Summary
The ubiquity of small RNAs (sRNAs) in bacteria is now well established. These transcripts are members of regulatory circuits involved in diverse processes ranging from stress adaptation to virulence to metabolism. Recent large-scale searches suggest that there exist many times more sRNAs than previously predicted even in the best studied of bacterial transcriptomes. Based on these and other recent findings of regulatory sRNAs that do not function in a “classical” manner, we propose that the working definition of sRNAs be broadened.
Introduction
It has become abundantly evident that small RNAs (sRNAs) can act as regulators of gene expression in all organisms in which they have been investigated. In the past decade, there has been an explosion in the number of sRNAs identified in bacteria [1-5]. Although still a young field, the study of bacterial sRNAs has already greatly extended our knowledge of genetic regulatory circuits in bacteria [6-13]. Historically, the bacterial sRNA field has focused on trans-encoded sRNAs, which differ from the cis-encoded antisense RNAs of plasmids, bacteriophages and chromosomes in that they have only imperfect complementarity with their RNA targets [14]. Small RNAs are often defined as short non-coding transcripts that, together with the RNA chaperone, Hfq, act in trans to control the translation or stability of target mRNAs. Indeed, many of the sRNAs originally identified, primarily in the model organism Escherichia coli, function in this manner (Figure 1A,B) [15-20].
Figure 1. Different types of sRNAs based on their mechanism of action.
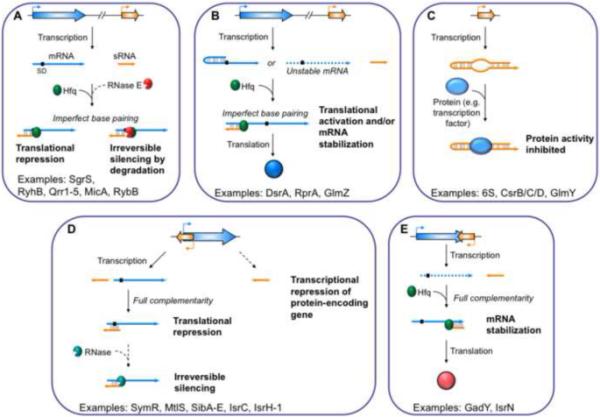
Trans-encoded sRNAs can base-pair imperfectly with mRNA targets and either (A) repress or (B) activate translation. Alternatively, (C) some trans-encoded sRNAs interact with proteins, including transcription factors, and inhibit their activity. Cis-encoded antisense sRNAs can also either (D) activate or (E) repress protein expression. Colored arrows represent RNA transcripts; black boxes indicate Shine-Dalgarno (SD) sequences. Dashed colored arrows represent unstable transcripts. Dashed black arrows represent hypothetical mechanistic steps of sRNA-mediated regulatory pathways.
Recent reports have revealed that there are more types of sRNA in bacteria than were predicted [21-24]. Additionally, it is becoming increasingly apparent that the established sRNA paradigm does not describe all or even most sRNAs in bacteria. Many cis-acting, chromosomally-encoded antisense sRNAs have now been identified, and Hfq-independent sRNAs have also been described. In addition, the known functions of sRNAs are broader than just the control of translation or mRNA stability. For example, the 6S and CsrB/C/D sRNAs directly bind protein transcription factors to affect downstream gene expression (Figure 1C) [25,26]. There are even sRNAs that serve both as regulatory RNAs and translated messenger RNAs. These observations are compelling reasons for broadening the working definition of sRNAs to accommodate all of these examples, as well as to leave room for the inevitable additional types of sRNAs yet to be reported.
sRNAs that do not fit the mold
Classically, sRNAs that regulate mRNA expression were known as non-coding transcripts that regulate gene expression by binding Hfq and an mRNA expressed in trans. Below, we provide examples from the recent literature that suggest that there are many aspects of this definition that must be relaxed or redefined.
