Summary
Ubiquitinylation of proteins is a critical mechanism in regulating numerous eukaryotic cellular processes including cell cycle progression, inflammatory response, and vesicular trafficking. Given the importance of ubiquitinylation, it is not surprising that several pathogenic bacteria have developed strategies to exploit various stages of the ubiquitin pathway for their own benefit. One such strategy is the delivery of bacterial ‘effector’ proteins into the host cell cytosol, which mimic the activities of components of the host ubiquitin pathway. Recent studies have highlighted a number of bacterial effectors that functionally mimic the activity of eukaryotic E3 ubiquitin ligases, including a novel structural class of bacterial E3 ligases that provides a striking example of convergent evolution.
Introduction
Several pathogenic bacteria utilize specialized type III or type IV secretion systems to deliver bacterial proteins, called effectors, into host cells to modulate a variety of cellular pathways. There are a growing number of effectors that usurp the host ubiquitin pathway by functionally mimicking components of the pathway. Ubiquitinylation results in the covalent attachment of ubiquitin to a lysine residue on a target protein [1]. Following the initial conjugation, subsequent ubiquitin molecules can be ligated to one of seven lysines in the previously attached ubiquitin molecule, resulting in polyubiquitinylation of various linkages. Therefore, a substrate can be monoubiquitinylated at a single lysine residue, multi-ubiquitinylated at multiple lysine residues, or polyubiquitinylated at one or more lysine residues. The type of ubiquitinylation and the topology of the ubiquitin chains formed direct substrate fate [2]. Ubiquitinylation can signal for proteasome-dependent degradation or function as non-proteolytic signals important for DNA repair, signal transduction and vesicular trafficking [3–7].
Ubiquitinylation involves an enzymatic cascade resulting in the formation of an isopeptide bond between ubiquitin and internal lysine residues of a substrate protein [8]. This process involves an ubiquitin-activating enzyme (E1), which forms a thioester bond between a catalytic cysteine and the carboxy terminal glycine residue of ubiquitin. The ubiquitin is then transferred to an ubiquitin-conjugating enzyme (E2). Finally, an ubiquitin ligase (E3) facilitates the covalent conjugation of ubiquitin from an ubiquitin-loaded E2 to one or more lysine residues in the substrate. Therefore, E3 ubiquitin ligases confer specificity to the reaction through substrate binding. E3 ubiquitin ligases are defined by their ability to facilitate the transfer of ubiquitin from a cognate E2 to a specific substrate. There are two major known types of E3 ubiquitin ligases in eukaryotes, which possess distinct structural and mechanistic properties: the RING (really interesting new gene)/U-box domain and the HECT (homologous to E6-associated protein C terminus) domain [9,10]. In this review, we focus on the structures features used by bacterial effectors to mimic the activity of eukaryotic E3 ubiquitin ligases.
RING/U-box-like E3 Ubiquitin Ligases
Eukaryotic RING/U-box E3s mainly function as scaffolds to facilitate the transfer of ubiquitin directly from an E2 to a substrate. RING domains are defined by the consensus sequence Cx2Cx9 – 39Cx1–3Hx2–3C/Hx2Cx4–48Cx2C, which forms a ‘cross-brace’ motif [11]. This motif coordinates the binding of two zinc ions between alternating Cys and His residues. In the U-box variant, the Cys and His residues are replaced with charged and polar residues that mediate salt bridges and hydrogen bonds resulting in a similar overall structure [12]. RING/U-box domains also include a concave surface consisting of a three amino acid hydrophobic patch that binds to a conserved sequence present on most E2s [11,13]. RING domains are often part of a multidomain protein or a multisubunit complex. While the RING/U-box domain recruits ubiquitin-loaded E2s, an additional domain or subunit that encodes a protein-protein interaction motif, such as scr homology-2 or leucine-rich repeat (LRR) domain, is required for substrate recognition.
