Abstract
Membrane curvature has emerged as a key regulatory factor in endocytic vesicle formation. From a theoretical perspective, we summarize recent progress in understanding how membrane curvature and biochemical pathways are coupled and orchestrated during the coherent process of endocytic vesicle formation. We mainly focus on clathrin- and actin- mediated endocytosis in yeast and in mammalian cells. We further speculate on how mechanochemical feedback could modulate other membrane-remodeling processes.
Introduction
Recent experimental efforts have greatly advanced our understanding of clathrin- and actin- dependent endocytosis. Endocytic vesicle formation typically takes ~ 1–2 minutes, and is highly robust in vivo, [1,2]. Despite differences in details, general themes are emerging for different endocytic vesicle formation pathways, both across and within species.
Endocytic vesicle formation involves a sequence of dramatic membrane shape changes. From a mechanical standpoint, endocytic vesicle formation can be divided into two stages. First, the cargo-laden membrane at the endocytic site is intruded to form a tubular or spherical vesicle. Second, the vesicle is pinched off into the interior cytoplasm. The large energy penalty for bending the cell plasma membrane (~ 100's kBT) makes it resist deforming to high curvatures [3,4]. Therefore, substantial forces must be generated to power endocytic vesicle formation. Many proteins and lipids have been identified as part of the endocytic machinery [1,5], some of which conspire to generate the forces that deform the membrane and pinch off the vesicle. Thus, finding the link between force generation and biochemical reactions on endocytic membranes is key to understanding the mechanism of endocytic vesicle formation.
Endocytic proteins often bind to each other via specific motifs [6], and many bind directly to the membrane [7]. However, in the noisy cellular environment, specific protein-protein and/or protein-lipid interactions alone are not sufficient to achieve rapid and robust endocytic internalization. For this reason, self-accelerating feedback mechanisms—which are widely utilized in many cellular processes—must facilitate endocytic vesicle formation. Specifically, biochemical pathways generate forces that remodel the membrane, and membrane shape changes in turn regulate biochemical pathways, forming a closed mechanochemical feedback loop. Testing this hypothesis in vivo is difficult because the process takes but a few seconds. A theoretical model, however, can establish whether such feedback loops can account for the observed robust dynamics.
We first summarize recent experimental evidence for a mechanochemical feedback loop during endocytic vesicle formation. Clathrin- and actin-dependent endocytosis will be used as an example to address how the mechanochemical model can account for the sequential dynamics of the endocytic machinery. Finally, we will discuss the implications of the model for other membrane trafficking processes.
Properties of clathrin- and actin-dependent endocytosis
Forty years of in vivo and in vitro studies have revealed the mechanochemical nature of the biochemical and biophysical activities of endocytic proteins and lipids. Here we briefly summarize these properties.
Actin/myosin force
Actin polymerization is required for yeast endocytic vesicle formation to overcome the resistance of the cell's turgor pressure (the detailed roles of actin function in endocytosis are described in [8]) [9]. FRAP (fluorescence recovery after photobleaching) experiments in yeast show that during endocytosis actin filaments, which are anchored to the bud region of the endocytic membrane via coat proteins, polymerize with their barbed-ends facing the plasma membrane/cell cortex [10••]. Theoretical analysis has predicted that actin polymerization can generate mechanical force via a Brownian ratchet mechanism [11,12]. The magnitude of this force has been predicted and subsequently confirmed to be ~ 1–2 pN per actin filament [13]. At this magnitude, theory suggests actin polymerization could provide the pulling force to overcome the turgor pressure, and drive membrane invagination [14]. Evidence also suggests a role for actin in endocytic internalization in mammalian cells [15].
In addition, type I myosin localizes at the endocytic site in both mammalian and yeast cells [16,17•]. Both actin nucleating and motor activities of type I myosin are required for actin's endocytic function in yeast [17•]. Recent single-molecule experiments demonstrate that the duty ratio of type I myosin increases with load [18]. Thus type I myosin could act as a mechanosensor. When actin assembles, its polymerization rate decreases exponentially with load [12]. The bending resistance of cell membranes always opposes actin polymerization during invagination. Type I myosin's motor activity could push the filaments away from the plasma membrane allowing intercalation of new monomers. Therefore, the combined effects of actin assembly and type I myosin motor activity can invaginate the endocytic membrane despite the high load due to the high membrane bending resistance at early stages.
Curvature sensing and curvature sculpting of BDPs (Bin/Amphiphysin/Rvs Domain Proteins)
BAR (Bin/Amphiphysin/Rvs) domains were first identified as conserved motifs in yeast Rvs167/161p and mammalian amphiphysin. In recent years, crystal structure studies have facilitated the discovery of many new BAR domains. Growing evidence shows that BAR domains function in membrane-remodeling (see [19] for details). In yeast, Rvs167/161p are proposed to play important roles in endocytic vesicle fission [20•].
Dimers of BAR-domain proteins (BDPs) have a preferred banana-like shape and bind to PIP2 via a positively-charged concave surface [21]. Experiments suggest that local membrane curvature can act as a guiding signal for BDP assembly, and BDPs bind more tightly to more highly curved membranes [22•]. Conversely, BDPs, as well as related F-BAR proteins, can deform membranes into tubular shapes with diameters ranging from 20–100 nm [23•,24•,25•,26]. In order for membranes to change shape the driving force must overcome the energy penalty for membrane deformation. As BDPs have their own preferred shape, upon binding to the membrane BDPs can `sculpt' the local membrane into the preferred shape for their own binding. Molecular dynamics simulations support this mechanism [27,28]. Thus, the curvature sensing and curvature producing properties of BAR domains form a positive feedback loop. Note that existence of multiple binding sites spanning the entire inner concave surface of BDPs can deform membranes more effectively than a single site could because they mutually stabilize one another's binding to the membrane while forcibly curving it [29].
Dynamin
The key differences between clathrin-mediated endocytosis in yeast and mammals are in the relative functional importance of dynamin, BDPs and actin [1,30••]. Actin and BDPs—but not dynamin homologues—are important for yeast endocytic vesicle formation [5,20•]. However, in mammalian cells, dynamin accumulates at the scission site and is crucial for efficient endocytic vesicle scission [31••,32,33••,34,35], while actin plays an auxiliary role. The spatial localization of dynamin and the phenotypes of dynamic GTPase mutants imply that dynamin can act as a `pinchase', squeezing the membrane tubule to directly pinch off the vesicle [34]. However, dynamin can only squeeze membrane tubules down to ~ 20 nm in diameter [36••]. As a result, the opposing membrane leaflets are still ~ 10 nm apart, too far to spontaneously fuse [37]. Indeed, recent in vitro and in vivo experiments suggest that dynamin always falls off the membrane before fission [38••,39••]. These observations indicate that dynamin may not directly pinch off the vesicle, and may in fact need to release its grip on the membrane before the opposing leaflets can get close enough to fuse.
