Abstract
Background
Computational methods are needed to design multivalent vaccines against flaviviruses such as West Nile (WNV) or Dengue (DENV).
Objective
To use physico-chemical property consensus (PCP-consensus) sequences of Flavivirus strains to delineate conserved motifs, areas of maximum variability, and specific loci that correlate with arthropod vector, serotype and disease severity.
Methods
PCP-consensus sequences for 27 species were prepared from 928 annotated sequences catalogued in Flavitrack. Alignments of these correlated well with the known structures of the NS3 protease domain and envelope (E) proteins. The PCPMer suite was used to identify motifs common to all Flaviviruses. Areas of PCP variability that correlated with phenotype were plotted on the structures.
Results
Despite considerable diversity at the amino acid level, PCPs for both proteins were well conserved throughout the Flaviviruses. A series of insertions in E separated tick- from mosquito-borne viruses, and all arthropod-borne from isolates with no known vector or directly from insects. Comparison of a PCP-consensus sequence of E derived from 600 DENV strains (DENV600) with individual ones for DENV1-DENV4 showed most major serotype specific variation occurs near these insertions. The DENV600 differed from one prepared from 8 hemorrhagic or fatal strains from four DENV serotypes at only 3 positions, two of which overlap known escape mutant sites
Conclusions
Comparing consensus sequences showed substantial changes occur in only a few areas of the E protein. PCP-consensus sequences can contribute to the design of multivalent vaccines.
Keywords: vaccine design, arthropod-borne virus, Dengue hemorrhagic disease, West nile encephalitis, functional motifs
Introduction
Flaviviruses (FV) are important human and animal pathogens 1; 2; 3; 4; 5; 6, which typically require insect vectors to infect mammalian hosts 7. Perhaps due to expansion in the growth zone of their vectors or intermediate hosts1, many FV, such as West Nile virus (WNV) and Dengue (DENV1 to DENV4) are now spreading into new areas. There were 1356 cases of WNV reported to the CDC in 2008 in the United States, and 44 fatalities (http://www.cdc.gov/ncidod/dvbid/westnile/surv&controlCaseCount08_detailed.htm). Billions of people throughout the world are at risk for DENV8. While mosquito control can be effective, antiviral agents and wide-spectrum vaccines are being sought to aid in stemming the tide of infections9; 10; 11; 12; 13; 14; 15. To design effective vaccines, we need to know which areas of proteins are required for virus function or infectivity, and thus should be targeted by antibodies. Flaviviruses are variable, with many sequence variants found even in single virus isolates from the same patient (so-called “quasispecies”)16. Within groups of related viruses, a certain amount of variation will occur. We need better methods to determine where variance can indicate a more severe disease, or alter phenotype significantly.
The current way to classify variants is to compare them to “reference sequences”, which are primarily historical isolates. Here we present a more generally useful method, to use consensus sequences to filter noise due to random amino-acid variations within strains or subtypes. The consensus approach can simplify vaccine design by highlighting true variance that could influence the properties of viral proteins. Variant positions would be obvious by comparing a novel sequence to consensus sequences that represent the common properties of each group of FV. We tested a new method for defining consensus sequences, based on conservation of physicochemical properties (PCPs) at each position of correctly aligned sequences (where the alignment was checked to insure that it was consistent with the tertiary structure of the protein). The basis of our work was the cross-referenced database of annotated FV sequences, Flavitrack (http://carnot.utmb.edu/flavitrack) 17; 18. Using this information resource, we aligned and analyzed 928 naturally occurring (i.e, we excluded vaccine strains or lab-made chimeras) strains of 27 flavivirus species. To insure that the alignments were correct, we restricted the analysis to two proteins whose structures were known: the NS3 protease, a target for design of anti-viral drugs 19, and E, for which there is considerable additional information on epitopes and interacting areas 7; 20; 21; 22; 23; 24; 25; 26. To avoid prejudicing the results toward the FV species with the largest number of representatives, such as WNV or DENV, each species was separately analyzed with our programs for determining conservation according to physicochemical properties (PCP) of the amino acid side chains (summarized in the PCPMer suite (http://landau.utmb.edu:8080/WebPCPMer/; 20; 27; 28). We also demonstrated that the method used to derive PCP-consensus sequences was robust, by comparing sequences generated from a few diverse DENV sequences to one based on approximately 600 DENV strains.
Our overall goal was to decompose the sequences of the Flaviviruses to distinguish conserved areas that probably are associated with common function (i.e., needed for growth by similar viruses) and variable areas that correlated with the disease type or vector phenotype of closely related viruses. The challenge in multivalent vaccine design is to determine a sequence that will generate antibodies to block both the common and specific regions. Alignments of the PCP-consensus sequences easily distinguished mosquito-borne viruses from those spread by ticks 29; 30, and the mosquito-borne flaviviruses according to disease phenotype. The alignments were used to identify, in a statistically significant way, PCP-motifs of areas common to all the FV. The most striking results of the diversity analysis were a series of insertions and residue interchanges in the E protein that correlated with the ability to grow in either ticks or mosquitoes, distinguished mosquito-borne viruses according to their disease phenotype (hemorrhagic or encephalitic), and selected a few positions that could correlate with disease severity in DENV infections.
Materials and Methods
A. FV sequences and structures; alignment tools
The Flavitrack database 17; 18 contains more than 2000 annotated, complete flavivirus genomes, of which 928 natural isolates were used in this study. The E and NS3 sequences were obtained for 10 mosquito-borne, 13 tick-borne and 4 no-known-vector (NKV; sequences of viruses isolated from bats or rodents that have not been found in insects) species; annotated according to CDC abbreviations (http://www.ncid.cdc.gov/arbocat/browse.asp). Multiple sequence alignments were generated with Clustalw 2.0.3, or MUSCLE, for very large alignments, with default parameters.
Structures for the proteins were from PDB files 1OK8 (the DENV2 envelope protein in the postfusion conformation), 1URZ (the TBE envelope protein in the membrane fusion conformation) and 1BEF (the DENV2 NS3 protease). Models of DENV consensus sequences were prepared using 1OK8 as a template, and our MPACK modeling suite.31; 32; 33 The structural alignments and figures were prepared using the PyMol program 34.
B. PCP-Consensus Sequences
Because FV exhibit considerable subtype variability, methods that compare randomly selected representatives from each of the virus groups can give misleading results. Here, ClustalW alignments of all available sequences for each group were separately analyzed to determine conservation of physicochemical properties (PCP). In this approach, the 20 naturally occurring amino acids are represented as points in a five dimensional space, where the five dimensions roughly correspond to hydrophobicity/hydrophylicity (E1); size (E2); alpha-helix propensity (E3); the property E4 is partially related to the partial specific volume, number of codons and relative abundance of the amino acids; and E5 correlates weakly with beta-strand propensity 35. To obtain PCP-consensus sequences, one amino acid was chosen that best approximated the average value of the PCPs at each position of the multiple alignment 17. The average PCP was calculated for each vector p=E1,..,E5:
where N is the number of amino acids in the given column of the alignment; and are the five quantitative descriptors of the amino acid at that column of the j-th sequence. The consensus amino acid (Aa) was chosen from those occurring naturally at that position with the least Euclidean distance from the average:
The scale factors bp make for each column more significant the vectors with higher relative entropies and were calculated as described elsewhere 17.
C. Conserved residues and PCP-motifs were identified with our PCPMer suite 27; 28; 36, with modifications as described in this section (see also 17). For each column of a multiple sequence alignment we determined a “physicochemical distance” D as
where , p=E1..E5, are the five quantitative descriptors of i-th amino acid and N is the number of sequences in the multiple alignment. Thus D is the average of all pairs’ physicochemical distances. To visualize the 3D-relationships of conserved or variable residues, stereophysicochemical variability plots (SVP) display the physicochemical distances using a color scale for “similarity”, defined as
where Nno gaps is the number of sequences not containing a gap in the given column. The similarity is 1 for absolutely conserved (identical) columns and 0 for the most diverse. The definition of similarity also contains a term which lowers its value when gaps are present in the column of the alignment.