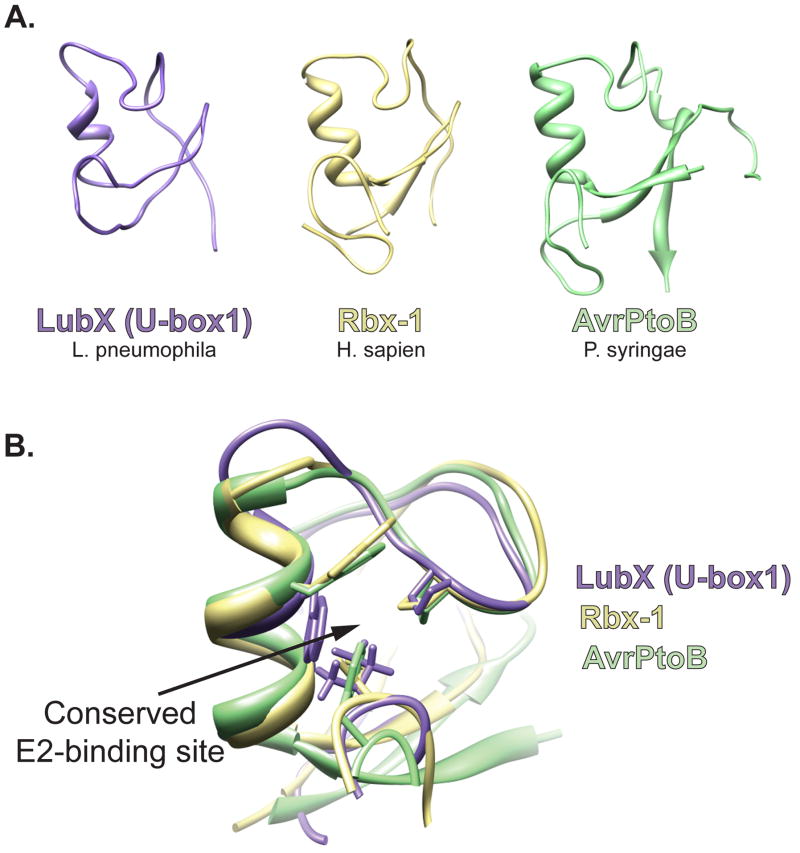
The type IV effector LubX of Legionella pheumophila has two domains both with striking sequence similarity to eukaryotic E3 U-box domains. U-box1 of LubX was shown to have ligase activity, while U-box2, which lacks a hydrophobic residue critical for interaction with E2s, is inactive [14]. When the three-dimensional structure of the U-box1 of LubX is modeled using the PHYRE threading program, the structure and positioning of key residues is very similar to known RING/U-box proteins [15] (Fig. 1 A and B). As predicted, mutations disrupting the putative E2 binding domain of U-box1 abolished ubiquitin ligase activity of LubX. Interestingly, U-box2 functions to bind host Cdc2-like kinase 1 (Clk1) and targets it for ubiquitinylation by U-box1 in vitro. Therefore, Legionella likely modulates Clk1 function during infection by translocating LubX, a RING-like E3 ubiquitin ligase. The consequence of Clk1 ubiquitinylation during Legionella infection remains to be determined.
Figure 1.
Bacterial mimics of eukaryotic RING/U-box E3 ligases. (A). Using the Phyre threading program, the sequence of U-box1 of L. pneumophila LubX was aligned to known structures and the structure was modeled to its best fit, human E3 traf6 (E-value of 2.6e−11; estimated precision of 100%); the RING/U-box structure of H. sapien, Rbx-1 (PDB ID 3DPL); the core fold of P. syringae, AvrPtoB (PDB ID 2FD4). (B) Visualization of the E2-binding site residues of Rbx-1 with homologous regions in LubX and AvrPtoB. The three putative E2-binding residues are shown.
However unlike LubX, most known bacterial E3 ligases do not share sequence similarity with eukaryotic E3s. The Pseudomonas syringae effector AvrPtoB is an example of a structural mimic of the RING/U-box family of E3 ligases. Pseudomonas syringae pathovar tomato causes bacterial speck disease on tomato and Arabidopsis [16]. In susceptible plants, Pseudomonas syringae injects the type III effector AvrPtoB, which suppresses programmed cell death (PCD) [17]. Structural studies revealed that the C-terminal domain of AvrPtoB adopts a ‘core fold’ nearly identical to the RING/U-box domain of the human E3, Rbx1, consisting of a three-stranded sheet with a single helix and two extended loops (Figure 1A). Similar to LubX, AvrPtoB also encodes a highly-conserved binding site for host E2s, where the positioning of critical residues is preserved (Figure 1B) [18,19]. Experiments confirmed that AvrPtoB possesses ubiquitin ligase activity and this activity is required to suppress PCD during Pseudomonas syringae infection. Therefore, while undetectable at the sequence level, the potential biochemical activity of AvrPtoB was revealed by its structural homology to RING/U-box E3 ligases. Subsequent studies have shown that the N-terminal domain of AvrPtoB mediates the recruitment of substrates and directs the ubiquitin ligase activity of the C-terminal domain. The N-terminal domain of AvrPtoB binds to several host kinases important in plant immunity, such as tomato Fen kinase, Flagellin-sensing receptor kinase 2, and the Chitin elicitor receptor kinase 1, resulting in their ubiquitinylation and proteosomal degradation [20–22]. Therefore, AvrPtoB promotes Pseudomonas syringae virulence by targeting signaling proteins important for plant immunity for ubiquitin-mediated degradation.
