SUMMARY
Stress-induced mutagenesis describes the accumulation of mutations that occur in non-growing cells, in contrast to mutagenesis that occurs in actively dividing populations, and has been referred to as stationary phase or adaptive mutagenesis. The most widely studied system for stress-induced mutagenesis involves monitoring the appearance of Lac+ revertants of the strain FC40 under starvation conditions in Escherichia coli [1]. The SOS inducible translesion DNA polymerase DinB [2–4], plays a particularly important role in this phenomenon. Loss of DinB (DNA pol IV) function results in a severe reduction of Lac+ revertants [5, 6]. We previously reported that NusA, an essential component of elongating RNA polymerases, interacts with DinB [7]. Here we report our unexpected observation that wild-type NusA function is required for stress-induced mutagenesis. We present evidence that this effect is unlikely to be due to defects in transcription of lac genes, but rather due to an inability to adapt and mutate in response to environmental stress. Furthermore, we extended our analysis to the formation of stress-induced mutants in response to antibiotic treatment, observing the same striking abolition of mutagenesis under entirely different conditions. Our results are the first to implicate NusA as a crucial participant in the phenomenon of stress-induced mutagenesis, in addition to its roles in transcription elongation and termination.
RESULTS AND DISCUSSION
nusA+ function is indispensible for the viability of bacteria and its roles in transcription elongation and termination have been appreciated for many years, however our recent data suggest that NusA may also be involved in the process of DNA damage repair/tolerance. We previously reported that the temperature sensitivity of the nusA11(ts) allele [8] can be suppressed by overexpression of the TLS (translesion synthesis) polymerase DinB, in a manner that requires the catalytic, and specifically the translesion DNA synthesis, capabilities of DinB [7]. Intrigued by these observations, we looked to see if the DinB dependent phenomenon of stress-induced mutagenesis was altered in a nusA11 background.
The widely used system devised by Cairns and Foster, utilizes a strain (FC40) which has lac deleted on the chromosome and carries an F'128 episome containing a lacI33-lacZ fusion with a +1 frameshift in a run of G:C base pairs [1]. When E. coli are starved for a carbon source and plated onto minimal lactose medium, Lac+ mutants occur at a rate of approximately 10−7 per cell per day for about 7 days [1]. Furthermore, it has been found that stress-induced Lac+ mutations mainly result from a −1 frameshift mutation in a run of consecutive bases [9, 10]. DinB in both prokaryotes and eukaryotes is proficient for the formation of −1 frameshift mutations [11, 12]. The analysis of dinB’s role in stress-induced mutagenesis is complicated by the fact that dinB+ is present on the F' episome as well as on the chromosome. However, depending on the dinB alleles used loss of DinB function results in a 2–4 fold reduction in the appearance of Lac+ revertants [5, 6]. Additionally, it has recently been found that dinB+ is the only SOS regulated gene required at induced levels for stress-induced mutagenesis [13].
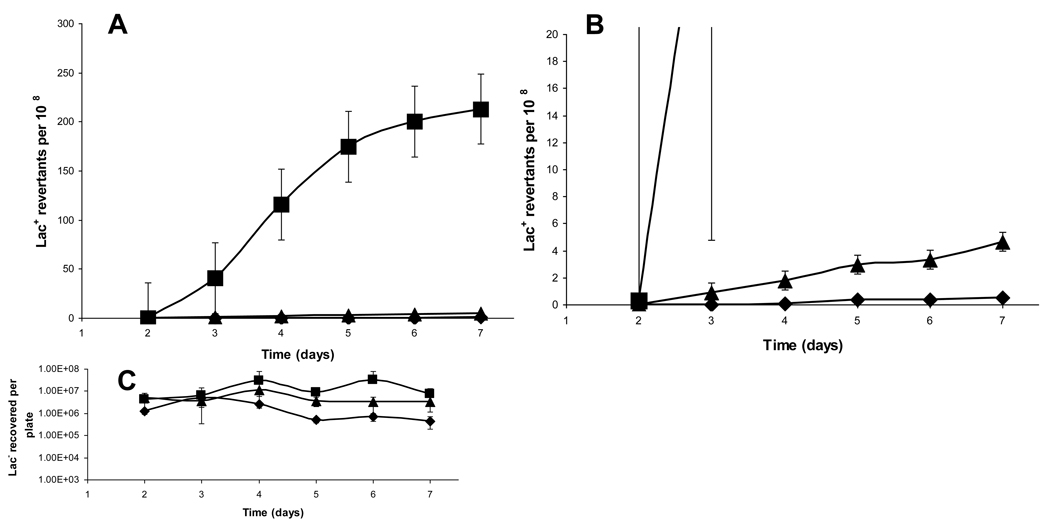
Using the lacI33-lacZ fusion system described above, we found that FC40 cells carrying the nusA11 mutation display an approximately 470-fold reduction of the rate of mutation to Lac+ at the permissive temperature (30°C), with reversion to Lac+ occurring at a rate of 0.14 ± 0.03/ 108 cells/day compared to the wild-type (67 ± 4.5/108 cells/day) (Figure 1A–B). The reduction in stress-induced mutations observed in a nusA11 background is even more extreme than the reduction that is seen in cells which are deleted for both the chromosomal and episomal copies of dinB (1 ± 0.05 Lac+ revertants/108 cells/day) (Figure 1A–B). These results suggest that the role of nusA+ in stress-induced mutagenesis may extend beyond an interaction with DinB and the formation of −1 frameshift mutations. Additionally we observe that the effect of nusA11 on stress-induced mutagenesis is independent of the cells’ dinB status, as FC40 nusA11 derivatives lacking the episomal copy of dinB, the chromosomal copy, or both show the same low level of stress-induced mutagenesis as the nusA11 dinB+ parent strain (data not shown). Daily measurements of viable Lac− cells on the plates show that no significant net growth or death during the course of the experiments and indicate that the nusA11 cells were not dying upon incubation on minimal lactose plates, but rather were incapable of mutating (Figure 1C).
