Abstract
The calcium-triggered neurotransmitter release requires three SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) proteins: synaptobrevin 2 (or VAMP2) on the synaptic vesicle and syntaxin 1 and SNAP-25 at the presynaptic plasmas membrane. This minimal fusion machinery is believed to drive fusion of the vesicle to the presynaptic membrane. Complexin, also known as synaphin, is a neuronal cytosolic protein that acts as a major regulator of synaptic vesicle exocytosis. Stimulatory and inhibitory effects of complexin have both been reported, suggesting the duality of its function. To shed light on the molecular basis of the complexin’s dual function, we have performed an EPR investigation of the complexin-SNARE quaternary complex. We found that the accessory α-helix (amino acids 27–48) by itself has the capacity to replace the C-terminus of the SNARE motif of VAMP2 in the four-helix bundle and makes the SNARE complex weaker when the N-terminal region of complexin I (amino acids 1–26) is removed. However, the accessory α-helix remains detached from the SNARE core when the N-terminal region of complexin I is present. Thus, our data show the possibility that the balance between the activities of the accessory α-helix and the N-terminal domain might determine the final outcome of the complexin function, either stimulatory or inhibitory.
Keywords: site-directed spin labeling, EPR spectroscopy, membrane fusion, SNAREs, Complexin I
Introduction
Synaptic vesicle fusion is driven by assembly of the trans-SNARE complex between the vesicle and the plasma membrane, which facilitates merging of the opposite bilayers.1–7 The physiological trigger for this process is the spike in the local concentration of calcium. As Ca2+ enters the presynaptic cytoplasm, the Ca2+-sensor synaptotagmin I, 8–11 which is localized on synaptic vesicles, interacts with the SNARE complex and phospholipids simultaneously,12–14 leading to the fast opening of the fusion pore.15–18
In addition to synaptotagmin I, complexins also participate in the fast Ca2+-dependent neuronal exocytosis.19–22 Complexins are small soluble cytoplasmic proteins (15- to 18-kDa) that are mainly found in the presynaptic part of neuronal cells23–25 and the central region (amino acids 48–70) binds to the SNARE core as an anti-parallel α-helix, which attaches complexin to the SNARE complex.26,27
Complexin is believed to have a dual function20,28,29 in synaptic membrane fusion, both stimulation19,26,27 and inhibition.21,30–35 Although the exact molecular mechanism is unknown, the N-terminal region of complexin is shown to play an important role in promoting the neurotransmitter release.28,29,36 It is proposed that the N-terminal region interacts with the membrane-proximal region of VAMP2 and functions as a force transducer.29 However, under different conditions, complexin exhibits the inhibitory function,28 which is the basis for the recent “complexin-clamp model”.34 Two possible molecular-level mechanistic models have been proposed to explain the inhibition by complexin. Firstly, complexin is found to have the µM affinity to the t-SNARE complex in addition to its sub µM affinity to the SNARE complex.20,26 Thus, under favorable conditions complexin could compete with the VAMP2 binding to the t-SNARE complex, which would competitively inhibit SNARE assembly and membrane fusion. Secondly, when bound to the SNARE core the accessory α-helix of complexin, which franks the central SNARE-anchor domain and the stimulatory N-terminal region, might replace the C-terminal portion of the VAMP2 helix to form an alternative four-helix bundle with the helices from t-SNAREs.28,34,37 Such structural replacement would prevent formation of the complete SNARE four helix bundle, whereby inhibiting membrane fusion.
In this work, we performed the EPR investigation of the conformational changes in the v-SNARE VAMP2 after complexin binding to the SNARE complex. The results show that there is no major conformational change, such as the replacement of VAMP2 by complexin, occurring at the membrane-proximal region of VAMP2 upon binding of the full-length complexin to the SNARE complex. However, when the stimulatory N-terminal region of complexin is removed, the accessory α-helix replaces the C-terminal region of VAMP2 partially. Thus, our results show that the balance between the stimulatory and inhibitory activities of the accessory α-helix and the adjacent N-terminal region, respectively, might determine the exact function of the particular complexin.