For the pairwise percentage identity calculations, the length of the shorter consensus sequence was used in the denominator.
Results
A. Generating unbiased sequence alignments of FV species and vector-specific groups
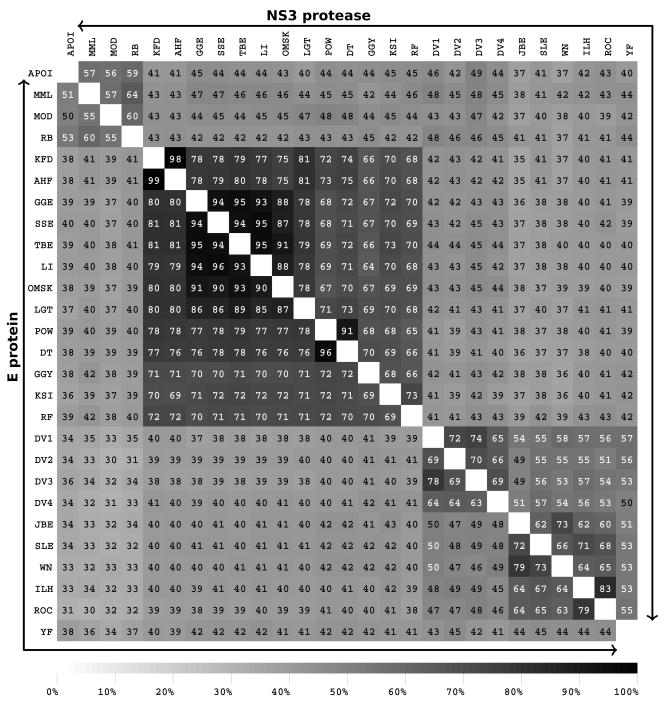
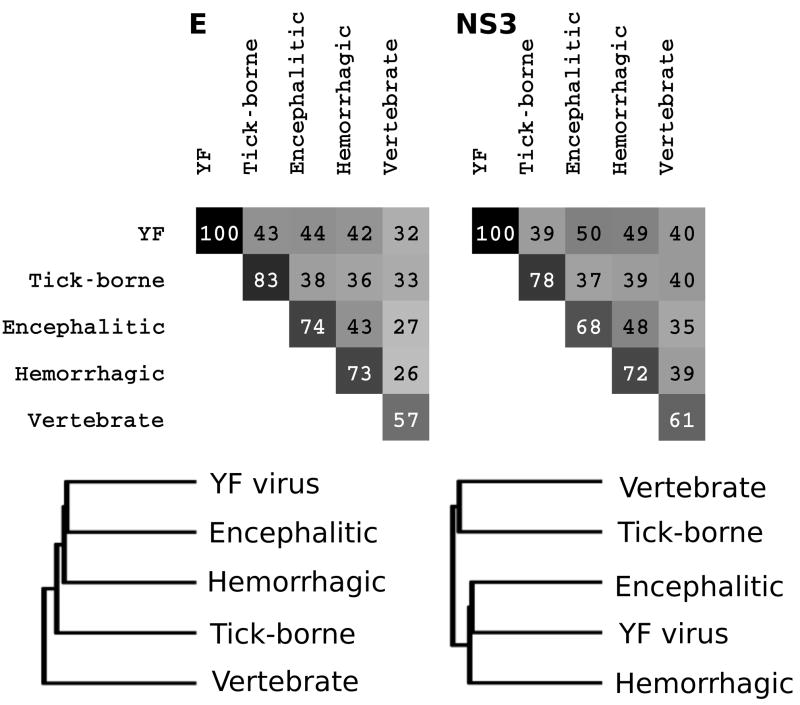
We chose to use consensus sequences to compare the FV groups for several reasons. First, trying to view 928 sequences simultaneously emphasized the need for unbiased data reduction methods. Secondly, consensus sequences should allow better discrimination of residues changes that fall outside the expected group variance. Because there are a many more sequences for certain mosquito-borne viruses than for any of the tick-borne or no-known-vector (NKV) groups, variability calculations that included all the natural sequences in Flavitrack would be biased toward species with the largest number of representatives (WNV and DENV). PCP-consensus sequences were created using alignments of all the sequences of each virus species (Figure S1 and S2). Alignments of the resulting PCP-consensus sequences were used to calculate interspecies identities (Figure 1), insertions and deletions, PCP-motifs and areas of diversity that distinguished the tick- or mosquito-borne species from the NKV-FV. The final set of PCP-consensus sequences may still be somewhat biased, as, for example, several of the tick-borne virus species, such as KFDV/AHFV and DTV/POWV differ from one another by only a few residues. This may lead to overestimating the similarity and underestimating the group variability of the tick-borne FV.
Figure 1.
Interspecies conservation of the NS3 protease (the upper triangular matrix) and the envelope protein (the lower triangular matrix) in terms of sequence identity between PCP-consensus sequences. The tick-borne (KFD-RF), mosquito-borne (DENV1-YF), no-vector (APOI-RB), mosquito-borne encephalitic (JBE-ROC), mosquito-borne hemorrhagic (DENV1-DENV4), and YF form distinct groups of FV. See the Flavitrack website for virus abbreviations (http://carnot.utmb.edu/flavitrack).
The +-strand RNA genome of a flavivirus is translated as a poly protein that is cleaved into three structural (C, prM, and E) and seven nonstructural (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) proteins. The NS5 polymerase is the most conserved throughout all the FV (78% in tick-borne and 59% in mosquito-borne FV) while the NS2a protein, especially in the C-terminal half, is the most variable 17. The E and NS3 proteases show considerable diversity at the amino acid level, with pairwise sequence identities between tick- and mosquito-borne FV of 37–43% for E and 35–45% for NS3 protease (Figure 1). However, the colored alignments (Supplementary Figure S1 and S2) show that despite differences at the amino acid level, both proteins maintain a similar pattern of charged, aromatic and aliphatic amino acids, consistent with them all having a 3D-structure similar to that determined for representative sequences for each protein. The alignments were further analyzed to define PCP-based motifs that were common to all FV (Table 1 and 2). The motifs for E and NS3 protease correspond to those we previously identified using reference sequences for 8 species 20. With a larger number of sequences, plus avoiding the bias arising from different number of representatives, the current motifs are generally a few residues shorter. Such areas of high conservation are likely to be involved in maintaining protein stability or function37.
Table 1.
Motifs in the envelope protein as recognized by the PCPMer program, common to both mosquito- and tick-borne flaviviruses. A variable relative entropy cutoff was used, ranging from 0.35 (least significant) to 0.6 (most significant). The residues in bold are surface exposed on the virus particle. The minimum length of a motif was 5, and the largest allowed gap (a continuous stretch of variable residues) in motifs was 2. The numbering corresponds to the sequence DENV2h64THmXxX_U87411.
| Entropy | Start | Sequence | End | |
|---|---|---|---|---|
| 0.55 | 9 | RDFVEGVSG | 17 | 1 |
| 0.50 | 26 | EHGSCVTTMA | 35 | |
| 0.55 | 54 | ATLRKYCIEA | 63 | 2 |
| 0.50 | 73 | RCPTQGEP | 80 | 3 |
| 0.60 | 99 | RGWGNGCGLF | 108 | 4 |
| 0.40 | 130 | VQPENLEYTIVITPH | 144 | |
| 0.35 | 159 | GKEIKITPQSS | 169 | 3 |
| 0.40 | 190 | GLDFNEMVLLQM | 201 | |
| 0.55 | 206 | WLVHRQWFLD | 215 | 3, 5 |
| 0.50 | 239 | TFKNPHA | 245 | |
| 0.45 | 250 | VVVLGSQEG | 258 | |
| 0.50 | 295 | KGMSYSMC | 302 | 6, 7 |
| 0.45 | 312 | IAETQHGTIVIRVQYEG | 328 | 7, 8 |
| 0.40 | 332 | PCKIP | 336 | 8 |
| 0.40 | 349 | GRLITVNP | 356 | |
| 0.40 | 366 | NIEAEPPFGDSYIIIG | 381 | 6, 7, 8, 9 |
| 0.45 | 391 | WFKKGSSIG | 399 | 7, 8, 10 |
| 0.55 | 416 | GDTAWDFGSLGG | 427 | |
| 0.40 | 431 | SIGKALHQVFGAIY | 444 | 2 |
| 0.45 | 448 | FSGVSW | 453 | |
| 0.40 | 460 | GVIITWIGMNSRS | 472 |
Part of a cytotoxic T-cell epitope identified for YF 69.