Figure 1.
Stress-induced mutation in response to starvation A) nusA11 strains are defective for stress-induced mutagenesis. Wild-type FC40 (squares), ΔdinB (SEC1414) (triangles) deleted on both chromosome and episome, and nusA11 (SEC182) (diamonds). B) Magnification of A). C) Recovery of viable Lac− cells from agar plugs reveals that no significant growth or death occurring during the duration of the experiment. Error bars represent the standard deviation from five independent experiments.
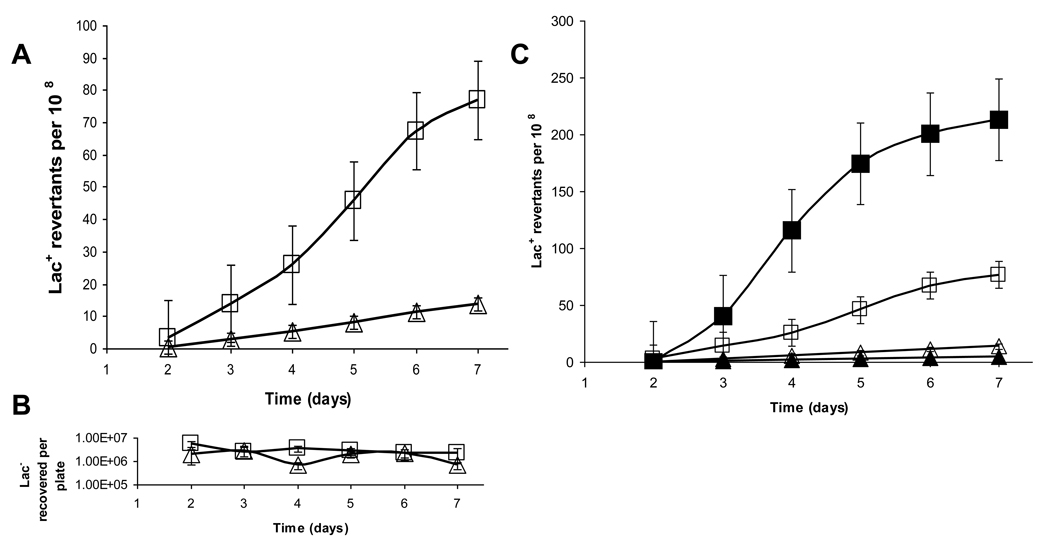
Strikingly, we observe that dinB deletion strains display an approximately 75-fold reduction in Lac+ revertants at 30°C (Figure 1A–B), which is significantly greater than the reported 2–4-fold reduction observed at 37°C [5, 6]. This discrepancy could be due to either the dinB deletion allele used or the temperature at which experiments were performed. We therefore performed these experiments at both temperatures utilizing the same doubly deleted ΔdinB strain. Compared to wild-type (FC40) the ΔdinB strain displays an approximately 6-fold reduction at 37°C and 75-fold reduction at 30°C (Figure 2 A–C). Most notable, is that while the number of Lac+ revertants in the doubly deleted ΔdinB strain is similar at the two temperatures, there are more Lac+ revertants produced at 30°C in the FC40 background than at 37°C, indicating that there are more dinB-dependent mutants at 30°C. To our knowledge, this is the first report of the temperature dependence of stress-induced mutagenesis in the lacI33-lacZ system.
Figure 2.
Stress-induced mutation in response to starvation at 30° and 37°C. A) Reversion to Lac+ of wild-type FC40 (open squares), ΔdinB (SEC1414) (open triangles) deleted on both chromosome and episome, at 37 °C. B) Overlay of graphs from Fig. 1A and Fig. 2A. Open symbols represent experiments carried out at 37°C, and closed symbols at 30°C. Symbols as in A). C) Recovery of viable Lac− cells from agar plugs reveals that no significant growth or death occurring during the duration of the experiment. Error bars represent the standard deviation from five independent experiments.