Results
Complexin-SNARE quaternary complex formation
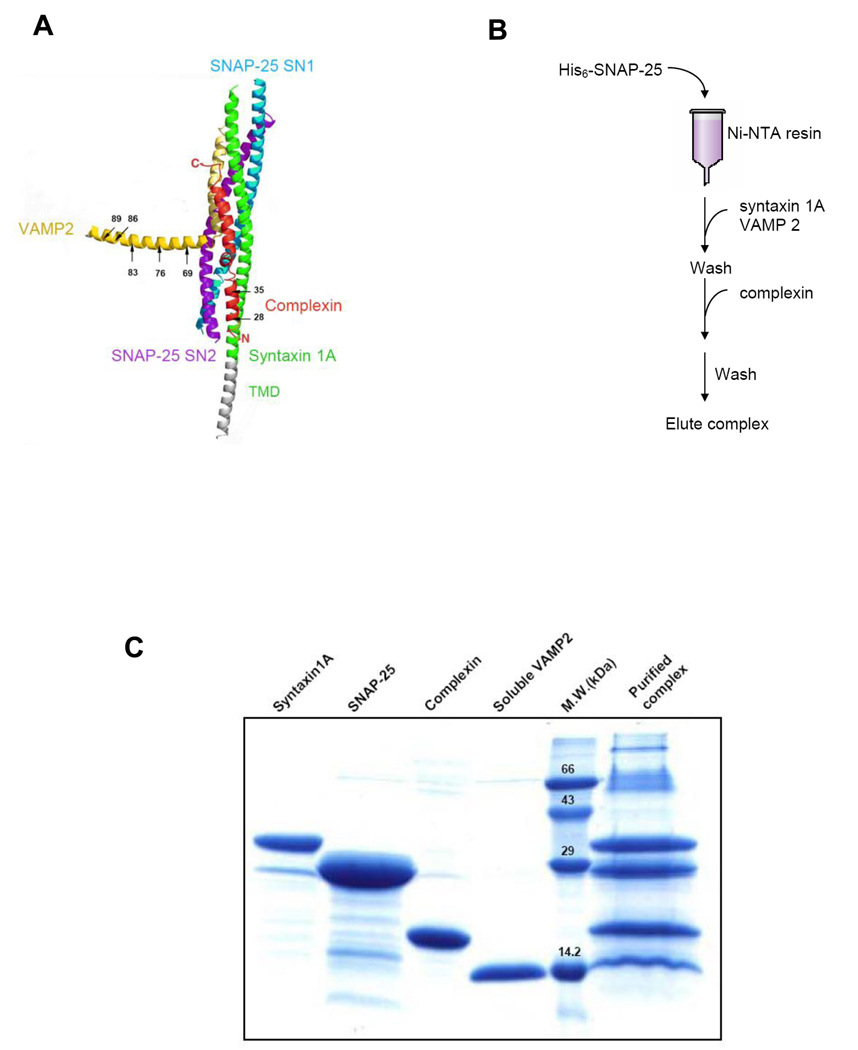
The recent study by Rothman and coworkers suggests that the accessory α-helix of complexin replace VAMP2 and form an alternative four-helix bundle with t-SNAREs near the membrane, thereby inhibiting complete SNARE complex formation and membrane fusion.34 According to this proposed model, the displaced sequence of VAMP2 includes both the cleavage site (between residues 76 and 77) and recognition site (residues 62 to 71) for BoNT/B toxin. Therefore, our EPR investigation was focused on the structure of the C-terminal region of VAMP2. We prepared five single cysteine mutants of VAMP2 (Fig 1A), including A69C within the recognition site of BoNT/B and Q76C on its cleavage site, and K83C, R86C, and W89C in the membrane-proximal region. The cysteine mutants were derivatized with methanethoisulfonate spin label (MTSSL) for EPR measurements. Next, we used the pull-down method (Fig 1B) to prepare the complexin-SNARE quaternary complex that includes the spin-labeled soluble VAMP2, full-length syntaxin 1A, SNAP-25, and complexin. As the bait, His6-tagged SNAP-25 was attached to the Ni-NTA beads and the SNARE complex was first formed by flowing syntaxin 1A and soluble VAMP2 into the bead solution. Complexin was next added to the column to form the complexin-SNARE quaternary complex. The transmembrane domain (TMD) of VAMP2 was not included in order to avoid the interaction between two TMDs in one membrane and to best mimic the trans-SNARE complex. For individual mutants we analyzed formation of the complexin-SNARE quaternary complex by the SDS-PAGE gel after heating. We found that the immobilized His6-tagged SNAP-25 co-purified with stoichiometric amounts of syntaxin 1A, soluble VAMP2 and complexin (Fig 1C).
Figure 1.
Characterization of the complexin-SNARE quaternary complex. A. The fusion clamp model proposes that complexin can replace the C terminus of VAMP2 upon binding to the trans-SNARE complex (adapted from ref. 34). Positions 69, 76, 83, 86 and 89 are the spin-labeled sites of the soluble VAMP2. Positions 35 and 28 of complexin have also been shown in the model. Positions 42, 25 and 22, which are located at the N-terminal of complexin, have not been shown. B. The flow diagram of purification of the complexin-SNARE quaternary complex. C. SDS-PAGE analysis of the purified complexin-SNARE quaternary complex.
