Abstract
tRNA isodecoders share the same anticodon but have differences in their body sequence. An unexpected result from genome sequencing projects is the identification of a large number of tRNA isodecoder genes in mammalian genomes. In the reference human genome, over 270 isodecoder genes are present among the approximately 450 tRNA genes distributed among 49 isoacceptor families. Whether sequence diversity among isodecoder tRNA genes reflects functional variability is an open question. To address this, we developed a method to quantify the efficiency of tRNA isodecoders in stop-codon suppression in human cell lines. First, a GFP (Green Fluorescent Protein) gene is introduced that contains a single UAG stop codon at two distinct locations. GFP is only produced when a tRNA suppressor containing CUA anticodon is co-transfected with the GFP gene. The suppression efficiency is examined for 31 tRNA isodecoders (all contain CUA anticodon), 21 derived from four isoacceptor families of tRNASer genes, 7 from five families of tRNALeu genes and 3 from three families of tRNAAla genes. We found that isodecoder tRNAs display a large difference in their suppression efficiency. Among those with above background suppression activity, differences of up to 20-fold were observed. We were able to tune tRNA suppression efficiency by subtly adjusting the tRNA sequence, and inter-convert poor suppressors into potent ones. We also demonstrate that isodecoder tRNAs with varying suppression efficiencies have similar stability and exhibit similar levels of aminoacylation in vivo. Our results indicate that naturally occurring tRNA isodecoders can have large functional variations, and suggest that some tRNA isodecoders may perform a function distinct from translation.
Keywords: tRNA, isodecoder, suppression, translation, GFP
Introduction
A remarkable finding among mammalian genome sequences is the presence of a large number of tRNA genes that have the same anticodon, but different body sequences [1]. In the NIH reference human genome, there are over 270 unique tRNA genes distributed among 49 isoacceptor families. The number of tRNA genes is only ~450, representing a gene/sequence ratio of ~1.7. For comparison, Saccharomyces cerevisiae has ~270 tRNA genes, but only 51 different sequences, representing a gene/sequence ratio of 5.4 [1]. For convenience, tRNA genes that share the same anticodon but have different body sequences have been termed as “isodecoders”, implying that they all read the same codon in translation [2].
At first glance, the presence of tRNA isodecoders may simply be an evolutionary relic. In general, isodecoder sequences vary in ways that still allow these tRNAs to fold into the same secondary and tertiary structure. It is possible that isodecoders are derived from neutral genetic drift, and they could translate the same codons at similar efficiencies. However, a large body of work showed that the body sequences of a tRNA are fine tuned for aminoacylation and interaction with the ribosome [3,4]. A sequence change in the tRNA body may therefore lead to differences in their efficiencies in translation. On the other hand, certain tRNAs are known to participate in extra-translational functions where they interact with proteins not involved in translation [5–7]. A sequence change in the tRNA body may lead to improved efficiency for these processes because of the reduced competition from the translation machinery. For these reasons, a rudimentary test of translational efficiency of tRNA isodecoders in vivo would be informative regarding their significance and considerations on their evolutionary conservation.
To our knowledge, no simple test exists that allows functional studies of tRNA isodecoders in mammalian cells. In single cell organisms such as E. coli and yeast, such tests may be carried out by deleting the chromosomal genes of one tRNA isoacceptor family, followed by introduction of a single isodecoder species to the cell [8,9]. In mammalian cells, the chromosomal copies of each isoacceptor family are numerous and cannot all be removed from the chromosome. Therefore, any assay designed to test isodecoder functions must separate the effects derived from the endogenous tRNA genes. Here, we develop a suppression based assay to study tRNA isodecoder function in human cell lines. This assay is based on the ability of transfected tRNA to read through a single stop codon in the GFP coding region. We use this assay to compare the efficiencies of 31 isodecoder tRNAs in stop-codon suppression using sequences derived from the human acceptor families of serine, leucine and alanine tRNA genes. Our results show a large difference in functional efficiency among isodecoders, and this difference is attributed to the steps downstream of aminoacylation.
Results
Translational suppression by tRNA transcripts
Exogenous tRNA can be introduced into cells either as DNA or directly as RNA [10,11]. In the case of isodecoder genes, some sequence differences reside in the D and T stem loop regions which are part of RNA polymerase III promoter and may affect transcription efficiency [12]. We therefore choose to directly use tRNA transcripts in this work to avoid possible complications derived from sequence effects of isodecoder genes on RNA polIII transcription.
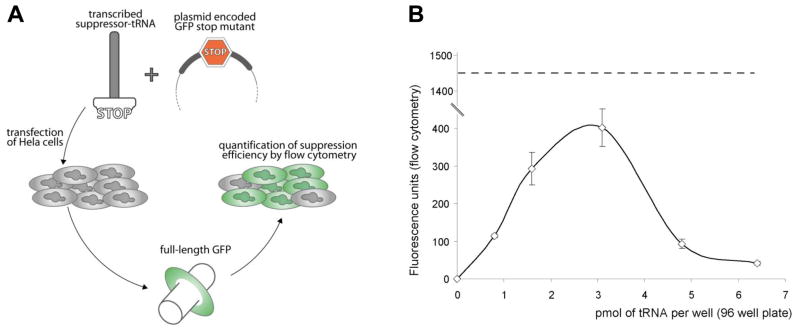
In vitro transcribed tRNA have been reported to be active in translation when transfected into eukaryotic cells [13]. Suppression of the UAG stop codon has also been used previously to map identity elements on tRNA [14] or to encode non-natural amino acids to generate proteins with new properties [15]. We use UAG suppression as a way to gauge the translational efficiency of different natural tRNA scaffolds (Fig. 1A). First, in vitro tRNA transcripts are co-transfected into HeLa cells with a plasmid DNA encoding a GFP mutant containing a single UAG stop codon at either residue S29 or S65. Serine at position 29 does not participate in the fluorescence activity of GFP; consequently position 29 can accommodate any amino acid without disturbing light emission [16]. The S29 mutant construct was used here to test isodecoders derived from isoacceptor families from three different amino acids: leucine, serine and alanine. Serine at position 65 accommodates only hydrophilic residues such as serine and threonine without perturbing GFP fluorescence [17] (Ribas de Pouplana, personal communication). The S65 mutant construct was used to test isodecoders derived from isoacceptor families of serine. The S65 reporter allows simultaneous evaluation of both charging specificity and ranking of suppression efficiency.
Figure 1. The experimental set-up and dosage optimization.
