Abstract
Alkyne modified phospholipids can be unambiguously identified and differentiated from native species in complex mixtures by formation of dicobalthexacarbonyl complexes. This reaction is specific for alkynes and is unaffected by other glycerophospholipid related moieties. Enrichment of cells with alkyne-derivatized fatty acids or glycerophospholipids followed by solid phase sequestration and release is a promising new method for unequivocally monitoring individual glycerophospholipids following incorporation and facilitates lipidomic analysis of substrates and products.
Tracking changes in the cellular composition of lipids is essential to understanding metabolic and signaling processes. Most of the studies to date have utilized radioactive 3H-, 14C-, or 32P-labeled compounds to monitor changes in lipids during cellular processes.1–5 While these procedures are widely used, they provide limited information about the lipid molecular species involved in signaling processes. As an example, glycerophospholipids labeled with 32P are typically used to monitor changes for an entire lipid class (such as PC or PI). Furthermore, even advanced methods of mass spectrometry have limitations with regards to discriminating very complex mixtures of isobaric species.6 More recently, naturally occurring as well as stable-isotope labeled lipids have been used to follow changes in lipid patterns.7–8 These methods, while an improvement, still provide limited information on changes in the fatty acid content of individual lipid species, or are only feasible for a limited number of lipid classes. The main drawback for all of these methods is the inability to discriminate between labeled and naturally occurring lipids in very complex mixtures that contain in excess of 1000 species of phospholipids.
The use of alkyne tags for substrates has become popular with recent advances in click chemistry. Alkyne-modified substrates, in conjunction with click chemistry, offer an approach to capture the substrate through selective modification of the alkyne moiety. While this approach has been successfully used in a wide variety of applications, it has some potential drawbacks as a technique for tracking the cellular incorporation of lipids. The alkyne tag alone is not sufficient for identifying the modified lipid because the alkynyl-lipid is isobaric with naturally occurring compounds. Modification of the alkynyl-lipid through click chemistry would allow one to identify the alkynyl-lipid in a complex mixture. However, quantification of dozens of lipid species simultaneously would not be practical since standards for these heavily derivatized phospholipids are not commercially available. Finally, conventional click chemistry results in the permanent modification of the phospholipid of interest.
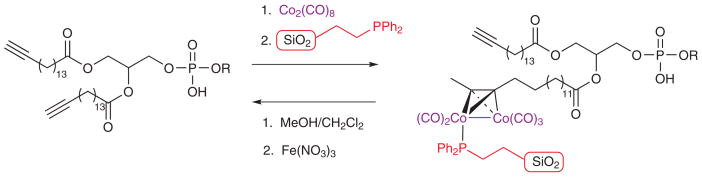
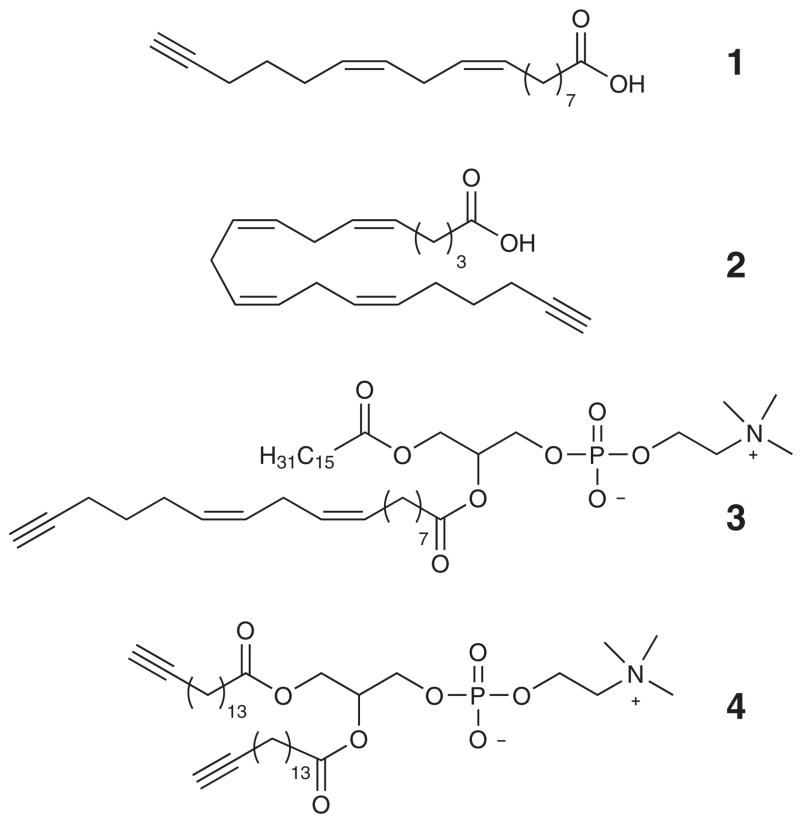
The selective reaction of alkynes with dicobalt octacarbonyl has been extensively described in many publications since the early 1970s. This well-established reaction has been successfully utilized in such diverse organic synthetic procedures as alkyne protections9–11, the Nicholas reaction12, and the Pauson-Khand cyclization13–16. We envisioned using cobalt complexation to quickly and unambiguously identify alkyne-modified lipids in complex mixtures. The incorporation of fatty acids and phospholipids into cells and their subsequent reaction with dicobaltoctacarbonyl was carried out with a variety of analogs containing an ω-alkyne functionality (Scheme 1). The terminal alkyne serves as an affinity tag in these fatty acids and their derived phospholipids. (see Supplemental Methods for synthetic details).
