Abstract
More than 40 species of primates and over 20 species of cats harbor antibodies that sero-react to lentiviral antigens. In nearly all cases where viral genetic analysis has been conducted, each host species is infected with a unique lentivirus. Though lentivirus clades within a species can be substantially divergent, they are typically monophyletic within that species. A notable significant departure from this observation is apparent cross-species transmission of FIV between bobcats (Lynx rufus) and pumas (Puma concolor) in southern California that has occurred at least three times; evidence from one bobcat sequence suggests this cross-over may have also occurred in Florida between bobcats and the endangered Florida panther. Several other isolated reports demonstrate cross-species transmission of FIV isolates among captive animals housed in close proximity, and it is well established that HIV-1 and HIV-2 arose from human contact with SIV-infected nonhuman primates. Using an experimental model, we have determined that domestic cats (Felis catus) are susceptible to FIVs originating from pumas or lions. While infections are initially replicative, and animals seroconvert, within a relatively short period of time circulating virus is reduced to nearly undetectable levels in a majority of animals. This diminution of viral load is proportional to initial viral peak. Although viral reservoirs can be identified in gastrointestinal tissues, most viral genomes recovered peripherally are highly mutated, suggesting that the non-adapted host successfully inhibits normal viral replication, leading to replication incompetent viral progeny. Mechanisms possible for such restriction of cross-species infections in natural settings include: 1. Lack of contact conducive to lentiviral transmission between infected and shedding animals of different species; 2. Lack of suitable receptor repertoire to allow viral entry to susceptible cells of a new species; 3. Cellular machinery in the new host sufficiently divergent from the primary host to support viral replication (ie passive unfacilitated viral replication); 4. Intracellular restriction mechanisms present in the new host that is able to limit viral replication (i.e. active interrupted viral replication. These include factors that limit uncoating, replication, packaging, and virion release); 5. Unique ability of new host to raise sterilizing adaptive immunity, resulting in aborted infection and inability to spread infections among con-specifics; or, 6. Production of defective or non-infectious viral progeny that lack cellular cofactors to render them infectious to conspecifics (i.e. particles lacking appropriate cellular components in viral Env to render them infectious to other animals of the same species). Data to support or refute the relative importance of each of these possibilities is described in this review. Insights based on our in vivo cross-species model suggest intracellular restriction mechanisms effectively inhibit rapid inter-specific transmission of lentiviruses. Further, limited contact both within and between species in natural populations is highly relevant to limiting the opportunity for spread of FIV strains. Studies of naturally-occurring SIV and innate host restriction systems suggest these same two mechanisms are significant factors inhibiting widespread cross-species transmission of lentiviruses among primate species as well.
Keywords: FIV, cross-species infection, feline, lentivirus, pathogenesis
Introduction
FIV infection has many analogies to HIV infection; the virus has emerged relatively recently, targets similar cell types, and the disease state demonstrates a similar time course, clinical signs and outcome (Bendinelli et al., 1995; Burkhard and Dean, 2003; Dias et al., 2006; Dunham, 2006; Kanzaki and Looney, 2004; Siebelink et al., 1990). Concurrently, FIVs which are endemic to the non-domestic cat population parallel SIVagm and other naturally occurring SIVs in that they are minimally pathogenic in their natural host (VandeWoude and Apetrei, 2006). Species-specificity of lentiviral infection, viral and host factors leading to establishment of naturally-occurring virulent versus avirulent infections, and emergence of new strains are areas with high relevance for understanding the current HIV epidemic. Studies of FIV have contributed to elucidating possible mechanisms for restriction of cross-species infections in natural settings as described below.
Lack of contact conducive to lentiviral transmission between infected and shedding animals of different species
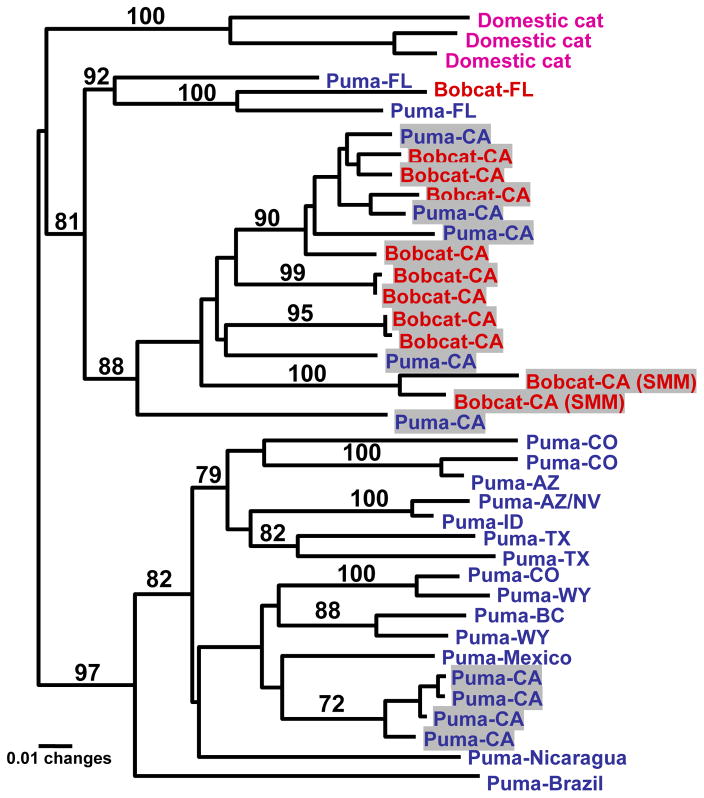
Lentiviruses that naturally occur in pumas (Puma concolor, also known as the cougar, mountain lion, Florida panther) are among the most widely studied in nondomestic feline species. Several large surveys of conserved regions of the puma lentivirus genome (FIVPco ) identify two unique viral clades. The most common, FIVPco clade B, is detected in hundreds of isolates ranging throughout North and South America and is highly diverse, whereas FIVPco clade A was originally identified as a divergent strain found only in Florida panthers, who did not seem to harbor clade B, and in one animal in California (Biek et al., 2003; Brown et al., 1994; Carpenter et al., 1996; Troyer et al., 2005). Lentiviral proviral sequences isolated from blood cells of bobcats from Southern California demonstrate strong phylogenic identity to FIVPco clade A in a 500-bp region of pol. All bobcat FIV (FIVLru) sequences fell within this clade, along with 5 of 12 proviral sequences isolated from pumas in three geographically distinct populations from Southern California (Fig. 1). An historical bobcat FIV sequence obtained from an animal captured in Florida in 1984 also clustered with FIVPco clade A sequences. Interestingly, FIVPco clade A of bobcats and pumas clustered most closely when geographic proximity of the hosts were considered—in other words, FIVPco clade A sequences isolated from bobcats and pumas north of Los Angeles (Ventura County) were more closely related than FIVPco Clade A isolates from pumas and bobcats south of Los Angeles, and the bobcat isolate from Florida was more closely related to Florida panther FIVPco clade A than to bobcat isolates from California (Franklin et al., 2007). Seven of twelve pumas from California harbored FIVPco Clade B that also clustered based upon geographic proximity of hosts; to date no bobcat has harbored an FIVPco Clade B isolate. These findings strongly suggest that FIV has been transmitted between bobcats and pumas in Southern California and in Florida on at least three occasions, likely in the direction of transmission from bobcats to pumas.