sRNAs that code
There are now several examples of sRNAs that have regulatory functions as RNA but are also translated into small proteins. One of the first sRNAs to be identified, RNAIII, a cis-acting regulatory RNA in Staphylococcus aureus involved in quorum-sensing, also encodes a 26-amino acid peptide that may be involved in biofilm integrity [27]. In E. coli, the 227-nucleotide (nt) sRNA SgrS, which is expressed when phosphosugar intermediates reach high, toxic levels, is a translational repressor of ptsG encoding the glucose transporter component of the phosphoenolpyruvate phosphotransferase system (PTS) [28]. SgrS also codes for the 43-amino acid SgrT that inhibits glucose transport through PtsG [29]. Finally, transcriptional profiling of Listeria monocytogenes recently revealed, among many putative sRNAs, five that are also predicted to encode small proteins ranging from 28–64 amino acids [24]. These findings collectively suggest that sRNAs are not necessarily non-coding, but can have dual functions as both messenger RNA and regulatory RNA.
Hfq is not always required
Originally identified as a host factor for replication of the RNA phage Qβ in E. coli [30], Hfq is an RNA binding protein that has been studied extensively for its role in sRNA-mediated gene regulation. Many of the best-characterized sRNAs require Hfq for stability and/or activity [15,19,31]. In several cases, Hfq has been shown to aid activity by enhancing the rate of duplex formation between sRNA and target RNA [32,33], which is often followed by hydrolysis of target, sRNA, or both, by ribonucleases such as RNase E [34,35] (Figure 1A).
The presumed central role of Hfq in sRNA biology has led to the use of this protein in the identification of new sRNA species [5,23,36]. Most recently, this approach was used towards the identification of 31 putative new sRNAs in Salmonella enterica serovar Typhimurium [23]. RNA sequences that co-immunoprecipitated with Hfq were cloned and sequenced en masse [23]. In addition to identifying candidate sRNAs, an advantage of this method is that 727 mRNAs that are Hfq-bound in vivo were also identified [23]. Thus, this method provides potential targets for each of the newly identified sRNAs. Through this analysis, the total number of known and putative S. Typhimurium sRNAs was increased to 64, and the authors were able to identify post-transcriptional regulons involving Hfq and sRNAs.
Obviously, sRNAs that do not bind Hfq will be missed in any Hfq-based sRNA identification strategy. There are several examples of Hfq-independent sRNAs, and we anticipate that there are additional such sRNAs remaining to be discovered and characterized. For example, a recently characterized sRNA in V. cholerae, VrrA, modulates colonization of the host small intestine and is involved in regulating outer membrane vesicle formation, but is not dependent upon Hfq for these activities [37]. In E. coli, the cis-acting sRNA, SymR, represses the translation of symE, encoding a toxic protein, in an Hfq-independent manner [38]. Additionally, using purified components, it was recently demonstrated that SgrS does not require Hfq for translational silencing of ptsG, though it does enhance the rate of duplex formation, which, in this case, still supports the role of Hfq as a chaperone [39]. Finally, multiple searches in both Streptomyces [22,40,41] and Mycobacterium tuberculosis [42] have identified numerous candidate sRNAs. However, an Hfq homolog has yet to be identified in these bacteria [40]. While it remains possible that these bacteria employ a novel RNA chaperone that is functionally equivalent to Hfq, it is also possible that these newly identified sRNAs do not require a protein chaperone for activity.
A multitude of cis-encoded sRNAs
The emphasis on sRNAs being expressed in trans to their targets needs to be reevaluated, for there is growing evidence that bacterial transcriptomes contain numerous chromosomally-encoded cis-acting sRNAs [2,42-44]. Though this class of sRNA is not yet fully characterized, it appears that most cis-encoded sRNAs are transcribed antisense to their target RNA. As a result, the sRNA shares perfect complementarity over at least a portion of its length with its target, resulting in highly specific, high affinity binding (Figure 1 D,E). Following are several recently described examples of chromosomally cis-encoded sRNAs.
Several cis-acting sRNAs in E. coli (SibA-E, SymR) have been characterized as antitoxins belonging to type I toxin-antitoxin pairs [38,45]. These sRNAs are transcribed antisense to, and repress the expression of, ORFs encoding toxic proteins. This mode of regulation is similar to the well-characterized plasmid sRNAs that are involved in maintenance and replication [46,47]. For example, SymR is transcribed in cis to the 5′ end of symE, which encodes an RNA-associating protein with toxin-like properties (Figure 1D) [38].