Scheme 1.
Alkyne analogs of fatty acids and phospholipids.
To explore this possibility, we first needed to confirm the cellular incorporation of the modified lipids. Enrichment of RAW 264.7 cells with alkyne-derivatized linoleic acid (1), or alkyne-derivatized arachidonic acid (2) was carried out using approximately 3 million cells per plate. After incubating in fatty acid-enriched media for 20 hours, samples were extracted using a modified Bligh and Dyer protocol.17–19 Cells treated with 1, 2, and untreated control samples were analyzed by direct infusion mass spectrometry in both positive and negative ionization modes. 17–19 Analysis showed that the alkyne fatty acids were incorporated into dozens of lipid species throughout all glycerophospholipid classes. Not only were phospholipids with these fatty acids detected, lipids were also observed that corresponded to species composed of alkyne fatty acids elongated by 2 carbons. The detection of normal analogs, as well as elongation products, was confirmed by extensive MSn analysis (see Supplemental Fig. 1 for a partial list of identified alkyne-containing phospholipids). As examples, MS/MS analysis of 38:4 PE from RAW 264.7 cells enriched with 1 showed that a fraction of this lipid species had a fatty acid combination of 20:0/alkyne18:4; analysis of 38:6 PI from RAW 264.7 cells enriched with 2 showed that this lipid was entirely due to a fatty acid combination of 18:0/alkyne20:6. It is important to note that routine MS analysis of lipid extracts containing alkyne-tagged phospholipids would not be possible due to the complex mixture of isobaric species which could not be identified by precursor ion scan analysis or completely resolved by LC retention time. As an example, the parent mass and MS/MS fragmentation patterns of alkyne 38:4 PE (containing the two carbon elongated alkyne fatty acid 20:4) and naturally occurring 38:4 PE would be identical.
Incubating RAW 264.7 cells for four hours with PC 3, or 4 supplemented media yielded cells enriched in these alkyne analog phospholipids (Fig. 1). For example, LC/MS analysis of the RAW 264.7 cell extracts indicated that a substantial amount of lipid 3 had incorporated by quantitative comparison to other naturally occurring PC lipids (Fig 1b). Quantification of the PC lipids was accomplished with the aid of four odd-carbon internal standards (25:0, 31:1, 37:4, and 43:6) and standard curves generated using even-carbon PC lipid standards.19 In the case of PC analog 3, 370 pg (500 pmol) lipid/1×106 cells were incorporated after the four hour incubation. By comparison, typical native PCs (e.g. 34:1 PC) are 200 to 400 pmol/1 × 106 untreated RAW 264.7 cells. Lipid identification was confirmed by comparing MS/MS spectra of enriched cell extracts with the fragmentation patterns of pure standards (Supplemental Fig. 2). Cellular enrichment with alkyne PC 4 yielded an incorporation of 466 pmol lipid/1 × 106 cells.
