Abstract
Generation of motile force is one of the main functions of the eukaryotic kinetochore during cell division. In recent years, the KMN network of proteins (Ndc80 complex, Mis12 complex and KNL-1 complex) has emerged as a highly conserved core microtubule-binding complex at the kinetochore. It plays a major role in coupling force generation to microtubule plus-end polymerization and depolymerization. In this review, we discuss current theoretical mechanisms of force generation, and then focus on emerging information about mechanistic contributions from the Ndc80 complex in eukaryotes, and the microtubule-binding Dam1/DASH complex from fungi. New information has also become available from super-resolution light microscopy on the protein architecture of the kinetochore-microtubule attachment site in both budding yeast and humans, which provides further insight into the mechanism of force generation. We briefly discuss potential contributions of motors, other microtubule-associated proteins, and microtubule depolymerases. Using the above evidence, we present speculative models of force generation at the kinetochore.
Introduction
The kinetochore is a unique motor essential to the segregation of chromosomes during cell division. It is a macromolecular protein complex that assembles on a specialized locus on each DNA molecule, known as the centromere. It attaches to the plus-ends of one or more spindle microtubules (MTs) forming kinetochore-microtubules (kMTs), and tightly couples chromosome movement to kMT polymerization and depolymerization. More than 40 different proteins are necessary for kMT formation and persistent attachment (see [1] for an in-depth review of kinetochore composition, organization, and regulation). Many of these proteins are conserved in all eukaryotes.
The goal of this review is to discuss the biophysical mechanisms of force generation coupled to MT polymerization and depolymerization, and their relevance to the kinetochore. An exciting confluence of results from diverse fields has brought us closer to understanding this problem. Investigations utilizing structural biology, single molecule biophysics, high-resolution microscopy, and cell biology seek to examine two fundamental aspects of kinetochore function: the nature and characteristics of the force generation mechanism, and the processes that control transitions between polymerization and depolymerization of kMT plus-ends. We first describe the principles of force generation at the MT plus-end. We review available biophysical data on the kinetochore-MT attachment. Next, results from cell biology for the kinetochore complexes necessary for end-on attachment: Ndc80 complex and the Dam1/DASH complex (in fungi), are emphasized. These two complexes have been studied extensively in vitro. We summarize these results, and discuss their implications in vivo. We then examine new data on the protein architecture of the kinetochore and its predictions in force generation mechanisms. We also briefly examine the general contributions of motors, depolymerases, and MT associated proteins (MAPs) to controlling force generation and stable maintenance of the kinetochore-MT attachment. Kinetochore functions in attachment error correction and in the spindle assembly checkpoint are also vital for accurate segregation. These topics have been reviewed elsewhere [2,3].
Theoretical mechanisms of force generation at the plus-end
Discovery of polymeric structure of MTs, which grows and shrinks in length by adding or losing subunits only from its two ends, prompted inquiries into general MT end-based force generation mechanisms [4]. Details of these mechanisms depend on the nature of kinetochore-MT attachment, and several molecular arrangements were proposed for force generation at the kinetochore that is coupled with MT disassembly (reviewed in [5]). During growth, a MT plus-end adds GTP-tubulin dimers, which have a relatively straight conformation along the long axis. GTP bound to β-tubulin is hydrolyzed after incorporation, but with some time delay. GDP-tubulin dimers have a bent conformation, but within the MT lattice relaxation to this state is not allowed because of lateral interactions. The energy of GTP-hydrolysis is stored in the lattice as a strain instead, which manifests itself in the flared geometry of depolymerizing plus-ends (Fig. 1A) [6]. A depolymerizing plus-end thus provides two modes of force generation. The MT lattice immediately behind the plus-end provides regular array of tubulin-dimers as attachment sites. Alternatively, the strain energy associated with bending of GDP-tubulin dimers during MT disassembly can be converted into work. A polymerizing plus-end can also generate forces. The mechanisms include differential binding energies of a kinetochore protein for GTP- and GDP-tubulin or thermal ratcheting against a rigid barrier.
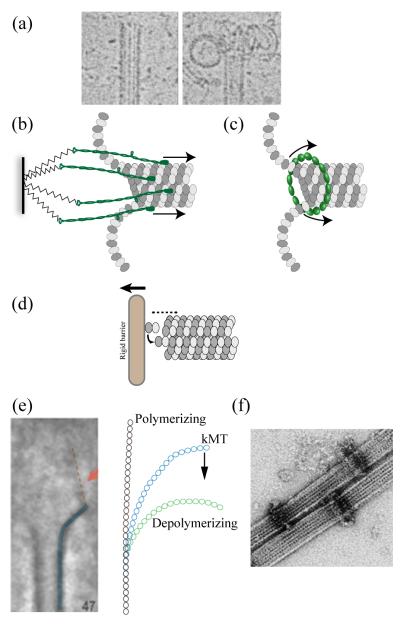
Figure 1.
(a) Morphology of polymerizing and depolymerizing MT plus-ends in vitro. Disassembling MT plus-ends allow two mechanisms of force generation coupled to the disassembly. (b) Biased diffusion models - proteins weakly bound to the MT lattice diffuse along the lattice in the absence of external force. However, the shortening plus-end acts as a moving boundary and biases the diffusive movement towards the minus end. (c) Forced walk models – A ring around the lattice serves as a barrier to the relaxation of curling protofilaments. Continuous outwards curling pushes the ring towards the minus end. (d) A plus-end growing against a rigid barrier generates force by thermal ratchet mechanism. Two types of forced walk couplers based on EM and in vitro data have been proposed (e and f). (e) Morphology of kMT plus-ends observed in ultrathin sections. Averaging over many protofilaments reveals the presence of fibrillar structures (orange) of unknown identity that bind on the inside of the curling protofilament (blue). kMT protofilaments (blue) display an intermediate curvature that may be indicative of incomplete relaxation of strain. If the fibrils bound to each protofilament are responsible for this curvature, then they can convert the strain into a minus-end directed force. (f) Dam1/DASH rings in vitro act as force couplers that convert the strain energy efficiently into a pushing force or work.
The first class of models for MT-disassembly coupled force generation uses the MT lattice as an array of binding sites (Fig. 1B). These models propose that the kinetochore-MT attachment is achieved by multiple, relatively weak linkages that are dispersed over a stretch of MT lattice behind the plus-end. The weak linkages can be broken and reform under the influence of thermal fluctuations allowing the attachment site to diffuse over the MT lattice. Loss of dimers at the plus-end creates a moving boundary, and biases the diffusion of the coupler towards the minus end of the MT (hence the term “biased diffusion”). For this general mechanism, persistence of attachment is an important characteristic, as the reliance on multiple, weak linkages near a shortening plus-end would make the kinetochore susceptible to complete detachment. The persistence of attachment depends on the rate of MT depolymerization, energy of individual linkages, and the distribution of linkages over the MT lattice. To avoid complete detachment, linkages must be established away from the plus-end. Therefore, explicit mechanisms that bias the formation of linkages away from the plus-end are an important requirement of the biased diffusion models. The inside-out curved protofilament geometry at the disassembling plus-end may also bias movement of the binding proteins away from the plus-end. The maximum amount of force generated by biased diffusion depends on the energy of individual interactions and the number of such linkages. For example, the sleeve mechanism as originally proposed by Hill can generate up to 13 pN of force, although the persistence of attachment diminishes with higher forces [7,8].