Pinching force from spatial heterogeneity in lipid composition
At higher membrane curvatures, lipid head groups on the outside of the forming vesicle are more splayed, and therefore more exposed to lipid-binding and modifying enzymes. Indeed, several experiments suggest that such curvature-sensitivity of lipid-modifying enzyme activities may be a general phenomenon. Phosphoinositide 3-kinase (PI 3-kinase) activity can be enhanced by higher membrane curvature [40], and PI-Specific Phospholipase C activity is also modulated by membrane curvature [41].
At endocytic sites, PIP2 phosphatases regulate PIP2 turnover [42•,43•]. The endocytic membrane is composed of bud and tubule/neck regions, each with different local curvatures. Curvature of the membrane neck increases greatly as it is squeezed during vesicle scission: if PIP2 phosphatase activity is regulated by curvature, PIP2 levels should distribute non-uniformly along curved endocytic membranes. Several independent experiments support this notion. In vitro experiments showed that hydrolysis rates of PIP2 phosphatase are critically dependent on the radius of liposomes (Chang and Di Paolo, personal communication). In vivo fluorescence experiments suggest that the PIP2 phosphatase activity concentrates at the bud region of the endocytic membrane invagination in yeast [42•]. Furthermore, PIP2-binding endocytic proteins show different localization along curved endocytic membranes [44••]. ANTH- and ENTH- domain proteins are confined to the bud region, while BAR-domain proteins are centered along the tubule region of endocytic membranes [44••]. Interestingly, the PIP2 binding affinity of BAR-domain proteins is much higher than that of ANTH- and ENTH- domain proteins [45,46]. The different PIP2 binding affinities not only dictate different abilities to accumulate local PIP2, but also should differentially protect PIP2 from hydrolysis. Due to technical limitations, definitive experimental proof for this effect has not yet been achieved; nevertheless, the above evidence suggests that spatial heterogeneity of PIP2 exists along the curved endocytic membrane.
Many in vitro experiments [47–51•] and theoretical studies [14•,48,52,53•] have shown that interfacial force can arise from lipid composition heterogeneity, and can directly squeeze the lipid domain boundary, triggering membrane fission (see Box 1 for detailed discussion of the physical mechanism of interfacial force generation). Thus the spatial heterogeneity of PIP2 along the curved endocytic membrane could give rise to a circumferential interfacial force at the bud-tubule boundary. Subsequently, the more the interfacial force squeezes the membrane neck, the more the increased curvature would promote PIP2 phosphatase activity, leading to further local depletion of PIP2. As the interfacial force is proportional to the lipid composition difference across the lipid domain boundary, the increased PIP2 level depletion at the bud-tubule boundary would induce an even larger interfacial force, forming a positive feedback loop.
An integrated theoretical model
Despite detailed knowledge of the molecular repertoire of the endocytic machinery, the interplay between endocytic proteins and lipids in space and time is largely obscure. Recently, we constructed the first coherent theoretical model of endocytic vesicle formation [54••]. This model can serve as a unified framework for investigating different types of endocytosis.
Mechanochemical feedback
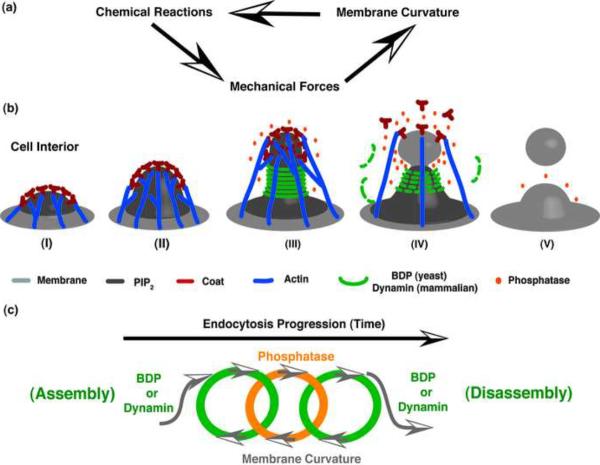
The central idea of the model is that the local membrane curvature is both slave to, and master of, the accompanying biochemical reactions involved in endocytosis (Figure 1a). Such curvature-mediated mechanochemical feedback provides a mechanism for orchestrating the process of endocytosis. The model shows that mechanochemical feedback can produce an interfacial force sufficient to pinch off a vesicle, and ensure that the process of endocytic vesicle formation is robust.
Mechanochemical feedback mechanism for endocytosis. (a) A general scheme for mechanochemical feedback during membrane vesicle formation. (b) Mechanochemical coupling drives the progression of yeast endocytic vesicle formation. The process can be divided into 5 steps. Step (I): PIP2 accumulates at the presumptive endocytic site, recruiting coat proteins to the bud region and initiating membrane invagination. The coat proteins nucleate and anchor F-actin. Step (II): Actin polymerization drives membrane invagination. Step (III): The resulting membrane curvature at the tubule region matches the preferred curvature for BDPs, and positive feedback between BDP binding and membrane shape change results in rapid local BDP recruitment. The membrane tubule is further extended by the curvature sculpting activity of BDPs. This high curvature allows the PIP2 phosphatase to access PIP2 at the endocytic site. Step (IV): BDP protection of the tubule PIP2 from phosphatase action induces PIP2 lipid phase segregation. The resulting interfacial force at the bud-tubule boundary squeezes the neck, while the coat in bud region starts to disassemble. The line tension squeezing increases the local curvature so that the phosphatase further depletes the PIP2 at the bud-tubule boundary, and this increases the interfacial force. This positive feedback loop rapidly pinches off the vesicle. Step (V): As the vesicle is pinched off, BDPs dissociate from the endocytic membrane as the local curvature deviates from the preferred curvature for BDP binding. The decreasing BDP protection concurrently increases the PIP2 phosphatase activity all over the endocytic site, and the endocytic machinery is eventually fully disassembled. (c) Endocytosis chain reactions. In both yeast and mammalian endocytosis, intertwined positive feedback loops between local endocytic membrane curvature and recruitment of endocytic proteins drives the progression of vesicle formation.