(A) Schematic description of the method used for the evaluation of the different tRNA scaffolds. (B) Fluorescence recovery versus the amount of suppressor tRNASer transfected. Identical amount of reporter plasmid encoding GFP S29stop is used in all transfections. Fluorescent values reported here are obtained from the average of 4 independent transfections. The dashed line indicates the observed fluorescence when equivalent amount of plasmid encoding wild type GFP is used.
We tested this approach using the human tRNASer(AGA) isodecoder encoded by 6 identical gene copies in the reference human genome (Fig. 1B). Cells transfected with the reporter plasmid alone were not fluorescent. After 24 h, fluorescence recovery depended on the amount of synthetic suppressor tRNA introduced into human cells. Suppression activity follows a bell-shaped profile with a maximum response around 3 pmol of tRNA per well transfected (~0.1 fmol/cell). Higher doses of tRNA transcript appear to be detrimental for suppression, with no reduction of cell growth in the short term (data not shown). This result may be explained by the activation of the GCN2 kinase [18]. GCN2 is part of the general amino acid control pathway and is activated upon binding to uncharged tRNA. When activated, GCN2 phosphorylates the translational initiation factor eIF2α leading to down-regulation of general translation. Massive and rapid introduction of synthetic tRNA into the intracellular environment can potentially overwhelm the cellular seryl-tRNA synthetases, leaving a significant fraction of tRNA uncharged and resulting in GCN2 activation. Regardless, this result established an optimally useful amount of tRNA transcript for transfection at 2 pmol and we used this amount of tRNA in all subsequent experiments. At this level, the suppression efficiency is ~20% when compared to the fluorescent intensity of cells transfected with the wild-type GFP gene.
Isodecoder tRNAs have a wide range of suppression activity
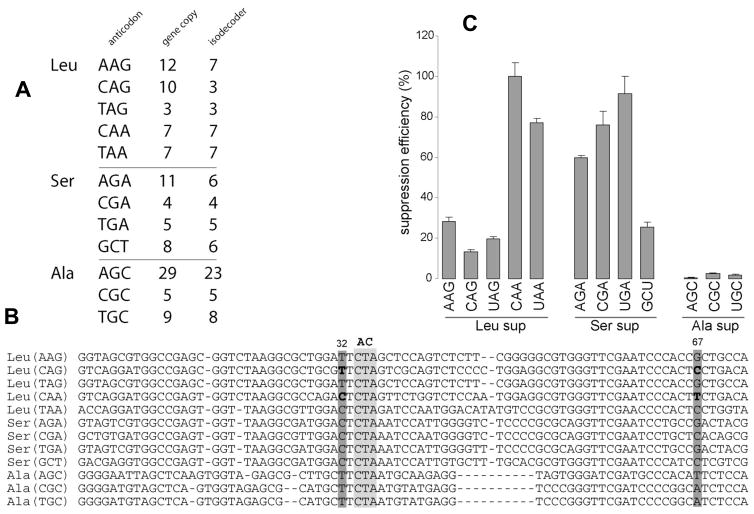
tRNAs specific to leucine, serine and alanine are organized in 5, 4 and 3 isoacceptor families in the reference human genome respectively (Fig. 2A). tRNAs for these three amino acids are unique because their anticodons are not a primary determinant for aminoacylation. The primary determinant for aminoacylation for leucine and serine tRNAs is a long variable arm that contains a short hairpin loop [19,20], whereas the determinant for alanine tRNA is the G3U70 base pair in the acceptor stem [21]. Since the suppression assay requires changing the anticodon to CUA, these tRNA species when converted to suppressors are the best candidates to become charged in the cell. We selected the tRNA species most represented in the genome per isoacceptor family and converted their anticodons to CUA, thus generating 12 isodecoder tRNAs with an identical anticodon (Fig. 2B).
Figure 2. Suppression analysis of isodecoders derived from isoacceptor families of leucine, serine and alanine.
(A) Detailed organization of tRNA isoacceptor families. Number of genes and isodecoders in each isoacceptor family is indicated. (B) Structural alignment of representative isodecoders of each isoacceptor family. Representatives correspond to tRNA encoded by the highest number of gene copies. Light grey box indicates position of the anticodon. Dark grey box highlights position 32 and 67 where mutations have been made. (C) Suppression efficiency for the 12 Leu, Ser and Ala-tRNA specific scaffolds tested. 100% corresponds to the most active scaffold (Leu(CAA)). Efficiency is proportional to fluorescence recovery. GFP S29stop is used as a reporter.
The suppression efficiencies of the 12 isodecoders vary widely (Fig. 2C). All tRNAAla derived isodecoders are poor scaffolds for suppression and their suppression activity is near background levels. All leucine and serine tRNA derived isodecoders show suppression efficiency significantly above background. These results suggest that the long variable arm, present in the tRNALeu and tRNASer, but lacking in the tRNAAla, is important for suppression. There is significant difference in suppression efficiency among Leu and Ser-tRNA derived isodecoders: the tRNALeu(CAA) scaffold was 6-fold more efficient compared to tRNALeu(CAG); and the tRNASer(UGA) scaffold was 4-fold more efficient compared to tRNASer(GCU).
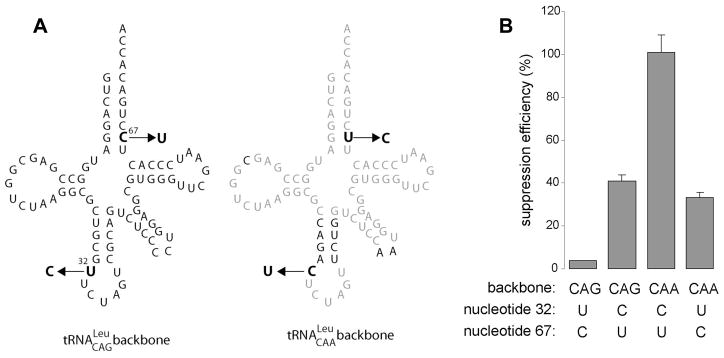
The family of leucine tRNAs can be organized in two subgroups according to their suppression efficiencies. The Leu(CAA) and (UAA) scaffold are ~5 times more active compared to Leu(AAG), (CAG) and (UAG). We noticed that the highly efficient Leu-tRNA scaffolds contain C32 in their body sequence, whereas the poor tRNALeu scaffolds contain U32. To test the influence of position 32 on suppression, U32 in Leu(CAG) was mutated to C and C32 in Leu(CAA) was mutated to U. Transplantation of U32 in Leu(CAA) reduces its suppression efficiency by 3-fold, whereas grafting C32 into Leu(CAG) increased its suppression efficiency by ~10-fold (Fig. 3). Although U67 was inadvertently changed to C67 (changing a GU pair to GC) in this experiment, this change unlikely affected suppression, as this base pair is not conserved among the high efficiency tRNALeu derived suppressors (Fig. 2B). This result indicates that the precise sequence of the anticodon loop can be crucial for suppression efficiency, a finding that has been demonstrated repeatedly [22–25].