HECT-like E3 Ubiquitin Ligases
HECT-type E3 ubiquitin ligases are modular proteins consisting of an N-terminal substrate binding domain and a HECT domain characterized by a ~350 residue C-terminal domain with a conserved catalytic cysteine residue ~35 residues from the C terminus [23]. Unlike RING/U-box E3 ubiquitin ligases, HECT E3s participates directly in the catalysis of ubiquitinylation by forming a covalent thioester intermediate between the catalytic Cys of the HECT domains and the C-terminus of ubiquitin prior to transferring ubiquitin to the target protein. The HECT domain consists of two subdomains: the N-terminal lobe (N lobe), which binds E2s, and the C-terminal lobe (C lobe), which possesses the catalytic Cys. The N and C lobes are connected by a flexible loop, and while the overall structure of HECT domains is similar, the orientation and position of the lobes can vary [24]. The structural organization of most HECT domains requires that a substantial conformational change occurs to bring the catalytic Cys of the HECT domain close enough to the E2 active site for ubiquitin transfer [25,26]. Mutations that restrict the rotation of the N and C lobes about the flexible loop reduce ubiquitin ligase activity, supporting the requirement for such a conformational change for catalytic activity [25]. This conformational flexibility is thought to enable the movement of ubiquitin from the E2 to the catalytic cysteine of the HECT E3 and then finally to the substrate.
The Salmonella enterica type III effector protein SopA structurally mimics HECT E3 ubiquitin ligases despite lacking significant amino acid similarity. SopA regulates host inflammatory responses, which leads to the transepithelial migration of polymorphonuclear (PMN) cells during Salmonella infection [27]. Salmonella strains expressing an E3 ligase-defective mutant of SopA induced less PMN transepithelial migration indicating that the E3 ligase activity of SopA is important for SopA function [27].
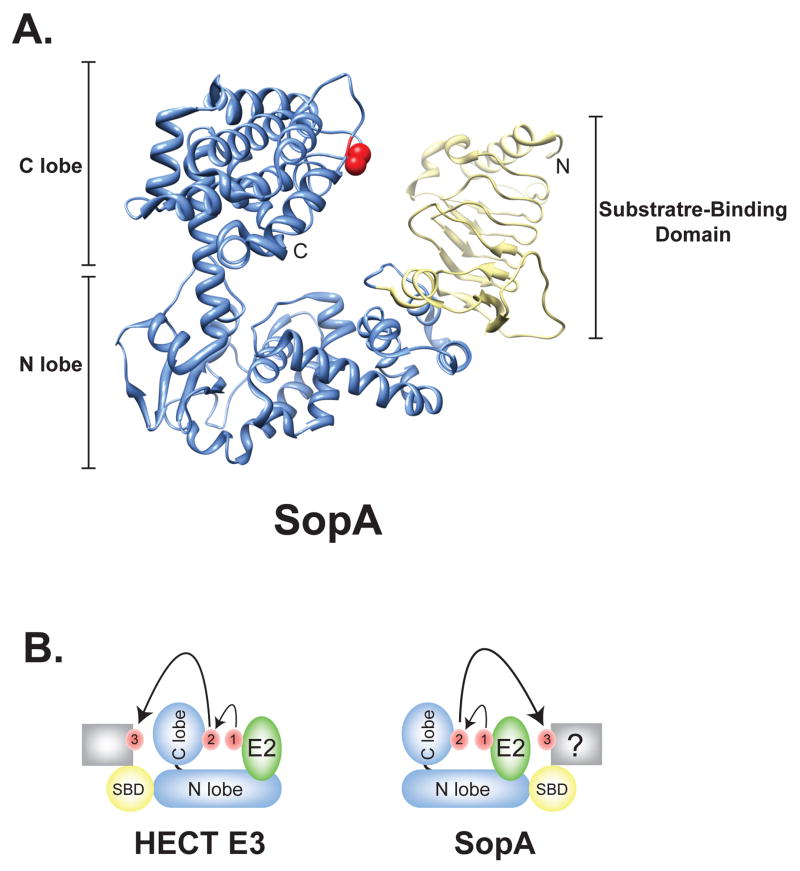
SopA is organized into an N-terminal putative substrate binding domain and a C-terminal HECT-like domain (Figure 2A). Similar to the human HECT E3 ligase E6-AP, the HECT domain of SopA is comprised of N and C lobes that are orientated in a L-shape [28]. The N lobe of SopA forms the long arm while the C lobe, which contains a catalytic Cys located 30 residues from the C-terminus, forms the short arm of the L structure. The lobes of SopA are connected by a flexible linker helix, which likely allows for the conformational changes required for ubiquitin transfer.