Figure 1.
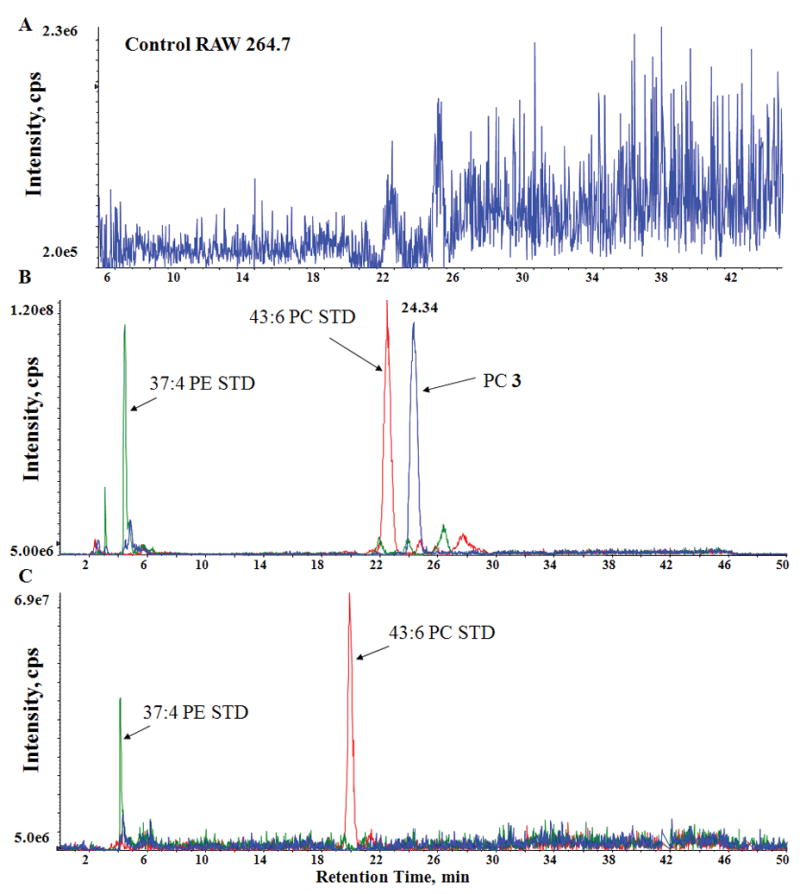
LC/MS results from PC (3) enriched RAW 264.7 cell extracts. (a) Extracted ion chromatogram (34:4 PC, 738 m/z) from a control RAW 264.7 cell extract. No detectable amount of 34:4 PC was observed in these samples. (b) 738 m/z (blue trace) extracted ion chromatogram corresponding to [PC (3)-15]− from cells treated with PC (3), and extracted ion chromatograms for 43:6 PC (red trace) and 37:4 PE (green trace) internal standards (c) 738 m/z extracted ion chromatogram (blue trace) from cells treated with PC (3), followed by reaction with Co2(CO)8. Note that the lack of a peak at this mass confirms that PC (3) was completely consumed. Extracted ion chromatograms corresponding to the 43:6 PC (red trace) and 37:4 PE (green trace) internal standards are also shown. Treatment with Co2(CO)8 had no detectable effect on these or any other non-alkyne lipid.
As an initial demonstration of the cobalt-alkyne reaction, a pure sample of PC (3) was treated with Co2(CO)8 under an atmosphere of N2. The resulting solution was analyzed by direct infusion MS which showed that the alkyne PC (3) had been completely consumed by the reaction and that a new peak corresponding to the dicobalthexacarbonyl complex 3a was detected at 1038 m/z. MS/MS analysis of this peak showed the loss of six consecutive 28 m/z units attributable to the loss of the six CO ligands (Supplemental Fig. 3), further confirming the identity of the complex. Under routine MS conditions, the CO ligands are quite labile.