Full-length complexin does not replace VAMP2
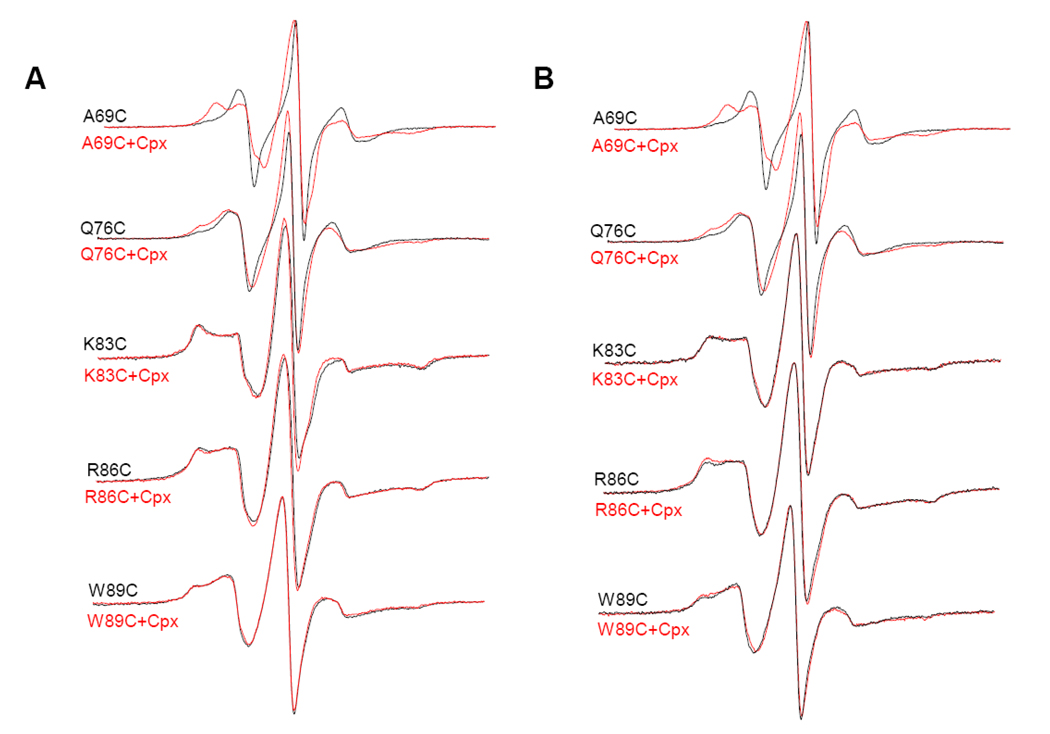
The EPR line shape is a sensitive function of the motional rate. The slower the motion of the nitroxide side chain, the broader the EPR spectrum. Therefore, if the proposed structure of the quaternary complex in Fig 1A were to be true, we expect the narrowing of the EPR spectra for the spin labeled positions when compared with the EPR spectra from the SNARE complex only. EPR spectra of the SNARE complex before and after binding to full-length complexin I were shown in Figure 2. The spectra for positions 69 and 76 of the complexin-SNARE complex became much broader than those from the SNARE complex only. This tendency was more obvious at 69 indicating that there was strong tertiary interaction at this site. Our data were also consistent with the deuterium exchange results which showed that position 69 was highly protected after complexin binding.27 Thus, the EPR results for positions 69 and 76 were not consistent with the proposed structure in Fig 1A. Instead, the results suggest that complexin bind on the surface of the SNARE four helix bundle, as shown in the crystal structure.27
Figure 2.
EPR spectra of the spin-labeled soluble VAMP2 in the full-length complexin-SNARE quaternary complex at room temperature. First-derivative mode EPR spectra for the complex in the detergent (A) and in the membrane (B) are shown.
For positions 83, 86, and 89, which are located in the membrane-proximal region, there was no obvious narrowing due to the replacement of VAMP2 by complexin either. Unlike the cases with positions 69 and 76, we did not observe extra line broadening due to the complexin binding, suggesting complexin might not even interact with the SNARE complex locally in the region. Further addition of complexin to the complexin-SNARE quaternary complex did not change the EPR lineshape either (Fig S1), suggesting that the reason we did not observe complexin-induced line broadening was neither due to the insufficient complexin binding nor due to the intermolecular complexin-binding equilibrium. Therefore, the EPR analysis of the quaternary complex suggested that full-length complexin I does not replace VAMP2 when bound to the SNARE complex.
The accessory α-helix of complexin has no obvious interaction with the SNARE core
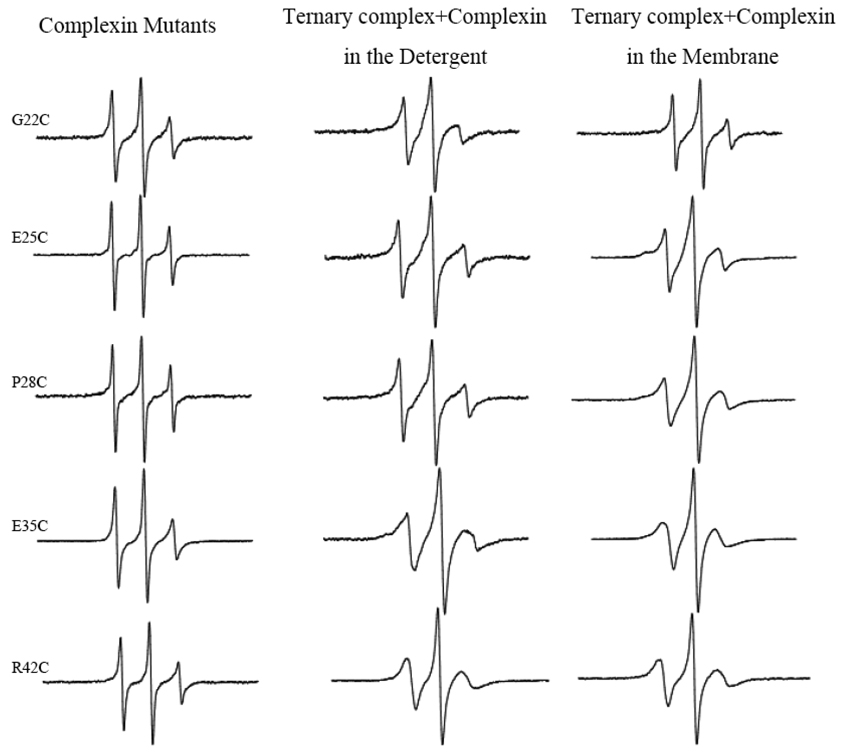
We now examined the conformation of complexin in the compelxin-SNARE quaternary complex using spin-labeled complexin. We aligned the sequences of complexin with VAMP2 in an antiparallel orientation as they were shown in the crystal structure27,34 and selected five positions R42C, E35, P28, E25, and G22 in the accessory α-helix as the spin labeling sites. EPR spectra for these sites in the complexin-SNARE complex were shown in Figure 3. The line shapes for spin labeled complexin alone were all very sharp, reflecting the freely diffusing random coil. When the spin-labeled mutants were bound to the SNARE complex, the spectra became broader, reflecting the slower motion of the nitroxide, most likely due to formation of α-helix. However, the spectra were still not so broad with no indication of any tertiary contact. In fact, the spectra are sharper than those for the solvent exposed positions on the surface helix,38,39 indicative of substantial motion of the accessory α-helix. Therefore, our EPR analysis shows that the accessory α-helix is most likely has a little or no tertiary interaction with the SNARE core.