Figure 2.
SopA is a HECT-like E3 ligase. (A) Overall structure of SopA163–782 (PDB ID 2QYU); the HECT domain of SopA is shown in blue and the N-terminal β-helix domain in yellow. The catalytic cysteine is signified in red. N, NH2 terminus; C, COOH terminus. (B) Schematic diagram of the transfer of ubiquitin. Ubiquitin is shown in red. The C lobe of a generic HECT E3 or SopA is positioned similarly to accept ubiquitin following binding of Ub-charged E2 (green). However, the placement of the substrate binding (SBD; yellow) domain of SopA on the opposite end of the N lobe as found in eukaryotic HECT E3s likely requires a significantly different conformational change to facilitate ubiquitin transfer to a target substrate (grey box).
While there is minimal sequence similarity around the active site of SopA in comparison to other HECT E3 ligases, alignment of the active site loop sequence revealed that Leu747 and Thr752 in addition to catalytic Cys753 of SopA are conserved in all known HECT domains [28]. Alanine replacement of either of these residues reduced ubiquitin ligase activity.
The N-terminal putative substrate-binding domain of SopA forms a parallel -helix structure, similar to those found in the pectin lyase-like structural superfamily, which is involved in carbohydrate binding [28]. While the presence of a β-helix motif suggests that SopA may ubiquitinylate carbohydrate-modified substrates, the substrates for SopA area still unknown. Interestingly, the substrate-binding domain of SopA extends from the opposite end of the N lobe structure of SopA as compared to the structure of the HECT-type E3 ligase E6-AP (Figure 2B). Yet Diao et al. showed that placement of the β-helix domain near the putative E2 binding region neither obstructed nor contributed to E2 binding of SopA [28]. The placement of the putative substrate-binding domain of SopA is markedly different from that observed in eukaryotic HECT E3s and will require additional co-structures of SopA with its cognate E2 and/or substrates are needed to help characterize the conformational changes required for ubiquitin transfer from SopA to its substrate (Figure 2B).
NEL E3 Ubiquitin Ligases
Recently, a new family of E3 ubiquitin ligases has been described that possesses a structural domain (termed NEL for Novel E3 Ligase), which is distinct from either the RING or HECT domains. NEL E3 ligases comprise a large family of a bacterial effector proteins encoded by a subset of pathogenic bacteria (Table 1). Prior to being recognized as ubiquitin ligases, NEL E3 ligases were classified as leucine-rich repeat (LRR) effector proteins due to the presence of an N-terminal LRR domain. In 2007, Rohdes et al. discovered that two family members, Shigella spp. IpaH9.8, and Salmonella, SspH1, possessed E3 ubiquitin ligases activity despite lacking sequence similarity to any known E3 ubiquitin ligases [29]. Since uncovering the ubiquitin ligase activity of this family of effector proteins, three structural studies have shed light on this new class of E3 ubiquitin ligases.