The concept of `Functional Modules'
At least 60 proteins are involved in yeast endocytosis, and an even larger number are likely involved in mammalian cells [1,2]. These endocytic proteins can be grouped into modules according to their function [20•]. The modules we used were: (1) coat proteins, (2) actin/myosin, (3) BDPs, (4) PIP2, and (5) PIP2 phosphatases. The simplification provided by the module concept allows us to take a step back and look beyond detailed molecular interactions, and to focus instead on the system level to see how endocytic membrane dynamics and local biochemical pathways might be coupled.
The mechanochemistry of yeast endocytosis
Here we use yeast as an example to show how different modules can work together in a mechanochemical feedback system to ensure robust and rapid endocytic vesicle formation (Figure 1(b); see [54••] for details).
In yeast, coat proteins first deform the membrane into a shallow dome-shaped invagination. Actin then drives of the dome deeper into the cytoplasm to form the endocytic membrane tubule. During this process the membrane is shaped by BDPs that bind at the right location (i.e. curvature) and time. Due to the curvature positive feedback, the more BDPs that bind, the faster they assemble, which in turn further tubulates the membrane. The highly curved tubule shape also activates PIP2 phosphatase activity. This gives rise to a PIP2 concentration difference across the boundary between the tubule, where BDPs protect PIP2 from hydrolysis, and the vesicle bud, which has less protection. The phase boundary thus created generates an interfacial force that squeezes the neck. As the neck constricts, its curvature increases and provides even better access for hydrolysis. Thus, the squeezing creates a positive feedback loop via the curvature-dependent PIP2 hydrolysis at the interface. The culmination of this positive feedback is the rapid evolution of a pinching force sufficient to drive vesicle scission. Eventually, as the neck constricts, the tubule curvature increases and so deviates from the shape preferred by BDPs, and this triggers rapid BDP disassembly. In this way, endocytosis proceeds like an autocatalytic chain reaction. One step paves the way for the next to drive the progression of events in which membrane curvature feedback is coupled to protein assembly and disassembly (Figure 1c). This model quantitatively recapitulates the key features of yeast endocytosis, including the temporal and spatial progression of endocytic vesicle formation.
Implications of the model for mammalian endocytosis
Since the model describes endocytosis at the level of functional modules rather than specific proteins, it allows us to `re-wire' the interaction diagram to one that applies to mammalian endocytosis. The model captures the key aspects of mammalian endocytosis, and makes predictions about specific functions of specific endocytic proteins [54••].
The model suggests that dynamin can affect the local PIP2 lipid composition, which can then give rise to an interfacial force [54••]. The novel aspect is that, rather than dynamin itself, it is the interfacial force that directly pinches off the vesicle. Dynamin binding acts as a component of a positive feedback loop with membrane shape changes. During squeezing, the local membrane curvature at the scission site deviates from the shape preferred for optimal dynamin binding, which triggers its rapid disassembly before fission. Moreover, we predict that dynamin persistence would hold the membrane tubule at its preferred diameter (~ 20nm), thus working against any further squeezing by interfacial forces. Thus dynamin disassembly is necessary for vesicle scission. This analysis predicts that dynamin must disassemble before membrane fission and provides a mechanism and function for the experimentally observed disassembly [38••,39••].
The model further predicts that endocytosis will be impaired if dynamin or actin activities are elevated. This is because in addition to the inhibitory effects of dynamin persistence, excessive actin pulling forces would raise membrane tension, thus opposing pinching and scission. Interestingly, dynamin was shown to negatively regulate actin polymerization during mammalian endocytosis [55]. It is tempting to speculate that such dynamin-actin interactions could be a safety measure evolved to ensure successful endocytosis.
Interactions between BDPs, actin and dynamin in mammalian endocytosis
In contrast to yeast, in mammalian endocytosis there is an extra level of regulation by BDPs that affects dynamin's curvature-mediated feedback loop [56–58]. It would be interesting to compare and dissect endocytosis across organisms from an evolutionary perspective; in fact, this has been initiated in [59]. Moreover, BDPs in yeast must rely on actin to tubulate the endocytic membrane in order to initiate their own assembly. In mammalian cells, however, dynamin alone can make full use of the positive feedback loop between curvature sensing and sculpting activities to deform the endocytic membrane. This could be due to dynamin's utilization of GTPase hydrolysis energy [33••]. Without membrane tubulation driven by actin polymerization, the mammalian endocytic membrane is shaped like a constricted coated vesicle with a very short neck, rather than the elongated tubule seen in yeast. Thus actin may play a more passive role in mammalian endocytosis; it may simply follow the membrane invagination driven by dynamin, instead of creating the membrane shape for BDP assembly as in yeast. Roles for dynamin and actin underscore a key difference between mammalian and yeast endocytosis.
Implications for other endomembrane trafficking processes
Despite their diverse molecular components, mechanochemical feedback may be a general theme shared by different vesicle formation processes. In this section, we summarize our current knowledge of other membrane remodeling systems in this regard.
In the ER (endoplasmic reticulum), PI (phosphoinositide) is the major component of phosphoinositol lipids, and PIP2 phosphatases are essential for ER function. The curvature-sensitive Sar1 GTPase is crucial for recruiting the COPII complex that packages cargo into vesicles [60]. Sar1 is capable of generating membrane curvature [61]. As with the positive feedback loop proposed in our model, Sar1 is likely to be a curvature sensor. However, Sar1 alone cannot trigger membrane fission in a purified in vitro system [62]. In this sense, Sar1 might function similarly to BDPs, or like dynamin since it consumes GTP. We predict that Sar1 may need to collaborate with other proteins, such as phosphoinositol lipid-modifying enzymes, to regulate the local phosphoinositol lipid levels to complete vesicle scission. In the Golgi, the Arf1 GTPase helps organize the COPI complex, which in turn recruits cargo to promote vesicle formation [60]. Arf1 GAP activates Arf GTPases and, hence, releases Arf GDP from the cargo/vesicle. The release of Arf GDP is essential for vesicle fission, and membrane curvature greatly stimulates Arf1 GTPase activity [63•]. The activities of phospholipase C and PIP2 phosphatases are both essential for vesicle scission in the Golgi, and both are curvature-sensitive [41].