Residues corresponding to 60–68, 431–440 are part of T-cell epitopes in JBE70.
Residues corresponding to 75, 76, 81, 83, 86, 170, 234 are part of non-neutralizing epitopes in WNV 71.
Mutations at W101, L107, F108 block cross-reactive antibody recognition in DENV 21.
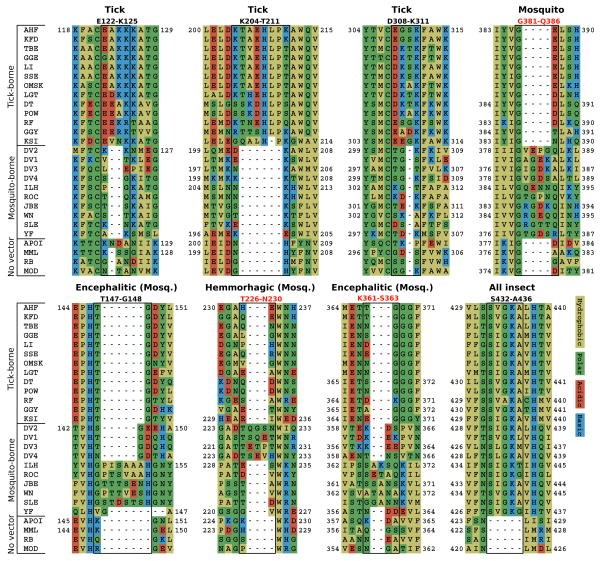
Adjacent to the K204-T211 insertion specific to tick-borne viruses (D203-K204 in DENV2).
Residues 291, 301–307, 381–383 form part of serotype specific epitopes 23.
Residues corresponding to 307, 308, 310–312, 325, 383, 384, 386, 388, 389, 391, 393 neutralizing epitope in DENV3 72.
Residues 325–331, 335–342, 368–398 in JBE are part of B-cell epitopes 70.
Adjacent to the insertion V382-G385 specific to mosquito-borne viruses.
Residue 390. which immediately precedes this region, is primarily Asn (N) in Asian strains of DENV2, while it is Glu (E) in American strains. Mutants of the Asian virus with E390 replicate more slowly in moncyte-derived macrophages than the wild type with N390.48
Table 2.
Motifs in the NS3 protease common to both mosquito- and tick-borne flaviviruses. The residues in bold are part of the peptide binding site. The same parameters were used as for the envelope protein. The numbering corresponds to the reference sequence DENV2h64THmXxX_U87411.
B. 3D-analysis of the NS3 protease shows limited group specific variation
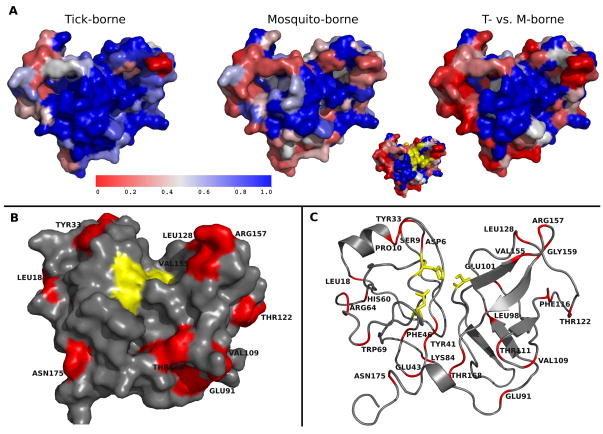
The N-terminal region of NS3 encodes a trypsin-like serine protease that, together with its cofactor, NS2B, cleaves four sites of the viral polyprotein (NS2A-NS2B, NS2B-NS3, NS3-NS4A, and NS4B-NS5). The cleavage sites are formed by the KR-, RK-, RR- or QR- residues downstream and a small amino acid S, G, or A upstream of the cleavage site 38. The physicochemical properties of the cleavage sites are well conserved in both tick- and mosquito-borne FV (Figure S3). As Figure 2 shows, the active site region of NS3 (near residues Q35, D75, S135) is completely conserved, but is surrounded by more variable residues (Figure 2c). There are only a few one residue insertions or deletions that distinguish the tick- from mosquito-borne NS3 proteins (at G29, G91, T156, and G179).
Figure 2.
Stereophysicochemical variability plots of the NS3 protease from DENV-2, colored to (A) show conserved (blue) and variable (red) residues within the tick-borne, mosquito-borne, or both (far right). The active site (yellow in the insert) is well conserved in all FV. The NS2B cofactor, required for activity, binds to NS3 from the back in the orientation shown. The residues colored red in B and C are conserved in a different fashion in tick- and mosquito-borne FV.
C. Insertions in E distinguish five different groups of FV
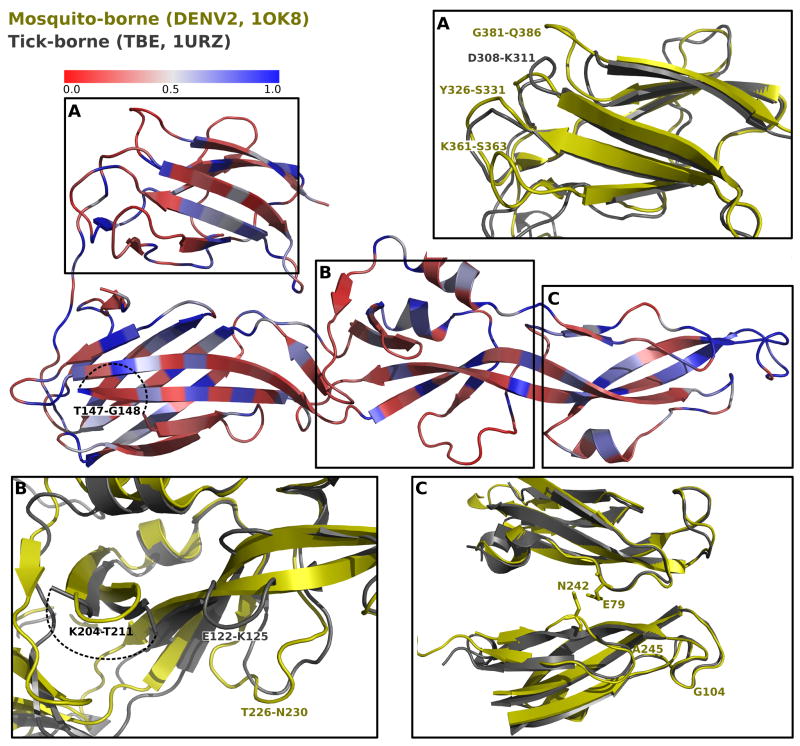
In contrast, the alignment of the E protein sequences demarcates eight well defined insertions that distinguish four FV groups (Figure 3, and illustrated pictorially in Figure 4); an additional insertion is found only in YF (see below). While the motifs common to both tick- and mosquito-borne FV are within the protein core or the surface that faces the viral membrane (surface exposure, from the GETAREA program, is annotated in Table 1), the group specific insertions, primarily residues with charged and polar sidechains, occur primarily in loops on the face of E that is free to interact with the host cell surface during cell entry (Figure 4). With one exception, all insertions are not present in the four NKV viruses, indicating either that they only are required for growth in insect cells, or that their elimination aids in establishing a chronic infection in vertebrates. The insertions occur in all three domains of E (Figure 4); one insertion (T431-L437), which distinguishes all the arthropod borne from the NKV-viruses, occurs in the stem, not included in the crystal structures. Three (D308-K311, K361-S363, and G381-Q386) are part of a ridge that forms part of a serotype-specific neutralization epitope which is know to contain residues important for receptor binding in vertebrate cells 26; 39; 40; 41. Two tick specific insertions, E122-K125 and K204-T211, are near one another in domain II (Figure 4b), near the insertion T226-N230 that is characteristic of mosquito-borne FV causing hemorrhagic disease. This region may contribute to binding to specific insect cells, as it lies near domain III in dimer structures and is probably on the mature virion particle surface (c.f. Figure 1A in 42). The two insertions that correlate with encephalitic phenotype, T147-G148 and K361-S363, are linked by a salt bridge.