Figure 3.
EPR spectra of the spin-labeled complexin in the full-length complexin-SNARE quaternary complex at room temperature. First-derivative mode EPR spectra for the complex in the detergent and in the membrane are shown. Positions 22, 25, 28, 35 and 42 of complexin are corresponding to the positions 89, 86, 83, 76 and 69 of VAMP2 respectively.
The accessory α-helix of complexin can partially displace VAMP2 locally when the N-terminal region is absent
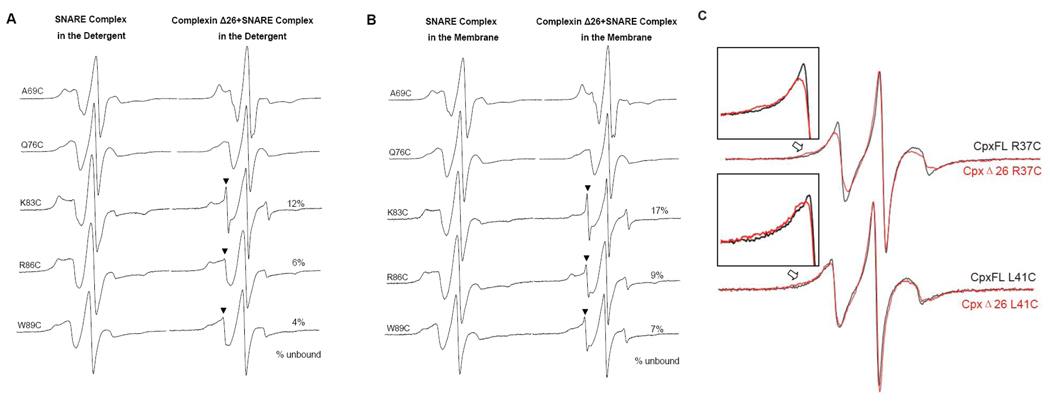
When complexins are deleted from the neuron, the Ca2+-triggered fast release is severely reduced. In contrast, the frequency of the Ca2+-independent spontaneous release is dramatically enhanced. However, when the N-terminal 26 residues are removed from complexin, both the evoked and the spontaneous releases are reduced when compared with those of complexin-null mutant.29 Thus, these results show that the shortened complexin mutant lacking the stimulatory N-terminal sequence has an inhibitory function for the spontaneous release. In this case, it is possible that the accessory α-helix displaces VAMP2, thereby inhibiting membrane fusion. To test this possibility, we prepared the shortened construct of complexin lacking the N-terminal 26 residues (amino acids 27–134), and investigate the structure of the complexin Δ26-SNARE quaternary complex employing the spin labeled VAMP2 mutants (Fig 4A and B). For nitroxides attached to positions 69 and 76, the EPR spectra were very similar to those obtained from the full-length complexin-SNARE quaternary complex (Fig 2). However, for C-terminal three spin-labeled positions 83, 86 and 89, the EPR spectra show a small but distinctly sharp spectral component (Fig 4B, see arrows, 7–17%), which is most likely from the frayed VAMP2 chain that is freely diffusing and fast moving in solution. Thus, the EPR analysis show that the accessory α-helix of complexin has the capacity to replace VAMP2 and make the SNARE complex somewhat weaker in the absence of the N-terminal region. It appears though that the sharp spectral component for N-terminal position 83 (17%) is larger than those for C-terminal positions 86 and 89 (9 and 7%, respectively). One however must be cautious in interpreting these numbers when taking into account the perturbation by nitroxides and the uncertainty of the spectral subtraction analysis. Therefore, given the experimental uncertainty we would not want to put much weight on the order of the extent of displacement among the spin labeled positions.
Figure 4.
(A) and (B), EPR spectra of the spin-labeled soluble VAMP2 in the truncated complexin Δ 26-SNARE quaternary complex at room temperature. First-derivative mode EPR spectra for the complex in the detergent (A) and in the membrane (B) are shown. The arrowheads indicate the sharp component from the locally displaced VAMP2. The percentage of the sharp component was calculated using the spectral subtraction method (ref. 42). (C), EPR spectra of the spin-labeled full-length and truncated complexin upon binding with the ternary SNARE complex at room temperature. The inset figures show the difference between full-length and truncated complexin involved in the quaternary complex clearly.