In early endosomes, in addition to Arf6 GTPase, the protein ctBP/BARS appears to be essential for vesicle fission. ctBP/BARS has been demonstrated to tubulate membranes, and phospholipase D appears to be essential for membrane fission in early endosomes [64,65]. In late endosomes, the ESCRT III complex is important for vesicle budding [66,67] and recently it has been shown by in vitro assays that ESCRT III can trigger membrane fission in a fashion similar to dynamin [68•], and like dynamin, it falls off the membrane before fission. Lastly, in mitochondria, dynamin-related GTPases are essential for fission of both outer and inner membranes [69,70].
Conclusions
Our theoretical model provides insights into the mechanochemistry of endocytic vesicle formation. We suggest that during endocytosis local membrane curvature is both slave to, and master of, the accompanying biochemical reactions. We expect this mechanochemical feedback mechanism to be a general feature of membrane trafficking processes. The challenge for biologists is to determine how each mechanochemical feedback mechanism operates at the molecular level. We believe that a close synergy between theory and experiments will be key to achieving this daunting task.
Box 1. Physical mechanism of interfacial force generation.
Cell membranes consist of many different lipids. Experiments have demonstrated the existence of lipid domains (e.g., `lipid rafts') in cell membranes in vivo [71–74], although controversies regarding their size, composition, lifetime and functional roles remain [75].
From a thermodynamics perspective, spatial organization of lipids within a membrane results from the competition between entropy and enthalpy [4]. On the one hand, lipid molecules are highly diffusive, which is driven by entropy and tends to smear out any non-uniform lipid distribution. On the other hand, the interactions amongst different lipids can be very different: some are more attractive to one another while others are repulsive. Such energetic differences are enthalpic in nature, and have several origins. They could be due to different electrostatic properties of lipids, particularly dipolar interactions [76,77]. They could also originate from lipid height mismatches [78–81]; that is, when longer lipids mix with shorter ones to form membranes, the longer ones expose more of their hydrophobic core to the water, which is energetically unfavorable. This effect would tend to cluster longer lipids together. Similarly, energy differences could stem from differences in hydrogen bonds that can form between lipids, e.g., PIP2 and non-PIP2 lipids [82,83].
When enthalpy dominates over entropy, the lipids with favorable interactions will form domains enriched in some lipid species. Within a domain, energetically unfavorable interactions still exist along the domain boundary. Here an `interfacial force' develops proportional to the lipid composition difference across the lipid domain boundary [4]. As with oil droplets in water, the interfacial force provides the thermodynamic driving force to minimize the lipid domain boundary, causing the lipid domain to bulge out-of-plane, and potentially to pinch off as a vesicle [14,52,53•]. In the case of a lipid bilayer, one can theoretically estimate the interfacial force arising from a lipid height mismatch using the lipid-water surface tension energy (~ 50mN/m) [37]. A lipid height mismatch of 0.5–1 nm can produce an interfacial force of ~25–50 pN, sufficient to pinch off lipid vesicles[4].
The idea that an interfacial force can drive vesicle scission is supported by many in vitro experiments, in which lipid domains form on liposomes or lipid bilayers when the temperature is lowered [47–51•]. Interestingly, the interfacial forces derived from these experiments are only about one pN, much smaller than theoretical estimates. How can we reconcile this discrepancy? One explanation is as follows. The large interfacial force of 10's of pNs is the initial driving force before lipid re-arrangement, which can proceed in at least two ways. (1) An out-of-plane membrane shape change is driven by the initial interfacial force, which could remain constant throughout the process. (2) An in-plane tilting of the lipids that shields their hydrophobic core from water. This minimizes the initial interfacial force. Process (1) is a collective behavior that is relatively slow (e.g., the relaxation time of out-of-plane membrane fluctuation is ~ 1s) [84]. Process (2) is much faster (e.g., the relaxation time of lipid wobbling and self-rotation is ~1–100 ns)[85]. Thus, it is possible that the measured interfacial force is simply the resulting residual energy penalty for exposing the hydrophobic core to water after the lipids have already optimized their configuration via process (2). To balance the bending energy of the lipid bilayer, this residual energy penalty from which the interfacial force is derived, has to be comparable to thermal fluctuations at equilibrium (1 kBT per lipid; 1 kBT ~ 4 pN·nm). Since the length scale of each lipid head group is ~ 1 nm, the effective force from thermal fluctuation is ~ 4 pN per lipid, exactly in the range of the interfacial force determined by in vitro measurements. Thus, the measured interfacial force may be the result of lipid domain formation, rather than the driving force for it.
Cells maintain constant temperature, and so lipid-protein interactions may drive lipid domain formation. Many proteins, including BDPs and actin filaments [86•,87•], can not only bind (directly or indirectly) with high affinity to PIP2-containing lipid bilayers. They can also induce PIP2 lipid domain formation. Thus, lipid domain formation can result from lipid-protein interactions. In this case, a theoretical estimate of the interfacial force induced by lipid-protein interaction can be derived. The compressibility modulus of a lipid bilayer is typically ~10–100 mN/m [88–90]. Inserting a typical protein into the membrane (e.g., with diameter ~ 1 nm) would incur an interfacial force of 10–100pN from the surrounding lipids [91]. As lipid tilting is more restricted due to lipid-protein binding [91], the initial interfacial force of 10's pNs could persist throughout the process of vesicle scission in vivo.