Figure 3.
Regions of the PCP-consensus sequence alignment, illustrating the insertions that characterize the different groups of FV: tick-borne (AHF-KSI), mosquito-borne (DENV2-YF), No-Known-Vector (NKV; APOI-MOD), mosquito-borne hemorrhagic (DENV2-DENV4), and mosquito-borne encephalitic (ILH-SLE). One insertion in the C-terminus of the protein distinguishes all arthropod borne viruses from the endogenous ones.
Figure 4.
Stereophysicochemical variability plots showing the 3D-relationships between variable (red) and conserved (blue) residues of the ectodomain of the E- protein of mosquito- (DENV2 in fusion conformation, 1OK8, yellow) and tick-borne (TBE; 1URZ, gray) viruses. The insertions common to the different FV groups (see Figure 3) are shown in the expanded figures. Dotted lines indicate the position of insertions that are too flexible to be discerned from the crystal data.
D. Distinguishing features of YFV-E
Consistent with its mosquito-borne phenotype, YFV contains the G381-Q386 mosquito-specific insertion and lacks all three tick-specific insertions (Figure 3). However, it also lacks all three insertions that characterize the encephalitic or hemorrhagic phenotypes of mosquito-borne FV. YF can be further distinguished by an extended “deletion” around the T147-G148 loop common to the encephalitic strains, and one strain contains a DNN insertion at position 270. In addition to these, many individual residues of YF differ from all the other FV (~ 70/493 residues, or 14%), and YF appears to be equidistant from any of the other arthropod-borne viruses (Figure 5). Among the most striking of these distinguishing residues are a conserved tyrosine (Y326 in DENV2)43 that may be included in a “tyrosine corner motif” 44 in domain III of the E protein. This tyrosine is conserved as an aromatic residue (Y or F) in all FV, but is M in YF. The Y326 is involved in orbital overlap with F306 20, a residue that is absolutely conserved in all the FV except YF (where it is V).
Figure 5.
Pairwise distances (normalized to 100; calculated from the number of positions conserved within both groups and similar in physicochemical properties) and phylogenetic trees based on the envelope protein and the NS3 protease illustrate that the “outlier” position of YF depends on the protein taken as reference. While the E protein is equidistant from all groups, the NS3 protein lies between the encephalitic and hemorrhagic mosquito borne viruses.
E. Highly variable regions in DENV serotypes
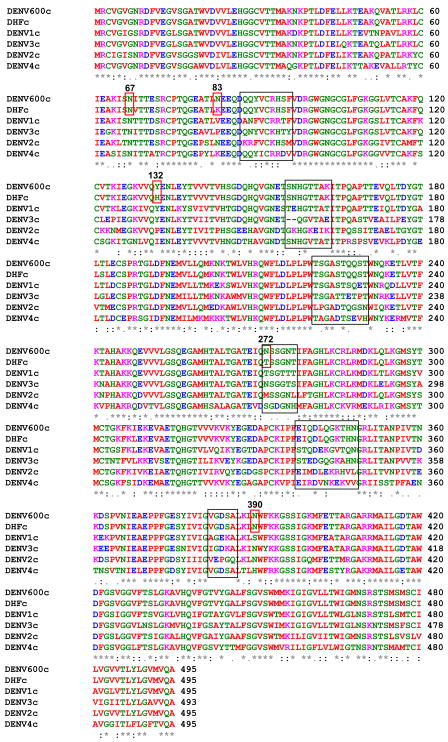
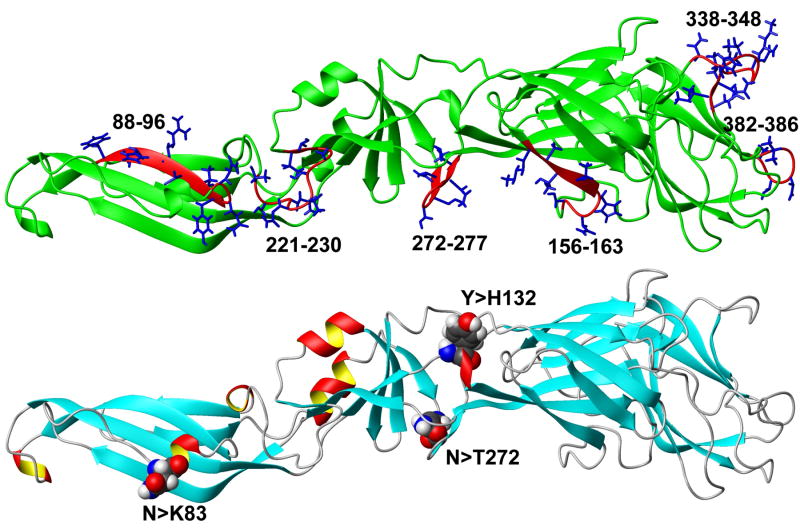
One use of PCP-consensus sequences is to distinguish viral areas that represent functional or serotype specific differences, rather than random variability that arises from error-prone RNA synthesis. For example, aligning a PCP-consensus sequence derived from 600 DENV strains (DENV600) in Flavitrack with a consensus sequences of DENV serotypes (Figure 6a) only showed major differences (i.e., the PCPs of the sequences differed significantly) in discrete areas (made up of residues 88–96, 156–163, 221–230, 272–277, 338–348, 382–386), which coincide for the most part with the areas where insertions occur (Figures 3 and 4). Indeed, 3D-mapping of the 6 regions where the consensus sequences from all 4 DENV serotypes vary (black boxes in Figure 6a) shows that the variants group to the same 4 areas, in all 3 domains, in the 3D structure of the protein. The variable region between 156 and 163 surrounds a 2 residue deletion that is characteristic of DENV3 strains.
Figure 6.
Determining areas of variability by comparing PCP-consensus sequences for each DENV serotype to one prepared from 600 DENV strains or 8-DHF strains from all 4 types (DENV600 and DENVDHF)
A) Alignment of the PCP-consensus sequences for each DENV serotype (DENV1–4) with DENV600 and DENVDHF. Areas of significant difference from the consensus sequences are blocked (88–96, 156–163, 221–230, 272–277, 338–348, 382–386), the three residues that differ (83, 132, 272) in the DENVDHF consensus sequence as compared to the DENV600, as well as 67 and 390, identified in other studies, are boxed in red. The amino acids are colored according to ClustalW2. Red: small and hydrophobic; Blue: acidic; Magenta: basic; Green: Hydroxyl, Amine and Basic.
B) Top: Ribbon diagram of a homology model of the DENV600 sequence, illustrating the positions of the regions of variability (from 6A). Only the variable sidechains in each residue group are illustrated; the 156–163 variable region includes a 2 amino acid deletion that is found only in DENV3 strains. Bottom: the same model, illustrating only positions where a significant change is found in the PCPs between the DENV600 and DENVDHF consensus sequences.