The second class of models can be termed as “forced walk” mechanisms. These models rely on the peeling of protofilaments at depolymerizing plus-ends [6,9]. A force coupler that resists the outward bending of protofilaments at the MT plus-end effectively transforms the bending of individual GDP-tubulin dimers as they lose their lateral lattice attachments into a power stroke (Fig. 1C). Explicit mechanical and mechanochemical models of MT depolymerization [10,11] enabled characterization of two types of forced walk models that are relevant to force generation at the kinetochore: rings that encircle the MT lattice just behind a flared plus-end, and fibrils that bind at the top of a curling protofilament [12,13]. By design, ring couplers can track a flared plus-end indefinitely. Theoretically, the energy of GTP hydrolysis is stored as strain energy in the MT lattice, and it is available for performing work [14]. An optimally designed ring (an oligomeric Dam1/DASH ring, see below) acts as a highly efficient coupler converting up to 70% of the strain energy into work generating 40-60 pN of force per MT [13, 14].
Growing MT plus-ends can also provide force generation via two different modes. The first mechanism depends on the presence of GTP-tubulin dimers at the MT plus-end. A kinetochore protein that binds preferentially to GTP-tubulin over GDP-tubulin is biased in its thermal movement over the MT lattice towards the plus-end. The second mechanism is commonly known as “thermal ratcheting”, wherein the plus-end is made to grow against an impenetrable barrier (Fig. 1D). Bacterial plasmid segregation machinery involving parC and parM provides an excellent example of thermal ratchets. The free energy that can be converted into work comes in form of chemical potential for GTP-tubulin addition at the plus-end [15]. Physiological values predict forces as high as 3 pN per dimer added at plus-end, which was confirmed by in vitro force measurements [16].
Biophysical characterization of the kinetochore in vivo
The in vivo mechanism of kinetochore force generation remains uncharacterized largely due to a dearth of in vivo biophysical data. There are models based on microtubule-motors, on non-motor mechanisms involving microtubule depolymerization-dependent mechanisms as discussed above and more conceptual mechanisms involving hypothetical long range electrostatic forces [17]. Current lack of quantitative information on the molecular interactions with the MT plus-end however, necessitates major assumptions regarding MT plus-end behavior at the kinetochore to force these models to mimic observed chromosome dynamics [7,18,19].
Some biophysical characteristics of the kinetochore-MT attachment are well characterized. MT-depolymerization coupled kinetochore movement occurs in vitro in the absence of ATP, demonstrating that the core kinetochore attachment machinery acts as a passive motor [9,20]. Kinetochores tethered to the spindle pole by kMTs move towards and away from the spindle pole with similar speeds, which suggests that the speed of movement does not critically depend on the opposing force. Direct measurement of force generated by a kinetochore in anaphase was obtained by the classic experiments conducted by Nicklas in grasshopper spermatocytes [21]. These experiments estimated the attachment force per kMT to be >10 pN. However, it should be noted that poleward kMT flux (poleward movement of MTs coupled with depolymerization at the minus-end) is a significant component of kinetochore motility in meiotic systems including grasshopper spermatocytes. The measured rate of poleward flux is similar to the speed of chromosome movement implying that the kMTs must polymerize at the plus-end even as the kinetochore is moving poleward [22]. The force measured in the Nicklas experiments indicate the maximum forces that can be transmitted by the kinetochore machinery including motors and MAPs. MT-depolymerization coupled force generated by the core kinetochore machinery then remains an unknown quantity. Thus, persistent attachment and force generation may involve the same molecular mechanism or involve two different mechanisms.
The morphology of kMT plus-ends could potentially provide valuable information regarding the coupling between kMT state and the direction of movement, and point to mechanisms of modulating plus-end polymerization dynamics. For example, sister kinetochore pairs in HeLa cells can oscillate about the spindle equator with excursions of ~ 2 μm in either direction. During such oscillations, kMTs attached to the leading kinetochore must lose tubulin subunits (depolymerize), whereas its trailing sister kinetochore must gain subunits (polymerize). Surprisingly, morphology of kMT plus-ends determined in ultrastructural studies however, failed to find significant differences in protofilament curvature of kMT populations at leading and trailing kinetochores [12] [23]. These studies found that both kinetochores in a sister kinetochore pair have ~ 70% of the kMT plus-ends with a flared geometry suggesting that they likely depolymerize (Fig. 1E), whereas the remaining plus-ends have a blunt morphology suggesting that they polymerize. This finding implies that the kMTs can on average add and lose subunits without significant changes in plus-end structure, and that mechanochemical transitions from one state to the other may be fluid and reversible. Comparison of protofilament morphology reveals that the curvature of kMT protofilaments is significantly less than the curvature at the ends of unattached, depolymerizing MT protofilaments (Fig. 1E). This observation implies that the release of lattice strain is incomplete, and possibly opposed by kinetochore proteins. Indeed, this study finds that fibrils of unknown identity consistently attach to a specific region of the protofilaments, where such fibrils can impart a minus-end directed force to the kinetochore (Fig. 1E). It is important to establish what fraction of the strain energy in these protofilaments is converted into useful work, or if the observed geometry is a consequence of strong modulation of kMT dynamics [24]. Finally, polymerization force by kMTs is thought to be small, since the centromere of a bioriented chromosome is rarely compressed [25].
Cell biology: Ndc80 and Dam1/DASH complex
Ndc80 and Dam1/DASH complexes in particular have emerged as the principle MT-binding kinetochore proteins that organize end-on kinetochore-MT attachment. The critical role of Ndc80 in vertebrate cells is vividly demonstrated by the observation that the deletion of an 80 amino acid long N-terminal tail of Ndc80 molecule completely abrogates end on kinetochore-MT attachments [26-28]. Although Dam1/DASH has been discovered only in fungi, its ability to form rings around a MT lattice in vitro and preferential binding to GTP-tubulin set it apart from a number of other MAPs studied so far. KNL-1/Blinkin/AF15q14 is another MT-binding conserved kinetochore protein that may be as important for MT attachment [29,30]. So far, its ability to act as a force generator has not been investigated in detail.