Acknowledgements
We thank Dr. Helen Stimpson for her critical reading of the manuscript and helpful suggestions. This work was supported by National Institutes of General Medical Sciences grants 1 R01 GM 42759, 1 R01 GM 50399 and 1 R01 GM 65462 to DGD. GFO was supported by National Science Foundation grant DMS 0414039.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest published within the period of review have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Perrais D, Merrifield CJ. Dynamics of Endocytic Vesicle Creation. Developmental Cell. 2005;9:581–592. doi: 10.1016/j.devcel.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 2.Toret CP, Drubin DG. The budding yeast endocytic pathway. J Cell Sci. 2006;119:4585–4587. doi: 10.1242/jcs.03251. [DOI] [PubMed] [Google Scholar]
- 3.Janmey PA, Kinnunen PKJ. Biophysical properties of lipids and dynamic membranes. Trends in Cell Biology. 2006;16:538–546. doi: 10.1016/j.tcb.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 4.Lipowski R, Sackmann E. Structure and Dynamics of Membranes. Amsterdam; North–Holland: 1995. [Google Scholar]
- 5.Munn AL. Molecular requirements for the internalisation step of endocytosis: insights from yeast. Biochim Biophys Acta. 2001;1535:236–257. doi: 10.1016/s0925-4439(01)00028-x. [DOI] [PubMed] [Google Scholar]
- 6.Engqvist-Goldstein AE, Drubin DG. Actin assembly and endocytosis: from yeast to mammals. Annu Rev Cell Dev Biol. 2003;19:287–332. doi: 10.1146/annurev.cellbio.19.111401.093127. [DOI] [PubMed] [Google Scholar]
- 7.Lemmon MA. Phosphoinositide recognition domains. Traffic. 2003;4:201–213. doi: 10.1034/j.1600-0854.2004.00071.x. [DOI] [PubMed] [Google Scholar]
- 8.Robertson AS, Smythe E, Ayscough KR. Functions of actin in endocytosis. Cell Mol Life Sci. 2009;66:2049–2065. doi: 10.1007/s00018-009-0001-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aghamohammadzadeh S, Ayscough KR. Differential requirements for actin during yeast and mammalian endocytosis. Nat Cell Biol. 2009;11:1039–1042. doi: 10.1038/ncb1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••10.Kaksonen M, Sun Y, Drubin DG. A Pathway for Association of Receptors, Adaptors, and Actin during Endocytic Internalization. Cell. 2003;115:475–487. doi: 10.1016/s0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]; This work, together with ref [17, 20, 42, 43], provides the first direct evidence that endocytic proteins are sequentially recruited to endocytic sites during clathrin-dependent endocytosis of budding yeast. It shows that such sequential recruitment is very robust and regular.
- 11.Mogilner A, Oster G. Force Generation by Actin Polymerization II: The Elastic Ratchet and Tethered Filaments. Vol. 84. 2003. pp. 1591–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mogilner A, Oster G. Cell motility driven by actin polymerization. Vol. 71. 1996. pp. 3030–3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kovar DR, Pollard TD. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proceedings of the National Academy of Sciences. 2004;101:14725–14730. doi: 10.1073/pnas.0405902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J, Kaksonen M, Drubin DG, Oster G. Endocytic vesicle scission by lipid phase boundary forces. Proceedings of the National Academy of Sciences. 2006;103:10277–10282. doi: 10.1073/pnas.0601045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doherty GJ, McMahon HT. Mechanisms of endocytosis. Annu Rev Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- 16.Krendel M, Osterweil EK, Mooseker MS. Myosin 1E interacts with synaptojanin-1 and dynamin and is involved in endocytosis. FEBS Lett. 2007;581:644–650. doi: 10.1016/j.febslet.2007.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •17.Sun Y, Martin AC, Drubin DG. Endocytic Internalization in Budding Yeast Requires Coordinated Actin Nucleation and Myosin Motor Activity. Developmental Cell. 2006;11:33–46. doi: 10.1016/j.devcel.2006.05.008. [DOI] [PubMed] [Google Scholar]; This work illustrates that both the actin nucleation and motor activities of type I myosin are essential for endocytic vesicle formation, and that these roles are separable.
- 18.Laakso JM, Lewis JH, Shuman H, Ostap EM. Myosin I Can Act As a Molecular Force Sensor. Science. 2008;321:133–136. doi: 10.1126/science.1159419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frost A, Unger VM, De Camilli P. The BAR domain superfamily: membrane-molding macromolecules. Cell. 2009;137:191–196. doi: 10.1016/j.cell.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •20.Kaksonen M, Toret CP, Drubin DG. A Modular Design for the Clathrin- and Actin-Mediated Endocytosis Machinery. Cell. 2005;123:305–320. doi: 10.1016/j.cell.2005.09.024. [DOI] [PubMed] [Google Scholar]; This work provides evidence for functional modules in yeast endocytosis. It also demonstrates a role for BAR-domain proteins in vesicle scission.
- 21.Zimmerberg J, McLaughlin S. Membrane Curvature: How BAR Domains Bend Bilayers. Current Biology. 2004;14:R250–R252. doi: 10.1016/j.cub.2004.02.060. [DOI] [PubMed] [Google Scholar]
- •22.Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJG, Evans PR, McMahon HT. BAR Domains as Sensors of Membrane Curvature: The Amphiphysin BAR Structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- •23.Gallop JL, Jao CC, Kent HM, Butler PJG, Evans PR, Langen R, McMahon HT. Mechanism of endophilin N-BAR domain-mediated membrane curvature. EMBO J. 2006;25:2898–2910. doi: 10.1038/sj.emboj.7601174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •24.Frost A, Perera R, Roux A, Spasov K, Destaing O, Egelman EH, De Camilli P, Unger VM. Structural Basis of Membrane Invagination by F-BAR Domains. Cell. 2008;132:807–817. doi: 10.1016/j.cell.2007.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •25.Henne WM, Kent HM, Ford MGJ, Hegde BG, Daumke O, Butler PJG, Mittal R, Langen R, Evans PR, McMahon HT. Structure and Analysis of FCHo2 F-BAR Domain: A Dimerizing and Membrane Recruitment Module that Effects Membrane Curvature. Structure. 2007;15:839–852. doi: 10.1016/j.str.2007.05.002. [DOI] [PubMed] [Google Scholar]; The work in ref [22–25] collectively show that many BAR-domain and F-BAR proteins share a general characteristics: they are not only capable of curvature sensing, but also capable of curvature sculpting.
- 26.Itoh T, De Camilli P. BAR, F-BAR (EFC) and ENTH/ANTH domains in the regulation of membrane-cytosol interfaces and membrane curvature. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2006;1761:897–912. doi: 10.1016/j.bbalip.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 27.Blood PD, Swenson RD, Voth GA. Factors Influencing Local Membrane Curvature Induction by N-BAR Domains as Revealed by Molecular Dynamics Simulations. Biophys. J. 2008;95:1866–1876. doi: 10.1529/biophysj.107.121160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yin Y, Arkhipov A, Schulten K. Simulations of Membrane Tubulation by Lattices of Amphiphysin N-BAR Domains. Vol. 17. 2009. pp. 882–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernandes F, Loura LMS, Chichon FJ, Carrascosa JL, Fedorov A, Prieto M. Role of Helix 0 of the N-BAR Domain in Membrane Curvature Generation. Biophys. J. 2008;94:3065–3073. doi: 10.1529/biophysj.107.113118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••30.Merrifield CJ, Feldman ME, Wan L, Almers W. Imaging actin and dynamin recruitment during invagination of single clathrin-coated pits. Nat Cell Biol. 2002;4:691–698. doi: 10.1038/ncb837. [DOI] [PubMed] [Google Scholar]; This work provides the first direct evidence that endocytic proteins are sequentially recruited to endocytic sites during clathrin-dependent endocytosis of mammalian cells. It shows that dynamin accumulates at the endocytic site first, followed by actin.