A PCP consensus sequence (DENVDHF) was prepared from 8 viruses (2 from each DENV serotype) that that were isolated from patients with a severe form of Dengue, Dengue hemorrhagic fever (DHF). Of these, at least two were isolated from patients with a fatal outcome. Comparing the PCP consensus sequences DENV600 to DENVDHF showed remarkably little variation (first 2 lines of the alignment in Figure 6a) except at three positions, 83,132 and 272. When we expanded the analysis to include the occurrence of amino acids at these three positions for all the DENV sequences in Flavitrack, we found little variance outside of that expected for the serotype for the DHF strains (Table 3), except that only 32% of the 307 strains of DENV2 had Lysine at residue 83, while 64% of the DHF strains did. We also note that residues 132 and 272 map near one another in domain 1 (Figure 6b), and both are close to previously described neutralizing epitopes (as determined from the position of escape mutants45). We also included position 67, a reported marker of hemorrhagic disease, in our analysis46, but found Asn at this position, in all DENV strains, regardless of reported disease severity. We found Asn at position 390 in all DENV except DENV4, where His predominates. The D390 variant, which correlates with milder DENV2 infections and reduced growth in macrophages47; 48, is quite rare in the selected sequences. This residue is differentially conserved as a non-charged polar residue in the FV groups: predominantly Asn in DENV1–3, Gln in the Tick-borne and YFV, and His in DENV4 and the encephalitic mosquito borne viruses. We would be interested in further tests at these sites by groups more familiar with the sources of the DENV strains, as there is often no information in the NCBI headers or related references on disease severity.
Table 3.
Comparison of residue choice at positions 67, 83, 132, 272 and 390 according to serotype in 670 DENV strains in Flavitrack and in strains that were designated as isolated from DHF cases (serotype followed by h). The residue in the DENVDHF consensus is highlighted in green, while residue in DENV600 consensus is showed as blue. All DENV strains had N at position 67, making this meaningless for discrimination of severe phenotype46.
| 67 | 83 | 132 | 272 | 390 | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | N | L | S | A | S | M | N | T | K | V | I | P | Y | H | S | M | N | T | H | D | S | N | |
| DENV1 | 215 | 215 | 0 | 0 | 0 | 0 | 3 | 0 | 212 | 0 | 0 | 0 | 1 | 213 | 1 | 0 | 3 | 0 | 212 | 0 | 0 | 214 | 1 |
| DENV2 | 307 | 307 | 0 | 3 | 1 | 0 | 305 | 0 | 2 | 97 | 0 | 0 | 307 | 0 | 0 | 0 | 305 | 0 | 2 | 0 | 5 | 16 | 286 |
| DENV3 | 136 | 136 | 1 | 0 | 0 | 0 | 0 | 98 | 38 | 0 | 0 | 0 | 0 | 83 | 53 | 0 | 0 | 98 | 38 | 0 | 0 | 0 | 136 |
| DENV4 | 12 | 12 | 0 | 0 | 0 | 12 | 0 | 0 | 0 | 12 | 1 | 11 | 0 | 0 | 0 | 12 | 0 | 0 | 0 | 12 | 0 | 0 | 0 |
| Total | 670 | 670 | 1 | 3 | 1 | 12 | 308 | 98 | 252 | 109 | 1 | 11 | 308 | 296 | 54 | 12 | 308 | 98 | 252 | 12 | 5 | 230 | 423 |
| DENV1h | 7 | 7 | 0 | 0 | 0 | 0 | 1 | 0 | 6 | 0 | 0 | 0 | 0 | 7 | 0 | 0 | 1 | 0 | 6 | 0 | 0 | 7 | 0 |
| DENV2h | 25 | 25 | 0 | 0 | 0 | 0 | 24 | 0 | 1 | 16 | 0 | 0 | 25 | 0 | 0 | 0 | 24 | 0 | 1 | 0 | 0 | 0 | 25 |
| DENV3h | 9 | 9 | 0 | 0 | 0 | 0 | 0 | 3 | 6 | 0 | 0 | 0 | 0 | 2 | 7 | 0 | 0 | 3 | 6 | 0 | 0 | 0 | 9 |
| DENV4h | 3 | 3 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | 0 | 0 |
| Total | 44 | 44 | 0 | 0 | 0 | 3 | 25 | 3 | 13 | 19 | 0 | 3 | 25 | 9 | 7 | 3 | 25 | 3 | 13 | 3 | 0 | 7 | 34 |
Discussion
Our results demonstrate the power of using PCP-consensus sequence alignments to discriminate constant regions and areas of significant diversity in a sea of variation. Protein decomposition, using statistical sequence analysis and 3D structure comparison, delineated common motifs (Tables 1 and 2), group specific insertions (Figures 2–5), and zones of variability (Figure 6). Comparison of individual PCP-consensus sequences for E proteins of DENV serotypes indicated that real diversity (i.e, areas with significant changes in physicochemical properties) was limited to a few regions (Figure 6), that generally coincided with areas where the observed group specific insertions throughout the FV family also occurred (Figures 3 and 4). Our comparison of a DHF-consensus from 8 strains with that obtained from comparing 600 sequences of DENV showed differences only in three residues. This highlights that our method for extracting PCP-consensus sequences is extremely robust, generating similar results for only a small subsection of the overall data, and is consistent with earlier studies that indicated the sequences of DHF strains were similar to those isolated from milder disease cases16; 49. In light of this, others have suggested that the severity of DENV infection may be more related to the immune state of the patient and preexisting antibodies than to the exact sequence of the infecting virus.21; 50; 51; 52; 53 Although the three positions where the DENVDHF differs from the overall DENV consensus cannot be related to disease severity with our current data, two lie near binding sites of previously identified escape mutations (i.e., mutations that allow viruses to “escape” from growth inhibition by neutralizing antibodies)45. This data, including the online alignment and supplementary data, provide a framework for determining variance that lies outside that anticipated for a given FV group.
The significance of variable regions
In our previous work, we isolated areas of maximum conservation, in order to identify functional motifs20; 28; 36. Here, we also wanted to highlight diversity among the sequences, especially as this applied to specific properties of the viruses. The full alignment (online in Flavitrack), or the more easily viewed, reduced representation of 27 PCP-consensus sequences (Figures S1 and S2), showed 8 insertions in the E protein (Figure 3, 4), that clearly distinguished 4 groups of arthropod-borne FV from the NKV-viruses isolated from mammals. An additional deletion in a flexible loop in domain II of the E protein distinguishes DENV3 from the other serotypes of DENV.
Insertions that distinguish tick from mosquito borne FV
There are many differences in the life cycle of ticks and mosquitoes that could account for the pronounced differences in the sequences of FV that replicate in them. 54 The non-protein encoding regions of the genomes of tick- and mosquito-borne FV have different complementary 5′- and 3′-cyclization elements 55; 56; 57; 58; 59 that are required for viral replication 60. Our results clearly delineated E protein regions that distinguish tick from mosquito borne FV. Other insertions separated the mosquito-borne FV into three subgroups: those causing hemorrhagic disease (DENV1-DENV4), encephalitic disease (JBE-ROC) and YF. These insertions include two previously characterized insertions in the DE and FG loops (K361-S363 and G381-Q386) in the domain III of DENV2 and WNV 61; 62; 63 and one in the glycosylated E0F0 loop (T147-G148) domain I of WNV 62. The tick specific insertions have not been previously reported.
It is possible that these insertion regions could be used to design anti-viral peptides. For example, a peptide based on one of the insertions common to mosquito borne FV, between residues 380–389, prevented DENV2 from binding to mosquito, but not mammalian cells 61. We plan to test other peptides, based on both the wild type and PCP-consensus sequences of these insertions, to see if they can inhibit the growth of WNV in mammalian and insect cells.