Our EPR analysis show that the displacement of VAMP2 by complexin Δ26 happens locally in the membrane-proximal region. If true, we expect that the accessory α-helical region of complexin Δ26 would interact with the t-SNARE helices in replacement of VAMP2. Indeed, for spin labeled complexin Δ26 mutants R37C and L41C we observe small but distinct line broadening (Fig 4C) most likely due to the tertiary interaction between the accessory α-helix and the t-SNARE core. Although positions 37 and 41 are not in register with VAMP2 residues 83–89, the results show that the tertiary interaction between complexin Δ26 and the t-SNARE core might occur in the part preceding this region.
Discussion
The EPR analysis of the complexin-SNARE quaternary complex revealed that the accessory α-helix has small but distinct capacity to displace the C-terminal part of VAMP2 from the SNARE four-helix bundle. However, in full-length complexin I such an inhibitory function of the accessory α-helix against SNARE complex formation is nearly completely suppressed by the presence of the N-terminal sequence. It is unclear how the N-terminal sequence might override the inhibition by the accessory α-helix. One possible scenario is that the N-terminal region directly interacts with the accessory α-helix to pull it out from its interaction with the SNARE complex. An alternative scenario is that the N-terminal region itself binds to the SNARE complex in a yet unknown site in such a way that it destabilizes the binding of accessory α-helix to the three helix bundle made of t-SNAREs syntaxin 1A and SNAP-25 non-competitively.
In the previous NMR study of complexin-SNARE quaternary complex using an N-terminally truncated fragment (Ref. 27), such displacement of the membrane-proximal region of VAMP2 was not observed. A fundamental difference between EPR and NMR is in the time scales of measurement. The EPR time scale is at least two orders of magnitude faster than that of NMR and it can pick up fast exchanging events. We speculate that the equilibrium exchange between the accessory α-helix and the VAMP2 is sufficiently fast for NMR not to be able to detect. Further, only ~10% insertion exacerbates the situation for the detection by NMR.
Although complexin’s ability to displace VAMP2 in the quaternary complex appears to be small, it could actually be much bigger for the trans-SNARE complex in which the interaction between membrane-anchored VAMP2 and t-SNARE on the opposite membrane is expected to be much weaker than that in our model system. In our system, the transmembrane domain of VAMP2 was deleted to avoid the situation of the cis-SNARE complex in which the transmembrane domains of v- and t-SNAREs are in the same membrane, imposing no repulsion of the bilaye-bilayer interaction to the SNARE core.
The modes of interaction between complexin and the SNARE complex appear to be quite diverse. So far, three different interaction modes are discovered. Firstly, complexin has the capacity to interact with the SNARE complex additively without changing the basic coiled coil structure of the SNARE core.27 Such an interaction is expected to work favorably for the stabilization of the SNARE complex and believed to play a stimulatory role in SNARE-dependent membrane fusion. Secondly, the accessory α-helix displaces the C-terminal part of VAMP2 to form an alternative four-helix bundle locally while the four plus one helix bundle structure is maintained in the middle.28,34 Our EPR results however show that only a small fraction of the quaternary complex have such a structure. It is most likely that the first and the second modes are inter-convertible and in equilibrium. Third, complexin interacts with the t-SNARE complex, which acts as the competitive inhibitor for VAMP2 binding to the t-SNARE complex.20,40 The binding constant for the complexin/t-SNARE complex is reported to be somewhat higher than its affinity to the ternary SNARE complex.20 It is highly likely that all three interaction modes exist in neuronal cells and the delicate balance between these three modes of interaction produces the inhibitory-stimulatory control of the neurotransmitter release. For example, the knockout experiments reveals that complexin from drosophila melanogaster is highly inhibitory,22,32 while complexin from rat is stimulatory.19 We speculate that such species specific function of complexin stems from the balance between the functional strengths between the inhibitory accessory α-helix and the stimulatory N-terminal sequence.
The interaction mode in which the accessory α-helix replaces the VAMP2 while bound to the SNARE core constitutes the structural basis for the fusion clamp model.34 Indeed, in the absence of the N-terminal sequence the accessory α-helix has the capacity of replacing VAMP2 partially and locally from the SNARE core while the other part of the complex is fully engaged. Such local structural inhibition may be advantageous over the fully competitive inhibition relying on the t-SNARE/complexin binding because the system could react more quickly in responding to the Ca2+ signal. Paradoxically, however, for full-length complexin, where the N-terminal sequence is present, such replacement was not seen. In pure speculation, if another protein factor such as synaptotagmin I could sequester the N-terminal sequence, the inhibition of SNARE assembly might be possible and the inhibition could be removed by the influx of Ca2+, which might produce synchronized membrane fusion. The results from the present analysis show that such a mechanistic model is structurally possible.
Materials and Methods
Plasmid construction and site-directed mutagenesis
DNA sequences encoding full-length syntaxin 1A (amino acids 4–288), the soluble VAMP2 (amino acids 1–94), full-length complexin I (amino acids 1–134 with one native cysteine C105 replaced by alanine), and the truncated complexin I Δ26 (amino acids 27–134) were cloned into the pGEX-KG vector and expressed as the N-terminal glutathione S-transferase (GST) fusion protein. SNAP-25 (amino acids 1–206 with four native cysteines C85, 88, 90 and 92 replaced by alanines) was cloned into pET-28b vector and expressed as N-terminal His6-tagged protein. All cysteine mutants were generated by QuikChange site-directed mutagenesis kit (Stratagene), and they were confirmed by DNA sequencing (Iowa State University DNA Sequencing Facility).