- ••31.Sever S, Damke H, Schmid SL. Dynamin:GTP Controls the Formation of Constricted Coated Pits, the Rate Limiting Step in Clathrin-mediated Endocytosis. J. Cell Biol. 2000;150:1137–1148. doi: 10.1083/jcb.150.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Damke H, Baba T, Warnock DE, Schmid SL. Induction of mutant dynamin specifically blocks endocytic coated vesicle formation. J. Cell Biol. 1994;127:915–934. doi: 10.1083/jcb.127.4.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••33.Marks B, Stowell MHB, Vallis Y, Mills IG, Gibson A, Hopkins CR, McMahon HT. GTPase activity of dynamin and resulting conformation change are essential for endocytosis. Nature. 2001;410:231–235. doi: 10.1038/35065645. [DOI] [PubMed] [Google Scholar]
- 34.Praefcke GJK, McMahon HT. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol. 2004;5:133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- 35.Song BD, Schmid SL. A Molecular Motor or a Regulator? Dynamin's in a Class of Its Own. Biochemistry. 2003;42:1369–1376. doi: 10.1021/bi027062h. [DOI] [PubMed] [Google Scholar]
- •36.Danino D, Moon K-H, Hinshaw JE. Rapid constriction of lipid bilayers by the mechanochemical enzyme dynamin. Journal of Structural Biology. 2004;147:259–267. doi: 10.1016/j.jsb.2004.04.005. [DOI] [PubMed] [Google Scholar]; Ref [31, 33, 36] demonstrate that dynamin is not only essential for clathrin-dependent endocytosis in mammalian cells, but also that dynamin GTPase activity is necessary for the constriction of membrane neck and, hence, endocytic vesicle formation.
- 37.Israelachvili . Intermolecular and Suface forces. Academic press; 1992. [Google Scholar]
- ••38.Pucadyil TJ, Schmid SL. Real-Time Visualization of Dynamin-Catalyzed Membrane Fission and Vesicle Release. Cell. 2008;135:1263–1275. doi: 10.1016/j.cell.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••39.Bashkirov PV, Akimov SA, Evseev AI, Schmid SL, Zimmerberg J, Frolov VA. GTPase Cycle of Dynamin Is Coupled to Membrane Squeeze and Release, Leading to Spontaneous Fission. Cell. 2008;135:1276–1286. doi: 10.1016/j.cell.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]; Using very innovative in vitro experimental techniques, Ref [38, 39] not only demonstrate that dynamin alone can trigger membrane fission under the right conditions, but also provide the key evidence that dynamin falls off the membrane before fission. This result suggests that dynamin may not directly pinch off the vesicle; it points to the possibility that dynamin could affect the local lipid composition or configuration to trigger membrane fission.
- 40.Hubner S, Couvillon AD, Kas JA, Bankaitis VA, Vegners R, Carpenter CL, Janmey PA. Enhancement of phosphoinositide 3-kinase (PI 3-kinase) activity by membrane curvature and inositol-phospholipid-binding peptides. Eur J Biochem. 1998;258:846–853. doi: 10.1046/j.1432-1327.1998.2580846.x. [DOI] [PubMed] [Google Scholar]
- 41.Ahyayauch H, Villar AV, Alonso A, Goni FM. Modulation of PI-specific phospholipase C by membrane curvature and molecular order. Biochemistry. 2005;44:11592–11600. doi: 10.1021/bi050715k. [DOI] [PubMed] [Google Scholar]
- •42.Sun Y, Carroll S, Kaksonen M, Toshima JY, Drubin DG. PtdIns(4,5)P2 turnover is required for multiple stages during clathrin- and actin-dependent endocytic internalization. J. Cell Biol. 2007;177:355–367. doi: 10.1083/jcb.200611011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •43.Perera RM, Zoncu R, Lucast L, De Camilli P, Toomre D. Two synaptojanin 1 isoforms are recruited to clathrin-coated pits at different stages. Proceedings of the National Academy of Sciences. 2006;103:19332–19337. doi: 10.1073/pnas.0609795104. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ref [42, 43] provides key evidence that not only PIP2 hydrolysis is essential for uncoating the endocytic vesicle, but also of critical for endocytic vesicle formation.
- ••44.Idrissi F-Z, Grotsch H, Fernandez-Golbano IM, Presciatto-Baschong C, Riezman H, Geli M-I. Distinct acto/myosin-I structures associate with endocytic profiles at the plasma membrane. J. Cell Biol. 2008;180:1219–1232. doi: 10.1083/jcb.200708060. [DOI] [PMC free article] [PubMed] [Google Scholar]; Using Immuno-EM, this work provides the clearest picture of the differential distribution of essential endocytic proteins along endocytic membrane invaginations. Specifically, it shows that coat proteins concentrate at the bud region, BAR-domain proteins accumulate at the tubule region, and actin filaments are all over the endocytic site. It provides definitive support for conclusions from fluorescence microscopy studies [10, 17, 20].
