium. Biotechnol. Bioeng. 1998;58:325–328. [PubMed] [Google Scholar]