There exist additional classes of cis-encoded sRNAs besides antitoxins. In E. coli, the GadY sRNA base-pairs with the 3′ UTR of gadX mRNA, which encodes a transcriptional activator of the acid response system. The pairing of the two RNAs significantly increases the stability of the message and ultimately results in increased expression of downstream acid resistance genes (Figure 1E) [48]. Working with V. cholerae, our lab recently identified MtlS, an sRNA transcribed in cis to, and in antisense orientation relative to the 5′ UTR of mtlA, encoding the mannitol-specific transporter of the PTS [21]. MtlS and mtlA mRNA share 70 nt of perfect complementarity, and their expression, which is dependent upon carbon source, is inversely correlated. MtlS is also able to repress mtlA expression when expressed in trans. This suggests that MtlS represses the translation and/or the stability of mtlA mRNA by direct base-pairing (Figure 1D), as opposed to cis affects on mtlA transcription, such as changes in DNA supercoiling caused by transcription of mtlS.
A recent computational screen for sRNAs encoded within the pathogenicity islands of S. Typhimurium identified 19 candidate sRNA genes, 11 of which are complementary to a flanking gene [49]. Subsequent work examining the expression patterns of these sRNAs suggested that they play a role during invasion and virulence. Interestingly, three of the sRNAs (IsrC, IsrH-1 IsrN) that overlap with the 3′ end of a flanking gene each appear to affect the cis-encoded gene in a distinct manner (Figure 1D,E). The expression pattern of IsrC sRNA and msgA mRNA is the same, and the two transcripts undergo mutual degradation when expressed in cis from a plasmid [49]. In contrast, IsrH-1 sRNA appears to inhibit the expression of the cis-encoded gene, glpC, and the two transcripts have inverse expression patterns [49]. Lastly, overexpression of sRNA IsrN from a plasmid results in accumulation of the cis-encoded STM2765 mRNA. Thus, not only are cis-acting sRNAs common genetic elements in S. Typhimurium, but they appear to function through several different mechanisms. These and other studies not highlighted here show that cis-acting sRNAs are encoded in the genomes of numerous bacteria. Furthermore, these regulatory RNAs should not be considered exceptions but, instead, consistent with a common property of sRNAs as regulators of gene expression.
Experimental identification of sRNAs
One of the dangers of limiting the definition of bacterial sRNAs is that experimental approaches designed to identify new sRNAs may provide incomplete catalogues. For example, the search for new sRNAs continues to rely heavily on computer programs that often search exclusively within intergenic regions (IGRs), and that define sRNAs based on a combination of sequence conservation, specific promoters and Rho-independent terminators [22,40,43,49]. Although many sRNAs have been successfully identified in this manner, sRNAs that are transcribed within or antisense to ORFs [45,49] will be missed. Moreover, sRNAs whose 3′ ends are formed by Rho-dependent termination and/or processing by RNases or ribozymes will be missed by computational methods that rely on the presence of Rho-independent terminators. Likewise, experimental sRNA searches that use Hfq-binding as a prerequisite will miss Hfq-independent sRNAs. Thus, there is a need to supplement these approaches with additional, less biased approaches for the discovery of bacteria sRNAs. One such approach, mentioned above, used transcriptional profiling with whole genome tiling microarrays to reveal many new candidate sRNAs in L. monocytogenes, including ones that are antisense to ORFs [24]. Several of these candidate sRNAs exhibit the same expression patterns as virulence genes, suggesting their involvement in regulating pathogenic processes. Below, we review two additional experimental approaches for discovering sRNAs that do not depend on the classical definition of sRNAs.