Figure 3. Sequence swap between two tRNALeu suppressors.
(A) Cloverleaf representation (CAG and CAA scaffolds). Common nucleotides are in light grey. Mutations are indicated by the arrows. (B) Suppression efficiency of the four corresponding suppressors. Backbone and nucleotides 32 and 67 are indicated below.
Isodecoders derived from serine tRNAs
The human genome contains 28 tRNASer genes encoding 21 different tRNA sequences (Fig. 4). The AGA and UGA isoacceptor families share two tRNA scaffolds, i.e. two sets of tRNASer differ only in their anticodon nucleotide at position 34, so the total tRNASer scaffolds is 19. All 19 tRNASer sequences have identical anticodon loops (except S17) and especially nucleotide C32. Therefore, analysis of tRNASer derived isodecoders should illuminate functional effects of sequence differences away from the anticodon loop. The 19 corresponding tRNASer genes are converted to CUA suppressors, tRNAs are transcribed in vitro and tested in translation in HeLa cells.
Figure 4. Sequence alignment of isodecoders from all tRNASer genes.
(A) Detailed organization of the complete set of tRNASer species in the reference human genome. The number of gene copies and the sequence of the original anticodon are indicated. (B) Sequence alignment, ordered according to their suppression efficiency (data in figure 5). Anticodon triplet is boxed. Sequence S19 is not aligned due to its non-canonical folding.
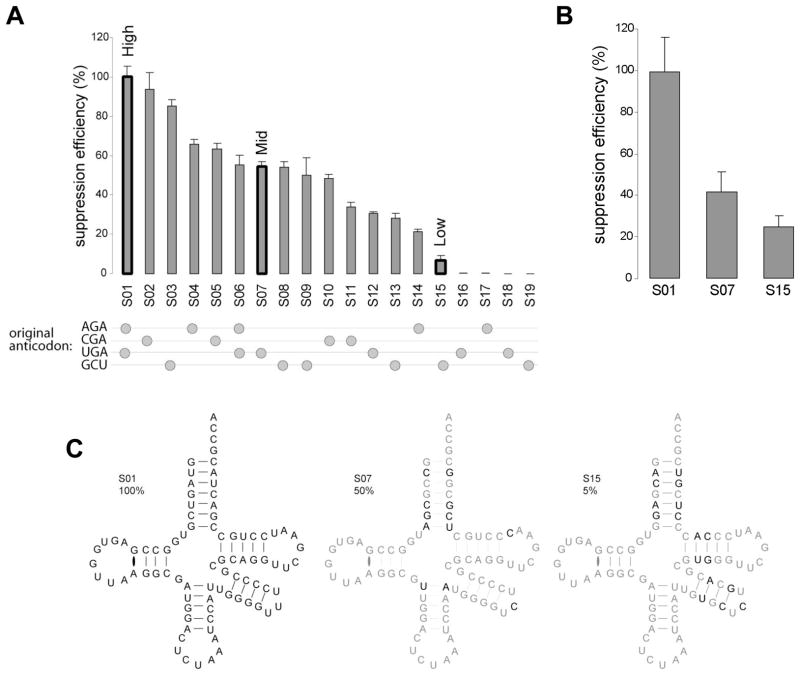
We observe a wide distribution of suppression efficiencies among the 19 isodecoders (Fig. 5A). Four isodecoders show no suppression activity, all four lack the long variable loop, supporting the importance of the variable loop in suppression. Among the 15 isodecoders that show suppression activity above background, the efficiency differs by 20-fold. Noticeably, all 15 isodecoders share the same anticodon loop and the bottom four base pairs of the anticodon stem and maintain the same canonical secondary structure (Fig. 4C), indicating that sequences outside of the anticodon stem-loop play a role in suppression efficiency.
Figure 5. Suppression analysis of isodecoders derived from tRNASer genes.
(A) Suppression efficiency for the 19 tRNASer specific scaffolds tested. 100% corresponds to the most active scaffold (S01). GFP S65stop is used as a reporter for simultaneous evaluation of both charging specificity and ranking of suppression efficiency. S01, S07 and S15 were chosen as representatives of high, medium and low suppressors respectively and used for all further studies. The beads on strings indicate the sequence of the original anticodon before substitution by suppressor CUA triplet. (B) Suppression efficiency for previously tested S01, S07 and S15 suppressors. GFP S29stop is used as a reporter. Position 29 can accommodate any amino acid without disturbing light emission. (C) Cloverleaf diagram of S01, S07 and S15. Sequence changes between S01 (high efficiency suppressor) and the mid (S07) and low (S15) efficiency suppressor are in black (common nucleotides appear in grey).
In previous studies [26–28], suppression efficiency has been shown to be influenced by mRNA nucleotides directly surrounding the stop codon. The context of S65stop is CUG UAG UAC (Leu-stop-Tyr), whereas the context of S29stop is UUC UAG GUG (Phe-stop-Val). Isodecoders derived from scaffolds 01, 07 and 15 (Fig. 4B) show the same rank order and similar magnitude of suppression activity with S29stop (Fig. 5B), indicating that the suppression activity of these isodecoders reflects the ability of each scaffold to interact with the translation machinery.