Table 1.
Examples of Bacterial E3 Ligase Mimics
| Bacteria | Effector | E3 Structural Family | Host Targets | Refs |
|---|---|---|---|---|
| Bradyrhizobium spp | Blr1676 | NEL | Unknown | [31] |
| Blr1904 | NEL | Unknown | [31,32] | |
| Escherichia coli ssp | Ecol5_01000486 | NEL | Unknown | [34] |
| Ecol5_01001536 | NEL | Unknown | [34] | |
| Ecol5_01001967 | Unknown | [34] | ||
| Ecol5_01003958 | NEL | Unknown | [34] | |
| Ecol5_01004202 | NEL | Unknown | [34] | |
| Ecol5_01004539 | NEL | Unknown | [34] | |
| Ecol5_01004764 | NEL | Unknown | [34] | |
| Ecol5_01004830 | NEL | Unknown | [34] | |
| Ecol5_01004885 | NEL | Unknown | [34] | |
| NleG2–3 | RING/U-box | Unknown | [35] | |
| Legionella spp | LubX | RING/U-box | Clk1? | [15] |
| Pseudomonas spp | AvrPtoB | RING/U-box | Fen, CERK1, FSL2 | [18–22] |
| PflO1_4099 | NEL | Unknown | [31] | |
| PflO1_4565 | NEL | Unknown | [31] | |
| PP_2212 | NEL | Unknown | [32] | |
| PP_2394 | NEL | Unknown | [32] | |
| PSPTO_1492 | NEL | Unknown | [31] | |
| PSPTO_4093 | NEL | Unknown | [31] | |
| Rhizobium spp | Y4fR | NEL | Unknown | [31,32] |
| Salmonella ssp | Slrp | NEL | Thioredoxin? | [30,33] |
| SopA | HECT-like | Unknown | [27,28] | |
| SspH1 | NEL | PKN-1? | [29,30] | |
| SspH2 | NEL | Unknown | [30] | |
| Shigella ssp | IpaH1 | NEL | Unknown | [34] |
| IpaH1.4 | NEL | Unknown | [32] | |
| IpaH2 | NEL | Unknown | [34] | |
| IpaH2.5 | NEL | Unknown | [34] | |
| IpaH3 | NEL | Unknown | [31] | |
| IpaH4 | NEL | Unknown | [34] | |
| IpaH4.5 | NEL | Unknown | [31] | |
| IpaH5 | NEL | Unknown | [34] | |
| IpaH6 | NEL | Unknown | [34] | |
| IpaH7 | NEL | Unknown | [34] | |
| IpaH7.8 | NEL | Unknown | [31] | |
| IpaH9.8 | NEL | STE7? | [32] | |
| Yersinia spp | YPA_3361 1 | NEL | Unknown | [31,34] |
| YPA_3364 1 | NEL | Unknown | [31,34] | |
these are representative open reading frames from Yersinia pestis Angola that are found in most Yersinia pestis and Yersinia pseudotuberculosis strains
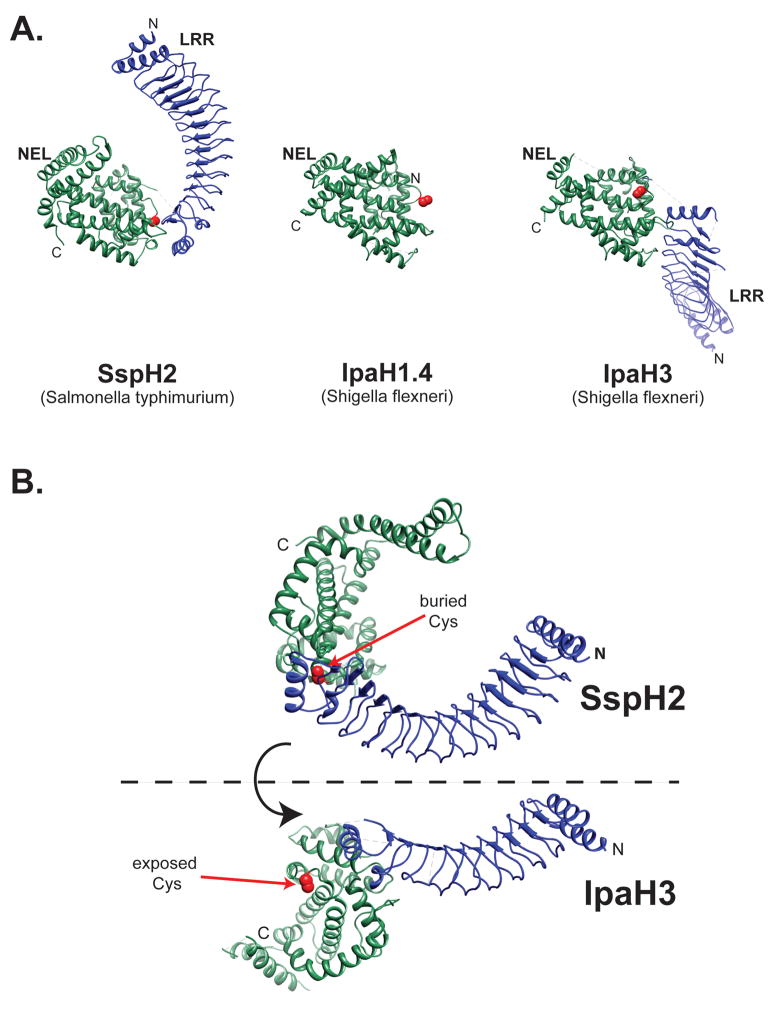
The nearly full-length structures of two NEL E3 ubiquitin ligases, Salmonella SspH2 and Shigella IpaH3, reveal similar folds composed of two well-defined structural elements: a N-terminal LRR domain linked by a short stretch of residues to a novel C-terminal helical domain with ubiquitin ligase activity (Figure 3A) [30,31]. Both LRR domains resemble a curving solenoid made of either 12 (SspH2) or 9 (IpaH3) LRR motif repeats capped at the N- and C-terminal regions with α-helices. The inner, concave surfaces of both LRR domains are lined with hydrophobic residues consistent with their putative involvement in protein-protein interactions. LRR domains provide a versatile platform for protein-protein interactions whereby simply varying the number of repeats, the curvature of the structure or the surface characteristics can alter binding specificity.