Figure 1.
Unrooted maximum likelihood (ML) tree of a 500bp region in polRT demonstrating the relationship between FIV sequences from pumas and bobcats. Minimum evolution (ME), maximum parsimony (MP), and Bayesian analyses (random starting trees; burn-in values set at 45,000 generations; two simultaneous runs with four Markov chains each were run for one million generations and sampled every 20 generations) produced trees with similar topologies. Bootstrap values and posterior probabilities are included for nodes discussed in the text (ML/MP/ME/Bayesian). Colors represent the species of origin (blue=bobcat, red=puma). Viral sequences isolated from animals in California (CA) are highlighted and each sequence is identified by a location consisting of a country name or a state abbreviation followed by a specific site in parentheses for the CA samples (OC=Orange County, VC=Ventura County, SDRC=San Diego and Riverside Counties). Analyses used empirical base frequencies, an estimated shape parameter of 0.7495, and an estimated substitution matrix as follows: A/C = 2.8733, A/G = 12.0716, A/T = 0.8769, C/G = 4.8937, C/T = 16.8913, and an estimated proportion of invariant sites of 0.3419.
Why this phenomenon has been observed in isolated geographic regions when habitat overlap between pumas and bobcats occurs across most of western North America and in Florida is unknown. One possibility is that interactions between these species are heightened in Southern California and Florida. This is plausible in that both of these regions have been under intense development pressure over the last century, which has had substantial impact on habitat connectivity for species with large home range requirements (Dobson et al., 1997; Harris and Atkins, 1991; Hoctor et al., 2000; Myers, 1990). Competition between these two species for remaining resources may result in increased inter-specific interactions leading to increased opportunities for transmission. Another contributing factor is that, at least in Southern California, bobcat FIVLru infection rates appear to be significantly higher than other cohorts whereby nearly 60% of bobcats in Orange County and 30% of animals from Ventura County are seropositive (Franklin et al., 2007). In contrast, seroprevalence reported in other bobcat populations are less than 10% (Barr et al., 1989) (Olmsted et al., 1992), including a report of 0% in a population of 25 animals from Northern California (Riley et al., 2004). It is plausible that habitat degradation has therefore also lead to increased intra-specific interactions among bobcats in Southern California, resulting in higher local rates of FIVLru, which therefore may be more likely to be transmitted to pumas in disrupted patches of habitat.
While the example presented above document cross-species transmission events between bobcats and pumas, several other reports suggests that sustained contact between felid species, typically in a zoo setting, may result in cross-species transmission of FIV (Table 1). This includes domestic cat FIVFca infection of a captive puma in an Argentinian zoo (Carpenter et al., 1996) and in a Tsushima Japan leopard cat (Nishimura et al., 1999), and African lion (Panthera leo) FIV infection (FIVPle) in a captive tiger and snow leopard in Asian zoos (Troyer et al., 2005).
Table 1.
Naturally occurring lentiviral transmissions between adapted and nonadapted/new species reported in the literature.
| “Adapted” Host | Nonadapted/New Host | Lentivirus Transmitted | Comment and Citation |
|---|---|---|---|
| Bobcat, Lynx rufus | Puma, Puma concolor | FIVPco Clade A, (FIVLru) | It is hypothesized that the route of transmission is from bobcat to puma, though this has not been definitively proven. (Franklin et al., 2007) |
| Domestic cat, Felis catus | Puma, Puma concolor | FIVFca (domestic cat FIV) | Recorded in one captive puma housed in a zoo in Argentina. (Carpenter et al., 1996) |
| Domestic cat, Felis catus | Tsushima leopard cat, Felis bengalensis euptilura | FIVFca (domestic cat FIV) | (Nishimura et al., 1999) |
| African lion, Panthera leo | Tiger, (Panthera tigris) Snow Leopard, (Uncia uncia) | FIVPle (African lion FIV) | All three animals (lion, tiger and snow leopard) were in the same zoo in Asia. (Troyer et al., 2005) |
| Sheep, Ovis aries | Goat, Capra hirus | Maedi-Visna Virus; Caprine Arthritis Encephalitis | (Pisoni et al., 2007; Ravazzolo et al., 2001; Rolland et al., 2002; Zanoni, 1998) |
| Chimpanzee, Pan troglodytes | Human, Homo sapiens | SIVCpz; HIV-1 | (Gao et al., 1999; Hahn et al., 2000); for review see (VandeWoude and Apetrei, 2006) |
| Sooty mangabey, Cercocebus atys | Human, Homo sapiens | SIVSmm; HIV-2 | (Chen et al., 1997); for review see (VandeWoude and Apetrei, 2006) |
Other instances of enhanced inter-specific contacts resulting in lentiviral cross species transmission have been noted to occur in ruminants and primates. Sheep and goats are each infected with lentiviruses, Maedi Visna Virus (MVV), and Caprine Arthritis Encephalitis Virus (CAEV). Serologic and genetic evidence suggests that transmission of species-specific strains has occurred on multiple occasions under natural conditions (Table 1). Dual infection of goats from a large herd were naturally infected with both MVV and CAEV (Pisoni et al., 2007). Control measures were not taken to prevent transmission of lentiviral infections in this herd, and it is likely high population density and management practices contributed to cross-species transmissions. Both of the contemporary human lentivirus infections, HIV-1 and HIV-2, arose following cross-species infection events (Apetrei et al., 2004; Hahn et al., 2000; Korber et al., 2000). Genetic diversity within circulating strains of HIV-1, and to a lesser extent HIV-2, suggest several cross-species transmission events from the respective primate hosts, the chimpanzee (Pan troglodytes), and sooty mangebey (Cercocebus atys) (Chen et al., 1997; Gao et al., 1999; Hahn et al., 2000). It has been postulated that the emergence of HIV-1 occurred as a result of convergence of anthropological factors including: 1. Increased human populations in areas inhabited by infected chimpanzees in the Democratic Republic of Congo, associated with development of commerce in rubber trade, and the general explosive population growth in this region; 2. The prevalence of bushmeat as an alternative food source and the availability of guns to facilitate ape capture; and, 3. Enhanced viral adaptation and accelerated spread during vaccine campaigns that may have exposed otherwise low risk individuals via use of practices such as re-used needles (Chitnis et al., 2000) (Drucker et al., 2001; Hahn et al., 2000; Korber et al., 2000).