Isodecoders show similar stability and charging level in vivo
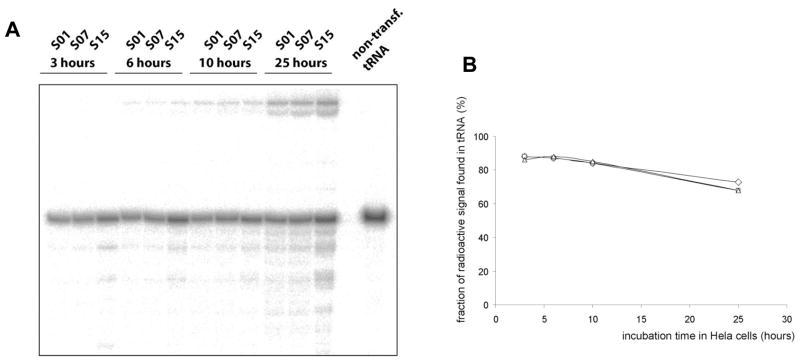
We chose 3 isodecoders derived from scaffolds for high (S01), medium (S07) and low (S15) suppression efficiency for further characterization. Foreign RNAs can be rapidly degraded when introduced into human cells [29]. tRNA modification enzymes are generally confined inside the nucleus [30]. It was shown that lipofected nucleic acids remain in the cytoplasm [31,32]. However, some lipofected material can cross the nuclear membrane when the cell is dividing and nucleus is more permeable. HeLa cells in our experiments double in ~24h, consequently it can be assumed that most of our tRNA transcripts remain unmodified within this time frame. To determine whether the suppression efficiency is attributed to differential stability of tRNA isodecoders, we directly transfected 32P-labeled tRNAs into HeLa cells. In this experiment, tRNAs are labeled at the phosphate of the A76 nucleotide using CCA-adding enzyme and α-32P-ATP [33]. Total RNAs were extracted at different time points and directly analyzed on denaturating PAGE (Fig. 6A). tRNA isodecoders derived from all three scaffolds show essentially the same stability in HeLa cells (Fig. 6B). Recycling of the 32P-label occurs at the same rate, and the radioactive A76 is slowly incorporated into higher molecular weight RNA. This result shows that tRNA isodecoder stability in vivo is not a contributing factor in the observed suppression efficiency.
Figure 6. Comparing in vivo stability of high, medium and low efficiency suppressors.
(A) S01, S07 and S15 scaffolds show equivalent stability when incubated in human cells. The 3′ terminal A of corresponding tRNAs has been 32P-labeled using CCA-adding enzyme and lipofected in HeLa cells. Total RNAs have been extracted at different time points and separated on denaturating PAGE. (B) Fraction of radioactive signals for the tRNA band at different time points shows similar half-lives for these suppressor-tRNAs. It corresponds to the ratio of the radioactive signal specific to full-length tRNA over the signal of the entire lane.
We next tested whether the charging level of isodecoders derived from these three scaffolds contributes to their differences in suppression efficiency. The three isodecoders were radiolabeled as described above and transfected into HeLa cells. After 12–24 hours, total charged tRNA was isolated under mildly acidic conditions. The total charged tRNA was then treated with nuclease P1 which cleaved charged tRNA to produce [32P]-pA and [32P]-pA-ser which were separated by thin-layer chromatography (Fig. 7). All three isodecoders show identical charging levels at both time points (around 5%). Wild type tRNASer transcripts displaying native AGA anticodon show equivalent charging level when treated the same way suggesting that the modest aminoacylation level of the suppressors is most likely due to the lack of post-transcriptional modifications rather than the presence of a non-cognate anticodon. Low in vivo aminoacylation levels is apparently idiosyncratic to tRNA transcripts. Indeed, Köhrer and collaborators have observed very similar aminoacylation values (around 8%) with synthetic tRNA in COS1 cells [13].
Figure 7. Comparing in vivo charging levels of high, medium and low efficiency suppressors.
32P-labeled S01, S07 and S15 scaffolds (label at 3′A) are incubated in HeLa cells, extracted under acidic conditions, digested by nuclease P1 and separated by TLC. Spots corresponding to free and acylated [32P]-pA are indicated. Right panel shows control with Wild Type (WT) transcripts of tRNASer (AGA) treated the same way. Suppressors and native tRNASer show comparable aminoacylation levels in vivo.
Comparative structural mapping of synthetic tRNA suppressors
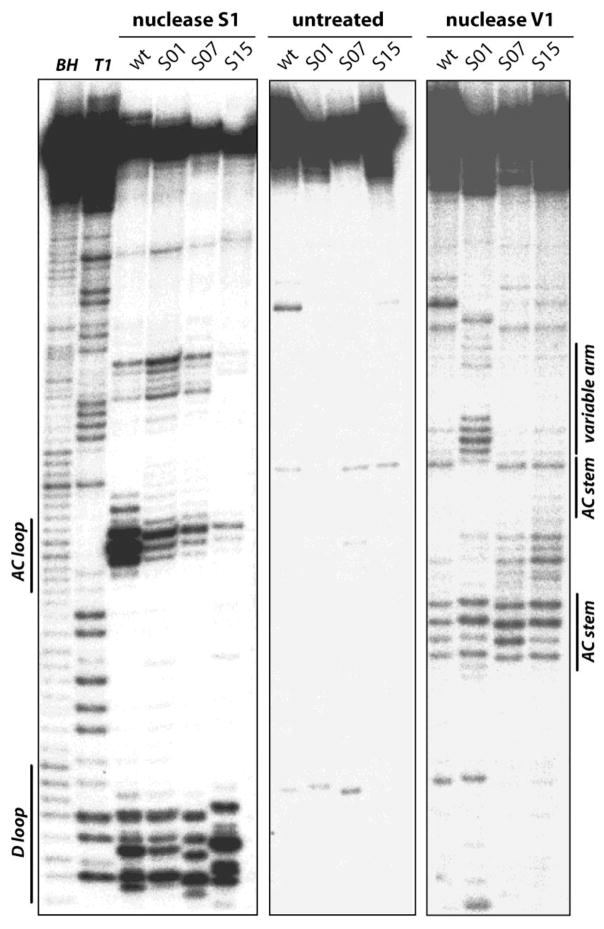
Nucleotide sequence changes can modulate the folding of tRNA. The sequences of all 15 tRNASer isodecoders that show suppression efficiency above background are consistent with the canonical secondary and tertiary structure of tRNA. However, none of these 15 transcripts has their native anticodon, a sequence change that may differentially affect the folding of these tRNAs. To investigate and compare the folding properties of tRNASer isodecoders, we used enzymatic probing in vitro to investigate the folding of scaffold S01, S07, S15 plus a tRNASer containing its wild-type anticodon sequence (Fig. 8). In this experiment, 5′ 32P-labeled tRNA transcript was refolded and the unpaired regions probed by nuclease S1 and the paired and stacked regions probed by nuclease V1 [34]. The structural mapping result is consistent with the formation of the D, anticodon and variable stem-loops in all four tRNAs, although the detailed extent of these stem-loop formation varies under this particular solution condition (20 mM tris, pH 7.5, 10 mM MgCl2, 0.1 M KCl).
Figure 8. Comparing folding in solution of high, medium and low efficiency suppressors.