- 45.Ford MGJ, Pearse BMF, Higgins MK, Vallis Y, Owen DJ, Gibson A, Hopkins CR, Evans PR, McMahon HT. Simultaneous Binding of PtdIns(4,5)P2 and Clathrin by AP180 in the Nucleation of Clathrin Lattices on Membranes. Science. 2001;291:1051–1055. doi: 10.1126/science.291.5506.1051. [DOI] [PubMed] [Google Scholar]
- 46.Itoh T, Koshiba S, Kigawa T, Kikuchi A, Yokoyama S, Takenawa T. Role of the ENTH Domain in Phosphatidylinositol-4,5-Bisphosphate Binding and Endocytosis. Science. 2001;291:1047–1051. doi: 10.1126/science.291.5506.1047. [DOI] [PubMed] [Google Scholar]
- •47.Baumgart T, Hess ST, Webb WW. Imaging coexisting fluid domains in biomembrane models coupling curvature and line tension. Nature. 2003;425:821–824. doi: 10.1038/nature02013. [DOI] [PubMed] [Google Scholar]
- •48.Allain JM, Storm C, Roux A, Amar MB, Joanny JF. Fission of a Multiphase Membrane Tube. Physical Review Letters. 2004;93:158104–158104. doi: 10.1103/PhysRevLett.93.158104. [DOI] [PubMed] [Google Scholar]
- •49.Rozovsky S, Kaizuka Y, Groves JT. Formation and Spatio-Temporal Evolution of Periodic Structures in Lipid Bilayers. Journal of the American Chemical Society. 2004;127:36–37. doi: 10.1021/ja046300o. [DOI] [PubMed] [Google Scholar]
- •50.Roux A, Cuvelier D, Nassoy P, Prost J, Bassereau P, Goud B. Role of curvature and phase transition in lipid sorting and fission of membrane tubules. The EMBO Journal. 2005;24:1537–1545. doi: 10.1038/sj.emboj.7600631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •51.Veatch SL, Keller SL. Separation of Liquid Phases in Giant Vesicles of Ternary Mixtures of Phospholipids and Cholesterol. Vol. 85. 2003. pp. 3074–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]; Using liposomes, supported bilayers or membrane tubules consisting of different kinds of lipids, ref [47–51] provide definitive evidence that interfacial force at lipid phase boundaries is capable of inducing membrane fission.
- 52.Julicher F, Lipowsky R. Domain-induced budding of vesicles. Physical Review Letters. 1993;70:2964. doi: 10.1103/PhysRevLett.70.2964. [DOI] [PubMed] [Google Scholar]
- •53.Lipowsky R. Budding of membranes induced by intramembrane domains. J Phys. II France. 1992;2:1825–1840. [Google Scholar]; This is the first theoretical model to show how the antagonism between the interfacial force from lipid phase segregation and the membrane bending modulus in determines the final membrane shape.
- ••54.Liu J, Sun Y, Drubin D, Oster G. The mechanochemistry of endocytosis. PLoS Biology. 2009 doi: 10.1371/journal.pbio.1000204. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is the first theoretical model that recapitulates all of the key features of clathrin- and actin-dependent endocytosis in coherent manner for both yeast and mammalian cells. It also provides the framework to understand many other vesicle formation processes.
- 55.Schafer DA, Weed SA, Binns D, Karginov AV, Parsons JT, Cooper JA. Dynamin2 and Cortactin Regulate Actin Assembly and Filament Organization. Current Biology. 2002;12:1852–1857. doi: 10.1016/s0960-9822(02)01228-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shin N, Ahn N, Chang-Ileto B, Park J, Takei K, Ahn S-G, Kim S-A, Di Paolo G, Chang S. SNX9 regulates tubular invagination of the plasma membrane through interaction with actin cytoskeleton and dynamin 2. J Cell Sci. 2008;121:1252–1263. doi: 10.1242/jcs.016709. [DOI] [PubMed] [Google Scholar]
- 57.Yarar D, Waterman-Storer CM, Schmid SL. SNX9 Couples Actin Assembly to Phosphoinositide Signals and Is Required for Membrane Remodeling during Endocytosis. Vol. 13. 2007. pp. 43–56. [DOI] [PubMed] [Google Scholar]
- 58.Soulet F, Yarar D, Leonard M, Schmid SL. SNX9 Regulates Dynamin Assembly and Is Required for Efficient Clathrin-mediated Endocytosis. Mol. Biol. Cell. 2005;16:2058–2067. doi: 10.1091/mbc.E04-11-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Elde N, Morgan G, Winey M, Sperling L, Turkewitz A. Elucidation of Clathrin-Mediated Endocytosis in Tetrahymena Reveals an Evolutionarily Convergent Recruitment of Dynamin. PLoS Genetics. 2005;1:e52. doi: 10.1371/journal.pgen.0010052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee MCS, Miller EA, Goldberg J, Orci L, Schekman R. BI-DIRECTIONAL PROTEIN TRANSPORT BETWEEN THE ER AND GOLGI. Annual Review of Cell and Developmental Biology. 2004;20:87. doi: 10.1146/annurev.cellbio.20.010403.105307. [DOI] [PubMed] [Google Scholar]
- 61.Lee MCS, Orci L, Hamamoto S, Futai E, Ravazzola M, Schekman R. Sar1p N-Terminal Helix Initiates Membrane Curvature and Completes the Fission of a COPII Vesicle. Cell. 2005;122:605–617. doi: 10.1016/j.cell.2005.07.025. [DOI] [PubMed] [Google Scholar]
- 62.Futai E, Hamamoto S, Orci L, Schekman R. GTP/GDP exchange by Sec12p enables COPII vesicle bud formation on synthetic liposomes. EMBO J. 2004;23:4286–4296. doi: 10.1038/sj.emboj.7600428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •63.Bigay J, Gounon P, Robineau S, Antonny B. Lipid packing sensed by ArfGAP1 couples COPI coat disassembly to membrane bilayer curvature. Nature. 2003;426:563–566. doi: 10.1038/nature02108. [DOI] [PubMed] [Google Scholar]; Using utilizing liposomes of different sizes, this study demonstrates that ArfGTPase hydrolysis activity critically depends on the local membrane curvature. This result suggests that membrane curvature could be an important regulatory factor in governing vesicle formation on Golgi membranes.
- 64.Yang J-S, Gad H, Lee SY, Mironov A, Zhang L, Beznoussenko GV, Valente C, Turacchio G, Bonsra AN, Du G, et al. A role for phosphatidic acid in COPI vesicle fission yields insights into Golgi maintenance. Nat Cell Biol. 2008;10:1146–1153. doi: 10.1038/ncb1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Haga Y, Miwa N, Jahangeer S, Okada T, Nakamura S-i. CtBP1/BARS is an activator of phospholipase D1 necessary for agonist-induced macropinocytosis. EMBO J. 2009;28:1197–1207. doi: 10.1038/emboj.2009.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saksena S, Sun J, Chu T, Emr SD. ESCRTing proteins in the endocytic pathway. Trends in Biochemical Sciences. 2007;32:561–573. doi: 10.1016/j.tibs.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 67.Williams RL, Urbe S. The emerging shape of the ESCRT machinery. Nat Rev Mol Cell Biol. 2007;8:355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- •68.Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. Membrane scission by the ESCRT-III complex. Nature. 2009;458:172–177. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]; Using an in vitro set-up, this study demonstrates for the first time that the ESCRT-III complex alone is sufficient to trigger membrane fission.