These observations suggest that opportunities for cross-species transmission of lentiviruses are very likely enhanced by environmental and anthropogenic influences, and in instances when such transmissions have occurred, it is useful to evaluate population structure and ecological influences to determine underlying predisposing factors for development of new epidemics.
Lack of suitable receptor repertoire to allow viral entry into susceptible cells of a new species
Though the previous section outlines instances where lentiviral cross-species transmissions have occurred successfully, such observations are rare relative to the number of inter-species exposures that are likely to occur. Epidemiologic surveys of 1800 persons from nine villages in Cameroon suggested very high (>60%) exposure to primate blood and body fluids and demonstrated that 1% of exposed individuals were seropositive for Simian Foamy Virus of three different nonhuman primate origins (Wolfe et al., 2004). Despite the fact that these events clearly demonstrate that human-primate contact occurs commonly, and can result in primate to human retroviral transmissions, human exposure to SIVs resulting in patent infections has been extremely rare. Therefore, exposure of humans to SIVs does not a priori result in successful cross-species infection as would be the case for a classic zoonotic disease transmission such as rabies (Apetrei and Marx, 2004). Mechanisms underlying the low cross-species transmission rate are explored further below.
An initial barrier that must be overcome when an individual is exposed to a mature lentiviral particle originating from a different host species is efficient Env binding to the cellular receptor of a new host. While FIVs of lion and puma origin can productively infect domestic cats (Terwee et al., 2008; Terwee et al., 2005; VandeWoude et al., 2003; VandeWoude et al., 1997a), or domestic cat cells (Brown et al., 1994; VandeWoude et al., 1997b), receptor blocking studies have indicated that different receptor repertoires are used by FIVPco clade B and FIVPle Clade B viruses (Smirnova et al., 2005). Lion subtypes B and E are monophyletic across all gene regions except env, where the SU and TM regions vary substantially (Pecon-Slattery et al., 2008). Subtype E env is somewhat more related to domestic cat sequences than to lion subtype B, likely reflecting a recombination event during superinfection with two FIV strains of highly divergent origin resulting in a mosaic virus able to propagate successfully in lion cells (Pecon-Slattery et al., 2008). The recombination event leading to subtype heterogeneity has resulted in divergent receptor usage between clades; clade E subtype uses a CD134/CXCR4 receptor repertoire, similar to domestic cat FIV, whereas as noted above, subtype B uses an as yet undefined cellular entry mechanism (McEwan et al., 2008).
Primate lentiviruses primarily bind CD4 via gp120, triggering conformational rearrangements that allow binding to a co-receptor, typically one of the seven-transmembrane chemokine receptors CCR5 or CXCR4. It is well-established that initial HIV infection occurs via CCR5 binding, but that end-stage isolates are more likely to use CXCR4 as a co-receptor. SIVs typically utilize CCR5 binding, and correlates between CCR5 expression on CD4+T cells suggest that expression patterns of CCR5 on these cells can influence the pathogenicity of SIV for the host. Certain HIV and SIV isolates, particularly those isolated from non-lymphoid tissues, or host-adapted strains, may lose the requirement for either primary or co-receptor presence to infect tissues (Reeves et al., 1999; Willey et al., 2003). This is also true for FIV (Elder et al., 2008; Willett and Hosie, 2008), though FIV strain pathogenicity is not clearly related to receptor binding affinity (de Rozieres et al., 2008). The apparent high rate of adaptation of lentiviruses to utilize alternative receptor repertoires depending upon host presentation suggest that discordant virus-receptor interactions are relatively easily overcome, and that this factor is likely not a primary determinant limiting cross-species transmission.
Cellular machinery in the new host sufficiently divergent from the primary host to support viral replication (ie passive unfacilitated viral replication)
While no published studies have reported that species-specificity of host cellular enzymes limit lentiviral replication, it is possible that this factor may limit completion of the viral lifecycle within a new host. This phenomenon has been most well studied in the family Polyomaviridae. SV40 viral replication occurs efficiently in vitro in the presence of human, but not murine, DNA polymerase α-primase, as the human enzyme is specifically required for initiation complex formation (Schneider et al., 1994; Stadlbauer et al., 1996); (Fanning and Zhoa, 2009; Pipas, 2009; Smith and Nasheuer, 2003). The fact that lentiviral integration, LTR-mediated gene transcription, viral transport and assembly are all influenced or mediated by cellular factors suggests that host- and cell-specific expression patterns or functional differences between these enzymes in different species could substantially impact the rate and success of viral replication. However, differences in the structure and function of essential cellular machinery between cat species are likely to be minimal, since the order Felidae represents a recent and rapid evolutionary radiation (on the order of 10 MYA in contrast to the 80 million years that separate mice and men; Johnson et al., 2006). Highly conserved genes involved in cell maintenance are unlikely candidates for species-specific evolution on such a short timescale without extreme selective pressure. In contrast, cell factors involved in viral restriction and innate and adaptive immunity may evolve much more rapidly resulting in species-specific restriction.