Structural mapping of in vitro transcribed wild-type tRNASer(AGA) and suppressors S01, S07 and S15 by limited nuclease S1 and V1 digestion. Cleavage products of 5′-labeled transcripts are separated by electrophoresis on a 10% polyacrylamide-8M urea gel and visualized by phosphorimaging. The two first lines correspond to alkaline (BH) and RNase T1 hydrolysis ladder of transcripts S01. Transcripts and the nature of treatment are indicated on top of each line. Some tRNA structural units are indicated on the side of the gel.
We conclude from the above results that the large difference in the suppression efficiency observed in vivo cannot be solely accounted for by differences in tRNA folding. Rather, steps downstream of aminoacylation, e.g. interaction with the ribosome must play a very important role in adjusting the suppression efficiency of tRNA isodecoders.
Effect of variable loop on suppression efficiency
Unexpectedly, the structural probing pattern for the variable stem-loop is different between the most efficient suppressor S01 and the three other tRNAs tested (Fig. 8). The sequence of S01 and S04 (which is derived from the wild-type tRNASer used in structural mapping) differs by a single nucleotide in the tip of the variable stem-loop: the loop sequence for S01 is UUU and for S04 is UCU. Yet, the suppression efficiency between these two tRNAs differ by almost 2-fold. This loop sequence for both S07 and S15 is also UCU. In addition, S15 has two base pair switches in the variable stem. Hence, the feature of the variable arm may play an important role in suppression efficiency.
In addition to these sequence changes in the variable loop, a major sequence differences between the high (S01), mid (S07) and low (S15) isodecoders are four base pair switches in the acceptor stem (Fig. 5C). The suppression efficiency between the high (S01) and mid (S07) isodecoders is only 2-fold, whereas this difference is 20-fold between the high (S01) and low (S15) isodecoders. This comparison suggests that changes in the acceptor stem play only a minor role in changes in the suppression efficiency.
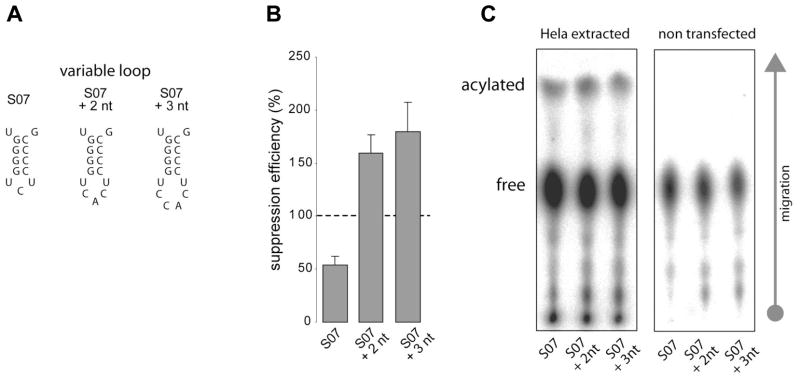
In an effort to derive suppressors that may be even more efficient than the best naturally derived e.g. S01, we enlarged the variable arm by 2–3 nucleotides, while still maintaining the same variable stem sequence (Fig. 9). The expansion is based on the observation that the human selenocysteine-tRNA has a variable loop 2 nucleotides larger than the variable loop of all tRNASer isodecoders [35]. This natural tRNA reads UGA stop codon and is accommodated in the A site of the ribosome with no apparent steric clash. Indeed, increasing the loop size in the variable arm in the context of the mid-level suppressor increases the suppression efficiency by up to 4-fold. The best suppressor obtained this way is 2-times more efficient than the best suppressor derived from natural tRNASer sequences.
Figure 9. Expansion of the loop size in the variable arm significantly increases suppression efficiency.
(A) Nucleotides have been grafted in the loop of the hairpin in the variable arm of S07 scaffold. (B) Suppression efficiency of wild type and two grafting mutants are indicated. GFP S65stop is used as a reporter. (C) In vivo charging analysis of these three transcripts shows comparable aminoacylation levels.
Discussion
This work describes the functional study of human tRNA isodecoders using UAG stop codon suppression as a proxy for translational efficiency. tRNA isodecoders share the same anticodon but have different body sequences. Isodecoder genes appear at relatively high frequencies in mammalian genomes, and mammalian isodecoder genes were present before the evolutionary divergence of mice and man [2]. The vast majority of mammalian isodecoders have sequence changes that preserve the canonical secondary and tertiary structure of tRNA.
Prior to this work, functional evaluation of mammalian tRNA isodecoders had not been carried out systematically. While the tRNA suppressor system is only a proxy for translational efficiency, it is at present the best tool available in vivo and offers some insight into translational disparities among tRNA isodecoders.
The suppression efficiency combines several cellular mechanisms. The first step of this chain of events is the internalization of synthetic tRNAs and their ability to be released into the cytoplasm. Lipofectamine has been used to transfect virtually all kind of nucleic acids without discrimination. Isodecoders are very similar molecules sharing equivalent secondary structures. As a consequence the subtle sequence variations are expected to have little or no influence on transfection efficiency. The fluorescence values we report in this study are the average of four independent transfections. Standard deviation is typically below 10% of the average value reflecting the consistency of the transfection procedure. It can be assumed that the different suppressors tested here are introduced at the same amount into human cell.
We have shown here that synthetic tRNA transfected into human cells are charged at ~5% level and this level of aminoacylation is sufficient to support up to 20% of suppression for the best suppressors. Interestingly, Kohrer and collaborators observed similar suppression efficiencies using plasmid encoded suppressors on luciferase reporter; however, the plasmid-derived suppressors are fully charged in vivo [25]. These results indicate that the suppression efficiency can be modulated by numerous parameters such as: (i) the source of the suppressor tRNA (transcripts versus plasmid encoded); (ii) the nature of the reporter genes (GFP versus luciferase) and the context of non-sense mutations; and (iii) the precise scaffold used to build the tRNA suppressor. Our work clearly illustrates the importance of tRNA scaffolds: equally charged suppressor tRNAs can differ by up to 20-fold in suppression efficiency.
This work reveals some trends in translational efficiency in human cells. The affinity of a tRNA for the ribosome is dominated by the interaction of the anticodon stem-loop with the small ribosomal subunit. More precisely nucleotide 32 together with 38 serves to weaken or strengthen ribosome affinity [36–38]. It was shown in E. coli that the efficiency of tRNAAla-derived suppressors depends on the 32–38 base pair, which increasingly promote suppression in the order UU < UC < CC < CA [37]. This rule could be relevant to human tRNAs as well. All three alanine isoacceptors tested here have the weak U32-U38 pair, which may contribute to the very low activity of the corresponding suppressors. For leucine tRNA isoacceptors, the 32–38 base pair promotes suppression as follows: UU < UC < CA < CU. We confirmed that a single substitution at position 32 (from U→C or C→U) can drastically modify the suppression efficiency and largely negate contribution of nucleotides elsewhere on the scaffold.