The Ndc80 complex is the principle MT-attachment site at the kinetochore in all eukaryotes. It also organizes key components of the kinetochore-based spindle assembly checkpoint machinery [31]. Structural studies of the Ndc80 complex reveal that it is a 56 nm long rod-like molecule, with globular domains at either end of the rod [32,33]. The Ndc80/Nuf2 subunits contain a pair of globular, calponin-homology domains that are involved in MT attachment [27]. As mentioned above, the Ndc80 subunit (Hec1 in vertebrates) also carries an N-terminal unstructured tail with nine positively charged residues that primarily mediates MT-binding through electrostatic interactions with the negatively charged, C-terminal tail of β-tubulin [26,28]. In vitro binding studies have shown that the Ndc80 complex binds the sides of microtubules by its N-terminal end and the rod domain extends toward the microtubule plus end at a 30-45° angle. At low concentrations, binding is cooperative by mechanisms that are not understood [32]. Surprisingly, this tail is not essential in budding yeast [34]. The CH-domains are thought to participate in MT-binding and these domains are also required for activating the spindle assembly checkpoint [26,28]. Initial structural studies and point mutations in amino acids suggested that the adjacent CH-domains of Ndc80 and Nuf2 (the “heal” of the foot-like conformation of the N-terminal end both make contacts with the microtubule lattice [32]. However, the binding geometry is uncertain since molecular microscopy by cryo-EM reveal the presence of alternating strong and weak densities protruding from tubulin monomers, with the heavier density (likely the “toe” of the CH-domain of Ndc80 with the CH-domain of Nuf2 situated behind) binding in the ridge between two tubulin heterodimers [35]. Structural analysis and EM images of isolated Ndc80 complexes have shown that the alpha helical coiled-coil rod domain of the Ndc80/Nuf2 dimer contains a loop in the Ndc80 polypeptide that produces a flexible domain about 16 nm behind the globular CH-domains [36]. The critical role of Ndc80 in regulating kMT attachment and in plus-end coupled force generation was first demonstrated by antibody injection experiments that disrupt Ndc80 phosphorylation [37]. This perturbation led to a strong suppression of kMT plus-end dynamics and turnover at bioriented chromosomes. kMT depolymerization at the minus-end by the flux machinery led to hyper-stretched centromeres and short kMTs. A study of kinetochore composition using GFP-fusion proteins and in vivo fluorescence microscopy found that there are 7-8 molecules of Ndc80 per MT in budding and fission yeast [38,39]. In light of the critical function of the Ndc80 complex, this number plays an important role in determining the properties of the kinetochore as a force generator.
The discovery that oligomeric Dam1/DASH complex forms rings around the MT lattice in vitro generated a lot of excitement [40,41]. This heterodecameric MAP has been found only in fungi so far, and in budding yeast, it is necessary for kinetochore-MT attachment and spindle integrity [42]. In vitro studies of the Dam1/DASH rings revealed that each ring contains 16-23 monomers (Fig. 1F). has an inside diameter of ~35 nm and thus there is a gap between it and the outside of the 25 nm diameter microtubule. Interactions occur through Dam1/DASH protein projections that extend to the lattice [40,43]. The data that indirectly supports the existence of rings comes from fluorescence measurements of the number of copies of the complex per kinetochore on average [39]. Although the Dam1/DASH rings have been well-characterized in vitro, such structures have not been discovered in vivo. Dam1/DASH is loaded onto the kinetochore only after kinetochore-MT attachment is established [44,45]. Dam1/DASH complex molecules assemble into rings autonomously. Once assembled, these rings dissociate from the MT mostly when they encounter a depolymerizing end in vitro [46]. These observations raise important issues. What process/factors then regulate the assembly of rings in vivo to ensure that each kinetochore acquires one ring? In vivo observation of Dam1/DASH recruitment during the retrieval of unattached kinetochores in budding yeast cells show that the kinetochore gradually acquires Dam1/DASH molecules after it establishes end-on attachment [45]. These observations lead us to two alternative scenarios for the possible recruitment of Dam1/DASH rings to the kinetochore: either end-on kinetochore-MT attachment is necessary for ring formation or a monitoring process distributes rings evenly. Finally, Dam1/DASH may also be important for establishing kinetochore coupling with growing kMTs [47]. Although Dam1/DASH is essential for survival in budding yeast, it is dispensable in fission yeast. The fission yeast kinetochore recruits a much smaller number of Dam1/DASH complex molecules, and this number is insufficient to form even one Dam1/DASH ring per kinetochore [38]. Interestingly, the fission yeast kinetochore supports 2-3 MT attachments as opposed to the single attachment in budding yeast.
Both Ndc80 and Dam1/DASH are regulatory targets of mitotic kinases. In vertebrates, Aurora B kinase suppress the binding affinity of Ndc80 mainly through the phosphorylation of positively charged residues in the N-terminal tail [26-28,33,37]. Phosphorylation is cell cycle dependent, and may be antagonized by the PP1 phosphatase. This phosphorylation is essential in the correction of erroneous kMT attachments in prometaphase cells (reviewed in [3,48]). Ipl1/Aurora B also targets several subunits of the Dam1/DASH. In vitro, phosphorylation affects the ability of Dam1/DASH to form rings and to associate with the Ndc80 complex [49].
In vitro interaction of Ndc80 and Dam1/DASH with a MT
Characteristics of Ndc80 complex interaction with a MT at the single molecule level were recently examined in vitro using bacterially reconstituted protein (no phosphorylation) [50]. Based on the residence times of Ndc80 complex molecules on stabilized MTs, this study found that the Ndc80 complex binds to the MT lattice with a relatively low affinity. Single Ndc80 molecules also diffuse along the MT lattice, although they cannot track a depolymerizing end. Instead, the low affinity necessitates an interlinked assemblage of at least 16 molecules for persistent depolymerization coupled movement with a lower diffusion coefficient. Beads coated with Ndc80 sustain depolymerization-driven movement against opposing forces as high as 3 pN. Tip-tracking behavior of Ndc80-coated microbeads is not surprising, as the relatively large diameter of the bead allows the Ncd80 molecules to attach over a significant length of the MT (~ 250 nm assuming a 40 nm long interaction zone between a 440 nm diameter bead surface and the MT lattice). Such motion has been seen for beads coated with a high enough number of a MT-binding protein [51,52]. The relatively low affinity of the Ndc80 molecule to the MT is also important, as it will allow diffusion of the molecule over the lattice instead of stable binding in one position. Together with the requirement of multiple Ndc80 molecules for tip-tracking ability, these observations lend support to biased-diffusion type of mechanism for force generation in vitro. It should be stressed that the geometry of binding between the Ndc80 complex aggregates and the MT tip, which is critical for reaching a definitive conclusion regarding its role in the mechanism of force generation at the kinetochore, was not characterized in this assay.
The Dam1/DASH complex exhibits complex behavior in vitro due to the two distinct oligomeric configurations: patches and bracelets, and rings containing 16-23 monomers. Grishchuck et al showed that the Dam1/DASH ring binding to the lattice is highly stable, and the strong binding allows very little thermal diffusion of the rings over the lattice [46]. This observation implies that rings must be moved over the lattice by an active mechanism significantly stronger than random thermal forces. In the in vitro assay, this pushing force comes from curling protofilaments on a depolymerizing plus-end, a forced-walk type of mechanism. Dam1/DASH rings harness a large portion of the strain energy to develop as much as 3-5 pN of force per protofilament (40-60 pN per MT) [14]. The authors also find that oligomeric Dam1/DASH patches also support tip-tracking behavior. Furthermore, these assemblies can act as force couplers, although the forces generated are lower on average. The diffusion coefficient of these assemblies depends inversely on the number of monomers. I This observation is consistent with concurrent studies demonstrating that rings are not necessary for persistent tip-tracking behavior of the Dam1/DASH molecules [53]. Interestingly, beads coated with Dam1/DASH complex also track growing plus-ends [54]. Such beads resist forces of up to 3 pN directed in the direction of growth. In contrast, a much lower force towards the minus-end is needed to dislodge the bead from the plus-end. Finally, tension applied to the MT-plus-end through beads coated with the Dam1/DASH complex increases the rescue rate and also suppresses the rate of catastrophe [55], a key feature of the kinetochore that has been proposed by cell biological observations and mathematical simulations [18,56]. This effect is negated by Aurora B phosphorylation. In conclusion, the Dam1/DASH complex can generate forces via both modes of MT-disassembly coupled force generation.