Figure 3.
NEL family of E3 ligases. (A) The structure of SspH2166–783 (PBD ID 3G06), IpaH1.4265–575 (PDB ID 3CKD), and Ipa325–561 (PDB ID 3CVR) is shown with leucine-rich repeat (LRR) domain in blue and the novel E3 ligase (NEL) domain in green. The catalytic cysteine residue is shown in red. N, NH2 terminus; C, COOH terminus. (B) Conformational changes likely activate the NEL family of E3 ligases. The structures of Shigella IpaH3 and Salmonella SspH2 suggests a dramatic hinge motion as the NEL domain rotates 180° from the closed conformation indicated by the structure of SspH2 to the open position represented by the IpaH3 structure. The catalytic cysteine residue is shown in red. N, NH2 terminus; C, COOH terminus.
The C-terminal NEL domain forms a unique fold that possesses ubiquitin ligase activity. It consists of 2 subdomains: an N-terminal globular region followed by two long finger-like helices extending from the globular fold [30–32]. The E3 ubiquitin ligase activity of the NEL domain requires a single conserved Cys residue. Similar to HECT E3s, this conserved Cys can act as a nucleophile forming a thioester bond with ubiquitin [31,32]. IpaH9.8 of Shigella contains several charged residues close to the catalytic Cys that are required for ligase activity, including Asp339, Arg340 and Asp387 [32]. These residues, as well as their positioning, are conserved in all NEL E3 effectors and likely participate directly in the ubiquitinylation reaction and/or substrate binding.
While the structures of both SspH2 and IpaH2 are composed of very similar N-terminal LRR domain and C-terminal NEL domains, there is a dramatic difference in the orientation of these domains with respect to each other (Figure 3). In SspH2, the concave surface of the LRR curves towards the NEL domain, partially occluding the putative protein-binding surface of the LRR domain and burying the catalytic Cys residue. In contrast, the LRR domain of IpaH3 is fully extended resulting in a solvent accessible catalytic Cys residue. We hypothesize that the structure of SspH2 depicts an autoinhibited or closed conformation, while the structure of IpaH3 represents an activated conformer in which the binding surface of the LRR is available and the catalytic cysteine accessible. A transition from the close conformation to the active conformation would result from a change in orientation of the NEL domain and the LRR domain translated through the linker region between the two domains (Figure 3B). We propose a model in which activation results in a relative reorientation of the two domains that exposes the catalytic site to the host ubiquitinylation machinery and substrate(s). Consistent with this model, mutations that destabilize the binding interface between the LRR and NEL domains of SspH2 dramatically increase the ubiquitin ligase activity of full-length SspH2 [30].
The role of NEL E3 ligase dependent ubiquitinylation during infection remains elusive. While overexpression of IpaH9.8 leads to the degradation of the mitogen-activated protein kinase (MAPK) kinase Ste7 in yeast, no decrease in components of the mammalian MAPK pathway was observed following Shigella infection [29]. In addition, Salmonella SspH1 and SlrP have been shown to target PKN1 and thioreodoxin respectively in vitro, but in vivo ubiquitinylation of either target has yet to be demostrated [29,33]. Therefore, in vivo targets of NEL E3 ligases during infection remain to be identified.
Conclusion
Several pathogenic bacteria have evolved effectors that have the capacity to exploit the host ubiquitin pathway by functionally mimicking eukaryotic E3 ubiquitin ligases (Table 1). Some bacteria accomplish this by expressing effectors, such as Legionella LubX, with significant amino acid sequence similarity to eukaryotic E3 ligases, which may have been acquired through horizontal gene transfer from a eukaryotic host. However, many effectors successfully mimic the activities of E3 ligases despite lacking amino acid sequence similarity. For example, Pseudomonas AvrPtoB and Salmonella SopA structurally mimic known eukaryotic E3 ligases by adopting homologous structures. While others, belonging to the NEL E3 ligase family, have evolved a novel structure to functionally mimic eukaryotic E3 ligases activity.
The discovery of the E3 ligase activity for these effectors is an exciting first step towards understanding the functional role they play during infection. However, much remains to be learned about these fascinating molecules. What are the types of ubiquitin conjugates that are formed by these different ligases? What are the host enzymes (i.e. E2s) they engage to carry out their function? And finally and arguably most important to understand their actual role in infection, what are the substrates they target? Understanding the mechanisms by which these enzymes restrict their activity to specific cellular locations would also help to clarify their role in host-pathogen interactions. In addition to broadening our understanding of bacterial pathogenesis, the study of bacterial E3 ubiquitin ligases will likely provide insights into the basic biology of the host ubiquitinylation pathway and may lead to the development of novel therapeutic strategies.