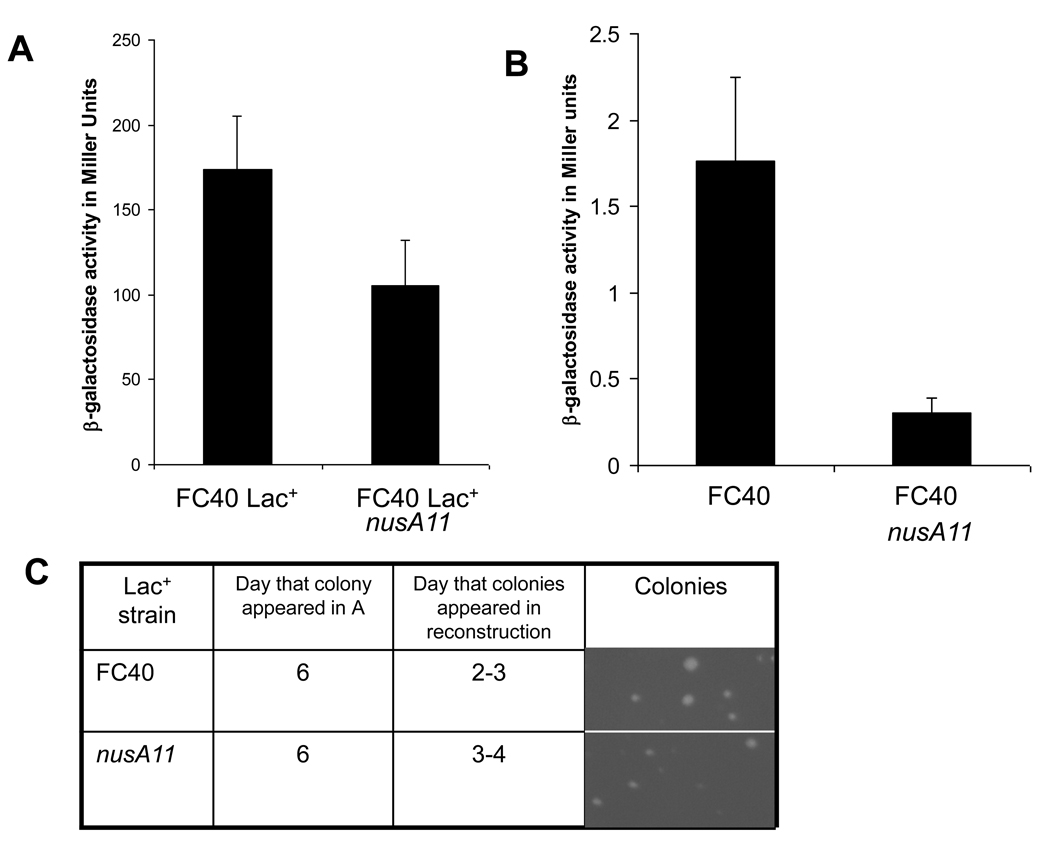
One caveat to our above results is that, in vitro, NusA is known to be required for β-galactosidase transcription [14]. In vivo, at the permissive temperature, the nusA11 mutation is reported to cause a 40 percent reduction in β-galactosidase activity [8], due to a Rho-dependent termination signal within the lacZ coding region. Could this defect in lacZ expression explain the elimination in stress-induced mutagenesis we are seeing? To test this, we isolated stress-induced mutants that arose from the wild type FC40 strain (FC40 Lac+) and nusA11 strains originally appearing at day 6 of selection and then monitored β-galactosidase activity as well as the ability to re-form colonies on lactose medium at 30°C in reconstruction experiments. We observe that a nusA11 derivative of a Lac+ strain shows an approximately 40 percent reduction in β-galactosidase activity from the episomal lacI33-lacZ reporter compared its nusA+ parent (Figure 3A). However a nusA11 derivative displays an approximately 75 percent reduction in residual β-galactosidase activity from the leaky lacI33-lacZ allele in a FC40 Lac− strain compared to its nusA+ parent (Figure 3B). Potential implications of this difference are discussed later.
Figure 3.
β-galactosidase activity and growth on lactose medium of nusA11 strains at 30°C. A) FC40 Lac+ nusA11 (SEC1429) strains show an approximately 40 percent reduction in β-galactosidase activity as compared to FC40 Lac+ nusA+ (SEC1419). B) FC40 nusA11 (SEC182) strains show an approximate 75 percent reduction in β-galactosidase activity as compared to FC40 nusA+. Error bars represent the standard deviation determined from three independent experiments. C) Reconstruction experiments showing that Lac+ revertants picked from day 6, take only two-three days for wild-type (91.3% ± 4.5 of colonies formed by day two) and three-four days for nusA11 strains (71.8% ± 13.8 of colonies formed by day three) to form colonies on lactose minimal media. Pictures of colonies taken on day 4 of reconstruction experiment. Data collected from the examination of 3–5 clones for each reconstruction.