Whole cell extracts from compound 3 enriched RAW 264.7 cells were also treated with Co2(CO)8. The alkyne-cobalt complex was not observed using a normal phase LC/MS protocol tailored towards glycerophospholipids (possibly due to the labile nature of the compound and a strong interaction with the silica support). However, analysis by LC/MS (Fig. 1c) showed that compound 3 had been completely consumed by the reaction with Co2(CO)8. No reaction with other non-alkyne lipids was observed during the course of these experiments. Odd-carbon internal standards (25:0, 31:1, 37:4, and 43:6) from the six major lipid classes (PA, PC, PE, PG, PI, and PS) were added to all of the LC/MS samples in concentrations much lower than that observed for PC (3). Whereas the entire signal corresponding to PC (3) was ablated during reaction with Co2(CO)8, no effect was observed on any of the internal standards, suggesting that this method is suitable for a variety of lipid classes with varying degrees of unsaturation.
We next sought to extend this technology by confirming that the alkynes could function in normal biological processes. We chose to follow the enzymatic conversion of an alkyne phospholipid substrate into its corresponding alkyne lipid product. To measure the flux from substrate to product a series of experiments were performed measuring products of the transphosphatidylation enzymatic reaction of phospholipase D. Primed RAW 264.7 cells enriched with PC (4) were treated with phorbol 12-myristate 13-acetate (PMA) to stimulate PLD in the presence of n-butanol, which serves as an alternate substrate.20–21 Analysis by LC/MS and LC/MS/MS indicated that the transphosphatidylation reaction product 32:4 alkynyl PtdBuOH (5) had been formed as expected (supplemental Fig. 4b). It was confirmed as the alkynyl product and not an isobaric phospholipid by complexation with Co2(CO)8, which resulted in complete disappearance of the peak (data not shown).
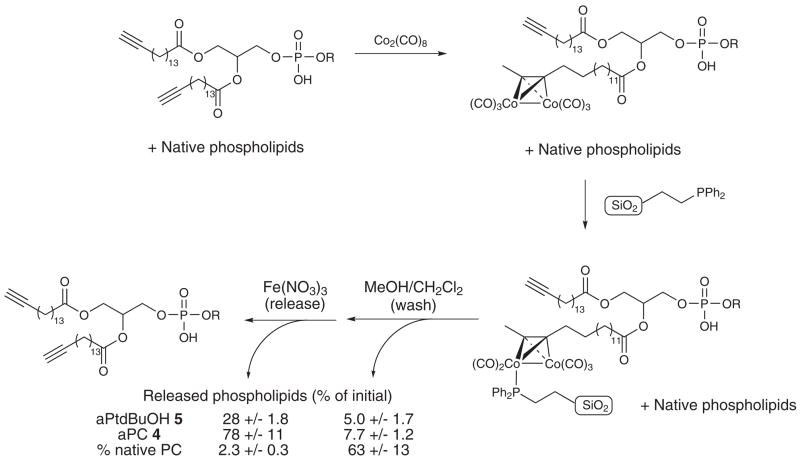
In addition to using cobalt complexation as a means of simply identifying alkyne-containing phospholipids, it can importantly also serve as a means to capture the alkynyl phospholipid from a complex mixture (Fig 2a). The first step involves cobalt complexation to the alkyne as described above. The alkyne-cobalt complex can be subsequently immobilized onto a solid-supported phosphine. It has been demonstrated that alkyne-cobalt complexes react selectively with triphenylphosphine (PPh3) and solid-supported PPh3.22–23 We envisioned using an approach similar to the latter case to selectively remove the alkyne from the vast majority of native phospholipids which are washed away. Finally, mild oxidation of the cobalt with Fe(NO3)3 releases the parent alkyne free of almost all non-alkynyl phospholipids, enabling the identification and quantification of the labeled lipid.
Figure 2.