Protein architecture of the kinetochore
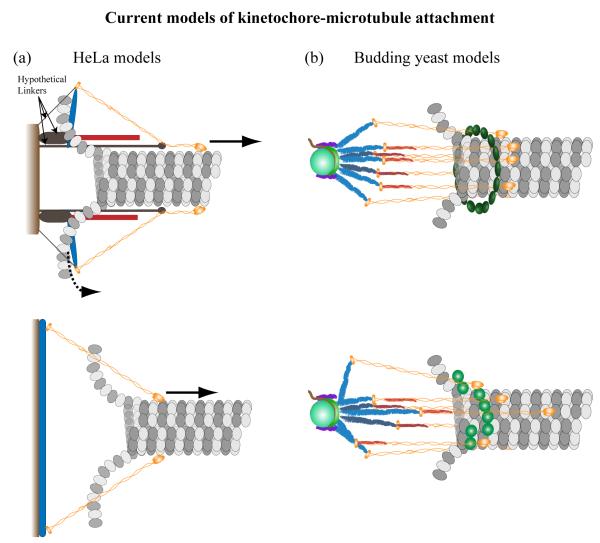
Both Ndc80 and Dam1/DASH operate in the context of the kinetochore as a macromolecular machine. Two recent studies made significant advances towards understanding the protein architecture of the kinetochore in yeast and in fixed HeLa cells [57,58]. Using super-resolution microscopy, these studies measured separations between a reference point on the kinetochore and known kinetochore proteins to assemble a map of kinetochore protein localization along the kinetochore-MT attachment. Significant similarities in the positions of kinetochore proteins emerge from the two maps, suggesting a high degree of conservation. There are significant differences as well. The length of the Ndc80 complex was measured to be 46 nm in metaphase in HeLa cells. This length is shorter than the 55 nm length of isolated Ndc80 complexes measured by structural studies [33]. Although the axis of a MT-bound Ndc80 molecule is tilted by 33° with respect to the MT axis in vitro, Wan et al. depicted Ndc80 molecules as constitutively bent at the hinge region to account for forces acting along the MT axis (Fig. 3A). Force along the kMT axis is transmitted by a hypothetical linker protein that attaches the hinge region of the Ndc80 complex to the inner centromere. Furthermore, they proposed that force from the curling protofilaments may also push the kinetochore complexes that link the Ndc80 complex to the centromere towards the minus-end (Fig. 3A). It is also possible that the measured length is smaller because of a small number of unattached Ndc80 molecules that lower the average length due to their random orientation. However, unattached Ndc80 molecules can activate the spindle assembly checkpoint, leading the authors to propose the bent Ndc80 molecule. Models of kinetochore architecture based on straight but tilted Ndc80 complex molecules require a rigid underlying foundation that resists the inwardly directed forces arising from the angle of the transmitted force (Fig. 3B) [1].
Budding yeast measurements reveal a simpler architecture. The 55 nm long Ndc80 complex binds to the kMT with its axis aligned with the MT axis, the alignment arising due to the force generated (Fig. 3A). Alignment of all Ndc80 molecules in metaphase raises two possibilities. The alignment arises because all Ndc80 molecules are constrained in this configuration even in the absence of kMT attachment. Alternatively, all eight Ndc80 molecules in a kinetochore mostly remain attached to the kMT. Interestingly, anaphase measurements in budding yeast revealed that the length of the Ndc80 molecule reduces to ~ 39 nm, which likely results from the bending of Ndc80 molecules in the hinge region. The Dam1/DASH complex localizes ~ 10 nm behind the Ndc80 head domains. At this location, a Dam1/DASH complex ring would encircle the kMT lattice as well as the coiled-coil domain of the Ndc80 complex. The 35 nm inner diameter of the Dam1/DASH ring makes this formally possible. This depiction of the kinetochore also suggests that the formation of the Dam1/DASH ring must follow the stable binding of a majority of the Ndc80 molecules to the kMT. Alternatively, the Dam1/DASH complex does not form rings in vivo, and instead contributes function as oligomeric patches or incomplete rings bound to the kinetochore.
Location of the MT plus-end within the kinetochore is critical for elucidating the mechanism of force generation. In Ptk1 cells, the end of a cold-stable fiber of kMTs extends ~ 60 nm beyond the MT-binding domain of the Ndc80 complex. Although this position likely marks the farthest that kMT plus-ends penetrate into the kinetochore, the measurement suggests that the Ndc80 complex binds the kMT lattice under the experimental conditions. The location of the Dam1/DASH complex within the yeast kinetochore implies that the kMT must extend beyond the MT-binding domains of the Ndc80 complex. For the Ndc80 complex to act as a forced-walk coupler, the position at which it binds to a curling protofilament decides the direction of the force being generated. Binding at the top of the ram’s horn is necessary for a minus-end directed force. If the kMT plus-end indeed extends beyond the MT-binding domains of the Ndc80 complex, action of the Ndc80 complex as a forced-walk coupler becomes difficult to envision.
Help in kMT attachment may also come in C. elegans from kinetochore protein KNL-1, which binds MTs in vitro [30]. Its fission yeast homolog Spc7 also bind MTs when over-expressed [59]. In HeLa cells, Blinkin/AF15q14 RNAi leads to the abrogation of kinetochore-MT attachment, although this phenotype may also arise from significant perturbation of kinetochore architecture [29]. The location of this protein ~ 30-40 nm inside the MT-binding head domains of the Ndc80 complex could provide another potential set of MT-binding sites. This arrangement of MT interactions sites distributed over 40 nm is well-suited for persistent attachment using biased diffusion. More studies are important to establish how KNL-1 and its homologs in vertebrates and fungi binds MTs in vivo, location of the MT-binding domain within the protein and within the kinetochore, and its role in force generation.
The observed architecture of the budding yeast kinetochore suggests a biased-diffusion mechanism for Ndc80 complex-mediated kinetochore motility (Fig. 3b). The Ndc80 complex connects to the inner kinetochore via a filamentous Mtw1 complex (blue bars in Fig. 3b). Such a mechanism may be possible, if one assumes that this complex is free to orient itself along the kMT axis, the in vitro length of this complex (up to 40 nm) combined with the 56 nm length of the Ndc80 complex can allow the MT-binding Ndc80 domains to reach significant distances (~ 100 nm) behind the MT plus-end. Such a model must invoke molecular strain or some form of position-dependent regulation that reduces the ability of the Ndc80 complex to bind close to the centromere, instead promoting binding to regions of the kMT lattice that are further away. Under this scenario, the role of the Dam1/DASH complex becomes completely dependent on the Ndc80 complex. Furthermore, Dam1/DASH molecules cannot form rings, but instead act as oligomeric patches to assist in MT-binding, and possibly influencing the polymerization dynamics of the kMT.
Contributions of motors, MT depolymerases, and MAPs
Motors, depolymerases, and MAPs are important regulators of MT polymerization dynamics in vivo. Assigning specific roles to each of these proteins at the kinetochore proves difficult for two reasons. First, many of these proteins have global and often overlapping roles in the regulation of MT polymerization dynamics. Observed effects on kinetochore function may be non-specific effects that arise from perturbations in MT polymerization dynamics or spindle structure. Second, spindle architectures possess different features and complexities in the various model organisms studied. Although many motors and MAPs are conserved, some of these proteins assume prominent, but system-specific roles, to maximize chromosome segregation accuracy. We therefore describe the general role of motors, depolymerases, and MAPs in HeLa cells, and highlight only a few intriguing results here.