Our suppression assay is much better suited for the analysis of tRNASer. All tRNASer based constructs have perfectly conserved anticodon arm and loop, a requirement for unbiased suppression analysis and dissection of the contribution of specific sequence features. Seryl-tRNA synthetase/tRNASer complexes are characterized by the lack of contact between the enzyme and tRNA anticodon loop [39,40]. Binding energy and specificity are compensated owing to interactions involving the enzyme coil-coiled region and the long variable arm of tRNA. This interaction mode has been shown to be structure dependent rather than sequence-specific. tRNASer scaffolds displaying short variable arms (constructs S16-S19) lack suppression activity, most likely because they are not aminoacylated. As for those constructs containing a long variable arm, the difference in suppression can be explained by the sequence difference of the helical stem in the variable arm where the more efficient suppressors have four consecutive G-C base pairs.
We were surprised to find that tRNAs with variable arms longer than the genomic encoded sequences are superior suppressors. In lengthening the variable arm, we were able to tune the suppression efficiency by up to 4-fold. It is commonly accepted that the different components of the translation machinery have been selected throughout evolution to optimize both velocity and accuracy of protein synthesis [41]. Our engineered, tRNASer based suppressors surpass genetically encoded isoacceptors, suggesting that hyperactive tRNA species may not necessarily confer a selective advantage. Super translators might perturb overall translational effectiveness by modifying the rate of mRNA decoding and favor local ribosomal slippage. This result also suggests that the variable arm contacts the ribosome directly. In the context of suppression, such extra contacts may allow tRNAs to better compete against release factor for the access to the stop codon by providing additional binding affinity. To increase the efficiency of suppression further, one could introduce additional changes in the variable arm sequence and size. Alternatively, the amount of the release factor eRF1 which reads the UAG stop codon [42] can be reduced by RNA interference. Overexpression of eRF1 increases termination efficiency whereas decreased eRF1 activity promotes stop codon read through in eukaryotic cells [43]. Hence, decreasing eRF1 activity would lessen the competition in favor of suppressor tRNAs for the access to the A site of the ribosome.
Due to the extensive secondary structure and post-transcriptional modifications, quantitative analysis of the tRNA isodecoders in human cells has been difficult at the genomic level [2]. A comparative analysis of several isodecoder pairs among Arg(UCG), Ala(CGC) and Pro(CGG) families showed that human tissues express varying amounts of isodecoders [2]. What would be a possible biological advantage of having a fleet of tRNA isodecoders with different efficiencies in translation? Aminoacylated tRNAs are known to be used for purposes other than translation. Arginine-tRNAArg is used in yeast and mammals as an amino acid donor by arginine-terminal transferase, an enzyme that tags proteins with arginine [7]. Lysine-tRNALys and Alanine-tRNAAla are used by some bacteria to remodel the membrane and enhance antibiotic resistance [44]. tRNAs that are inefficiently used in translation might serve as storage vehicles of free amino acid that could be rapidly re-injected into the translation machinery when amino acid availability is low. These tRNAs may be used to buffer small fluctuations in amino acid concentration and avoid the activation of energy demanding mechanisms like the GCN2 pathway.
Nonsense mutations are often responsible for a variety of human genetic diseases. Approaches that have been considered for treatment of these diseases include antibiotic induced read-through of the nonsense codon [45] and gene therapy involving suppressor tRNA genes [46,47]. Our findings that some tRNA scaffolds work better than others for suppression may provide the basis for the rational design of new and more efficient therapeutic suppressor tRNA.
Material and Methods
Cell work
Adherent Hela cells were maintained at 37°C, 5% CO2 in DMEM media supplemented with fetal calf serum and antibiotics. Transfections were performed using the Lipofectamine reagent and following the instructions provided by Invitrogen. Transfections were performed in 96 well plates using per well 2 pmol (~50 ng) of tRNA and 200 ng of reporter plasmid encoding GFP mutant. Transfected cells were grown 24h before resuspended and analyzed by flow cytometry. Fluorescence values are the average of four independent transfections.
In vitro transcription
tRNAs were in vitro transcribed using T7 RNA polymerase from template generated by primer extension of overlapping DNA oligonucleotides (IDT) and purified on 10% denaturating PAGE. RNA were extracted by soaking gel slices overnight at 4°C in 50 mM KOAc, 200 mM KCl, pH 7, precipitated and resuspended in H2O.
3′ tRNA labeling [48]
200 pmol of tRNA were incubated for 10 min at 37°C in 35 μl of 100 mM glycine pH 9, 20 mM MgCl2, 60 μM CTP, 20 μM ATP, 1 μg of purified E. coli CCA adding enzyme, 15 pmol of 3000 Ci/mmol [α-32P]ATP. Labeled tRNA were gel purified as describe above.
5′ tRNA labeling
25 pmol of dephosphorylated tRNA were incubated for 30 min at 37°C with 5U of T4 polynucleotide kinase (USB Affymetrix) and 25 pmol of 6000 Ci/mmol [γ-32P]ATP in 10μl of dedicated T4 PNK buffer. Labeled tRNA were gel purified as describe above.
RNA extraction
Transfected cells were rinsed in 1 ml of PBS and resuspended in 300 μl of 0.3 M NaOAc pH 4.5. Total RNA were purified by two consecutive phenol/chloroform extractions and ethanol precipitated. RNA pellet was resuspended in 20 μl of 10 mM NaOAc pH 4.5.
Nucleotides separation on TLC [48]
1 μl of total cellular RNA was digested in 4 μl of 0.3 units/μl of Nuclease P1 (Sigma-Aldrich) in 0.3 M of NaOAc/HOAc pH 4.5 for 10 min at room temperature. 3 μl of sample were spotted on TLC plate and migrated with 5% glacial acetic acid, 100 mM NH4Cl. TLC was exposed overnight on an imaging plate and quantification of radioactive spots was performed using a Fuji scanner.
S1 and V1 structural mapping [34]
5′ 32P -labeled tRNAs were subjected to nuclease cleavage under conditions that result in a limited scission of tRNA molecules to avoid secondary cleavages. Preceding the probing reactions, labeled tRNAs were diluted with unlabeled carrier tRNA of the same sequence. To renature tRNAs [49], RNA in buffer was first heated at 85°C for 2 min, followed by room temperature for 3 min, MgCl2 and KCl were added and the mixture incubated at 37°C for 5 min. The final condition contains 0.5 μM tRNA, 20 mM TrisHCl, pH 7.5, 10 mM MgCl2 and 0.1 M KCl. Nuclease S1 or V1 was added to a final concentration of 30 U/μl and 0.2 mU/μl, respectively. The nuclease reaction proceeded at 37°C for 5 min.