Identification of candidate sRNAs by massively parallel sequencing
The use of massively parallel sequencing as a tool for sRNA discovery has the advantages of precisely mapping the 5′ and 3′ ends of the sRNA transcripts and any sRNA identified is automatically known to be transcribed under the condition used. Our lab recently developed “sRNA-Seq” to identify the total population of sRNAs in any bacterial transcriptome with no assumptions a priori with respect to the nature of the sRNAs (e.g., genome location, promoter, terminator, Hfq-binding) [21]. Our method differs from previously reported direct cloning protocols [2,3] in that during the cloning, tRNAs and 5S rRNA are depleted from the sRNA population by using RNase H and a mixture of oligonucleotides complementary to the tRNAs and 5S rRNA. Applying this method to V. cholerae, we identified many hundreds of candidate sRNAs plus all 20 previously known sRNAs [21]. This population was enriched for candidate sRNAs encoded within IGRs, but we also observed many that were antisense to, or sense to annotated ORFs. The vast majority of these candidate sRNAs are of unknown function. Thus, a great challenge for the future will be deciphering the biological roles of candidate sRNAs identified by this and other methods.
Targeted identification of sRNAs
A recently reported genetic approach for sRNA discovery begins with the mRNA target, and then searches for sRNAs that regulate it [31]. DNA encoding the target transcript including its 5′ UTR is cloned under the control of an inducible promoter as a translational fusion to lacZ, thereby decoupling transcription from post-transcriptional control. The researchers applied this method to the E. coli dpiBA operon transcript encoding a two-component system involved in the SOS response to β-lactam antibiotics. The dpiBA-lacZ construct was introduced into the E. coli chromosome and a screen with a multicopy plasmid library successfully identified one trans-acting sRNA, RybC, as a direct negative regulator. Although RybC requires Hfq for stability, this method does not exclude Hfq-independent regulatory factors, nor is it specific to sRNAs since protein regulators of dpiBA may have been identified as well. This method should be broadly applicable to the search of sRNAs in any genetically tractable system.
Conclusions
The ubiquity of sRNAs in bacteria is now widely accepted. What remains is the challenge of identifying the complete repertoire of sRNAs, and the even greater challenge of deciphering the roles of these regulators within the context of bacterial regulatory circuits and perhaps other processes. We anticipate that many new signaling systems involving sRNAs will be uncovered, and even well known circuits may need to be revisited as the number of identified sRNAs continues to increase. The recent contributions to the field suggest that as we continue to identify and characterize sRNAs, we will benefit by maintaining a broad definition of what sRNAs are and how they function. We suggest that any bacterial RNA molecule <500 nt that functions in the regulation of gene expression or protein activity be considered an sRNA, regardless of its mechanism of action or other functions. The recently developed methods to identify new sRNAs in bacterial transcriptomes should facilitate the identification of additional “non-canonical” sRNAs and reveal how common these genetic elements actually are. It is likely that we are just beginning to uncover the vast repertoire of sRNAs, and that novel functions for sRNAs remain to be discovered.
Acknowledgements
We thank David Lazinski and Revati Masilamani for insightful discussion and suggestions. This work was supported by awards K12GM074869 from the National Institute of General Medical Sciences (J.M.L.) and AI45746 from the National Institutes of Health (A.C). A.C. is a Howard Hughes Medical Institute investigator. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EGH, Margalit H, Altuvia S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Current Biology. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 2.Kawano M, Reynolds AA, Miranda-Rios J, Storz G. Detection of 5′- and 3′-UTR-derived small RNAs and cis-encoded antisense RNAs in Escherichia coli. Nucleic Acids Research. 2005;33:1040–1050. doi: 10.1093/nar/gki256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vogel J, Bartels V, Tang TH, Churakov G, Slagter-Jager JG, Huttenhofer A, Wagner EGH. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Research. 2003;31:6435–6443. doi: 10.1093/nar/gkg867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Genes & Development. 2001;15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang AX, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S. Global analysis of small RNA and mRNA targets of Hfq. Molecular Microbiology. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
- 6.Andrade JM, Arraiano CM. PNPase is a key player in the regulation of small RNAs that control the expression of outer membrane proteins. RNA. 2008;14:543–551. doi: 10.1261/rna.683308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reichenbach B, Maes A, Kalamorz F, Hajnsdorf E, Gorke B. The small RNA GlmY acts upstream of the sRNA GlmZ in the activation of glmS expression and is subject to regulation by polyadenylation in Escherichia coli. Nucleic Acids Research. 2008;36:2570–2580. doi: 10.1093/nar/gkn091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Svenningsen SL, Tu KC, Bassler BL. Gene dosage compensation calibrates four regulatory RNAs to control Vibrio cholerae quorum sensing. EMBO Journal. 2009;28:429–439. doi: 10.1038/emboj.2008.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Svenningsen SL, Waters CM, Bassler BL. A negative feedback loop involving small RNAs accelerates Vibrio cholerae transition out of quorum-sensing mode. Genes & Development. 2008;22:226–238. doi: 10.1101/gad.1629908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tu KC, Waters CM, Svenningsen SL, Bassler BL. A small-RNA-mediated negative feedback loop controls quorum-sensing dynamics in Vibrio harveyi. Molecular Microbiology. 2008;70:896–907. doi: 10.1111/j.1365-2958.2008.06452.x.•This paper demonstrates that in V. harveyi LuxR directly activates transcription of Qrr sRNAs which, in turn, target and destabilize luxR mRNA. The resulting negative feedback loop is an example of a tightly controlled genetic circuit in which sRNA expression is itself tightly regulated.