Reconstruction experiments, taking Lac+ revertants and re-plating a known number of cells in the presence of ~109 scavenger cells to recapitulate lactose selection conditions, have been used to determine if mutations leading to Lac+ arose during selection or were preexisting in the population [15]. If the Lac+ colony originally appearing at day six after selection reappears after 6 days, it is presumed to be a slow growing preexisting mutant. If however, the number of days to reform colonies is less than the number of days when the original Lac+ colony arose, it is presumed to have occurred on the plate in response to stress. We observe that Lac+ colonies, from both the wild-type (FC40) and nusA11 strains take much less than 6 days to reform colonies at 30°C. FC40 nusA+ Lac+ colonies begin to appear at day 2, with the remaining colonies becoming visible at day 3. Consistent with their moderately reduced levels of β-galactosidase expression, nusA11 Lac+ colonies however, begin to appear on day 3, with remaining colonies developing on day 4, a delay of ca. 24 hours compared to the nusA+ parent (Figure 3C). These results suggest that, even under the conditions of reduced β-galactosidase activity in a nusA11 strain, there remains sufficient β-galactosidase function to grow when lactose is the only carbon source, albeit at a slower rate. Thus, the elimination of stress-induced mutagenesis seen in the nusA11 strain is likely not due to a defect in lacZ expression, but rather to an inability to adapt and mutate in response to environmental stress.
Although the above observations suggest that loss of stress-induced mutagenesis in a nusA11 background is not due to altered expression of the lacI33-lacZ fusion, we wanted to see if this observation would repeat in another context. In an effort to accomplish this, we thought we might take advantage of a pre-existing reporter that has been previously used to monitor unselected mutations in stress-induced mutagenesis experiments employing the lacI33-lacZ system. The strain FC722, carries an inactivated tetA gene due to a +1 frameshift mutation in a run of G:C base pairs within a Tn10 transposon on the episome (F'128), positioned approximately 5 kb away from the lac genes [16, 17], and has a reversion rate to tetracycline resistance of 3×10−9/cell/generation during exponential growth [18]. Tetracycline is a bacteriostatic antibiotic which functions through inhibition of the 30S ribosome [19]; resistance imparted by a Tn10 insertion is achieved by tetracycline efflux [20]. The tetracycline resistance (TetR) of a strain carrying a chromosomal Tn10 insertion was not affected in a nusA11 background, indirectly showing that tetA expression is not altered. In fact, the presence of the nusA11 allele even modestly improves tetracycline resistance at higher doses (Figure 4A). At the concentration of tetracycline used in the experiments described below (12 µg/mL), both the nusA+ and nusA11 TetR strains showed 100 percent survival. Stress-induced mutagenesis is defined as mutations that arise in non-growing bacteria held under non-lethal selection, allowing for growth [21]. Thus we hypothesized that, by plating FC722 cells on tetracycline medium, non-growing cells should accumulate mutations in the defective Tn10 that would then allow for growth on tetracycline medium, analogously to the Lac+ mutations that accumulate in starved FC40 cells plated on minimal lactose medium.
Figure 4.
Stress-induced mutagenesis in response to tetracycline at 30°C. A) Tetracycline resistance from chromosomal Tn10. Percent survival of nusA+ (SEC527) (squares) and nusA11 (SEC29) (diamonds) strains to tetracycline (0–50µg/mL) shows that nusA11 strains are not altered in their ability to express tetracycline resistance. Error bars represent the standard deviation determined from three independent experiments. B) Stress-induced mutagenesis of FC722 upon tetracycline treatment. Wild-type FC722 (SEC361) (closed squares), nusA11 (SEC369) (closed diamonds), ΔdinB on the chromosome and episome (SEC611) (closed triangles), and ΔruvC (SEC1466) (open triangles). C) Stress-induced TetR hypermutability. Hypermutation observed in FC722 ΔrecG mutants in response to tetracycline as is seen during lactose stress-induced mutagenesis of FC40. Wild-type FC722 (SEC361) (closed squares) and ΔrecG (SEC1464) (open squares). D) Reconstruction experiments showing that TetR revertants picked from day 5 or 6, take only two days to reform colonies on tetracycline medium. Data collected from the examination of 3–5 clones for each reconstruction, all colonies formed by day two.
When independent cultures were plated onto minimal glucose plates supplemented with tetracycline, we observed TetR colonies appearing over a period of several days (25 ± 4.6/109cells/day), an observation reminiscent of the appearance of Lac+ colonies over several days in standard lacI33-lacZ stress-induced mutagenesis (Figure 4B). Compared to the Lac system, the total number of mutants that are produced is lower when compared to FC40 strains plated on lactose minimal medium. Additionally, we were not able to isolate tetracycline sensitive (TetS) cells from the plates, by removing agar plugs from spaces in between colonies. However, if the plates were then overlaid with top agar containing glucose, growth of the lawn population was then visible (data not shown). These data indicate that although we were unable to quantify the TetS population over the course of the experiment, many (if not all) the cells remain viable upon continued exposure to tetracycline.