Acknowledgments
We thank Mariana Pavon-Eternod (University of Chicago, USA) and Dr. Eriani (CNRS, Strasbourg, France) for discussions and suggestions. We are grateful to Dr. Ribas de Pouplana (IRB, Barcelona, Spain) for the gift of the plasmids encoding wild type and variants of GFP. Cell culture was performed using Dr. Rosner’s facility (Ben May Institute, Chicago, USA). This work was supported by grants from the NIH and DoD (to T.P.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37:D93–7. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goodenbour JM, Pan T. Diversity of tRNA genes in eukaryotes. Nucleic Acids Res. 2006;34:6137–46. doi: 10.1093/nar/gkl725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dale T, Fahlman RP, Olejniczak M, Uhlenbeck OC. Specificity of the ribosomal A site for aminoacyl-tRNAs. Nucleic Acids Res. 2009;37:1202–10. doi: 10.1093/nar/gkn1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ledoux S, Olejniczak M, Uhlenbeck OC. A sequence element that tunes Escherichia coli tRNA(Ala)(GGC) to ensure accurate decoding. Nat Struct Mol Biol. 2009;16:359–64. doi: 10.1038/nsmb.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roy H, Ibba M. RNA-dependent lipid remodeling by bacterial multiple peptide resistance factors. Proc Natl Acad Sci U S A. 2008;105:4667–72. doi: 10.1073/pnas.0800006105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang DD, Wang WY. Chlorophyll biosynthesis in Chlamydomonas starts with the formation of glutamyl-tRNA. J Biol Chem. 1986;261:13451–5. [PubMed] [Google Scholar]
- 7.Kwon YT, Kashina AS, Davydov IV, Hu RG, An JY, Seo JW, Du F, Varshavsky A. An essential role of N-terminal arginylation in cardiovascular development. Science. 2002;297:96–9. doi: 10.1126/science.1069531. [DOI] [PubMed] [Google Scholar]
- 8.Geslain R, Martin R, Camasses A, Eriani G. A yeast knockout strain to discriminate between active and inactive tRNA molecules. Nucleic Acids Res. 2003;31:4729–4737. doi: 10.1093/nar/gkg685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McClain WH, Gabriel K. Construction of an Escherichia coli knockout strain for functional analysis of tRNA(Asp) J Mol Biol. 2001;310:537–542. doi: 10.1006/jmbi.2001.4785. [DOI] [PubMed] [Google Scholar]
- 10.Carbon P, Haumont E, Fournier M, deHenau S, Grosjean H. Site-directed in vitro replacement of nucleosides in the anticodon loop of tRNA: application to the study of structural requirements for queuine insertase activity. EMBO J. 1983;2:1093–1097. doi: 10.1002/j.1460-2075.1983.tb01551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buvoli M, Buvoli A, Leinwand LA. Suppression of nonsense mutations in cell culture and mice by multimerized suppressor tRNA genes. Mol Cell Biol. 2000;20:3116–24. doi: 10.1128/mcb.20.9.3116-3124.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geiduschek EP, Tocchini-Valentini GP. Transcription by RNA polymerase III. Annu Rev Biochem. 1988;57:873–914. doi: 10.1146/annurev.bi.57.070188.004301. [DOI] [PubMed] [Google Scholar]
- 13.Kohrer C, Xie L, Kellerer S, Varshney U, RajBhandary UL. Import of amber and ochre suppressor tRNAs into mammalian cells: a general approach to site-specific insertion of amino acid analogues into proteins. Proc Natl Acad Sci U S A. 2001;98:14310–5. doi: 10.1073/pnas.251438898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Normanly J, Masson JM, Kleina LG, Abelson J, Miller JH. Construction of two Escherichia coli amber suppressor genes: tRNAPhe/CUA and tRNACys/CUA. Proc Natl Acad Sci USA. 1986;83:6548–6552. doi: 10.1073/pnas.83.17.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu W, Brock A, Chen S, Schultz PG. Genetic incorporation of unnatural amino acids into proteins in mammalian cells. Nat Methods. 2007;4:239–44. doi: 10.1038/nmeth1016. [DOI] [PubMed] [Google Scholar]
- 16.Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–5. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 17.Ropp JD, Donahue CJ, Wolfgang-Kimball D, Hooley JJ, Chin JY, Hoffman RA, Cuthbertson RA, Bauer KD. Aequorea green fluorescent protein analysis by flow cytometry. Cytometry. 1995;21:309–17. doi: 10.1002/cyto.990210402. [DOI] [PubMed] [Google Scholar]
- 18.Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–50. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 19.Achsel T, Gross HJ. Identity determinants of human tRNASer: sequence elements necessary for serylation and mutation of a tRNA with a long extra arm. EMBO J. 1993;12:3333–3338. doi: 10.1002/j.1460-2075.1993.tb06003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Breitschopf K, Achsel T, Busch K, Gross HJ. Identity elements of human tRNA(Leu): structural requirements for converting human tRNA(Ser) into a leucine acceptor in vitro. Nucleic Acids Res. 1995;23:3633–3637. doi: 10.1093/nar/23.18.3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lovato MA, Chihade JW, Schimmel P. Translocation within the acceptor helix of a major tRNA identity determinant. Embo J. 2001;20:4846–53. doi: 10.1093/emboj/20.17.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bruce AG, Atkins JF, Wills N, Uhlenbeck O, Gesteland RF. Replacement of anticodon loop nucleotides to produce functional tRNAs: amber suppressors derived from yeast tRNAPhe. Proc Natl Acad Sci U S A. 1982;79:7127–31. doi: 10.1073/pnas.79.23.7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kleina LG, Masson JM, Normanly J, Abelson J, Miller JH. Construction of Escherichia coli amber suppressor tRNA genes. II. Synthesis of additional tRNA genes and improvement of suppressor efficiency. Journal of Molecular Biology. 1990;213:705–717. doi: 10.1016/S0022-2836(05)80257-8. [DOI] [PubMed] [Google Scholar]