- 11.Urban JH, Vogel J. Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation. PLOS Biology. 2008;6:631–642. doi: 10.1371/journal.pbio.0060064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Figueroa-Bossi N, Valentini M, Malleret L, Bossi L. Caught at its own game: regulatory small RNA inactivated by an inducible transcript mimicking its target. Genes & Development. 2009 doi: 10.1101/gad.541609.•This paper reports the identification of a regulatory circuit in Salmonella in which the sRNA ChiX is targeted for degradation by the sRNA Anti-ChiX, which is transcribed and processed when the bacteria are in the presence of chitooligosaccharides. As ChiX represses translation of chiP mRNA, expressing a chitoporin, the action of Anti-ChiX ultimately allows for the synthesis of ChiP and transport of chitooligosaccharides into the bacteria.
- 13.Overgaard M, Johansen J, Møller-Jensen J, Valentin-Hansen P. Switching off small RNA regulation with trap-mRNA. Molecular Microbiology. 2009;73:790–800. doi: 10.1111/j.1365-2958.2009.06807.x. [DOI] [PubMed] [Google Scholar]
- 14.Gottesman S. Micros for microbes: non-coding regulatory RNAs in bacteria. Trends in Genetics. 2005;21:399–404. doi: 10.1016/j.tig.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 15.Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 16.Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Majdalani N, Hernandez D, Gottesman S. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Molecular Microbiology. 2002;46:813–826. doi: 10.1046/j.1365-2958.2002.03203.x. [DOI] [PubMed] [Google Scholar]
- 18.Masse E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moller T, Franch T, Udesen C, Gerdes K, Valentin-Hansen P. Spot 42 RNA mediates discoordinate expression of the E. coli galactose operon. Genes & Development. 2002;16:1696–1706. doi: 10.1101/gad.231702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang AX, Altuvia S, Tiwari A, Argaman L, Hengge-Aronis R, Storz G. The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO Journal. 1998;17:6061–6068. doi: 10.1093/emboj/17.20.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu JM, Livny J, Lawrence MS, Kimball MD, Waldor MK, Camilli A. Experimental discovery of sRNAs in Vibrio cholerae by direct cloning, 5S/tRNA depletion and parallel sequencing. Nucleic Acids Research. 2009;37 doi: 10.1093/nar/gkp080.••This paper reports the identification of a large number of candidate sRNAs in the V. cholerae transcriptome. The authors used direct cloning along with massively parallel sequencing, a method that should be applicable to any organism of interest.
- 22.Panek J, Bobek J, Mikulik K, Basler M, Vohradsky J. Biocomputational prediction of small non-coding RNAs in Streptomyces. BMC Genomics. 2008;9 doi: 10.1186/1471-2164-9-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, Hinton JCD, Vogel J. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLOS Genetics. 2008;4 doi: 10.1371/journal.pgen.1000163.••This paper reports the identification of 31 candidate sRNAs in S. Typhimurium through a combined approach of co-immunoprecipitation with Hfq and massively parallel sequencing. Through sequence identification of mRNA that were also co-immunoprecipitated with Hfq, the authors were able to postulate genetic regulatory circuits involving sRNAs and target mRNAs.