- 69.Hoppins S, Lackner L, Nunnari J. The Machines that Divide and Fuse Mitochondria. Annual Review of Biochemistry. 2007;76:751. doi: 10.1146/annurev.biochem.76.071905.090048. [DOI] [PubMed] [Google Scholar]
- 70.Chan DC. Mitochondrial Fusion and Fission in Mammals. Annual Review of Cell and Developmental Biology. 2006;22:79. doi: 10.1146/annurev.cellbio.22.010305.104638. [DOI] [PubMed] [Google Scholar]
- 71.Brown DA, London E. FUNCTIONS OF LIPID RAFTS IN BIOLOGICAL MEMBRANES. Annual Review of Cell and Developmental Biology. 1998;14:111. doi: 10.1146/annurev.cellbio.14.1.111. [DOI] [PubMed] [Google Scholar]
- 72.Simons K, Toomre D. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 73.Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 74.Simons K, Vaz WLC. MODEL SYSTEMS, LIPID RAFTS, AND CELL MEMBRANES1. Annual Review of Biophysics and Biomolecular Structure. 2004;33:269. doi: 10.1146/annurev.biophys.32.110601.141803. [DOI] [PubMed] [Google Scholar]
- 75.Edidin M. Shrinking patches and slippery rafts: scales of domains in the plasma membrane. Trends in Cell Biology. 2001;11:492–496. doi: 10.1016/s0962-8924(01)02139-0. [DOI] [PubMed] [Google Scholar]
- 76.Leibler S, Andelman D. Ordered and curved meso-structures in membranes and amphiphilic films. Journal de Physique II. 1987;48:2013–2018. [Google Scholar]
- 77.Liu J, Qi S, Groves JT, Chakraborty AK. Phase Segregation on Different Length Scales in a Model Cell Membrane System. The Journal of Physical Chemistry B. 2005;109:19960–19969. doi: 10.1021/jp053562j. [DOI] [PubMed] [Google Scholar]
- 78.Garcia-Saez AJ, Chiantia S, Schwille P. Effect of Line Tension on the Lateral Organization of Lipid Membranes. J. Biol. Chem. 2007;282:33537–33544. doi: 10.1074/jbc.M706162200. [DOI] [PubMed] [Google Scholar]
- 79.Jorgensen K, Mouritsen OG. Phase separation dynamics and lateral organization of two-component lipid membranes. Biophysical Journal. 1995;69:942–954. doi: 10.1016/S0006-3495(95)79968-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wallace EJ, Hooper NM, Olmsted PD. Effect of Hydrophobic Mismatch on Phase Behavior of Lipid Membranes. Biophysical Journal. 2006;90:4104–4118. doi: 10.1529/biophysj.105.062778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Akimov SA, Chizmadzhev YA, Zimmerberg J, Cohen FS. Line Tension Of Membrane Domains Calculated From Chemical Interactions Betweem Lipids And Elastic Splay And Tilt. Biophysical Journal. 2009;96:607a–607a. doi: 10.1529/biophysj.104.048223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Levental I, Janmey PA, Cebers A. Electrostatic Contribution to the Surface Pressure of Charged Monolayers Containing Polyphosphoinositides. Biophys. J. 2008;95:1199–1205. doi: 10.1529/biophysj.107.126615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Levental I, Cebers A, Janmey PA. Combined Electrostatics and Hydrogen Bonding Determine Intermolecular Interactions Between Polyphosphoinositides. J. Am. Chem. Soc. 2008;130:9025–9030. doi: 10.1021/ja800948c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Groves JT. Bending Mechanics and Molecular Organization in Biological Membranes. Annual Review of Physical Chemistry. 2007;58:697. doi: 10.1146/annurev.physchem.56.092503.141216. [DOI] [PubMed] [Google Scholar]
- 85.Klauda JB, Roberts MF, Redfield AG, Brooks BR, Pastor RW. Rotation of Lipids in Membranes: Molecular Dynamics Simulation, 31P Spin-Lattice Relaxation, and Rigid-Body Dynamics. Biophys. J. 2008;94:3074–3083. doi: 10.1529/biophysj.107.121806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •86.Liu AP, Fletcher DA. Actin Polymerization Serves as a Membrane Domain Switch in Model Lipid Bilayers. Biophysical Journal. 2006;91:4064–4070. doi: 10.1529/biophysj.106.090852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •87.Saarikangas J, Zhao H, Pykäläinen A, Laurinmäki P, Mattila PK, Kinnunen PKJ, Butcher SJ, Lappalainen P. Molecular Mechanisms of Membrane Deformation by I-BAR Domain Proteins. Current Biology. 2009;19:95–107. doi: 10.1016/j.cub.2008.12.029. [DOI] [PubMed] [Google Scholar]; Using in vitro assays with purified proteins, Ref. [86] and [87] demonstrate that binding between PIP2 and endocytic proteins, such as actin and BAR-domain proteins, can trigger PIP2 lipid domain formation.
- 88.Rawicz W, Olbrich KC, McIntosh T, Needham D, Evans E. Effect of Chain Length and Unsaturation on Elasticity of Lipid Bilayers. Biophys. J. 2000;79:328–339. doi: 10.1016/S0006-3495(00)76295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lindahl E, Edholm O. Mesoscopic Undulations and Thickness Fluctuations in Lipid Bilayers from Molecular Dynamics Simulations. Biophys. J. 2000;79:426–433. doi: 10.1016/S0006-3495(00)76304-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Evans E, Rawicz W. Entropy-driven tension and bending elasticity in condensed-fluid membranes. Physical Review Letters. 1990;64:2094. doi: 10.1103/PhysRevLett.64.2094. [DOI] [PubMed] [Google Scholar]
- 91.Dan N, Safran SA. Effect of Lipid Characteristics on the Structure of Transmembrane Proteins. Biophys. J. 1998;75:1410–1414. doi: 10.1016/S0006-3495(98)74059-7. [DOI] [PMC free article] [PubMed] [Google Scholar]