Motors generate an auxiliary force at the kinetochore. Dynein and CENP-E provide prominent examples. The ability of some motors to depolymerize a MT plus-end or to induce catastrophe at a growing plus-end can significantly improve the persistence of kinetochore force generation coupled to kMT depolymerization. This function is especially important in chromosome segregation in anaphase, as in the case of the Kinesin-13 family proteins [60]. The ability of depolymerases to switch kMT plus-ends from growing to shrinking state also allows them to control coordination of motion of sister kinetochores. Both these functions are intriguingly on display in recent studies of Kinesin-8 and Kinesin-13 depolymerases [61,62]. Despite similar depolymerase activities, over-expression or depletion of these motors from the kinetochore has the opposite effects on the characteristics of chromosome oscillations in HeLa cells. Recruitment of additional MCAK to the centromeres increases chromosome speed, oscillation amplitude, and also increases the coordination between sister kinetochores. On the other hand, over-expression of Kif18A decreases the rate of chromosome movement as well as the amplitude of oscillations. These seemingly antagonistic functions likely arise due to the mode of recruitment of these proteins at the kinetochore and their location with respect to the plus-end. MCAK is bound to the interior of the kinetochore and in front of kMT plus-end, whereas Kif18A uses its motor activity to travel along the lattice to reach and act on kMT plus-ends. The location of MCAK within the kinetochore may provide a built-in negative feedback to its activity. MAPs yield an antagonistic influence on the state of kMT plus-ends, battling with the depolymerases to promote plus-end growth. A number of MAPs including EB1, CLIP170, XMAP215, CLASP, and Ska1/RAMA are important for kinetochore function [63-67]. The Ska1 complex has garnered attention in the recent year due to its role in maintaining stable end-on attachments at the kinetochore (see [68] for a review). It localizes to the kinetochores during mitosis, and depletion of multiple Ska1 subunits severely reduces the stability of kMT attachments. It can oligomerize in vitro, and it is subject to phosphoregulation. Furthermore, Ska1 complex coated beads were shown to track shortening plus-ends and slow down the rate of disassembly in a manner reminiscent of the activity of the Dam1/DASH complex [69]. A more recent study demonstrated that the Ska1 complex for the satisfaction of the spindle assembly checkpoint and for maintaining sister chromatid cohesion [70]. More studies are necessary to establish the role of this complex in force generation at the kinetochore.
The influence of motors and MAPs on kinetochore activity demonstrates that the leading and trailing kinetochores in a sister kinetochore pair must have distinct kMT populations, even if such populations cannot be discerned from their morphology apparent in ultrastructural studies. Indeed, EB1, which recognizes and tracks with polymerizing plus-ends, is seen at much higher concentrations at a trailing kinetochore where there is net kMT growth (polymerization) in comparison with its leading sister kinetochore where there is net kMT shrinkage (depolymerization) [71].
Conclusions and outlook
The kinetochore is a sophisticated macromolecular machine that has two critical functions in chromosome segregation: it must establish end-on attachments that enable the dividing cell to distinguish and selectively stabilize chromosome bi-orientation, and it must generate forces to move and segregate chromosomes in anaphase. Although we compare and contrast two different mechanisms of MT-depolymerization coupled force generation, both mechanisms may contribute to kinetochore force generation in vivo. A complete understanding of force generation and MT dynamics control mechanisms will require concerted efforts to obtain in vivo biophysical measurements of the kinetochore-MT attachment, and in vitro reconstitution of the entire kinetochore. Biophysical properties of interest are: magnitude of force generated, dynamics of the coupling between the kMT plus-end and the MT-binding proteins etc. The fluid control over the kMT plus-end dynamics exerted by the kinetochore is a critical aspect of its function. Understanding the mechanochemical events that allow a MT to polymerize and depolymerize will greatly facilitate elucidation of the kinetochore-MT interaction. Establishing the interfaces between different kinetochore protein complexes is a key objective of structural and biochemical studies of the kinetochore. Much more is understood about the protein architecture and mechanisms of attachment and force generation at budding yeast kinetochores than for higher eukaryotes. There is significant conservation in the core attachment site proteins, such as the KMN network, but so far, only the functions of the Ndc80 complex appear highly conserved, and much needs to be learned about the similarities and differences in the homologs of KNL-1 as well as other microtubule binding proteins important for attachment like the Ska1 complex. Functional contributions of motors and MAPs are essential for maximizing the accuracy of chromosome segregation. Single molecule studies of these proteins will greatly aid in assigning specific functions to these proteins in vivo. Finally, regulation of kinetochore activity by mitotic kinases and spindle assembly checkpoint machinery is a crucial factor in the accurate segregation of all the chromosomes in a cell. Exciting progress has been reported on this front recently [72]. Concerted efforts across multiple disciplines in the coming years will ultimately provide an integrative model of kinetochore function and regulation for accurate chromosome segregation.
Figure 2. Protein architecture and force generation in HeLa and yeast kinetochores.
(a) Proposed models for force generation at the vertebrate kinetochore. These schematics show the two possible arrangements of the Ndc80 complex binding to the MT lattice in HeLa kinetochores. The upper model is deduced from distance measurement data obtained by Wan et al., while the lower model is based on the tilted orientation of the Ndc80 bound to MT lattice observed in vitro. (b) Kinetochore architecture obtained from live budding yeast cells. Of particular interest are the positions of Dam1/DASH complex (green ellipsoids) and Ndc80 complex (orange fibrils). The first version depicts the simplest arrangement representing average distances. Other arrangements are also possible, and the second cartoon depicts a proposed model for persistent kMT attachment through the Ndc80 complex. The Mtw1 complex (blue fibrils) is assumed to attain various orientations with respect to the kinetochore-MT axis and allow the Ndc80 complex (orange fibrils) to reach significant distances downstream from the kMT plus-end.
Color key: NDC80 – ORANGE, KNL-1/Spc105 – RED, Mis12/MTW1 – BLUE, Dam1/DASH – GREEN
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Santaguida S, Musacchio A. The life and miracles of kinetochores. Embo J. 2009 doi: 10.1038/emboj.2009.173.A current and comprehensive review of kinetochore protein composition, its architecture, function, and regulation. Our review is narrowly focused on the mechanisms of force generation at the kinetochore. This review provides a good discussion of the core machinery of the kinetochore.
- 2.Cimini D. Merotelic kinetochore orientation, aneuploidy, and cancer. Biochim Biophys Acta. 2008;1786:32–40. doi: 10.1016/j.bbcan.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 3.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163.The spindle assembly checkpoint machinery and function is reviewed in depth.
- 4.Hill TL. Microfilament or microtubule assembly or disassembly against a force. Proc Natl Acad Sci U S A. 1981;78:5613–5617. doi: 10.1073/pnas.78.9.5613.This paper provides an excellent discussion of the thermodynamics of force generation coupled to polymerization and depolymerization at the ends of a polymer.