Protein expression, purification, and spin labeling
GST fusion proteins were expressed in E. coli Rosetta (DE3) pLysS (Novagene) and purified using glutathione-agarose beads (Sigma).41 Briefly, the cells were grown at 37 °C in LB with glucose (2 g/liter), ampicillin (100 µg/ml), and chloramphenicol (50 µg/ml) until A600 reached 0.6–0.8. After adding 0.3 mM isopropylthio-β-D-galactopyranoside (IPTG), the cells were further grown for 6 h at 22 °C for GST-soluble VAMP2, GST-complexin I, and GST-complexin I Δ26 but at 16 °C for GST-syntaxin 1A. Finally, the protein was cleaved to remove the GST tag by thrombin in the cleavage buffer (50 mM Tris-HCl, 150 mM NaCl, pH 8.0). We add 1% n-octyl-glucoside (OG) in the cleavage buffer for syntaxin 1A.
His6-tagged SNAP-25 was expressed in E. coli BL21 (DE3) Codon Plus RIL (Stratagene) and purified using Ni-NTA resin (Qiagen).41 The cells were grown at 37 °C in LB with glucose (2 g/liter), kanamycin (34 µg/ml), and chloramphenicol (50 µg/ml) until A600 reached 0.6–0.8. Protein expression was induced by 0.5 mM IPTG, and the cells were grown for an additional 6 h at 30 °C. Finally, the protein was eluted from the Ni-NTA resin by elution buffer (25mM HEPES, 100mM KCl with 250mM imidazole, pH 7.4).
The cysteine mutants of soluble VAMP2 and complexin I were reacted with (1-oxyl- 2,2,5,5-tetramethylpyrrolinyl-3-methyl) methanethiosulfonate (MTSSL) spin label at 4°C overnight while the proteins were bound to the GST-agarose beads. To remove free spin label, the beads with bound proteins were extensively washed with the cleavage buffer then cleaved by thrombin (Sigma). The spin-labeling efficiency was determined by comparing the spin concentrations with the 50 µM 2,2,6,6-tetramethyl-4-piperidine N-oxide (TEMPO) standard. For all samples, the efficiency was ~80%.
Preparation of the complexin-SNARE quaternary complex
Purified His6-tagged SNAP-25 was first added to the Ni-NTA resin solution and nutated for 1 h at room temperature. After washing out the free proteins, purified syntaxin 1A and soluble VAMP2 were mixed with His6-SNAP-25 to form the SNARE complex with the molar ratio of 2:5:1. The mixture was incubated at 4 °C overnight. After washing one time, 2-fold excess of purified complexin I or complexin I Δ26 was added to the solution and incubated at 4 °C overnight again. After extensive washing to remove the unbound proteins, the complexin-SNARE quaternary complex was eluted with a buffer containing 250 mM imidazole and 1% n-octyl-glucoside (OG). The formation of the complex was confirmed with SDS-PAGE gel. We obtained identical results when the quaternary complex was formed by adding all four components simultaneously.
EPR data collection
EPR spectra were obtained using a Bruker ESP 300 spectrometer (Bruker, Germany) equipped with a low noise microwave amplifier and a loop-gap resonator. The modulation amplitude was set at no greater than one-fourth of the line width. Spectra were collected at room temperature in the first-derivative mode with 1mW microwave power.
As for the complexin-SNARE quaternary complex in the detergent (1% OG), the samples were concentrated to the final concentration of 50–100 µM using a 5-kDa cutoff centrifugal filter (Millipore) before EPR spectra collection. For the purified complex in the membrane, the samples were reconstituted into vesicles at about 1:200 protein-to-lipid molar ratio using the published method.42 The large unilamellar vesicles (~100 nm in diameter) of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine (POPC) containing 15% 1,2-dioleoyl-sn-glycero-3-phosphatidylserine (DOPS) were prepared in a detergent-free buffer using an extruder (Avanti). The detergent was removed by treating the sample with the dialysis buffer (25 mM HEPES, 100mM KCl, pH 7.4) at 4 °C overnight. After dialysis, the sample solution was concentrated using a 100-kDa cutoff centrifugal filter (Millipore) before taking an EPR spectrum.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health grant GM51290 to Y.-K.S.