Supplementary Material
Acknowledgments
S.W.H is supported by the National Research Service Award postdoctoral fellowship, AI069704, from the National Institutes of Health (NIH). Work in the Galan laboratory on the subjects discussed in this article has also been supported by the NIH grant, AI055472 (to J.E.G.). We would like to thank Heather Scobie, Helena Hodak, Stefania Spanò and Matthew Lefebre for helpful comments and critical review of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
• of special interest
•• of outstanding interest
- 1.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 2.Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals. ‘Protein Modifications: Beyond the Usual Suspects’ review series. EMBO Rep. 2008;9:536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hicke L, Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol. 2003;19:141–172. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- 6.Huang TT, D’Andrea AD. Regulation of DNA repair by ubiquitylation. Nat Rev Mol Cell Biol. 2006;7 :323–334. doi: 10.1038/nrm1908. [DOI] [PubMed] [Google Scholar]
- 7.Haglund K, Dikic I. Ubiquitylation and cell signaling. EMBO J. 2005;24:3353–3359. doi: 10.1038/sj.emboj.7600808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Passmore LA, Barford D. Getting into position: the catalytic mechanisms of protein ubiquitylation. Biochem J. 2004;379:513–525. doi: 10.1042/BJ20040198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pavletich NP. Structural biology of ubiquitin-protein ligases. Harvey Lect. 2002;98:65–102. [PubMed] [Google Scholar]
- 10.Ardley HC, Robinson PA. E3 ubiquitin ligases. Essays Biochem. 2005;41:15–30. doi: 10.1042/EB0410015. [DOI] [PubMed] [Google Scholar]
- 11.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 12.Aravind L, Koonin EV. The U box is a modified RING finger - a common domain in ubiquitination. Curr Biol. 2000;10:R132–134. doi: 10.1016/s0960-9822(00)00398-5. [DOI] [PubMed] [Google Scholar]
- 13.Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 14.Kubori T, Hyakutake A, Nagai H. Legionella translocates an E3 ubiquitin ligase that has multiple U-boxes with distinct functions. Mol Microbiol. 2008;67:1307–1319. doi: 10.1111/j.1365-2958.2008.06124.x. [DOI] [PubMed] [Google Scholar]
- 15.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 16.Alfano JR, Collmer A. Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu Rev Phytopathol. 2004;42:385–414. doi: 10.1146/annurev.phyto.42.040103.110731. [DOI] [PubMed] [Google Scholar]
- 17.Abramovitch RB, Kim YJ, Chen S, Dickman MB, Martin GB. Pseudomonas type III effector AvrPtoB induces plant disease susceptibility by inhibition of host programmed cell death. EMBO J. 2003;22 :60–69. doi: 10.1093/emboj/cdg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18•.Janjusevic R, Abramovitch RB, Martin GB, Stebbins CE. A bacterial inhibitor of host programmed cell death defenses is an E3 ubiquitin ligase. Science. 2006;311:222–226. doi: 10.1126/science.1120131. These two studies [18•,19• ] describe the core fold of the C-terminal domain of AvrPtoB, revealing a structure similar to that of known RING/U-box E3 ubiquitin ligases. AvrPtoB residues required for E3 ligase activity are also required for AvrPtoB-dependent inhibition of programmed cell death during Pseudomonas springae infection. [DOI] [PubMed] [Google Scholar]
- 19•.Abramovitch RB, Janjusevic R, Stebbins CE, Martin GB. Type III effector AvrPtoB requires intrinsic E3 ubiquitin ligase activity to suppress plant cell death and immunity. Proc Natl Acad Sci U S A. 2006;103:2851–2856. doi: 10.1073/pnas.0507892103. See annotation for [18•] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gohre V, Spallek T, Haweker H, Mersmann S, Mentzel T, Boller T, de Torres M, Mansfield JW, Robatzek S. Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr Biol. 2008;18:1824–1832. doi: 10.1016/j.cub.2008.10.063. [DOI] [PubMed] [Google Scholar]
- 21.Rosebrock TR, Zeng L, Brady JJ, Abramovitch RB, Xiao F, Martin GB. A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature. 2007;448:370–374. doi: 10.1038/nature05966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gimenez-Ibanez S, Hann DR, Ntoukakis V, Petutschnig E, Lipka V, Rathjen JP. AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr Biol. 2009;19:423–429. doi: 10.1016/j.cub.2009.01.054. [DOI] [PubMed] [Google Scholar]