- 24.Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, Loh E, Gripenland J, Tiensuu T, Vaitkevicius K, et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature. 2009;459:950–956. doi: 10.1038/nature08080.••This paper applied tiling microarrays covering both strands of the L. monocytogenes chromosome to investigate the transcriptional profile of the bacteria in various conditions. 29 candidate sRNAs were identified in this manner, several of which demonstrate expression patterns suggesting a role in virulence.
- 25.Liu MY, Gui GJ, Wei BD, Preston JF, Oakford L, Yuksel U, Giedroc DP, Romeo T. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. Journal of Biological Chemistry. 1997;272:17502–17510. doi: 10.1074/jbc.272.28.17502. [DOI] [PubMed] [Google Scholar]
- 26.Wassarman KM, Storz G. 6S RNA regulates E. coli RNA polymerase activity. Cell. 2000;101:613–623. doi: 10.1016/s0092-8674(00)80873-9. [DOI] [PubMed] [Google Scholar]
- 27.Novick RP, Geisinger E. Quorum Sensing in Staphylococci. Annual Review of Genetics. 2008;42:541–564. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- 28.Vanderpool CK, Gottesman S. Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Molecular Microbiology. 2004;54:1076–1089. doi: 10.1111/j.1365-2958.2004.04348.x. [DOI] [PubMed] [Google Scholar]
- 29.Wadler CS, Vanderpool C. A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:20454–20459. doi: 10.1073/pnas.0708102104.•This paper demonstrates that the sRNA SgrS, which negatively regulates the translation of ptsG mRNA, is also translated into a 43-amino acid protein, SgrT. The authors provide evidence that SgrT negatively affects glucose uptake, possibly by inhibiting the activity of the PtsG transporter.
- 30.Franze de Fernandez MT, Eoyang L, August JT. Factor fraction required for the synthesis of bacteriophage Qbeta-RNA. Nature. 1968;219:588–590. doi: 10.1038/219588a0. [DOI] [PubMed] [Google Scholar]
- 31.Mandin P, Gottesman S. A genetic approach for finding small RNAs regulators of genes of interest identifies RybC as regulating the DpiA/DpiB two-component system. Molecular Microbiology. 2009;72:551–565. doi: 10.1111/j.1365-2958.2009.06665.x.••This work describes a genetic system in which an mRNA of interest is cloned, through reverse transcription, as a lacZ fusion, and the resulting construct may be screened along with a multicopy plasmid library to identify effectors of lacZ expression. Using this approach, the authors successfully identified an sRNA that negatively regulates the expression of a two component signal transduction system in E. coli.
- 32.Kawamoto H, Koide Y, Morita T, Aiba H. Base-pairing requirement for RNA silencing by a bacterial small RNA and acceleration of duplex formation by Hfq. Molecular Microbiology. 2006;61:1013–1022. doi: 10.1111/j.1365-2958.2006.05288.x. [DOI] [PubMed] [Google Scholar]
- 33.Rasmussen AA, Johansen J, Nielsen JS, Overgaard M, Kallipolitis B, Valentin-Hansen P. A conserved small RNA promotes silencing of the outer membrane protein YbfM. Molecular Microbiology. 2009;72:566–577. doi: 10.1111/j.1365-2958.2009.06688.x. [DOI] [PubMed] [Google Scholar]
- 34.Masse E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes & Development. 2003;17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morita T, Maki K, Aiba H. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes & Development. 2005;19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Christiansen JK, Nielsen JS, Ebersbach T, Valentin-Hansen P, Sogaard-Andersen L, Kallipolitis BH. Identification of small Hfq-binding RNAs in Listeria monocytogenes. RNA. 2006;12:1383–1396. doi: 10.1261/rna.49706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song T, Mika F, Lindmark B, Liu Z, Schild S, Bishop A, Zhu J, Camilli A, Johansson J, Vogel J, et al. A new Vibrio cholerae sRNA modulates colonization and affects release of outer membrane vesicles. Molecular Microbiology. 2008;70:100–111. doi: 10.1111/j.1365-2958.2008.06392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawano M, Aravind L, Storz G. An antisense RNA controls synthesis of an SOS-induced toxin evolved from an antitoxin. Molecular Microbiology. 2007;64:738–754. doi: 10.1111/j.1365-2958.2007.05688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maki K, Uno K, Morita T, Aiba H. RNA, but not protein partners, is directly responsible for translational silencing by a bacterial Hfq-binding small RNA. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:10332–10337. doi: 10.1073/pnas.0803106105.•This paper reports the use of a defined in vitro system to show that base pairing in the absence of Hfq beween the sRNA SgrS and its ptsG mRNA is sufficient for translational repression. Though not required, Hfq is shown to significantly enhance duplex formation.