We find that the genetic requirements for stress-induced mutations that occur after tetracycline treatment are similar to those that occur in response to starvation in that they require dinB+ and the recombination functions of ruvC+ [22, 23] (Figure 4B), with reversion to TetR occurring at 3 ± 0.5/109cells/day and 1 ± 0.2/109cells/day in the doubly deleted ΔdinB mutant and ΔruvC mutant respectively. Furthermore, stress-induced hypermutability after tetracycline treatment can be achieved through mutation of recG (125 ± 34 TetR revertants/109cells/day) (Figure 4C), as occurs in the Lac system [22, 23]. nusA+ function is also required for tetracycline adaptive mutagenesis, as the nusA11 mutation virtually eliminates the formation of stress-induced mutants and has a reversion rate to TetR of 0.1 ± 0.07/109cells/day (Figure 4B). Isolating TetR revertants, from both the wild-type and mutant strains, that originally appeared at day 5 or 6 allowed us to perform reconstruction experiments to test if these mutants are truly stress-induced. By plating a known number of cells, in the presence of ~109 scavenger cells (FC29) to mimic a lawn population of non-growing TetS cells, we observe that colonies appear after two days of incubation on minimal glucose plates supplemented with tetracycline with no notable difference in colony size between wild-type, nusA11, ΔdinB, and ΔruvC TetR strains (Figure 4D). These data imply that the TetR mutants that appear after tetracycline exposure are indeed stress-induced and further support the notion that the defect in stress-induced mutagenesis seen in the nusA11 strain is not due to improper transcription but rather to a disruption of a process that allows the cells to adapt and mutate in response to stress. Interestingly, E. coli cells plated on medium containing a bacteriostatic concentration of the gyrase inhibiting antibiotic, ciprofloxacin, acquired resistant mutants in non-growing cells over the course of a week [24], and display a dependence on the SOS response, homologous recombination and DinB as has been shown in the Lac system [25]. These data suggest that stress-induced mutagenesis in response to bacteriostatic antibiotic treatment may be a general phenomenon.
In summary, we have discovered a previously unrecognized role for the essential gene nusA+ in the process of stress-induced mutagenesis. Specifically a function of nusA altered by the nusA11(ts) mutation at the permissive temperature is critical for the formation of stress-induced mutations. The reduction in stress-induced mutagenesis caused by the nusA11 mutation is 470-fold, which is larger than the reductions caused by mutation of other genes whose products have been implicated in stress-induced mutagenesis, e.g. rpoS (10–20 fold) [26, 27], dinB (4–5 fold) [5, 6], recA (10–100 fold) [1, 28], groE (10–20 fold) [29], ppk (5 fold) [21], and ruvC (10 fold) [22, 23]. Furthermore, the nusA11 mutation does not affect the frequency of UV induced mutagenesis in a standard mutagenesis assay (Supplemental Table 1). Since nusA11 cells are viable at 30°C, the function of NusA altered by the nusA11 mutation is genetically separable from the essential roles of NusA in normal RNA polymerase elongation and termination/antitermination. Our finding that the reduction in stress-induced mutants seen in the nusA11 background is greater than that seen when dinB is deleted, leads us to hypothesize that NusA may play a role in stress-induced mutagenesis that extends beyond a potential interaction with DinB. Though future experiments will be required to elucidate what this role may be, we propose that NusA is required to link transcription to stress-induced mutation.
Our previous report suggested that a problem occurring in the nusA11 mutant, at the restrictive temperature, might be at the DNA level as overexpression of DinB can rescue growth in a manner requiring the catalytic and specifically the translesion DNA synthesis properties of DinB [7]. A possible role for NusA under stressed conditions could arise from a deficiency in DNA repair that results in unrepaired endogenous lesions being present in the transcribed strand. Alternatively, the very limited growth allowed by the leaky lacI33-lacZ allele might result in DNA replication intermediates that have more single stranded gaps than in exponentially growing cells. Either of these situations could stall an RNA polymerase if transcription is attempted. If this were the case, the function lost by the nusA11 mutation might be the ability to recruit DinB and potentially other cellular factors required for stress-induced mutagenesis to the stalled RNA polymerase. The process of transcription-coupled translesion synthesis we have hypothesized [7] might therefore be one mechanism that could contribute to the generation of stress-induced mutations. Below we suggest how this idea might fit into the proposed models to explain the phenomenon of stress-induced mutagenesis.