Intracellular restriction mechanisms present in the new host that is able to limit viral replication (ie active interrupted viral replication)
Apolipoprotein B mRNA-editing enzyme catalytic polypeptide (APOBEC) has been identified as a restriction factor limiting lentiviral replication that can be overcome by lentiviral vif in a species-specific manner (see review Munk et al this issue). The cytidine deamination activity of some APOBEC subtypes (APOBEC-3G in primates) results in C to U editing during single stranded DNA synthesis, resulting in G to A mutations in the positive sense DNA viral genome (Huthoff and Towers, 2008). Feline analogues of the APOBEC family have been identified in the cat (Lochelt et al., 2005; Munk et al., 2008).
Our previous work indicates that infection of domestic cats with a lentivirus native to the puma (FIVPco Strain PLV-1695, subsequently referred to as PLV) results in mild lymphadenopathy and only a slight, transient decrease in CD4+ T-cell count. PBMC viral burden gradually decreases over the course of several months of infection and is ultimately virtually eliminated by the host without evidence of adaptive immune activation that procedes viral diminution. In late infection, proviral burden persisted in gastro-intestinal versus lymphoid compartments (Terwee et al., 2005), and viral replication was not enhanced following immunosuppressive doses of corticosteroids. Full length proviral genome sequences recovered from circulating PBMC at various times post-infection revealed increased frequency of PLV G-to-A substitutions and accumulation of defective genomes following replication in a domestic cat host. Infection persisted despite the high frequency of errors in the viral genome. These findings provided evidence for DNA editing by a host cytidine deaminase (Poss et al., 2006). However, infection persisted despite the high frequency of errors in the viral genome. Five sites in PLV RT-pol were shown to be under positive selection; also significantly, virus recovered from cats that contained one of these selected sites in RT had measurable evolutionary rates and phylogenetic evidence of recombination (Poss et al., 2007). Thus, virus persistence in this cross-species infection correlates with selection on RT and enhanced viral recombination. Studies in primates have demonstrated that cytidine deamination, possibly in conjunction with cytokine activation, restricts nonadapted primate lentiviral replication in vitro, and one recent report documents construction of a ‘simian tropic’ HIV-1 by substitution of SIV vif on an HIV backbone (Hatziioannou et al., 2009).
TRIM5α is an intracellular factor that interferes with viral uncoating following binding and fusion events in the viral replication cycle in a species-dependent manner. The TRIM protein family consists of members with a “TRIpartate Motif” that are involved in a diverse set of cellar processes ranging from cell proliferation to apoptosis (Huthoff and Towers, 2008; Nisole et al., 2005)(see also Munk et al. this issue). TRIM5α of rhesus was shown to inhibit HIV-1 replication, and subsequently was shown to be equivalent to previously described Fv1 of mice and REF1 of humans—host genes that were associated with species-specific restriction of retroviral replication. The last intron of TRIM5α has been substituted with cyclophilin A by retrotransposition in several species, including owl monkeys and old world species. The resulting gene, called TRIMCyp has been shown to inhibit HIV replication, and to restrict viruses whose capsids bind CypA, including FIV (Huthoff and Towers, 2008). The FIV-Gag motif in this binding region is highly conserved among feline lentiviruses, demonstrating that it is a critical region for viral function, and potentially susceptible to TRIMCyp interference (Burkala and Poss, 2007). The mechanism by which TRIM5α inhibits viral replication is not completely understood, though capsid binding and interference with uncoating in the proteosome have been associated with TRIM5α anti-HIV activity. Analysis of gene loci reveal that TRIM5α is a pseudogene in the cat, and the SPRY domain associated with viral restriction in primates is truncated (Troyer et al., 2008). However, it is possible that a different TRIM family protein can restrict retroviral replication as has been determined in cattle. The role of Tripartite Motif proteins in restriction of feline lentiviral replication is therefore currently under investigation (Troyer et al., 2008).
Unique ability of new host to raise sterilizing adaptive immunity, resulting in aborted infection and inability to spread infections among con-specifics
In studies mentioned above, inoculation of cats with a strain of FIV that originated in pumas resulted in aborted infections. While evidence of cytidine deamination in proviral genomes was documented, measures of adaptive immune response (ie development of neutralizing antibodies or cytotoxic T cells) failed to detect protective responses (Terwee et al., 2005).
Co-infection with avirulent and virulent lentiviral strains further illucidates protective immunity developed during avirulent infection. When cats infected with PLV-1695 were challenged with a virulent strain of FIV, co-infection resulted; however, CD4+ T-cell depletion was prevented, and significant differences in cytokine profiles and immunocytes were noted over time (Terwee et al., 2008). By 24 days after FIVFca subtype C infection, the dual infected cats had a unique immunological profile compared to that of the other three groups; this profile was characterized by elevated levels of CD8, CD25, and FAS expressing cells, increased numbers of lymphocytes and neutrophils, and elevated IL4 and IFNγ (Poss, M. L., unpublished). However, evidence of adaptive immune responses (ie neutralizing antibody, cytotoxic T cell responses) against the virulent FIV strain were again lacking. This study demonstrates that co-infection with distinct strains of lentivirus results in dynamic differences in innate immune responses, resulting in distinct disease phenotypes and immunological profiles during dual vs. single infection, but sterilizing adaptive immunity to either strain is lacking.
While many studies have been conducted using an attenuated strain of HIV or SIV to afford protection against virulent SIV in nonhuman primates, the consequences of co-infection are not consistent. Typically, apathogenic infections with SHIV or SIV result in complete to partial protection against virulent strains, but it has been difficult to determine the mechanisms underlying these observations (Dunn et al., 1997; Petry et al., 1995; Putkonen et al., 1995; Stephens et al., 1997). A recent set of experiments have demonstrated that SIV isolated from the red-capped mangabey (SIVRcm), which has limited pathogenicity in rhesus macaques, affords partial protection against superinfection with pathogenic SIVmac251 by undefined mechanisms (Ling et al., 2008). These studies support observations made in feline infections described above, and further dissection of the mechanisms underlying partial protection is warranted.