- 40.Swiercz JP, Hindra, Bobek J, Haiser HJ, Di Berardo C, Tjaden B, Elliot MA. Small non-coding RNAs in Streptomyces coelicolor. Nucleic Acids Research. 2008;36:7240–7251. doi: 10.1093/nar/gkn898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tezuka T, Hara H, Ohnishi Y, Horinouchi S. Identification and gene disruption of small noncoding RNAs in Streptomyces griseus. Journal of Bacteriology. 2009;191:4896–4904. doi: 10.1128/JB.00087-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arnvig K, Young D. Identification of small RNAs in Mycobacterium tuberculosis. Molecular Microbiology. 2009;73:397–408. doi: 10.1111/j.1365-2958.2009.06777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiao B, Li W, Guo G, Li BS, Liu Z, Jia KR, Guo Y, Mao XH, Zou QM. Identification of small noncoding RNAs in Helicobacter pylori by a bioinformatics-based approach. Current Microbiology. 2009;58:258–263. doi: 10.1007/s00284-008-9318-2. [DOI] [PubMed] [Google Scholar]
- 44.Xiao B, Li W, Guo G, Li B, Liu Z, Tang B, Mao X, Zou Q. Screening and identification of natural antisense transcripts in Helicobacter pylori by a novel approach based on RNase I protection assay. Molecular Biology Reports. 2009;36:1853–1858. doi: 10.1007/s11033-008-9390-5. [DOI] [PubMed] [Google Scholar]
- 45.Fozo EM, Kawano M, Fontaine F, Kaya Y, Mendieta KS, Jones KL, Ocampo A, Rudd KE, Storz G. Repression of small toxic protein synthesis by the Sib and OhsC small RNAs. Molecular Microbiology. 2008;70:1076–1093. doi: 10.1111/j.1365-2958.2008.06394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thisted T, Gerdes K. Mechanism of post-segregational killing by the Hok Sok system of plasmid R1 - Sok antisense RNA regulates hok gene expression indirectly through the overlapping mok gene. Journal of Molecular Biology. 1992;223:41–54. doi: 10.1016/0022-2836(92)90714-u. [DOI] [PubMed] [Google Scholar]
- 47.Thisted T, Sorensen NS, Wagner EGH, Gerdes K. Mechanism of post-segregational killing - Sok antisense RNA interacts with hok messenger-RNA via its 5′-end single-stranded leader and competes with the 3′-End of hok messenger-RNA for binding to the mok translational initiation region. EMBO Journal. 1994;13:1960–1968. doi: 10.1002/j.1460-2075.1994.tb06465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Opdyke JA, Kang JG, Storz G. GadY, a small-RNA regulator of acid response genes in Escherichia coli. Journal of Bacteriology. 2004;186:6698–6705. doi: 10.1128/JB.186.20.6698-6705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Padalon-Brauch G, Hershberg R, Elgrably-Weiss M, Baruch K, Rosenshine I, Margalit H, Altuvia S. Small RNAs encoded within genetic islands of Salmonella Typhimurium show host-induced expression and role in virulence. Nucleic Acids Research. 2008;36:1913–1927. doi: 10.1093/nar/gkn050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen ZT, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]