A PCP-consensus sequence for DENV multivalent vaccine design
Our consensus sequence for 600 different DENV was generated as the first step in designing a multivalent vaccine against all 4 serotypes. We concentrated on the E protein for our analysis as others have shown that vaccines based on sections of the E protein64, especially domain III, may be sufficient to protect against WNV65 or DENV66; 67. Comparing our overall consensus sequence to individual consensus sequences for all 4 different DENV serotypes found significant changes in the PCPs primarily in 4 areas, which coincided with regions where group specific insertions occur across the vector borne FV. Our analysis also identified as variable a ridge formed from discontinuous regions of the E protein domain III, which had been previously identified as a binding site for neutralizing antibodies to WNV68 and for DENV-serotype-specific and complex-specific neutralizing monoclonal antibodies7; 25. Residue 390, Asn throughout all the DENV2 but Asp in five American strains reported to have milder phenotype47, lies near this ridge.
Our results indicate that the physicochemical properties of the E protein are highly conserved throughout the DENV serotypes. This suggests that a multivalent protein for vaccine use may involve optimization of only a few discrete regions, including the variable loop on the domain III of the E protein. Any vaccine protein should contain the motifs of Table 1, which are common to all the FV, as a stable scaffolding, and be altered to reflect the common properties of the other serotypes. The PCP consensus sequence, coupled with information on positions of neutralizing antibodies7; 25 and escape mutants, will be particularly valuable for modeling the variable areas. Our amino acid choice represents the amino acid most consistent with the physicochemical properties of all the sequences in the group, and should thus be most likely to offer multivalent protection against the different serotypes.
In conclusion, we present here a novel way to compare the amino acids sequences of the FV in a statistically significant fashion. The approach was used to define areas common to all Flaviviruses in two structurally characterized proteins, the NS3 protease domain and the E protein. We highlighted group specific insertions and selected areas of sequence variations that could be targeted in vaccine design. Optimal choice of residues in these variable areas will be important for the design of multivalent vaccines and inhibitors.
Supplementary Material
Acknowledgments
We thank Stephen Higgs and Werner Braun for a critical reading of the manuscript. Support for this project came from NIH grant AI064913. The computational resources of the Sealy Center for Structural Biology and Molecular Biophysics were also used in this project.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zell R, Krumbholz A, Wutzler P. Impact of global warming on viral diseases: what is the evidence? Current Opinion in Biotechnology. 2008;19:652–660. doi: 10.1016/j.copbio.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaunt MW, Gould EA. Rapid subgroup identification of the flaviviruses using degenerate primer E-gene RT-PCR and site specific restriction enzyme analysis. Journal of Virological Methods. 2005;128:113–127. doi: 10.1016/j.jviromet.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 3.Thiel HJ, Collett MS, Gould EA, Heinz FX, Houghton M, Meyers G, Purcell RH, Rice CM. Family Flaviviridae. Virus Taxonomy, Eight Report of the International Committee for the Taxonomy of Viruses. 2005:981–998. [Google Scholar]
- 4.Gaunt MW, Sall AA, de Lamballerie X, Falconar AK, Dzhivanian TI, Gould EA. Phylogenetic relationships of flaviviruses correlate with their epidemiology, disease association and biogeography. J Gen Virol. 2001;82:1867–1876. doi: 10.1099/0022-1317-82-8-1867. [DOI] [PubMed] [Google Scholar]
- 5.Mackenzie JS, Lindsay MD, Coelen RJ, Broom AK, Hall RA, Smith DW. Arboviruses causing human disease in the Australasian zoogeographic region. Archives of Virology. 1994;136:447–467. doi: 10.1007/BF01321074. [DOI] [PubMed] [Google Scholar]
- 6.Nisbet DJ, Lee KJ, van den Hurk AF, Johansen CA, Kuno G, Chang GJ, Mackenzie JS, Ritchie SA, Hall RA. Identification of new flaviviruses in the Kokobera virus complex. J Gen Virol. 2005;86:121–124. doi: 10.1099/vir.0.80381-0. [DOI] [PubMed] [Google Scholar]
- 7.Gromowski GD, Barrett ND, Barrett AD. Characterization of dengue virus complex-specific neutralizing epitopes on envelope protein domain III of dengue 2 virus. J Virol. 2008;82:8828–37. doi: 10.1128/JVI.00606-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monath TP, Kanesa-Thasan N, Guirakhoo F, Pugachev K, Almond J, Lang J, Quentin-Millet MJ, Barrett AD, Brinton MA, Cetron MS, Barwick RS, Chambers TJ, Halstead SB, Roehrig JT, Kinney RM, Rico-Hesse R, Strauss JH. Recombination and flavivirus vaccines: a commentary. Vaccine. 2005;23:2956–8. doi: 10.1016/j.vaccine.2004.11.069. [DOI] [PubMed] [Google Scholar]
- 9.Ishikawa T, Widman DG, Bourne N, Konishi E, Mason PW. Construction and evaluation of a chimeric pseudoinfectious virus vaccine to prevent Japanese encephalitis. Vaccine. 2008;26:2772–2781. doi: 10.1016/j.vaccine.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 10.Beasley DWC, Lewthwaite P, Solomon T. Current use and development of vaccines for Japanese encephalitis. Expert Opinion on Biological Therapy. 2008;8:95–106. doi: 10.1517/14712598.8.1.95. [DOI] [PubMed] [Google Scholar]
- 11.Widman DG, Ishikawa T, Fayzulin R, Bourne N, Mason PW. Construction and characterization of a second-generation pseudoinfectious West Nile virus vaccine propagated using a new cultivation system. Vaccine. 2008;26:2762–2771. doi: 10.1016/j.vaccine.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 12.Tang F, Zhang JS, Liu W, Zhao QM, Zhang F, Wu XM, Yang H, Ly H, Cao WC. Short report: failure of Japanese encephalitis vaccine and infection in inducing neutralizing antibodies against West Nile virus, People’s Republic of China. American Journal of Tropical Medicine and Hygiene. 2008;78:999–1001. [PubMed] [Google Scholar]
- 13.Barrett ADT. Flavivirus DNA vaccine with a kick. Nature Biotechnology. 2008;26:525–526. doi: 10.1038/nbt0508-525. [DOI] [PubMed] [Google Scholar]
- 14.Wiggan O, Huang CYH, Silengo SJ, Kinney RM, Osorio JE, Stinchcomb DT. Optimization of a chimeric DEN-2/west nile vaccine. American Journal of Tropical Medicine and Hygiene. 2007;77:135–135. [Google Scholar]
- 15.El Garch H, Minke JM, Rehder J, Richard S, Toulemonde CE, Dinic S, Andreoni C, Audonnet JC, Nordgren R, Juillard V. A West Nile virus (WNV) recombinant canarypox virus vaccine elicits WNV-specific neutralizing antibodies and cell-mediated immune responses in the horse. Veterinary Immunology and Immunopathology. 2008;123:230–239. doi: 10.1016/j.vetimm.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 16.Rico-Hesse R, Harrison LM, Nisalak A, Vaughn DW, Kalayanarooj S, Green S, Rothman AL, Ennis FA. Molecular evolution of dengue type 2 virus in Thailand. Am J Trop Med Hyg. 1998;58:96–101. doi: 10.4269/ajtmh.1998.58.96. [DOI] [PubMed] [Google Scholar]