- 5.Inoue S, Salmon ED. Force generation by microtubule assembly/disassembly in mitosis and related movements. Mol Biol Cell. 1995;6:1619–1640. doi: 10.1091/mbc.6.12.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mandelkow EM, Mandelkow E, Milligan RA. Microtubule dynamics and microtubule caps: a time-resolved cryo- electron microscopy study. J. Cell Biol. 1991;114:977–991. doi: 10.1083/jcb.114.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joglekar AP, Hunt AJ. A simple, mechanistic model for directional instability during mitotic chromosome movements. Biophys J. 2002;83:42–58. doi: 10.1016/S0006-3495(02)75148-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hill TL. Theoretical problems related to the attachment of microtubules to kinetochores. Proc Natl Acad Sci U S A. 1985;82:4404–4408. doi: 10.1073/pnas.82.13.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koshland DE, Mitchison TJ, Kirschner MW. Polewards chromosome movement driven by microtubule depolymerization in vitro. Nature. 1988;331:499–504. doi: 10.1038/331499a0. [DOI] [PubMed] [Google Scholar]
- 10.Molodtsov MI, Ermakova EA, Shnol EE, Grishchuk EL, McIntosh JR, Ataullakhanov FI. A molecular-mechanical model of the microtubule. Biophys J. 2005;88:3167–3179. doi: 10.1529/biophysj.104.051789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.VanBuren V, Cassimeris L, Odde DJ. Mechanochemical model of microtubule structure and self-assembly kinetics. Biophys J. 2005;89:2911–2926. doi: 10.1529/biophysj.105.060913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McIntosh JR, Grishchuk EL, Morphew MK, Efremov AK, Zhudenkov K, Volkov VA, Cheeseman IM, Desai A, Mastronarde DN, Ataullakhanov FI. Fibrils connect microtubule tips with kinetochores: a mechanism to couple tubulin dynamics to chromosome motion. Cell. 2008;135:322–333. doi: 10.1016/j.cell.2008.08.038.A quantitative study of the morphology of individual protofilaments in PtK1 kinetochores. The study reveals that the majority of kMTs at both sister kinetochores display flared plus-ends. Furthermore, the curvature of individual protofilaments is less than the curvature of protofilaments of MTs disassembling in vitro. The authors also discover fibrils that attach to a specific region on these peeling protofilaments, where they can convert the residual strain energy into a minus-end directed force. Finally, the authors also explore the requirements of such a force generation mechanism for driving persistent chromosome movement.
- 13.Efremov A, Grishchuk EL, McIntosh JR, Ataullakhanov FI. In search of an optimal ring to couple microtubule depolymerization to processive chromosome motions. Proc Natl Acad Sci U S A. 2007;104:19017–19022. doi: 10.1073/pnas.0709524104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grishchuk EL, Molodtsov MI, Ataullakhanov FI, McIntosh JR. Force production by disassembling microtubules. Nature. 2005;438:384–388. doi: 10.1038/nature04132. [DOI] [PubMed] [Google Scholar]
- 15.Dickinson RB, Caro L, Purich DL. Force generation by cytoskeletal filament end-tracking proteins. Biophys J. 2004;87:2838–2854. doi: 10.1529/biophysj.104.045211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dogterom M, Yurke B. Measurement of the force-velocity relation for growing microtubules. Science. 1997;278:856–860. doi: 10.1126/science.278.5339.856. [DOI] [PubMed] [Google Scholar]
- 17.Gagliardi LJ. Electrostatic force generation in chromosome motions during mitosis. Journal of Electrostatics. 2004;63:309–327. [Google Scholar]
- 18.Civelekoglu-Scholey G, Sharp DJ, Mogilner A, Scholey JM. Model of chromosome motility in Drosophila embryos: adaptation of a general mechanism for rapid mitosis. Biophys J. 2006;90:3966–3982. doi: 10.1529/biophysj.105.078691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sprague BL, Pearson CG, Maddox PS, Bloom KS, Salmon ED, Odde DJ. Mechanisms of microtubule-based kinetochore positioning in the yeast metaphase spindle. Biophys J. 2003;84:3529–3546. doi: 10.1016/S0006-3495(03)75087-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coue M, Lombillo VA, McIntosh JR. Microtubule depolymerization promotes particle and chromosome movement in vitro. J. Cell Biol. 1991;112:1165–1175. doi: 10.1083/jcb.112.6.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicklas RB. Measurements of the force produced by the mitotic spindle in anaphase. J Cell Biol. 1983;97:542–548. doi: 10.1083/jcb.97.2.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen W, Zhang D. Kinetochore fibre dynamics outside the context of the spindle during anaphase. Nat Cell Biol. 2004;6:227–231. doi: 10.1038/ncb1104. [DOI] [PubMed] [Google Scholar]
- 23.VandenBeldt KJ, Barnard RM, Hergert PJ, Meng X, Maiato H, McEwen BF. Kinetochores use a novel mechanism for coordinating the dynamics of individual microtubules. Curr Biol. 2006;16:1217–1223. doi: 10.1016/j.cub.2006.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salmon ED. Microtubules: a ring for the depolymerization motor. Curr Biol. 2005;15:R299–302. doi: 10.1016/j.cub.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 25.Skibbens RV, Skeen VP, Salmon ED. Directional instability of kinetochore motility during chromosome congression and segregation in mitotic newt lung cells: a push-pull mechanism. J Cell Biol. 1993;122:859–875. doi: 10.1083/jcb.122.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guimaraes GJ, Dong Y, McEwen BF, Deluca JG. Kinetochore-microtubule attachment relies on the disordered N-terminal tail domain of Hec1. Curr Biol. 2008;18:1778–1784. doi: 10.1016/j.cub.2008.08.012.This and the companion study [27] below vividly demonstrate the importance of the 80 amino acid N-terminal tail of the Ndc80 subunit in the Ndc80 complex. Deletion of this tail prevents binding of the Ndc80 complex with MTs in vitro, and completely abrogates end-on kinetochore-MT attachments in Ptk1 cells. Interestingly, the CH-domain is necessary for activating the spindle assembly checkpoint.
- 27.Wei RR, Al-Bassam J, Harrison SC. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat Struct Mol Biol. 2007;14:54–59. doi: 10.1038/nsmb1186. [DOI] [PubMed] [Google Scholar]
- 28.Miller SA, Johnson ML, Stukenberg PT. Kinetochore Attachments Require an Interaction between Unstructured Tails on Microtubules and Ndc80Hec1. Current Biology. 2008;18:1785–1791. doi: 10.1016/j.cub.2008.11.007.This study reports the same conclusions regarding the requirement of the N-terminal Ndc80 tail using HeLa cells. The authors also establish that Ndc80-MT interactions are established by electrostatic interactions between the N-terminal tail (net charge of +10) and the C-terminal tubulin tails or E-hooks (net charge of −6).
- 29.Kiyomitsu T, Obuse C, Yanagida M. Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev Cell. 2007;13:663–676. doi: 10.1016/j.devcel.2007.09.005.This study shows that depletion of the human homolog of KNL-1 abolishes kMT attachments, although Ndc80 is present at the kinetochore. The study does not investigate the impact of KNL-1 depletion on kinetochore architecture and regulation.
- 30.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039.This study uses purified/reconstituted KNL-1, Mis12/Mtw1 complex, and the Ndc80 complex from C. elegans to establish the MT-binding properties of the core MT attachment site within the kinetochore. It also obtains the first view of the Ndc80 complex bound to the MT lattice.