Production of defective or non-infectious viral progeny that lack cellular cofactors to render them infectious to conspecifics
Retroviral budding from the cell membrane to form intact virions results in the incorporation of host cell membranes into the viral envelope. MHC II may be ‘preferentially’ incorporated into HIV Env during budding as HIV interacts specifically at several points of viral replication with MHC II expression on the cell surface (Saifuddin et al., 2000a; Saifuddin et al., 2000b). Vaccination-induced sterilizing immunity has been achieved in rhesus macaques immunized with whole inactivated SIV (WIV) preparations (Warren and Dolatshahi, 1993); however, the mechanism of neutralization appeared to be antibodies raised against xenogeneic cell membrane proteins incorporated by the challenge SIV stocks grown in human cell culture systems, in particular MHC II antigens (Cranage et al., 1993; Goldstein et al., 1994; Mills et al., 1992; Stott, 1991). Recent reports suggest that the glycosphigolipid composition of HIV-Env can mediate interactions with immature and mature dendritic cell binding (Hatch et al., 2009), also demonstrating that the cell surface phenotype of a virus-producing host cell can influence the subsequent host range of progeny virions.
Other contributions of host cell constituents to lentiviral pathogenicity and/or species-specificity have not been examined, and may play an as yet unrecognized role in lentiviral pathogenicity. These components may be yet another mechanism that limits cross-species transmission of lentiviruses, particularly if highly immunogenic molecules such as MHC II affect the capacity for mature virions to infect susceptible cells.
Conclusions
This review of current literature suggests that the two most important factors resulting in opportunities for lentiviral cross-species transmission are enhanced contact rates between different species, and the ability to overcome intracellular restriction factors limiting viral replication. Viral Env-receptor specificity and the capacity of the new host species to develop a neutralizing adaptive immune response would appear to be less relevant to prevention of successful cross-species transmission events. Viral replication dependent upon host cellular machinery and incorporation of host components into mature virions are potential steps in the lentiviral life cycle that may be limited during cross-species transmission due to functional or structural incompatibilities, but these features have yet to be studied in feline or other lentivirus systems. Progress in understanding features of host-lentiviral adaptation and cross-species restriction has been greatly facilitated by studies of FIV.
Acknowledgments
We thank Sam Franklin for technical assistance in generating FIV sequences used for phylogenic analysis, and to Kevin Crooks, Seth Riley, Walter Boyce, Lisa Lyren, and Ray Langley (posthumously) for providing samples from these animals for genetic analysis. Thanks also to Sandra Quackenbush and Jill Pecon-Slattery for editorial comments and suggestions during the production of this manuscript.
Footnotes
Conflict of Interest
The authors state that here are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Sue VandeWoude, Email: suev@lamar.colostate.edu.
Jennifer Troyer, Email: jtroyer@ncifcrc.gov.
Mary Poss, Email: mposs@bx.psu.edu.
References Cited
- Apetrei C, Marx PA. Simian retroviral infections in human beings. The Lancet. 2004;364:137–138. doi: 10.1016/S0140-6736(04)16620-8. [DOI] [PubMed] [Google Scholar]
- Apetrei C, Robertson DL, Marx PA. The history of SIVS and AIDS: epidemiology, phylogeny and biology of isolates from naturally SIV infected non-human primates (NHP) in Africa. Front Biosci. 2004;9:225–254. doi: 10.2741/1154. [DOI] [PubMed] [Google Scholar]
- Barr MC, Calle PP, Roelke ME, Scott FW. Feline immunodeficiency virus-infection in nondomestic felids. J Zoo and Wildlife Medicine. 1989;20:265–272. [Google Scholar]
- Bendinelli M, Pistello M, Lombardi S, Poli A, Garzelli C, Matteucci D, Ceccherini-Nelli L, Malvaldi G, Tozzini F. Feline immunodeficiency virus: an interesting model for AIDS studies and an important cat pathogen. Clin Microbiol Rev. 1995;8:87–112. doi: 10.1128/cmr.8.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biek R, Rodrigo AG, Holley D, Drummond A, Anderson CR, Jr, Ross HA, Poss M. Epidemiology, genetic diversity, and evolution of endemic feline immunodeficiency virus in a population of wild cougars. J Virol. 2003;77:9578–9589. doi: 10.1128/JVI.77.17.9578-9589.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EW, Yuhki N, Packer C, O’Brien SJ. A lion lentivirus related to feline immunodeficiency virus: epidemiologic and phylogenetic aspects. J Virol. 1994;68:5953–5968. doi: 10.1128/jvi.68.9.5953-5968.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkala E, Poss M. Evolution of feline immunodeficiency virus Gag proteins. Virus Genes. 2007;35:251–264. doi: 10.1007/s11262-006-0058-8. [DOI] [PubMed] [Google Scholar]
- Burkhard MJ, Dean GA. Transmission and immunopathogenesis of FIV in cats as a model for HIV. Curr HIV Res. 2003;1:15–29. doi: 10.2174/1570162033352101. [DOI] [PubMed] [Google Scholar]
- Carpenter MA, Brown EW, Culver M, Johnson WE, Pecon-Slattery J, Brousset D, O’Brien SJ. Genetic and phylogenetic divergence of feline immunodeficiency virus in the puma (Puma concolor) J Virol. 1996;70:6682–6693. doi: 10.1128/jvi.70.10.6682-6693.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Luckay A, Sodora DL, Telfer P, Reed P, Gettie A, Kanu JM, Sadek RF, Yee J, Ho DD, Zhang L, Marx PA. Human immunodeficiency virus type 2 (HIV-2) seroprevalence and characterization of a distinct HIV-2 genetic subtype from the natural range of simian immunodeficiency virus-infected sooty mangabeys. J Virol. 1997;71:3953–3960. doi: 10.1128/jvi.71.5.3953-3960.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitnis A, Rawls D, Moore J. Origin of HIV type 1 in colonial French Equatorial Africa? AIDS Res Hum Retroviruses. 2000;16:5–8. doi: 10.1089/088922200309548. [DOI] [PubMed] [Google Scholar]