- 17.Danecek P, Schein CH. Flavitrack analysis of the structure and function of West Nile non-structural proteins. Int J of Bioinformatics Res Appl. 2009 doi: 10.1504/IJBRA.2010.032117. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Misra M, Schein CH. Flavitrack: an annotated database of flavivirus sequences. Bioinformatics. 2007;23:2645–2647. doi: 10.1093/bioinformatics/btm383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stoermer MJ, Chappell KJ, Liebscher S, Jensen CM, Gan CH, Gupta PK, Xu WJ, Young PR, Fairlie DP. Potent cationic inhibitors of West Nile Virus NS2B/NS3 protease with serum stability, cell permeability and antiviral activity. Journal of Medicinal Chemistry. 2008;51:5714–5721. doi: 10.1021/jm800503y. [DOI] [PubMed] [Google Scholar]
- 20.Schein CH, Zhou B, Braun W. Stereophysicochemical variability plots highlight conserved antigenic areas in Flaviviruses. Virol J. 2005;2:40. doi: 10.1186/1743-422X-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lai CY, Tsai WY, Lin SR, Kao CL, Hu HP, King CC, Wu HC, Chang GJ, Wang WK. Antibodies to envelope glycoprotein of dengue virus during the natural course of infection are predominantly cross-reactive and recognize epitopes containing highly conserved residues at the fusion loop of domain II. J Virol. 2008;82:6631–43. doi: 10.1128/JVI.00316-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ledizet M, Kar K, Foellmer HG, Bonafe N, Anthony KG, Gould LH, Bushmich SL, Fikrig E, Koski RA. Antibodies targeting linear determinants of the envelope protein protect mice against West Nile virus. J Infect Dis. 2007;196:1741–8. doi: 10.1086/523654. [DOI] [PubMed] [Google Scholar]
- 23.Modis Y, Ogata S, Clements D, Harrison SC. Variable surface epitopes in the crystal structure of dengue virus type 3 envelope glycoprotein. J Virol. 2005;79:1223–31. doi: 10.1128/JVI.79.2.1223-1231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oliphant T, Nybakken GE, Austin SK, Xu Q, Bramson J, Loeb M, Throsby M, Fremont DH, Pierson TC, Diamond MS. Induction of epitope-specific neutralizing antibodies against West Nile virus. J Virol. 2007;81:11828–39. doi: 10.1128/JVI.00643-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sukupolvi-Petty S, Austin SK, Purtha WE, Oliphant T, Nybakken GE, Schlesinger JJ, Roehrig JT, Gromowski GD, Barrett AD, Fremont DH, Diamond MS. Type- and subcomplex-specific neutralizing antibodies against domain III of dengue virus type 2 envelope protein recognize adjacent epitopes. J Virol. 2007;81:12816–26. doi: 10.1128/JVI.00432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rey FA. Dengue virus envelope glycoprotein structure: new insight into its interactions during viral entry. Proc Natl Acad Sci U S A. 2003;100:6899–901. doi: 10.1073/pnas.1332695100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mathura VS, Schein CH, Braun W. Identifying property based sequence motifs in protein families and superfamilies: application to DNase-1 related endonucleases. Bioinformatics. 2003;19:1381–1390. doi: 10.1093/bioinformatics/btg164. [DOI] [PubMed] [Google Scholar]
- 28.Schein CH, Zhou B, Oezguen N, Mathura VS, Braun W. Molego-based definition of the architecture and specificity of metal-binding sites. Proteins-Structure Function and Bioinformatics. 2005;58:200–210. doi: 10.1002/prot.20253. [DOI] [PubMed] [Google Scholar]
- 29.Kuno G. Host range specificity of flaviviruses: correlation with in vitro replication. J Med Entomol. 2007;44:93–101. doi: 10.1603/0022-2585(2007)44[93:hrsofc]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 30.Lawrie CH, Uzcategui NY, Armesto M, Bell-Sakyi L, Gould EA. Susceptibility of mosquito and tick cell lines to infection with various flaviviruses. Med Vet Entomol. 2004;18:268–74. doi: 10.1111/j.0269-283X.2004.00505.x. [DOI] [PubMed] [Google Scholar]
- 31.Oezguen N, Zhou B, Negi SS, Ivanciuc O, Schein CH, Labesse G, Braun W. Comprehensive 3D-modeling of allergenic proteins and amino acid composition of potential conformational IgE epitopes. Mol Immunol. 2008;45:3740–7. doi: 10.1016/j.molimm.2008.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schein CH, Nagle GT, Page JS, Sweedler JV, Xu Y, Painter SD, Braun W. Aplysia attractin: Biophysical characterization and modeling of a water-borne pheromone. Biophysical Journal. 2001;81:463–472. doi: 10.1016/S0006-3495(01)75714-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soman KV, Midoro-Horiuti T, Ferreon JC, Goldblum RM, Brooks EG, Kurosky A, Braun W, Schein CH. Homology modeling and characterization of IgE epitopes of mountain cedar allergen Jun a 3. Biophys J. 2000;79:1601–1609. doi: 10.1016/S0006-3495(00)76410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; Palo Alto, CA, USA: 2002. [Google Scholar]
- 35.Venkatarajan MS, Braun W. New quantitative descriptors of amino acids based on multidimensional scaling of a large number of physical-chemical properties. Journal of Molecular Modeling. 2001;7:445–453. [Google Scholar]
- 36.Schein CH, Ozgun N, Izumi T, Braun W. Total sequence decomposition distinguishes functional modules, “molegos” in apurinic/apyrimidinic endonucleases. Bmc Bioinformatics. 2002:3. doi: 10.1186/1471-2105-3-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kristensen DM, Ward RM, Lisewski AM, Erdin S, Chen BY, Fofanov VY, Kimmel M, Kavraki LE, Lichtarge O. Prediction of enzyme function based on 3D templates of evolutionarily important amino acids. Bmc Bioinformatics. 2008:9. doi: 10.1186/1471-2105-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murthy HM, Clum S, Padmanabhan R. Dengue virus NS3 serine protease. Crystal structure and insights into interaction of the active site with substrates by molecular modeling and structural analysis of mutational effects. J Biol Chem. 1999;274:5573–80. doi: 10.1074/jbc.274.9.5573. [DOI] [PubMed] [Google Scholar]
- 39.Chu JJ, Ng ML. Interaction of West Nile virus with alpha v beta 3 integrin mediates virus entry into cells. J Biol Chem. 2004;279:54533–41. doi: 10.1074/jbc.M410208200. [DOI] [PubMed] [Google Scholar]
- 40.Jindadamrongwech S, Thepparit C, Smith DR. Identification of GRP 78 (BiP) as a liver cell expressed receptor element for dengue virus serotype 2. Arch Virol. 2004;149:915–27. doi: 10.1007/s00705-003-0263-x. [DOI] [PubMed] [Google Scholar]
- 41.Lee JW, Chu JJ, Ng ML. Quantifying the specific binding between West Nile virus envelope domain III protein and the cellular receptor alphaVbeta3 integrin. J Biol Chem. 2006;281:1352–60. doi: 10.1074/jbc.M506614200. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y, Zhang W, Ogata S, Clements D, Strauss JH, Baker TS, Kuhn RJ, Rossmann MG. Conformational changes of the flavivirus E glycoprotein. Structure. 2004;12:1607–18. doi: 10.1016/j.str.2004.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maillard RA, Jordan M, Beasley DWC, Barrett ADT, Lee JC. Long range communication in the envelope protein domain III and its effect on the resistance of West Nile virus to antibody-mediated neutralization. Journal of Biological Chemistry. 2008;283:613–622. doi: 10.1074/jbc.M706031200. [DOI] [PubMed] [Google Scholar]
- 44.Hemmingsen JM, Gernert KM, Richardson JS, Richardson DC. The tyrosine corner: a feature of most Greek key beta-barrel proteins. Protein Sci. 1994;3:1927–37. doi: 10.1002/pro.5560031104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roehrig JT. Antigenic structure of flavivirus proteins. Adv Virus Res. 2003;59:141–75. doi: 10.1016/s0065-3527(03)59005-4. [DOI] [PubMed] [Google Scholar]