There are currently two alternative classes of models to explain how Lac+ mutations occur during lactose selection: models involving error-prone DNA synthesis during double-strand break repair and the amplification model [30–32]. In the models involving error-prone DNA synthesis during double-strand break repair, it is suggested that amplification results in an unstable Lac+ colony and is separate from, and does not lead to, mutation. The process through which actual mutation occurs is postulated to be initiated by a double-strand break and controlled by the SOS and RpoS stress responses. The process of repair of a double-strand break, which could be formed by replication encountering a nick generated at the episomal origin, could become error-prone through DNA replication by DinB. This switch to error-prone double-strand break repair by DinB is regulated by the SOS and RpoS stress responses [17, 30, 31, 33]. The limited DNA replication initiated at the episomal origin or during double-strand break repair could generate DNA substrates that could stall RNA polymerase and result in the recruitment of factors required for stress-induced mutagenesis by NusA.
The amplification model suggests that an amplified array of DNA of up to 100 copies containing the lac genes is an intermediate in the formation of Lac+ mutants. In this model it is suggested that the residual β-galactosidase activity, which is now amplified by up to 100 times, allows cells to grow very slowly on minimal lactose media and reversion to Lac+ occurs with a higher probability due to the repeated array [16, 32, 34–36]. Considering this model, it is possible that nusA11 cells, since they display a 75 percent reduction in residual β-galactosidase activity (Figure 3C), may be further restricting the slow growth of the Lac− cells thus inhibiting their ability to mutate to Lac+. Similar effects have been previously observed through the addition of LacZ inhibitors or through inactivation of the gal operon [36]. However, it is less clear how this type of explanation could account for the almost complete loss of stress-induced TetR mutants in a nusA11 strain. It is possible, however, that the nusA11 mutation could interfere with the creation of the illegitimate recombination event that creates the initial duplication or the subsequent events involved in generating the amplified arrays under stressed conditions.
The amplification model also proposes that the number of revertants depends highly on the pre-growth conditions in which the original duplication required for amplification is generated [32]. We observe that when an equivalent number of FC40 nusA+ and nusA11 cells are plated on minimal media with better carbon sources (glucose or glycerol) the nusA11 mutation does not affect colony formation with respect to either time of appearance or their size whereas, on poor carbon sources (acetate or pyruvate), the nusA11 mutation delays the appearance of colonies by ca. 24 hours, suggesting that nusA+ is important for growth under poor carbon sources. The replication involved in the generation of the amplified arrays under stressed conditions could generate the substrates we suggest might stall RNA polymerase and the recruitment of factors involved in stress-induced mutagenesis by NusA.
EXPERIMENTAL PROCEDURES
Mutagenesis Assays
All strains used are listed in Supplemental Table 2 and constructed using standard techniques. Lactose stress-induced mutagenesis assays were performed as previously described, except when noted incubation steps were performed at 30°C [37]. Briefly, ~108 cells, of at least five independent cultures of each strain were grown to saturation in M9 minimal media with either (0.1% Glycerol or Glucose), were mixed with ~109 cells of FC29 scavenger cells and 2.5mL minimal top agar and overlaid onto M9 minimal lactose (0.1%) plates. Viable Lac− cells were monitored as described in [1]. Tetracycline stress-induced mutagenesis assays were performed by growing each strain to saturation in M9 minimal media (0.2% Glucose) and diluted 105 fold into fresh medium (0.2% Glucose), divided into several cultures, and allowed to reach saturation, to produce at least five independent cultures. Approximately 109 cells from each independent culture is mixed with minimal top agar and overlaid onto M9 minimal glucose (0.2%) plates containing tetracycline (12 µg/mL). Viable cells were determined by dilution in M9 salts and plating cells on to M9 minimal glucose (0.2%) plates without tetracycline. Plates were incubated at 30°C and tetracycline resistant colonies are counted and marked every day. Reversion of argE3 allele in standard UV induced mutagenesis were performed according to the published method [38], all incubation steps were performed at 30°C.
β-galactosidase activity assays
Strains were grown at 30°C in minimal M9 lactose (0.2%) (Lac+ strains) or glycerol (0.2%) (Lac− strains) medium and β-galactosidase activity assays were performed as previously described [39].