- Cranage MP, Polyanskaya N, McBride B. Studies on the specificity of the vaccine effect elicited by inactivated simian immunodeficiency virus. AIDS Res Hum Retroviruses. 1993;9:13–22. doi: 10.1089/aid.1993.9.13. [DOI] [PubMed] [Google Scholar]
- de Rozieres S, Thompson J, Sundstrom M, Gruber J, Stump DS, de Parseval AP, VandeWoude S, Elder JH. Replication properties of clade A/C chimeric feline immunodeficiency viruses and evaluation of infection kinetics in the domestic cat. J Virol. 2008;82:7953–7963. doi: 10.1128/JVI.00337-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias AS, Bester MJ, Britz RF, Apostolides Z. Animal models used for the evaluation of antiretroviral therapies. Curr HIV Res. 2006;4:431–446. doi: 10.2174/157016206778560045. [DOI] [PubMed] [Google Scholar]
- Dobson AP, Rodriguez JP, Roberts WM, Wilcove DS. Geographic distribution of endangered species in the United States. Science. 1997;275:550–553. doi: 10.1126/science.275.5299.550. [DOI] [PubMed] [Google Scholar]
- Drucker E, Alcabes PG, Marx PA. The injection century: massive unsterile injections and the emergence of human pathogens. Lancet. 2001;358:1989–1992. doi: 10.1016/S0140-6736(01)06967-7. [DOI] [PubMed] [Google Scholar]
- Dunham SP. Lessons from the cat: development of vaccines against lentiviruses. Vet Immunol Immunopathol. 2006;112:67–77. doi: 10.1016/j.vetimm.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Dunn CS, Hurtrel B, Beyer C, Gloeckler L, Ledger TN, Moog C, Kieny MP, Mehtali M, Schmitt D, Gut JP, Kirn A, Aubertin AM. Protection of SIVmac-infected macaque monkeys against superinfection by a simian immunodeficiency virus expressing envelope glycoproteins of HIV type 1. AIDS Res Hum Retroviruses. 1997;13:913–922. doi: 10.1089/aid.1997.13.913. [DOI] [PubMed] [Google Scholar]
- Elder JH, Sundstrom M, de Rozieres S, de Parseval A, Grant CK, Lin YC. Molecular mechanisms of FIV infection. Vet Immunol Immunopathol. 2008;123:3–13. doi: 10.1016/j.vetimm.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning E, Zhoa K. SV40 DNA replication: From the A gene to a nanomachine. Virology. 2009;384:352–359. doi: 10.1016/j.virol.2008.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin SP, Troyer JL, Terwee JA, Lyren LM, Boyce WM, Riley SP, Roelke ME, Crooks KR, Vandewoude S. Frequent transmission of immunodeficiency viruses among bobcats and pumas. J Virol. 2007;81:10961–10969. doi: 10.1128/JVI.00997-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Bailes E, Robertson DL, Chen Y, Rodenburg CM, Michael SF, Cummins LB, Arthur LO, Peeters M, Shaw GM, Sharp PM, Hahn BH. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature. 1999;397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- Goldstein S, Elkins WR, London WT, et al. Immunization with whole inactivated vaccine protects from infection by SIV grown in human but not macaque cells. J Med Primatol. 1994;23:75–82. doi: 10.1111/j.1600-0684.1994.tb00105.x. [DOI] [PubMed] [Google Scholar]
- Hahn BH, Shaw GM, De Cock KM, Sharp PM. AIDS as a zoonosis: scientific and public health implications. Science. 2000;287:607–614. doi: 10.1126/science.287.5453.607. [DOI] [PubMed] [Google Scholar]
- Harris LD, Atkins A. Faunal movement corridors in Florida. Island Press and Defenders of Wildlife; Washington D.C: 1991. [Google Scholar]
- Hatch SC, Archer J, Gummuluru S. Glycosphingolipid composition of human immunodeficiency virus type 1 (HIV-1) particles is a crucial determinant for dendritic cell-mediated HIV-1 trans-infection. J Virol. 2009;83:3496–3506. doi: 10.1128/JVI.02249-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatziioannou T, Ambrose Z, Chung NP, Piatak M, Jr, Yuan F, Trubey CM, Coalter V, Kiser R, Schneider D, Smedley J, Pung R, Gathuka M, Estes JD, Veazey RS, KewalRamani VN, Lifson JD, Bieniasz PD. A macaque model of HIV-1 infection. Proc Natl Acad Sci U S A. 2009;106:4425–4429. doi: 10.1073/pnas.0812587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoctor TS, Carr MH, Zwick PD. Identifying a linked reserve system using a regional landscape approach: The Florida ecological network. Conservation Biology. 2000;14:984–1000. [Google Scholar]
- Huthoff H, Towers GJ. Restriction of retroviral replication by APOBEC3G/F and TRIM5alpha. Trends Microbiol. 2008;16:612–619. doi: 10.1016/j.tim.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Eizirik E, Pecon-Slattery J, Murphy WJ, Antunes A, Teeling E, O’Brien SJ. The late Miocene radiation of modern Felidae: a genetic assessment. Science. 2006;311:73–77. doi: 10.1126/science.1122277. [DOI] [PubMed] [Google Scholar]
- Kanzaki LI, Looney DJ. Feline immunodeficiency virus: a concise review. Front Biosci. 2004;9:370–377. doi: 10.2741/1235. [DOI] [PubMed] [Google Scholar]
- Korber B, Muldoon M, Theiler J, Gao F, Gupta R, Lapedes A, Hahn BH, Wolinsky S, Bhattacharya T. Timing the ancestor of the HIV-1 pandemic strains. Science. 2000;288:1789–1796. doi: 10.1126/science.288.5472.1789. [DOI] [PubMed] [Google Scholar]