Abbreviations used
- SNARE
soluble NSF attachment protein receptor
- NSF
N-ethylmaleimide-sensitive factor
- SNAP-25
synaptosome-associated protein of 25 kDa
- t-SNARE
target membrane SNARE
- v-SNARE
vesicle-associated SNARE
- VAMP2
vesicle-associated membrane protein 2
- Ni-NTA
nickel-nitrilotriacetic acid
- MTSSL
(1-oxyl-2,2,5,5-tetramethyl-D-pyrroline-3-methyl) methanethiosulfonate
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine
- DOPS
1,2-dioleoyl-sn-glycero-3-phosphatidylserine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sollner T, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman JE. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- 2.Weber T, Zemelman BV, McNew JA, Westermann B, Gmachl M, Parlati F, Sollner TH, Rothman JE. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 3.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 4.Poirier MA, Xiao W, Macosko JC, Chan C, Shin YK, Bennett MK. The synaptic SNARE complex is a parallel four-stranded helical bundle. Nat Struct Biol. 1998;5:765–769. doi: 10.1038/1799. [DOI] [PubMed] [Google Scholar]
- 5.Jahn R, Scheller RH. SNAREs--engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 6.Jahn R, Lang T, Sudhof TC. Membrane fusion. Cell. 2003;112:519–533. doi: 10.1016/s0092-8674(03)00112-0. [DOI] [PubMed] [Google Scholar]
- 7.Yoon TY, Shin YK. Progress in understanding the neuronal SNARE function and its regulation. Cell Mol Life Sci. 2009;66:460–469. doi: 10.1007/s00018-008-8372-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandez-Chacon R, Konigstorfer A, Gerber SH, Garcia J, Matos MF, Stevens CF, Brose N, Rizo J, Rosenmund C, Sudhof TC. Synaptotagmin I functions as a calcium regulator of release probability. Nature. 2001;410:41–49. doi: 10.1038/35065004. [DOI] [PubMed] [Google Scholar]
- 9.Chapman ER. Synaptotagmin: a Ca(2+) sensor that triggers exocytosis? Nat Rev Mol Cell Biol. 2002;3:498–508. doi: 10.1038/nrm855. [DOI] [PubMed] [Google Scholar]
- 10.Koh TW, Bellen HJ. Synaptotagmin I, a Ca2+ sensor for neurotransmitter release. Trends Neurosci. 2003;26:413–422. doi: 10.1016/S0166-2236(03)00195-4. [DOI] [PubMed] [Google Scholar]
- 11.Saraswati S, Adolfsen B, Littleton JT. Characterization of the role of the Synaptotagmin family as calcium sensors in facilitation and asynchronous neurotransmitter release. Proc Natl Acad Sci U S A. 2007;104:14122–14127. doi: 10.1073/pnas.0706711104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perin MS, Fried VA, Mignery GA, Jahn R, Sudhof TC. Phospholipid binding by a synaptic vesicle protein homologous to the regulatory region of protein kinase C. Nature. 1990;345:260–263. doi: 10.1038/345260a0. [DOI] [PubMed] [Google Scholar]
- 13.Petrenko AG, Perin MS, Davletov BA, Ushkaryov YA, Geppert M, Sudhof TC. Binding of synaptotagmin to the alpha-latrotoxin receptor implicates both in synaptic vesicle exocytosis. Nature. 1991;353:65–68. doi: 10.1038/353065a0. [DOI] [PubMed] [Google Scholar]
- 14.Davletov BA, Sudhof TC. A single C2 domain from synaptotagmin I is sufficient for high affinity Ca2+/phospholipid binding. J Biol Chem. 1993;268:26386–26390. [PubMed] [Google Scholar]
- 15.Chen YA, Scales SJ, Patel SM, Doung YC, Scheller RH. SNARE complex formation is triggered by Ca2+ and drives membrane fusion. Cell. 1999;97:165–174. doi: 10.1016/s0092-8674(00)80727-8. [DOI] [PubMed] [Google Scholar]
- 16.Dai H, Shen N, Arac D, Rizo J. A quaternary SNARE-synaptotagmin-Ca2+-phospholipid complex in neurotransmitter release. J Mol Biol. 2007;367:848–863. doi: 10.1016/j.jmb.2007.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tucker WC, Weber T, Chapman ER. Reconstitution of Ca2+-regulated membrane fusion by synaptotagmin and SNAREs. Science. 2004;304:435–438. doi: 10.1126/science.1097196. [DOI] [PubMed] [Google Scholar]
- 18.Wang CT, Bai J, Chang PY, Chapman ER, Jackson MB. Synaptotagmin-Ca2+ triggers two sequential steps in regulated exocytosis in rat PC12 cells: fusion pore opening and fusion pore dilation. J Physiol. 2006;570:295–307. doi: 10.1113/jphysiol.2005.097378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reim K, Mansour M, Varoqueaux F, McMahon HT, Sudhof TC, Brose N, Rosenmund C. Complexins regulate a late step in Ca2+-dependent neurotransmitter release. Cell. 2001;104:71–81. doi: 10.1016/s0092-8674(01)00192-1. [DOI] [PubMed] [Google Scholar]
- 20.Yoon TY, Lu X, Diao J, Lee SM, Ha T, Shin YK. Complexin and Ca2+ stimulate SNARE-mediated membrane fusion. Nat Struct Mol Biol. 2008;15:707–713. doi: 10.1038/nsmb.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang J, Maximov A, Shin OH, Dai H, Rizo J, Sudhof TC. A complexin/synaptotagmin 1 switch controls fast synaptic vesicle exocytosis. Cell. 2006;126:1175–1187. doi: 10.1016/j.cell.2006.08.030. [DOI] [PubMed] [Google Scholar]