- 46.Barker WC, Mazumder R, Vasudevan S, Sagripanti JL, Wu CH. Sequence signatures in envelope protein may determine whether flaviviruses produce hemorrhagic or encephalitic syndromes. Virus Genes. 2009;39:1–9. doi: 10.1007/s11262-009-0343-4. [DOI] [PubMed] [Google Scholar]
- 47.Leitmeyer KC, Vaughn DW, Watts DM, Salas R, Villalobos I, de C, Ramos C, Rico-Hesse R. Dengue Virus Structural Differences That Correlate with Pathogenesis. J Virol. 1999;73:4738–4747. doi: 10.1128/jvi.73.6.4738-4747.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pryor MJ, Carr JM, Hocking H, Davidson AD, Li P, Wright PJ. Replication of dengue virus type 2 in human monocyte-derived macrophages: comparisons of isolates and recombinant viruses with substitutions at amino acid 390 in the envelope glycoprotein. Am J Trop Med Hyg. 2001;65:427–434. doi: 10.4269/ajtmh.2001.65.427. [DOI] [PubMed] [Google Scholar]
- 49.Rico-Hesse R. Dengue virus evolution and virulence models. Clin Infect Dis. 2007;44:1462–6. doi: 10.1086/517587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gubler DJ. Epidemic dengue/dengue hemorrhagic fever as a public health, social and economic problem in the 21st century. TRENDS Microbiol. 2002;10:100–103. doi: 10.1016/s0966-842x(01)02288-0. [DOI] [PubMed] [Google Scholar]
- 51.Guzman MG, Kouri G. Dengue and dengue hemorrhagic fever in the Americas: lessions and challenges. J Clin Virol. 2003;27:1–13. doi: 10.1016/s1386-6532(03)00010-6. [DOI] [PubMed] [Google Scholar]
- 52.Loke H, Bethell DB, Phuong CXT, Dung M, Schneider J, White NJ, Day NP, Farrar J, Hill AVS. Strong HLA Class I-restricted T cell responses in dengue hemorrhagic fever: a double-edged sword? J Infect Dis. 2001;184:1369–1373. doi: 10.1086/324320. [DOI] [PubMed] [Google Scholar]
- 53.Mongkolsapaya J, Dejnirattisai W, Xu X, Vasanawathana S, Tangthawornchaikul N, Chairunsri A, Swasdivorn S, Duangchinda T, Dong T, Rowland-Jones S, Yenchitsomanus P, McMichael A, Malasit P, Screaton G. Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fevr. Nature Med. 2003;9:921–927. doi: 10.1038/nm887. [DOI] [PubMed] [Google Scholar]
- 54.Higgs S. Influences of arthropod vectors on encephalitic arboviruses. In: Reiss C, editor. Neurotropic Viral Infections. Cambridge University Press; New York: 2008. pp. 362–381. [Google Scholar]
- 55.Hahn CS, Hahn YS, Rice CM, Lee E, Dalgarno L, Strauss EG, Strauss JH. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J Mol Biol. 1987;198:33–41. doi: 10.1016/0022-2836(87)90455-4. [DOI] [PubMed] [Google Scholar]
- 56.Mandl CW, Holzmann H, Kunz C, Heinz FX. Complete genomic sequence of Powassan virus: evaluation of genetic elements in tick-borne versus mosquito-borne flaviviruses. Virology. 1993;194:173–84. doi: 10.1006/viro.1993.1247. [DOI] [PubMed] [Google Scholar]
- 57.Khromykh AA, Meka H, Guyatt KJ, Westaway EG. Essential role of cyclization sequences in flavivirus RNA replication. J Virol. 2001;75:6719–28. doi: 10.1128/JVI.75.14.6719-6728.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Markoff L. 5′- and 3′-noncoding regions in flavivirus RNA. Adv Virus Res. 2003;59:177–228. doi: 10.1016/S0065-3527(03)59006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thurner C, Witwer C, Hofacker IL, Stadler PF. Conserved RNA secondary structures in Flaviviridae genomes. J Gen Virol. 2004;85:1113–24. doi: 10.1099/vir.0.19462-0. [DOI] [PubMed] [Google Scholar]
- 60.Kofler RM, Hoenninger VM, Thurner C, Mandl CW. Functional analysis of the tick-borne encephalitis virus cyclization elements indicates major differences between mosquito-borne and tick-borne flaviviruses. J Virol. 2006;80:4099–113. doi: 10.1128/JVI.80.8.4099-4113.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hung JJ, Hsieh MT, Young MJ, Kao CL, King CC, Chang W. An external loop region of domain III of dengue virus type 2 envelope protein is involved in serotype-specific binding to mosquito but not mammalian cells. Journal of Virology. 2004;78:378–388. doi: 10.1128/JVI.78.1.378-388.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kanai R, Kar K, Anthony K, Gould LH, Ledizet M, Fikrig E, Marasco WA, Koski RA, Modis Y. Crystal structure of west nile virus envelope glycoprotein reveals viral surface epitopes. J Virol. 2006;80:11000–8. doi: 10.1128/JVI.01735-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature. 1995;375:291–8. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- 64.Lieberman MM, Nerurkar VR, Luo HY, Cropp B, Carrion R, de la Garza M, Coller BA, Clements D, Ogata S, Wong T, Martyak T, Weeks-Levy C. Immunogenicity and Protective Efficacy of a Recombinant Subunit West Nile Virus Vaccine in Rhesus Monkeys. Clinical and Vaccine Immunology. 2009;16:1332–1337. doi: 10.1128/CVI.00119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chu JHJ, Chiang CCS, Ng ML. Immunization of flavivirus West Nile recombinant envelope domain III protein induced specific immune response and protection against West Nile virus infection. Journal of Immunology. 2007;178:2699–2705. doi: 10.4049/jimmunol.178.5.2699. [DOI] [PubMed] [Google Scholar]
- 66.Chin JFL, Chu JJH, Ng ML. The envelope glycoprotein domain III of dengue virus serotypes 1 and 2 inhibit virus entry. Microbes and Infection. 2007;9:1–6. doi: 10.1016/j.micinf.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 67.Babu JP, Pattnaik P, Gupta N, Shrivastava A, Khan M, Rao PVL. Immunogenicity of a recombinant envelope domain III protein of dengue virus type-4 with various adjuvants in mice. Vaccine. 2008;26:4655–4663. doi: 10.1016/j.vaccine.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 68.Nybakken GE, Oliphant T, Johnson S, Burke S, Diamond MS, Fremont DH. Structural basis of West Nile virus neutralization by a therapeutic antibody. Nature. 2005;437:764–768. doi: 10.1038/nature03956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van der Most RG, Harrington LE, Giuggio V, Mahar PL, Ahmed R. Yellow fever virus 17D envelope and NS3 proteins are major targets of the antiviral T cell response in mice. Virology. 2002;296:117–24. doi: 10.1006/viro.2002.1432. [DOI] [PubMed] [Google Scholar]
- 70.Li P, Zheng QS, Wang Q, Li Y, Wang EX, Liu JJ, Cao RB, Chen PY. Immune responses of recombinant adenoviruses expressing immunodominant epitopes against Japanese encephalitis virus. Vaccine. 2008;26:5802–7. doi: 10.1016/j.vaccine.2008.08.035. [DOI] [PubMed] [Google Scholar]
- 71.Oliphant T, Nybakken GE, Engle M, Xu Q, Nelson CA, Sukupolvi-Petty S, Marri A, Lachmi BE, Olshevsky U, Fremont DH, Pierson TC, Diamond MS. Antibody recognition and neutralization determinants on domains I and II of West Nile Virus envelope protein. J Virol. 2006;80:12149–59. doi: 10.1128/JVI.01732-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Matsui K, Gromowski GD, Li L, Schuh AJ, Lee JC, Barrett AD. Characterization of dengue complex-reactive epitopes on dengue 3 virus envelope protein domain III. Virology. 2008 doi: 10.1016/j.virol.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 73.Zivny J, DeFronzo M, Jarry W, Jameson J, Cruz J, Ennis FA, Rothman AL. Partial agonist effect influences the CTL response to a heterologous dengue virus serotype. J Immunol. 1999;163:2754–60. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.