- Ling B, Veazey RS, Marx PA. Nonpathogenic CCR2-tropic SIVrcm after serial passage and its effect on SIVmac infection of Indian rhesus macaques. Virology. 2008;379:38–44. doi: 10.1016/j.virol.2008.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochelt M, Romen F, Bastone P, Muckenfuss H, Kirchner N, Kim YB, Truyen U, Rosler U, Battenberg M, Saib A, Flory E, Cichutek K, Munk C. The antiretroviral activity of APOBEC3 is inhibited by the foamy virus accessory Bet protein. Proc Natl Acad Sci U S A. 2005;102:7982–7987. doi: 10.1073/pnas.0501445102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan WA, McMonagle EL, Logan N, Serra RC, Kat P, Vandewoude S, Hosie MJ, Willett BJ. Genetically divergent strains of feline immunodeficiency virus from the domestic cat (Felis catus) and the African lion (Panthera leo) share usage of CD134 and CXCR4 as entry receptors. J Virol. 2008;82:10953–10958. doi: 10.1128/JVI.01312-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills KH, Page M, Chan WL, et al. Protection against SIV infection in macaques by immunization with inactivated virus from the BK28 molecular clone, but not with BK28-derived recombinant env and gag proteins. J Med Primatol. 1992;21:50–58. [PubMed] [Google Scholar]
- Munk C, Beck T, Zielonka J, Hotz-Wagenblatt A, Chareza S, Battenberg M, Thielebein J, Cichutek K, Bravo IG, O’Brien SJ, Lochelt M, Yuhki N. Functions, structure, and read-through alternative splicing of feline APOBEC3 genes. Genome Biol. 2008;9:R48. doi: 10.1186/gb-2008-9-3-r48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers N. The biodiversity challenge: expanded hot-spots analysis. The Environmentalist. 1990;10:243–256. doi: 10.1007/BF02239720. [DOI] [PubMed] [Google Scholar]
- Nishimura Y, Goto Y, Yoneda K, Endo Y, Mizuno T, Hamachi M, Maruyama H, Kinoshita H, Koga S, Komori M, Fushuku S, Ushinohama K, Akuzawa M, Watari T, Hasegawa A, Tsujimoto H. Interspecies transmission of feline immunodeficiency virus from the domestic cat to the Tsushima cat (Felis bengalensis euptilura) in the wild. J Virol. 1999;73:7916–7921. doi: 10.1128/jvi.73.9.7916-7921.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisole S, Stoye JP, Saib A. TRIM family proteins: retroviral restriction and antiviral defence. Nat Rev Microbiol. 2005;3:799–808. doi: 10.1038/nrmicro1248. [DOI] [PubMed] [Google Scholar]
- Olmsted RA, Langley R, Roelke ME, Goeken RM, Adger-Johnson D, Goff JP, Albert JP, Packer C, Laurenson MK, Caro TM, et al. Worldwide prevalence of lentivirus infection in wild feline species: epidemiologic and phylogenetic aspects. J Virol. 1992;66:6008–6018. doi: 10.1128/jvi.66.10.6008-6018.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecon-Slattery J, McCracken CL, Troyer JL, VandeWoude S, Roelke M, Sondgeroth K, Winterbach C, Winterbach H, O’Brien SJ. Genomic organization, sequence divergence, and recombination of feline immunodeficiency virus from lions in the wild. BMC Genomics. 2008;9:66. doi: 10.1186/1471-2164-9-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petry H, Dittmer U, Stahl-Hennig C, Coulibaly C, Makoschey B, Fuchs D, Wachter H, Tolle T, Morys-Wortmann C, Kaup FJ, et al. Reactivation of human immunodeficiency virus type 2 in macaques after simian immunodeficiency virus SIVmac superinfection. J Virol. 1995;69:1564–1574. doi: 10.1128/jvi.69.3.1564-1574.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipas JM. SV40: Cell transformation and tumorigenesis. Virology. 2009;384:294–303. doi: 10.1016/j.virol.2008.11.024. [DOI] [PubMed] [Google Scholar]
- Pisoni G, Bertoni G, Puricelli M, Maccalli M, Moroni P. Demonstration of coinfection with and recombination by caprine arthritis-encephalitis virus and maedi-visna virus in naturally infected goats. J Virol. 2007;81:4948–4955. doi: 10.1128/JVI.00126-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss M, Idoine A, Ross HA, Terwee JA, Vandewoude S, Rodrigo A. Recombination in feline lentiviral genomes during experimental cross-species infection. Virology. 2007;359:146–151. doi: 10.1016/j.virol.2006.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss M, Ross HA, Painter SL, Holley DC, Terwee JA, Vandewoude S, Rodrigo A. Feline lentivirus evolution in cross-species infection reveals extensive G-to-A mutation and selection on key residues in the viral polymerase. J Virol. 2006;80:2728–2737. doi: 10.1128/JVI.80.6.2728-2737.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putkonen P, Walther L, Zhang YJ, Li SL, Nilsson C, Albert J, Biberfeld P, Thorstensson R, Biberfeld G. Long-term protection against SIV-induced disease in macaques vaccinated with a live attenuated HIV-2 vaccine. Nat Med. 1995;1:914–918. doi: 10.1038/nm0995-914. [DOI] [PubMed] [Google Scholar]
- Ravazzolo AP, Reischak D, Peterhans E, Zanoni R. Phylogenetic analysis of small ruminant lentiviruses from Southern Brazil. Virus Res. 2001;79:117–123. doi: 10.1016/s0168-1702(01)00339-2. [DOI] [PubMed] [Google Scholar]
- Reeves JD, Hibbitts S, Simmons G, McKnight A, Azevedo-Pereira JM, Moniz-Pereira J, Clapham PR. Primary Human Immunodeficiency Virus Type 2 (HIV-2) isolates infect CD4-negative cells via CCR5 and CXCR4: comparison with HIV-1 and Simian Immunodeficiency Virus and relevance to cell tropism in vivo. J Virol. 1999;73:7795–7804. doi: 10.1128/jvi.73.9.7795-7804.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley SP, Foley J, Chomel B. Exposure to feline and canine pathogens in bobcats and gray foxes in urban and rural zones of a national park in California. J Wildl Dis. 2004;40:11–22. doi: 10.7589/0090-3558-40.1.11. [DOI] [PubMed] [Google Scholar]
- Rolland M, Mooney J, Valas S, Perrin G, Mamoun RZ. Characterisation of an Irish caprine lentivirus strain--SRLV phylogeny revisited. Virus Res. 2002;85:29–39. doi: 10.1016/s0168-1702(02)00015-1. [DOI] [PubMed] [Google Scholar]
- Saifuddin M, Roebuck KA, Chang C, Ting JP, Spear GT. Cutting edge: activation of HIV-1 transcription by the MHC class II transactivator. J Immunol. 2000a;164:3941–3945. doi: 10.4049/jimmunol.164.8.3941. [DOI] [PubMed] [Google Scholar]