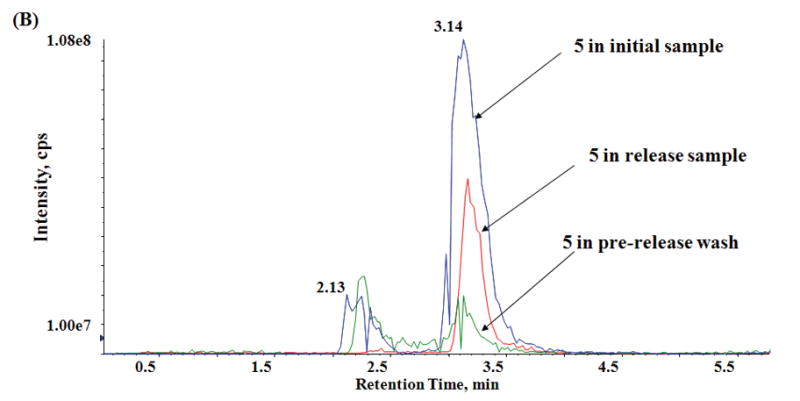
Capture and release of alkyne-modified lipids. (a) Functionalized silica gel and cobalt immobilization with subsequent release of alkynyl phospholipids. Lipid quantification shows results averaged from 3 independent sets of capture and release experiments. (b) LC/MS results from PMA treatment of RAW 264.7 cells enriched with alkyne PC (4). (Blue trace) 695 m/z extracted ion chromatogram, which corresponds to the 32:4 alkynyl PtdBuOH PLD reaction product 5 in the initial (pre capture and release) sample. (Red trace) Extracted ion chromatogram for 5 in the release sample. (Green trace) Extracted ion chromatogram for 5 in the post-capture combined washes.
We first conducted numerous experiments to determine a suitable solid support. We found that 2-diphenylphosphinoethyl-functionalized silica gel (Aldrich) yielded better results than supports containing polystyrene cores with or without PEG (polyethyleneglycol) linkers. Considerable method development was done utilizing triphenylphosphine Novagel (EMD chemicals). This class of solid support was found to be unsuitable due to solvent incompatibility (high mass spec background noise) and non-specific binding of native lipids to the support.
With the solid phase material optimized, we performed a full capture and release sequence (Fig 2, Supplemental Fig. 4). In the washes after the initial capture, the alkyne lipids 4 and 5 (Fig 2) were only present at 7 and 5% of initial respectively, whereas 63% of the natural PC was recovered. The opposite was true for the washes after release. Only 2% of natural phosphatidylcholine was present in the release samples. In contrast, the alkynyl phospholipids 4 and 5 (Fig. 2) have been enriched (78 and 28% of initial amounts were recovered respectively). We observed similar behavior for the other native glycerophospholipid classes in both the post-capture wash and release samples. In the latter case, many of these lipids were below the limit of absolute quantification. These combined values demonstrate that a negligible amount of native phospholipids from the PLD reaction cell extracts is present after the capture and release sequence, yielding a strongly enriched pool of modified lipids suitable for further study.
In summary, our results demonstrate that the alkyne lipids can incorporate into cellular membranes and enter into the substrate pool for enzymes such as PLD. Furthermore, the use of solid-supported phosphines, such as 2-diphenylphosphinoethyl functionalized silica gel, allows one to selectively remove the alkyne-containing lipids from the whole cell extracts by immobilizing the cobalt-alkyne complexes according to their unique chemistry. This ability to selectively purify reaction products, as well as potential binding partners, has tremendous potential for a wide range of lipidomic applications.
Supplementary Material
Acknowledgments
The authors thank Pavlina Ivanova and David Myers for technical assistance and many helpful conversations. This work was partially supported by the National Institutes of Health (P01-ES013125) (H.A.B., L.J.M, and N.A.P.), Vanderbilt Institute of Chemical Biology Pilot Program Grant (H.A.B, and N.A.P.), and National Institutes of Health Grant Lipid Maps U54 GM69338 (H.A.B).
Footnotes
AUTHOR CONTRIBUTIONS
S.B.M. analyzed cell extract samples by mass spectrometry. K.A.T. and R.S. performed chemical synthesis, purification, and analysis. S.B.M. and C.M.L. developed the alkyne lipid cobalt derivatization protocol. K.A.T. and S.B.M developed the capture and release methodology. M.D.A. and C.A.R. contributed to the lipid enrichment procedures. L.J.M., N.A.P., and H.A.B. were responsible for overall project direction and experimental design. All authors discussed the results and modified the manuscript.