- 23.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 24.Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol. 2009;10:398–409. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- 25.Verdecia MA, Joazeiro CA, Wells NJ, Ferrer JL, Bowman ME, Hunter T, Noel JP. Conformational flexibility underlies ubiquitin ligation mediated by the WWP1 HECT domain E3 ligase. Mol Cell. 2003;11:249–259. doi: 10.1016/s1097-2765(02)00774-8. [DOI] [PubMed] [Google Scholar]
- 26.Huang L, Kinnucan E, Wang G, Beaudenon S, Howley PM, Huibregtse JM, Pavletich NP. Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2–E3 enzyme cascade. Science. 1999;286:1321–1326. doi: 10.1126/science.286.5443.1321. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Higashide WM, McCormick BA, Chen J, Zhou D. The inflammation-associated Salmonella SopA is a HECT-like E3 ubiquitin ligase. Mol Microbiol. 2006;62:786–793. doi: 10.1111/j.1365-2958.2006.05407.x. [DOI] [PubMed] [Google Scholar]
- 28•.Diao J, Zhang Y, Huibregtse JM, Zhou D, Chen J. Crystal structure of SopA, a Salmonella effector protein mimicking a eukaryotic ubiquitin ligase. Nat Struct Mol Biol. 2008;15:65–70. doi: 10.1038/nsmb1346. Despite lacking amino acid sequence similarity, this study shows that Salmonella SopA adpots a similar bilobal structure as HECT-type E3 ubiquitin ligases. [DOI] [PubMed] [Google Scholar]
- 29•.Rohde JR, Breitkreutz A, Chenal A, Sansonetti PJ, Parsot C. Type III secretion effectors of the IpaH family are E3 ubiquitin ligases. Cell Host Microbe. 2007;1:77–83. doi: 10.1016/j.chom.2007.02.002. This study was the first to describe the E3 ubiquitin ligase activity of both Shigella IpaH9.8 and Salmonella SspH1. IpaH9.8 disrupts phermone response signaling in yeast by promoting the proteasomal-dependent degradation of MAPKK Ste7. The authors also show that SspH1 can ubiquitinylate PKN1 in vitro. [DOI] [PubMed] [Google Scholar]
- 30•.Quezada CM, Hicks SW, Galan JE, Stebbins CE. A family of Salmonella virulence factors functions as a distinct class of autoregulated E3 ubiquitin ligases. Proc Natl Acad Sci U S A. 2009;106:4864–4869. doi: 10.1073/pnas.0811058106. This work descibes the crystal structure of Salmonella SspH2, which possesses a canonical leucine-rich repeat domain that interacts with a C-terminal NEL E3 ligase domain. The interaction of these two domains regulates the E3 ligase activity of the protein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31•.Zhu Y, Li H, Hu L, Wang J, Zhou Y, Pang Z, Liu L, Shao F. Structure of a Shigella effector reveals a new class of ubiquitin ligases. Nat Struct Mol Biol. 2008;15:1302–1308. doi: 10.1038/nsmb.1517. These two studies [31•,32•] show that the Shigella IpaH family of effectors functionally mimics known E3 ligase activity through a novel C-terminal protein fold. The authors also show that a conserved CXD motif acts to catalyze ubiqutin transfer. [DOI] [PubMed] [Google Scholar]
- 32•.Singer AU, Rohde JR, Lam R, Skarina T, Kagan O, Dileo R, Chirgadze NY, Cuff ME, Joachimiak A, Tyers M, et al. Structure of the Shigella T3SS effector IpaH defines a new class of E3 ubiquitin ligases. Nat Struct Mol Biol. 2008;15:1293–1301. doi: 10.1038/nsmb.1511. See annotation for [31•] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernal-Bayard J, Ramos-Morales F. Salmonella type III secretion effector SlrP is an E3 ubiquitin ligase for mammalian thioredoxin. J Biol Chem. 2009;284:27587–27595. doi: 10.1074/jbc.M109.010363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaudhuri RR, Pallen MJ. xBASE, a collection of online databases for bacterial comparative genomics. Nucleic Acids Res. 2006;34:D335–337. doi: 10.1093/nar/gkj140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu B, Yee, A., Fares, C., Lemak, A., Semest, A., Claude, M., Coomes, B., Edwards, A., Singer, A., Arrowsmith, CH., T Montelione, G., Joachimiak, A., Savchenko, A. Pathogenic E. coli O157:H7 NleG family of Type 3 secretion effectors features a common C-terminal U-box domain and E3 ubiquitin ligase activity. PDN ID: 2KKX & 2KKY.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.