- 31.DeLuca JG, Howell BJ, Canman JC, Hickey JM, Fang G, Salmon ED. Nuf2 and Hec1 are required for retention of the checkpoint proteins Mad1 and Mad2 to kinetochores. Curr Biol. 2003;13:2103–2109. doi: 10.1016/j.cub.2003.10.056. [DOI] [PubMed] [Google Scholar]
- 32.Wei RR, Sorger PK, Harrison SC. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc Natl Acad Sci U S A. 2005;102:5363–5367. doi: 10.1073/pnas.0501168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ciferri C, Pasqualato S, Screpanti E, Varetti G, Santaguida S, Reis G Dos, Maiolica A, Polka J, De Luca JG, De Wulf P, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020.This study characterized the EM structure of the Ndc80 complex, and put forth the two-headed Ndc80-Nuf2 MT-binding model. This study also observed cooperative binding of the Ndc80 complex.
- 34.Kemmler S, Stach M, Knapp M, Ortiz J, Pfannstiel J, Ruppert T, Lechner J. Mimicking Ndc80 phosphorylation triggers spindle assembly checkpoint signalling. Embo J. 2009;28:1099–1110. doi: 10.1038/emboj.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson-Kubalek EM, Cheeseman IM, Yoshioka C, Desai A, Milligan RA. Orientation and structure of the Ndc80 complex on the microtubule lattice. J Cell Biol. 2008;182:1055–1061. doi: 10.1083/jcb.200804170.Through further EM analysis of the Ndc80 complex bound to the MT lattice, the authors proposed the toe-binding model for the Ndc80 complex wherein Ndc80 binds to either inter- or intra-dimer interface along the lattice.
- 36.Wang HW, Long S, Ciferri C, Westermann S, Drubin D, Barnes G, Nogales E. Architecture and flexibility of the yeast Ndc80 kinetochore complex. J Mol Biol. 2008;383:894–903. doi: 10.1016/j.jmb.2008.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 38.Joglekar AP, Bouck D, Finley K, Liu X, Wan Y, Berman J, He X, Salmon ED, Bloom KS. Molecular architecture of the kinetochore-microtubule attachment site is conserved between point and regional centromeres. J Cell Biol. 2008;181:587–594. doi: 10.1083/jcb.200803027.This study demonstrates that the stoichiometry of core kinetochore proteins determined for budding yeast is also conserved in fission yeast (Schizosaccharomyces pombe) and in Candida albicans. Both these fungi possess chromosomes with regional centromeres, and in the case of fission yeast, there are 2-3 kMT attachments per kinetochore. The study also finds that the number of kinetochore proteins per kMT at these kinetochores is nearly identical to the numbers found at the budding yeast kinetochore.
- 39.Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED. Molecular architecture of a kinetochore-microtubule attachment site. Nat Cell Biol. 2006;8:581–585. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Westermann S, Avila-Sakar A, Wang HW, Niederstrasser H, Wong J, Drubin DG, Nogales E, Barnes G. Formation of a dynamic kinetochore- microtubule interface through assembly of the Dam1 ring complex. Mol Cell. 2005;17:277–290. doi: 10.1016/j.molcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 41.Miranda JJ, De Wulf P, Sorger PK, Harrison SC. The yeast DASH complex forms closed rings on microtubules. Nat Struct Mol Biol. 2005;12:138–143. doi: 10.1038/nsmb896. [DOI] [PubMed] [Google Scholar]
- 42.Cheeseman IM, Enquist-Newman M, Muller-Reichert T, Drubin DG, Barnes G. Mitotic spindle integrity and kinetochore function linked by the Duo1p/Dam1p complex. J Cell Biol. 2001;152:197–212. doi: 10.1083/jcb.152.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang HW, Ramey VH, Westermann S, Leschziner AE, Welburn JP, Nakajima Y, Drubin DG, Barnes G, Nogales E. Architecture of the Dam1 kinetochore ring complex and implications for microtubule-driven assembly and force-coupling mechanisms. Nat Struct Mol Biol. 2007;14:721–726. doi: 10.1038/nsmb1274. [DOI] [PubMed] [Google Scholar]
- 44.Li Y, Bachant J, Alcasabas AA, Wang Y, Qin J, Elledge SJ. The mitotic spindle is required for loading of the DASH complex onto the kinetochore. Genes Dev. 2002;16:183–197. doi: 10.1101/gad.959402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanaka K, Kitamura E, Kitamura Y, Tanaka TU. Molecular mechanisms of microtubule-dependent kinetochore transport toward spindle poles. J Cell Biol. 2007;178:269–281. doi: 10.1083/jcb.200702141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grishchuk EL, Spiridonov IS, Volkov VA, Efremov A, Westermann S, Drubin D, Barnes G, Ataullakhanov FI, McIntosh JR. Different assemblies of the DAM1 complex follow shortening microtubules by distinct mechanisms. Proc Natl Acad Sci U S A. 2008;105:6918–6923. doi: 10.1073/pnas.0801811105.Using an innovative assay consisting of asymmetrically labeled microbeads and photobleaching experiments, the authors establish the presence of Dam1/DASH rings on MTs in their in vitro assay. The authors go on to measure the diffusion coefficient and force generation properties
- 47.Shimogawa MM, Graczyk B, Gardner MK, Francis SE, White EA, Ess M, Molk JN, Ruse C, Niessen S, Iii JR Yates, et al. Mps1 Phosphorylation of Dam1 Couples Kinetochores to Microtubule Plus Ends at Metaphase. Current Biology. 2006;16:1489–1501. doi: 10.1016/j.cub.2006.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cimini D. Detection and correction of merotelic kinetochore orientation by Aurora B and its partners. Cell Cycle. 2007;6:1558–1564. doi: 10.4161/cc.6.13.4452. [DOI] [PubMed] [Google Scholar]
- 49.Cheeseman IM, Anderson S, Jwa M, Green EM, Kang J, Yates JR, 3rd, Chan CS, Drubin DG, Barnes G. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 2002;111:163–172. doi: 10.1016/s0092-8674(02)00973-x. [DOI] [PubMed] [Google Scholar]
- 50.Powers AF, Franck AD, Gestaut DR, Cooper J, Gracyzk B, Wei RR, Wordeman L, Davis TN, Asbury CL. The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell. 2009;136:865–875. doi: 10.1016/j.cell.2008.12.045.A single molecule study of the MT-binding properties of the Ndc80 complex. The authors demonstrate that as few as 14 Ndc80 molecules on the surface of a bead can track the plus-ends of depolymerizing MTs. Although single Ndc80 molecules do not remain at the plus-ends of depolymerizing MT, aggregates of Ndc80 complex molecules assembled through antibody binding do achieve persistent tracking, demonstrating the need for multiple interactions for generating movement coupled to MT depolymerization. The authors also find that the properties of the yeast and human Ndc80 complex are very similar.
- 51.Lombillo VA, Stewart RJ, McIntosh JR. Minus-end-directed motion of kinesin-coated microspheres driven by microtubule depolymerization. Nature. 1995;373:161–164. doi: 10.1038/373161a0. [DOI] [PubMed] [Google Scholar]
- 52.Peskin CS, Oster GF. Force production by depolymerizing microtubules: load-velocity curves and run-pause statistics. Biophys J. 1995;69:2268–2276. doi: 10.1016/S0006-3495(95)80097-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gestaut DR, Graczyk B, Cooper J, Widlund PO, Zelter A, Wordeman L, Asbury CL, Davis TN. Phosphoregulation and depolymerization-driven movement of the Dam1 complex do not require ring formation. Nat Cell Biol. 2008;10:407–414. doi: 10.1038/ncb1702.This in vitro study looks at the behavior of fluorescently labeled Dam1/DASH complex at the plus-ends of disassembling MTs. The authors find that 1-4 Dam1/DASH molecules can track the plus-ends of depolymerizing MTs. The effect of Dam1/DASH complex phosphorylated by Ipl1 is also studied.