The authors declare they have no competing financial interests.
References
- 1.Walker SJ, Brown HA. Methods Mol Biol. 2004;237:89–97. doi: 10.1385/1-59259-430-1:89. [DOI] [PubMed] [Google Scholar]
- 2.Brown HA, Gutowski S, Moomaw CR, Slaughter C, Sternweis PC. Cell. 1993;75:1137–1144. doi: 10.1016/0092-8674(93)90323-i. [DOI] [PubMed] [Google Scholar]
- 3.Raetz CR. Phosphatidylserine synthetase mutants of Escherichia coli. J Biol Chem. 1976;251:3242–3249. [PubMed] [Google Scholar]
- 4.Ganong BR, Leonard JM, Raetz CR. J Biol Chem. 1980;255:1923–1929. [PubMed] [Google Scholar]
- 5.Serunian LA, Auger KR, Roberts TM, Cantley LC. J Virol. 1990;64:4718–4725. doi: 10.1128/jvi.64.10.4718-4725.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown HA, Murphy RC. Nat Chem Biol. 2009;5:602–606. doi: 10.1038/nchembio0909-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zemski Berry KA, Murphy RC. J Lipid Res. 2005;46:1038–1046. doi: 10.1194/jlr.M500014-JLR200. [DOI] [PubMed] [Google Scholar]
- 8.Ekroos K, Chernushevich IV, Simons K, Shevchenko A. Anal Chem. 2002;74:941–949. doi: 10.1021/ac015655c. [DOI] [PubMed] [Google Scholar]
- 9.Nicholas KM, Pettit R. Tetrahedron Lett. 1971:3475–3478. [Google Scholar]
- 10.Seyferth D, Nestle MO, Wehman AT. J Am Chem Soc. 1975;97:7417–7426. [Google Scholar]
- 11.Milgrom LR, Rees RD, Yahioglu G. Tetrahedron Lett. 1997:4905–4908. [Google Scholar]
- 12.Schreiber SL, Sammakia T, Crowe WE. J Am Chem Soc. 1986;108:3128–3130. [Google Scholar]
- 13.Khand IU, Knox GR, Pauson PL, Watts WE. J Chem Soc Chem Comm. 1971;36 [Google Scholar]
- 14.Khand IU, Graham RK, Pauson PL, Watts WE. J Chem Soc Perk T. 1973;1:975–977. [Google Scholar]
- 15.Khand IU, Knox GR, Pauson PL, Watts WE, Foreman MI. J Chem Soc Perk T. 1973;1:977–981. [Google Scholar]
- 16.Pauson PL. Tetrahedron. 1985;41:5855–5860. [Google Scholar]
- 17.Ivanova PT, Milne SB, Forrester JS, Brown HA. Mol Interv. 2004;4:84–94. doi: 10.1124/mi.4.2.6. [DOI] [PubMed] [Google Scholar]
- 18.Milne S, Ivanova P, Forrester J, Brown HA. Methods. 2006;39:92–103. doi: 10.1016/j.ymeth.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 19.Ivanova PT, Milne SB, Byrne MO, Xiang Y, Brown HA. Meth Enzymol. 2007;432:21–57. doi: 10.1016/S0076-6879(07)32002-8. [DOI] [PubMed] [Google Scholar]
- 20.Rouzer CA, et al. Biochemistry. 2006;45:14795–14808. doi: 10.1021/bi061723j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown HA, Henage LG, Preininger AM, Xiang Y, Exton JH. Meth Enzymol. 2007;434:49–87. doi: 10.1016/S0076-6879(07)34004-4. [DOI] [PubMed] [Google Scholar]
- 22.Comely AC, Gibson (née Thomas) SE, Hales NJ. Chem Commun. 1999:2075–2076. [Google Scholar]
- 23.Comely AC, Gibson SE, Hales NJ, Johnstone C, Stevenazzi A. Org Biomol Chem. 2003;1:1959–1968. doi: 10.1039/b303208f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.