Determination of nusA11 effects on reporter genes or growth on different carbon sources
TetA expression from Tn10. Strains harboring chromosomal Tn10 (SEC527, SEC29) were grown in LB medium, diluted in M9 salts and plated onto LB plates supplemented with tetracycline, CFU/mL were determined after incubation at 30°C. Reconstruction assays were performed by collecting Lac+ or TetR strains and plating a known number of cells with ~109 scavenger cells (FC29) onto either minimal lactose or tetracycline media and incubated at 30°C. Colony number and size were scored as a factor of time. When applicable tetracycline was used at 12 µg/mL. Growth on different carbon sources. FC40 nusA+ and nusA11 cells (SEC182) were diluted in M9 salts and plated onto M9 minimal plates containing glycerol, acetate or pyruvate at 0.2% and incubated at 30°C. Colonies number and size were scored as a factor of time.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Daniel F. Jarosz and anonymous reviewers for thoughtful comments on the manuscript. This work was supported by NIH grant CA21615 to G.C.W. as well as NIEHS Grant P30 ES002109 to the MIT Center for Environmental Health Sciences. G.C.W. is an American Cancer Society Research Professor.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Cairns J, Foster PL. Adaptive reversion of a frameshift mutation in Escherichia coli. Genetics. 1991;128:695–701. doi: 10.1093/genetics/128.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kenyon CJ, Walker GC. DNA-damaging agents stimulate gene expression at specific loci in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1980;77:2819–2823. doi: 10.1073/pnas.77.5.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis: Second Edition. Washington, DC: American Society for Microbiology; 2005. [Google Scholar]
- 4.Ohmori H, Friedberg EC, Fuchs RP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, Prakash L, Prakash S, Todo T, Walker GC, Wang Z, Woodgate R. The Y-family of DNA polymerases. Mol. Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- 5.McKenzie GJ, Lee PL, Lombardo MJ, Hastings PJ, Rosenberg SM. SOS mutator DNA polymerase IV functions in adaptive mutation and not adaptive amplification. Mol. Cell. 2001;7:571–579. doi: 10.1016/s1097-2765(01)00204-0. [DOI] [PubMed] [Google Scholar]
- 6.Foster PL. Adaptive mutation in Escherichia coli. Cold Spring Harb. Symp. Quant. Biol. 2000;65:21–29. doi: 10.1101/sqb.2000.65.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen SE, Godoy VG, Walker GC. Transcriptional modulator NusA interacts with translesion DNA polymerases in Escherichia coli. J Bacteriol. 2009;191:665–672. doi: 10.1128/JB.00941-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakamura Y, Mizusawa S, Court DL, Tsugawa A. Regulatory defects of a conditionally lethal nusAts mutant of Escherichia coli. Positive and negative modulator roles of NusA protein in vivo. J Mol Biol. 1986;189:103–111. doi: 10.1016/0022-2836(86)90384-0. [DOI] [PubMed] [Google Scholar]
- 9.Rosenberg SM, Longerich S, Gee P, Harris RS. Adaptive mutation by deletions in small mononucleotide repeats. Science. 1994;265:405–407. doi: 10.1126/science.8023163. [DOI] [PubMed] [Google Scholar]
- 10.Foster PL, Trimarchi JM. Adaptive reversion of a frameshift mutation in Escherichia coli by simple base deletions in homopolymeric runs. Science. 1994;265:407–409. doi: 10.1126/science.8023164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim SR, Matsui K, Yamada M, Gruz P, Nohmi T. Roles of chromosomal and episomal dinB genes encoding DNA pol IV in targeted and untargeted mutagenesis in Escherichia coli. Mol. Genet. Genomics. 2001;266:207–215. doi: 10.1007/s004380100541. [DOI] [PubMed] [Google Scholar]
- 12.Ohashi E, Bebenek K, Matsuda T, Feaver WJ, Gerlach VL, Friedberg EC, Ohmori H, Kunkel TA. Fidelity and processivity of DNA synthesis by DNA polymerase kappa, the product of the human DINB1 gene. J. Biol. Chem. 2000;275:39678–39684. doi: 10.1074/jbc.M005309200. [DOI] [PubMed] [Google Scholar]
- 13.Galhardo RS, Do R, Yamada M, Friedberg E, Hastings P, Nohmi T, Rosenberg S. DinB Up-Regulation Is the Sole Role of the SOS Response in Stress-Induced Mutagenesis in Escherichia coli. Genetics. 2009;182:55–68. doi: 10.1534/genetics.109.100735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenblatt J, Li J, Adhya S, Friedman DI, Baron LS, Redfield B, Kung HF, Weissbach H. L factor that is required for beta-galactosidase synthesis is the nusA gene product involved in transcription termination. Proc Natl Acad Sci U S A. 1980;77:1991–1994. doi: 10.1073/pnas.77.4.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prival MJ, Cebula TA. Adaptive mutation and slow-growing revertants of an Escherichia coli lacZ amber mutant. Genetics. 1996;144:1337–1341. doi: 10.1093/genetics/144.4.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hendrickson H, Slechta ES, Bergthorsson U, Andersson DI, Roth JR. Amplification-mutagenesis: evidence that "directed" adaptive mutation and general hypermutability result from growth with a selected gene amplification. Proc. Natl. Acad. Sci. U.S.A. 2002;99:2164–2169. doi: 10.1073/pnas.032680899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ponder RG, Fonville NC, Rosenberg SM. A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Mol Cell. 2005;19:791–804. doi: 10.1016/j.molcel.2005.07.025. [DOI] [PubMed] [Google Scholar]