- 24.Anderson JC, Schultz PG. Adaptation of an Orthogonal Archaeal Leucyl-tRNA and Synthetase Pair for Four-base, Amber, and Opal Suppression. Biochemistry. 2003;42:9598–9608. doi: 10.1021/bi034550w. [DOI] [PubMed] [Google Scholar]
- 25.Kohrer C, Sullivan EL, RajBhandary UL. Complete set of orthogonal 21st aminoacyl-tRNA synthetase-amber, ochre and opal suppressor tRNA pairs: concomitant suppression of three different termination codons in an mRNA in mammalian cells. Nucleic Acids Res. 2004;32:6200–6211. doi: 10.1093/nar/gkh959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller JH, AMA Effects of surrounding sequence on the suppression of nonsense codons. J Mol Biol. 1983;164:59–71. doi: 10.1016/0022-2836(83)90087-6. [DOI] [PubMed] [Google Scholar]
- 27.Phillips-Jones MK, Hill LS, Atkinson J, Martin R. Context effects on misreading and suppression at UAG codons in human cells. Mol Cell Biol. 1995;15:6593–600. doi: 10.1128/mcb.15.12.6593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kopelowitz J, Hampe C, Goldman R, Reches M, Engelberg-Kulka H. Influence of codon context on UGA suppression and readthrough. J Mol Biol. 1992;225:261–269. doi: 10.1016/0022-2836(92)90920-f. [DOI] [PubMed] [Google Scholar]
- 29.Banan M, Puri N. The ins and outs of RNAi in mammalian cells. Curr Pharm Biotechnol. 2004;5:441–50. doi: 10.2174/1389201043376643. [DOI] [PubMed] [Google Scholar]
- 30.Li JM, Hopper AK, Martin NC. N2, N2-dimethylguanosine-specific tRNA methyltransferase contains both nuclear and mitochondrial targeting signals in Saccharomyces cerevisiae. J Cell Biol. 1989;109:1411–9. doi: 10.1083/jcb.109.4.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Subramanian A, Ranganathan P, Diamond SL. Nuclear targeting peptide scaffolds for lipofection of nondividing mammalian cells. Nat Biotechnol. 1999;17:873–7. doi: 10.1038/12860. [DOI] [PubMed] [Google Scholar]
- 32.Yu Q, Morrow CD. Essential regions of the tRNA primer required for HIV-1 infectivity. Nucleic Acids Res. 2000;28:4783–9. doi: 10.1093/nar/28.23.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ledoux S, Uhlenbeck OC. [3′-32P]-labeling tRNA with nucleotidyltransferase for assaying aminoacylation and peptide bond formation. Methods. 2008;44:74–80. doi: 10.1016/j.ymeth.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ehresmann C, Baudin F, Mougel M, Romby P, Ebel JP, Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Research. 1987;15:9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palioura S, Sherrer RL, Steitz TA, Soll D, Simonovic M. The human SepSecS-tRNASec complex reveals the mechanism of selenocysteine formation. Science. 2009;325:321–5. doi: 10.1126/science.1173755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olejniczak M, Dale T, Fahlman RP, Uhlenbeck OC. Idiosyncratic tuning of tRNAs to achieve uniform ribosome binding. Nat Struct Mol Biol. 2005;21:21. doi: 10.1038/nsmb978. [DOI] [PubMed] [Google Scholar]
- 37.McClain WH, Schneider J, Bhattacharya S, Gabriel K. The importance of tRNA backbone-mediated interactions with synthetase for aminoacylation. Proc Natl Acad Sci U S A. 1998;95:460–5. doi: 10.1073/pnas.95.2.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Auffinger P, Westhof E. Singly and bifurcated hydrogen-bonded base-pairs in tRNA anticodon hairpins and ribozymes. J Mol Biol. 1999;292:467–83. doi: 10.1006/jmbi.1999.3080. [DOI] [PubMed] [Google Scholar]
- 39.Biou V, Yoremchuk M, Tukalo M, Cusack S. The 2, 9 Å crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNASer. Science. 1994;263:1404–1410. doi: 10.1126/science.8128220. [DOI] [PubMed] [Google Scholar]
- 40.Lenhard B, Orellana O, Ibba M, Weygand-Durasevic I. tRNA recognition and evolution of determinants in seryl-tRNA synthesis. Nucleic Acids Res. 1999;27:721–729. doi: 10.1093/nar/27.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ibba M. Protein synthesis. Discriminating right from wrong. Science. 2001;294:70–71. doi: 10.1126/science.1066106. [DOI] [PubMed] [Google Scholar]
- 42.Carnes J, Frolova L, Zinnen S, Drugeon G, Phillippe M, Justesen J, Haenni AL, Leinwand L, Kisselev LL, Yarus M. Suppression of eukaryotic translation termination by selected RNAs. Rna. 2000;6:1468–79. doi: 10.1017/s1355838200001242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ilegems E, Pick HM, Vogel H. Downregulation of eRF1 by RNA interference increases mis-acylated tRNA suppression efficiency in human cells. Protein Eng Des Sel. 2004;17:821–7. doi: 10.1093/protein/gzh096. [DOI] [PubMed] [Google Scholar]
- 44.Roy H, Dare K, Ibba M. Adaptation of the bacterial membrane to changing environments using aminoacylated phospholipids. Mol Microbiol. 2009;71:547–50. doi: 10.1111/j.1365-2958.2008.06563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barton-Davis ER, Cordier L, Shoturma DI, Leland SE, Sweeney HL. Aminoglycoside antibiotics restore dystrophin function to skeletal muscles of mdx mice. J Clin Invest. 1999;104:375–81. doi: 10.1172/JCI7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Temple GF, Dozy AM, Roy KL, Kan YW. Construction of a functional human suppressor tRNA gene: an approach to gene therapy for beta-thalassaemia. Nature. 1982;296:537–40. doi: 10.1038/296537a0. [DOI] [PubMed] [Google Scholar]
- 47.Panchal RG, Wang S, McDermott J, Link CJ., Jr Partial functional correction of xeroderma pigmentosum group A cells by suppressor tRNA. Hum Gene Ther. 1999;10:2209–19. doi: 10.1089/10430349950017194. [DOI] [PubMed] [Google Scholar]
- 48.Wolfson A, Pleiss J, Uhlenbeck O. A new assay for tRNA aminoacylation kinetics. RNA. 1998;4:1019–1023. doi: 10.1017/s1355838298980700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shelton VM, Sosnick TR, Pan T. Applicability of urea in the thermodynamic analysis of secondary and tertiary RNA folding. Biochemistry. 1999;38:16831–9. doi: 10.1021/bi991699s. [DOI] [PubMed] [Google Scholar]