- 54.Asbury CL, Gestaut DR, Powers AF, Franck AD, Davis TN. The Dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement. Proc Natl Acad Sci U S A. 2006;103:9873–9878. doi: 10.1073/pnas.0602249103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Franck AD, Powers AF, Gestaut DR, Gonen T, Davis TN, Asbury CL. Tension applied through the Dam1 complex promotes microtubule elongation providing a direct mechanism for length control in mitosis. Nat Cell Biol. 2007;9:832–837. doi: 10.1038/ncb1609.By exerting force on Dam1/DASH coated microbeads tracking disassembling plus-ends, the authors find that tension increases the rescue frequency and lowers catastrophe frequency for the MTs.
- 56.Gardner MK, Pearson CG, Sprague BL, Zarzar TR, Bloom K, Salmon ED, Odde DJ. Tension-dependent regulation of microtubule dynamics at kinetochores can explain metaphase congression in yeast. Mol Biol Cell. 2005;16:3764–3775. doi: 10.1091/mbc.E05-04-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wan X, O’Quinn RP, Pierce HL, Joglekar AP, Gall WE, DeLuca JG, Carroll CW, Liu ST, Yen TJ, McEwen BF, et al. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137:672–684. doi: 10.1016/j.cell.2009.03.035.The authors use two-color super-resolution microscopy on fixed HeLa cells stained with antibodies. The authors find that the projected length along the kMT axis of the Ndc80 complex is ~ 10 nm smaller than the length determined in EM studies of reconstituted complex. By comparing kinetochore architecture in metaphase cells and cells treated with taxol, the authors discover that the KNL-1 and Ndc80 complex show different movements with respect to each other. This observation suggests the presence of two “arms” within the MT attachment site that can move relative to each other in response to tension.
- 58.Joglekar AP, Bloom K, Salmon ED. In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr Biol. 2009;19:694–699. doi: 10.1016/j.cub.2009.02.056.This paper establishes the protein architecture of the budding yeast kinetochore in metaphase and anaphase using live cell super-resolution microscopy of fluorescently labeled kinetochore proteins. Of interest to this review are the more or less parallel orientation of the Ndc80 complex to the kMT axis, and the location of the Dam1/DASH complex 10 nm behind the MT-binding head domains of the Ndc80 complex. The length of the Ndc80 complex in anaphase also suggests that its front section likely bends at the hinge region.
- 59.Kerres A, Jakopec V, Fleig U. The conserved Spc7 protein is required for spindle integrity and links kinetochore complexes in fission yeast. Mol Biol Cell. 2007;18:2441–2454. doi: 10.1091/mbc.E06-08-0738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Manning AL, Ganem NJ, Bakhoum SF, Wagenbach M, Wordeman L, Compton DA. The kinesin-13 proteins Kif2a, Kif2b, and Kif2c/MCAK have distinct roles during mitosis in human cells. Mol Biol Cell. 2007;18:2970–2979. doi: 10.1091/mbc.E07-02-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stumpff J, von Dassow G, Wagenbach M, Asbury C, Wordeman L. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev Cell. 2008;14:252–262. doi: 10.1016/j.devcel.2007.11.014.This study finds that the motor activity of Kif18A is necessary for its concentration at metaphase kinetochores. Furthermore, the levels of Kif18A at a sister kinetochore pair are distinctly different. Surprisingly, over-expression of this MT depolymerase dampens chromosome oscillations, whereas its depletion increases chromosome oscillation speed and amplitude.
- 62.Wordeman L, Wagenbach M, von Dassow G. MCAK facilitates chromosome movement by promoting kinetochore microtubule turnover. J Cell Biol. 2007;179:869–879. doi: 10.1083/jcb.200707120.The authors use a CENP-B-MCAK fusion protein to recruit MCAK and its mutant alleles to the inner kinetochore and selectively deplete endogenous MCAK from the kinetochore. This innovative strategy allows the authors to study MCAK function specifically at the kinetochore. This paper along with the Kif18A study above [58] provides an insight into the importance of MT depolymerases in coordinating the function of sister kinetochores.
- 63.Draviam VM, Shapiro I, Aldridge B, Sorger PK. Misorientation and reduced stretching of aligned sister kinetochores promote chromosome missegregation in EB1- or APC-depleted cells. Embo J. 2006;25:2814–2827. doi: 10.1038/sj.emboj.7601168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cassimeris L, Becker B, Carney B. TOGp regulates microtubule assembly and density during mitosis and contributes to chromosome directional instability. Cell Motil Cytoskeleton. 2009;66:535–545. doi: 10.1002/cm.20359. [DOI] [PubMed] [Google Scholar]
- 65.Pereira AL, Pereira AJ, Maia ARR, Drabek K, Sayas CL, Hergert PJ, Lince-Faria M, Matos I, Duque C, Stepanova T, et al. Mammalian CLASP1 and CLASP2 Cooperate to Ensure Mitotic Fidelity by Regulating Spindle and Kinetochore Function. Mol. Biol. Cell. 2006;17:4526–4542. doi: 10.1091/mbc.E06-07-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cheeseman IM, MacLeod I, Yates JR, 3rd, Oegema K, Desai A. The CENP-F-like proteins HCP-1 and HCP-2 target CLASP to kinetochores to mediate chromosome segregation. Curr Biol. 2005;15:771–777. doi: 10.1016/j.cub.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 67.Maiato H, Khodjakov A, Rieder CL. Drosophila CLASP is required for the incorporation of microtubule subunits into fluxing kinetochore fibres. Nat Cell Biol. 2005;7:42–47. doi: 10.1038/ncb1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guimaraes GJ, Deluca JG. Connecting with Ska, a key complex at the kinetochore-microtubule interface. Embo J. 2009;28:1375–1377. doi: 10.1038/emboj.2009.124.A review of the recent reports on the importance of the Ska complex in maintaining stable end-on attachments in vertebrate cells. The review does not include observations published in a subsequent report that demonstrates that the Ska complex is also required for sister chromatid cohesion.
- 69.Welburn JPI, Grishchuk EL, Backer CB, Wilson-Kubalek EM, Iii JR Yates, Cheeseman IM. The Human Kinetochore Ska1 Complex Facilitates Microtubule Depolymerization-Coupled Motility. Developmental Cell. 2009;16:374–385. doi: 10.1016/j.devcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Daum JR, Wren JD, Daniel JJ, Sivakumar S, McAvoy JN, Potapova TA, Gorbsky GJ. Ska3 Is Required for Spindle Checkpoint Silencing and the Maintenance of Chromosome Cohesion in Mitosis. Curr Biol. 2009 doi: 10.1016/j.cub.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tirnauer JS, Canman JC, Salmon ED, Mitchison TJ. EB1 targets to kinetochores with attached, polymerizing microtubules. Mol Biol Cell. 2002;13:4308–4316. doi: 10.1091/mbc.E02-04-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu D, Vader G, Vromans MJ, Lampson MA, Lens SM. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 2009;323:1350–1353. doi: 10.1126/science.1167000. [DOI] [PMC free article] [PubMed] [Google Scholar]