- 18.Foster PL. Nonadaptive mutations occur on the F' episome during adaptive mutation conditions in Escherichia coli. J. Bacteriol. 1997;179:1550–1554. doi: 10.1128/jb.179.5.1550-1554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldman RA, Hasan T, Hall CC, Strycharz WA, Cooperman BS. Photoincorporation of tetracycline into Escherichia coli ribosomes. Identification of the major proteins photolabeled by native tetracycline and tetracycline photoproducts and implications for the inhibitory action of tetracycline on protein synthesis. Biochemistry. 1983;22:359–368. doi: 10.1021/bi00271a020. [DOI] [PubMed] [Google Scholar]
- 20.Hillen W, Berens C. Mechanisms underlying expression of Tn10 encoded tetracycline resistance. Annu Rev Microbiol. 1994;48:345–369. doi: 10.1146/annurev.mi.48.100194.002021. [DOI] [PubMed] [Google Scholar]
- 21.Stumpf JD, Foster PL. Polyphosphate kinase regulates error-prone replication by DNA polymerase IV in Escherichia coli. Mol Microbiol. 2005;57:751–761. doi: 10.1111/j.1365-2958.2005.04724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harris RS, Ross KJ, Rosenberg SM. Opposing roles of the Holliday junction processing systems of Escherichia coli in recombination-dependent adaptive mutation. Genetics. 1996;142:681–691. doi: 10.1093/genetics/142.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foster PL, Trimarchi JM, Maurer RA. Two enzymes, both of which process recombination intermediates, have opposite effects on adaptive mutation in Escherichia coli. Genetics. 1996;142:25–37. doi: 10.1093/genetics/142.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riesenfeld C, Everett M, Piddock LJ, Hall BG. Adaptive mutations produce resistance to ciprofloxacin. Antimicrob Agents Chemother. 1997;41:2059–2060. doi: 10.1128/aac.41.9.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cirz RT, Chin JK, Andes DR, de Crecy-Lagard V, Craig WA, Romesberg FE. Inhibition of mutation and combating the evolution of antibiotic resistance. PLoS Biol. 2005;3:e176. doi: 10.1371/journal.pbio.0030176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Layton JC, Foster PL. Error-prone DNA polymerase IV is controlled by the stress-response sigma factor, RpoS, in Escherichia coli. Mol. Microbiol. 2003;50:549–561. doi: 10.1046/j.1365-2958.2003.03704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lombardo MJ, Aponyi I, Rosenberg SM. General stress response regulator RpoS in adaptive mutation and amplification in Escherichia coli. Genetics. 2004;166:669–680. doi: 10.1534/genetics.166.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harris RS, Longerich S, Rosenberg SM. Recombination in adaptive mutation. Science. 1994;264:258–260. doi: 10.1126/science.8146657. [DOI] [PubMed] [Google Scholar]
- 29.Layton JC, Foster PL. Error-prone DNA polymerase IV is regulated by the heat shock chaperone GroE in Escherichia coli. J Bacteriol. 2005;187:449–457. doi: 10.1128/JB.187.2.449-457.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Foster PL. Stress-induced mutagenesis in bacteria. Crit Rev Biochem Mol Biol. 2007;42:373–397. doi: 10.1080/10409230701648494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galhardo RS, Hastings PJ, Rosenberg SM. Mutation as a stress response and the regulation of evolvability. Crit Rev Biochem Mol Biol. 2007;42:399–435. doi: 10.1080/10409230701648502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roth JR, Kugelberg E, Reams AB, Kofoid E, Andersson DI. Origin of mutations under selection: the adaptive mutation controversy. Annu Rev Microbiol. 2006;60:477–501. doi: 10.1146/annurev.micro.60.080805.142045. [DOI] [PubMed] [Google Scholar]
- 33.Rosenberg SM. Evolving responsively: adaptive mutation. Nat. Rev. Genet. 2001;2:504–515. doi: 10.1038/35080556. [DOI] [PubMed] [Google Scholar]
- 34.Kugelberg E, Kofoid E, Reams AB, Andersson DI, Roth JR. Multiple pathways of selected gene amplification during adaptive mutation. Proc Natl Acad Sci U S A. 2006;103:17319–17324. doi: 10.1073/pnas.0608309103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roth JR, Andersson DI. Adaptive mutation: how growth under selection stimulates Lac(+) reversion by increasing target copy number. J. Bacteriol. 2004;186:4855–4860. doi: 10.1128/JB.186.15.4855-4860.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andersson DI, Slechta ES, Roth JR. Evidence that gene amplification underlies adaptive mutability of the bacterial lac operon. Science. 1998;282:1133–1135. doi: 10.1126/science.282.5391.1133. [DOI] [PubMed] [Google Scholar]
- 37.Godoy VG, Gizatullin FS, Fox MS. Some features of the mutability of bacteria during nonlethal selection. Genetics. 2000;154:49–59. doi: 10.1093/genetics/154.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elledge SJ, Walker GC. Proteins required for ultraviolet light and chemical mutagenesis: Identification of the products of the umuC locus of Escherichia coli. J. Mol. Biol. 1983;164:175–192. doi: 10.1016/0022-2836(83)90074-8. [DOI] [PubMed] [Google Scholar]
- 39.Miller JH. A short course in bacterial genetics. A laboratory manual for Escherichia coli and related bacteria. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.