- Saifuddin M, Spear GT, Chang C, Roebuck KA. Expression of MHC class II in T cells is associated with increased HIV-1 expression. Clin Exp Immunol. 2000b;121:324–331. doi: 10.1046/j.1365-2249.2000.01290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C, Weisshart K, Guarino LA, Dornreiter I, Fanning E. Species-specific functional interactions of DNA polymerase alpha-primase with simian virus 40 (SV40) T antigen require SV40 origin DNA. Mol Cell Biol. 1994;14:3176–3185. doi: 10.1128/mcb.14.5.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebelink KH, Chu IH, Rimmelzwaan GF, Weijer K, van Herwijnen R, Knell P, Egberink HF, Bosch ML, Osterhaus AD. Feline immunodeficiency virus (FIV) infection in the cat as a model for HIV infection in man: FIV-induced impairment of immune function. AIDS Res Hum Retroviruses. 1990;6:1373–1378. doi: 10.1089/aid.1990.6.1373. [DOI] [PubMed] [Google Scholar]
- Smirnova N, Troyer JL, Schissler J, Terwee J, Poss M, VandeWoude S. Feline lentiviruses demonstrate differences in receptor repertoire and envelope structural elements. Virology. 2005;342:60–76. doi: 10.1016/j.virol.2005.07.024. [DOI] [PubMed] [Google Scholar]
- Smith RWP, Nasheuer H-P. Initiation of JC virus DNA replication invitro by human and mouse DNA polymerase a-primase. Eur J Biochem. 2003;270:2030–2037. doi: 10.1046/j.1432-1033.2003.03579.x. [DOI] [PubMed] [Google Scholar]
- Stadlbauer F, Voitenleitner C, Bruckner A, Fanning E, Nasheuer HP. Species-specific replication of simian virus 40 DNA in vitro requires the p180 subunit of human DNA polymerase alpha-primase. Mol Cell Biol. 1996;16:94–104. doi: 10.1128/mcb.16.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens EB, Joag SV, Atkinson B, Sahni M, Li Z, Foresman L, Adany I, Narayan O. Infected macaques that controlled replication of SIVmac or nonpathogenic SHIV developed sterilizing resistance against pathogenic SHIV(KU-1) Virology. 1997;234:328–339. doi: 10.1006/viro.1997.8662. [DOI] [PubMed] [Google Scholar]
- Stott EJ. Anti-cell antibody in macaques. Nature. 1991;353:393. doi: 10.1038/353393a0. [DOI] [PubMed] [Google Scholar]
- Terwee JA, Carlson JK, Sprague WS, Sondgeroth KS, Shropshire SB, Troyer JL, VandeWoude S. Prevention of immunodeficiency virus induced CD4+ T-cell depletion by prior infection with a non-pathogenic virus. Virology. 2008;377:63–70. doi: 10.1016/j.virol.2008.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwee JA, Yactor JK, Sondgeroth KS, Vandewoude S. Puma lentivirus is controlled in domestic cats after mucosal exposure in the absence of conventional indicators of immunity. J Virol. 2005;79:2797–2806. doi: 10.1128/JVI.79.5.2797-2806.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyer JL, Pecon-Slattery J, Roelke ME, Johnson W, VandeWoude S, Vazquez-Salat N, Brown M, Frank L, Woodroffe R, Winterbach C, Winterbach H, Hemson G, Bush M, Alexander KA, Revilla E, O’Brien SJ. Seroprevalence and genomic divergence of circulating strains of feline immunodeficiency virus among Felidae and Hyaenidae species. J Virol. 2005;79:8282–8294. doi: 10.1128/JVI.79.13.8282-8294.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyer JL, Vandewoude S, Pecon-Slattery J, McIntosh C, Franklin S, Antunes A, Johnson W, O’Brien SJ. FIV cross-species transmission: an evolutionary prospective. Vet Immunol Immunopathol. 2008;123:159–166. doi: 10.1016/j.vetimm.2008.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VandeWoude S, Apetrei C. Going wild: lessons from naturally occurring T-lymphotropic lentiviruses. Clin Microbiol Rev. 2006;19:728–762. doi: 10.1128/CMR.00009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VandeWoude S, Hageman CL, Hoover EA. Domestic cats infected with lion or puma lentivirus develop anti-feline immunodeficiency virus immune responses. J Acquir Immune Defic Syndr. 2003;34:20–31. doi: 10.1097/00126334-200309010-00003. [DOI] [PubMed] [Google Scholar]
- VandeWoude S, O’Brien S J, Hoover EA. Infectivity of lion and puma lentiviruses for domestic cats. J Gen Virol. 1997a;78:795–800. doi: 10.1099/0022-1317-78-4-795. [DOI] [PubMed] [Google Scholar]
- VandeWoude S, O’Brien SJ, Langelier K, Hardy WD, Slattery JP, Zuckerman EE, Hoover EA. Growth of lion and puma lentiviruses in domestic cat cells and comparisons with FIV. Virology. 1997b;233:185–192. doi: 10.1006/viro.1997.8587. [DOI] [PubMed] [Google Scholar]
- Warren JT, Dolatshahi M. First updated and revised survey of worldwide HIV and SIV vaccine challenge studies in nonhuman primates: progress in first and second order studies. J Med Primatol. 1993;22:203–235. [PubMed] [Google Scholar]
- Willett BJ, Hosie MJ. Chemokine receptors and co-stimulatory molecules: unravelling feline immunodeficiency virus infection. Vet Immunol Immunopathol. 2008;123:56–64. doi: 10.1016/j.vetimm.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willey SJ, Reeves JD, Hudson R, Miyake K, Dejucq N, Schols D, De Clercq E, Bell J, McKnight A, Clapham PR. Identification of a subset of Human Immunodeficiency Virus Type 1 (HIV-1), HIV-2, and Simian Immunodeficiency Virus strains able to exploit an alternative corecpetor on untransformed human brain and lymphoid cells. J Virol. 2003;77:6138–6152. doi: 10.1128/JVI.77.11.6138-6152.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe ND, Switzer WM, Carr JK, Bhullar VB, Shanmugam V, Tamoufe U, Prosser AT, Torimiro JN, Wright A, Mpoudi-Ngole E, McCutchan FE, Birx DL, Folks TM, Burke DS, Heneine W. Naturally acquired simian retrovirus infections in central African hunters. Lancet. 2004;363:932–937. doi: 10.1016/S0140-6736(04)15787-5. [DOI] [PubMed] [Google Scholar]
- Zanoni RG. Phylogenetic analysis of small ruminant lentiviruses. J Gen Virol. 1998;79:1951–1961. doi: 10.1099/0022-1317-79-8-1951. [DOI] [PubMed] [Google Scholar]