- 22.Huntwork S, Littleton JT. A complexin fusion clamp regulates spontaneous neurotransmitter release and synaptic growth. Nat Neurosci. 2007;10:1235–1237. doi: 10.1038/nn1980. [DOI] [PubMed] [Google Scholar]
- 23.McMahon HT, Missler M, Li C, Sudhof TC. Complexins: cytosolic proteins that regulate SNAP receptor function. Cell. 1995;83:111–119. doi: 10.1016/0092-8674(95)90239-2. [DOI] [PubMed] [Google Scholar]
- 24.Ishizuka T, Saisu H, Odani S, Abe T. Synaphin: a protein associated with the docking/fusion complex in presynaptic terminals. Biochem Biophys Res Commun. 1995;213:1107–1114. doi: 10.1006/bbrc.1995.2241. [DOI] [PubMed] [Google Scholar]
- 25.Reim K, Wegmeyer H, Brandstatter JH, Xue M, Rosenmund C, Dresbach T, Hofmann K, Brose N. Structurally and functionally unique complexins at retinal ribbon synapses. J Cell Biol. 2005;169:669–680. doi: 10.1083/jcb.200502115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pabst S, Margittai M, Vainius D, Langen R, Jahn R, Fasshauer D. Rapid and selective binding to the synaptic SNARE complex suggests a modulatory role of complexins in neuroexocytosis. J Biol Chem. 2002;277:7838–7848. doi: 10.1074/jbc.M109507200. [DOI] [PubMed] [Google Scholar]
- 27.Chen X, Tomchick DR, Kovrigin E, Arac D, Machius M, Sudhof TC, Rizo J. Three-dimensional structure of the complexin/SNARE complex. Neuron. 2002;33:397–409. doi: 10.1016/s0896-6273(02)00583-4. [DOI] [PubMed] [Google Scholar]
- 28.Xue M, Reim K, Chen X, Chao HT, Deng H, Rizo J, Brose N, Rosenmund C. Distinct domains of complexin I differentially regulate neurotransmitter release. Nat Struct Mol Biol. 2007;14:949–958. doi: 10.1038/nsmb1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maximov A, Tang J, Yang X, Pang ZP, Sudhof TC. Complexin controls the force transfer from SNARE complexes to membranes in fusion. Science. 2009;323:516–521. doi: 10.1126/science.1166505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Archer DA, Graham ME, Burgoyne RD. Complexin regulates the closure of the fusion pore during regulated vesicle exocytosis. J Biol Chem. 2002;277:18249–18252. doi: 10.1074/jbc.C200166200. [DOI] [PubMed] [Google Scholar]
- 31.Itakura M, Misawa H, Sekiguchi M, Takahashi S, Takahashi M. Transfection analysis of functional roles of complexin I and II in the exocytosis of two different types of secretory vesicles. Biochem Biophys Res Commun. 1999;265:691–696. doi: 10.1006/bbrc.1999.1756. [DOI] [PubMed] [Google Scholar]
- 32.Schaub JR, Lu X, Doneske B, Shin YK, McNew JA. Hemifusion arrest by complexin is relieved by Ca2+-synaptotagmin I. Nat Struct Mol Biol. 2006;13:748–750. doi: 10.1038/nsmb1124. [DOI] [PubMed] [Google Scholar]
- 33.Giraudo CG, Eng WS, Melia TJ, Rothman JE. A clamping mechanism involved in SNARE-dependent exocytosis. Science. 2006;313:676–680. doi: 10.1126/science.1129450. [DOI] [PubMed] [Google Scholar]
- 34.Giraudo CG, Garcia-Diaz A, Eng WS, Chen Y, Hendrickson WA, Melia TJ, Rothman JE. Alternative zippering as an on-off switch for SNARE-mediated fusion. Science. 2009;323:512–516. doi: 10.1126/science.1166500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melia TJ., Jr Putting the clamps on membrane fusion: how complexin sets the stage for calcium-mediated exocytosis. FEBS Lett. 2007;581:2131–2139. doi: 10.1016/j.febslet.2007.02.066. [DOI] [PubMed] [Google Scholar]
- 36.Xue M, Stradomska A, Chen H, Brose N, Zhang W, Rosenmund C, Reim K. Complexins facilitate neurotransmitter release at excitatory and inhibitory synapses in mammalian central nervous system. Proc Natl Acad Sci U S A. 2008;105:7875–7880. doi: 10.1073/pnas.0803012105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giraudo CG, Garcia-Diaz A, Eng WS, Yamamoto A, Melia TJ, Rothman JE. Distinct domains of complexins bind SNARE complexes and clamp fusion in vitro. J Biol Chem. 2008;283:21211–21219. doi: 10.1074/jbc.M803478200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McHaourab HS, Lietzow MA, Hideg K, Hubbell WL. Motion of spin-labeled side chains in T4 lysozyme. Correlation with protein structure and dynamics. Biochemistry. 1996;35:7692–7704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- 39.Kweon DH, Kim CS, Shin YK. Regulation of neuronal SNARE assembly by the membrane. Nat Struct Biol. 2003;10:440–447. doi: 10.1038/nsb928. [DOI] [PubMed] [Google Scholar]
- 40.Weninger K, Bowen ME, Choi UB, Chu S, Brunger AT. Accessory proteins stabilize the acceptor complex for synaptobrevin, the 1:1 syntaxin/SNAP-25 complex. Structure. 2008;16:308–320. doi: 10.1016/j.str.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Su Z, Zhang F, Chen Y, Shin YK. A partially zipped SNARE complex stabilized by the membrane. J Biol Chem. 2005;280:15595–15600. doi: 10.1074/jbc.M500736200. [DOI] [PubMed] [Google Scholar]
- 42.Chen Y, Xu Y, Zhang F, Shin YK. Constitutive versus regulated SNARE assembly: a structural basis. Embo J. 2004;23:681–689. doi: 10.1038/sj.emboj.7600083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.