Abstract
Filamentous fungal gene expression assays provide essential information for understanding systemic cellular regulation. To aid research on fungal gene expression, we constructed a novel, comprehensive, free database, the Filamentous Fungal Gene Expression Database (FFGED), available at http://bioinfo.townsend.yale.edu. FFGED features user-friendly management of gene expression data, which are assorted into experimental metadata, experimental design, raw data, normalized details, and analysis results. Data may be submitted in the process of an experiment, and any user can submit multiple experiments, thus classifying the FFGED as an “active experiment” database. Most importantly, FFGED functions as a collective and collaborative platform, by connecting each experiment with similar related experiments made public by other users, maximizing data sharing among different users, and correlating diverse gene expression levels under multiple experimental designs within different experiments. A clear and efficient web interface is provided with enhancement by AJAX (Asynchronous JavaScript and XML) and through a collection of tools to effectively facilitate data submission, sharing, retrieval and visualization.
Keywords: public experiments, public results, data submission, related experiments, experimental design, AJAX, microarray
1. Introduction
Due to their close relationship with Animalia and their experimental tractability, filamentous fungi are frequently used as model organisms for understanding diverse aspects of basic cellular regulation, including cell cycle progression, gene expression, circadian timing, light sensing, recombination, secretion, multicellular development (Davis 1966; Ballance 1986; Dunlap et al. 2007), and evolution (Bruns et al. 1991; Kohn 2005). For instance, the multicellular filamentous fungus Neurospora crassa (Galagan et al. 2003; Wu et al. 2007) has been studied in research laboratories for decades (Beadle and Tatum 1941; Davis and Perkins 2002; Dong et al. 2008), primarily owing to its interesting and diverse biology, ease of culture, facile genetics, and rapid growth rate (Borkovich et al. 2004; Merrow and Roenneberg 2007). A number of other fungi, such as Aspergillus nidulans (Galagan et al. 2005a), A. fumigatus (Schoberle and May 2007), Fusarium graminearum (Cuomo et al. 2007), Magnaporthe grisea (Soderlund et al. 2006), have become important fungal models for animal and plant pathogenesis (Gow et al. 2002; Tunlid and Talbot 2002). Increasingly, gene expression data for filamentous fungi provide a rich resource for the identification of candidate genes and the characterization of systemic biological responses to growth conditions and environmental stresses (Soderlund et al. 2006; Breakspear and Momany 2007). However, few resources have been made available for storage and access to diverse yet interrelated filamentous fungal gene expression studies.
A number of databases have been constructed to provide storage and management of gene expression data, several (Demeter et al. 2007; Hong et al. 2008; Barrett et al. 2009; Parkinson et al. 2009). The Stanford Microarray Database (SMD) (Demeter et al. 2007) serves as a research tool that is free of charge for Stanford researchers and their collaborators. The Saccharomyces Genome Database (SGD) (Hong et al. 2008) is a central repository for the budding yeast Saccharomyces cerevisiae, allowing the retrieval of microarray expression data for Saccharomyces (Ball et al. 2001).
Among them, the Gene Expression Omnibus (GEO) database (Barrett et al. 2009) at the US National Center for Biotechnology Information (NCBI) and the ArrayExpress repository (Parkinson et al. 2009) at the European Bioinformatics Institute (EBI) are major databases for public gene expression data. However, these databases simply collect user-submitted results and lack a clear classification of gene expression data. Gene expression metadata include a variety of information from wet-lab experiment to in-silico procedure, which may be classified into different sections. Considering the burgeoning collection of gene expression data, a necessary next step is to make full use of classified expression data to automatically correlate relevant experiments and discover some correlations with different experimental conditions. However, such an operation is not well-supported by these generalized unclassified databases. Moreover, none of these databases provide specific resources to facilitate well-annotated deposition or facile comparison of filamentous fungal gene expression studies.
To simplify data submission and management and to facilitate discovery of correlations among diverse gene expression experiments in filamentous fungi, we constructed a novel, comprehensive, free database, the Filamentous Fungal Gene Expression Database (FFGED; http://bioinfo.townsend.yale.edu), to serve as a collective and collaborative platform for filamentous fungi. Unlike existing related databases, FFGED focuses on comparisons of diverse experimental designs and correlations of different experimental results for filamentous fungi. The aim of FFGED is not only to provide users with friendly web interfaces for facilitating data submission and management, but also to rapidly share gene expression data within the scientific community at large, to automatically identify related gene expression experiments to assist users in connecting their experiments with similar experiments, and to effectively ease virtual comparisons of each gene’s expression under multiple experimental designs within different experiments across diverse fungi.
2. Materials and Methods
2.1. Database architecture
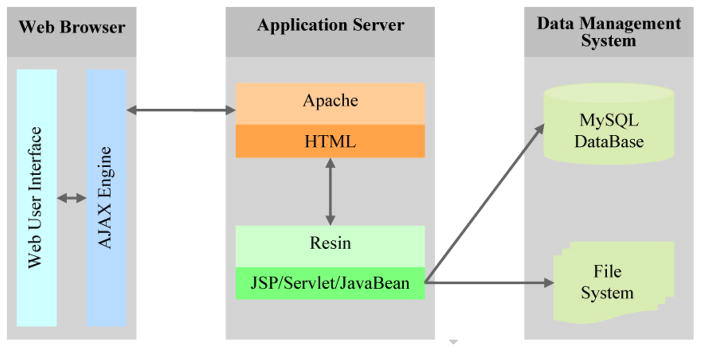
To develop FFGED, we used MySQL (a free and popular relational database management system, Version 5.1.30; http://www.mysql.org) on a Mac OS X Server. To provide efficient access to the database and to simplify data submission and management, we provide user-friendly graphical user interfaces (GUI) implemented with HTML, JSP (Java Server Pages), JavaBean, Servlet, and AJAX (Asynchronous JavaScript and XML, a collection of web development technologies for creating highly interactive web applications) with two web service applications, Apache and Resin (Figure 1). Apache serves static content (like HTML) and Resin serves dynamic content (like JSP, JavaBean, and Servlet). AJAX is used to transfer data between server and browser, based on JQuery (an open source, free, and lightweight framework for interaction between JavaScript and HTML; http://www.jquery.com).
Figure 1.
Database architecture of FFGED, including three tiers, web browser, application server and data management system. These tiers facilitate ease of use, efficient data processing, and reliable archiving of data.
2.2. Annotation
To provide gene annotations to result files from raw or statistical analyses of microarray data, we retrieved annotations for N. crassa from three databases: PEDANT (Protein Extraction, Description and Analysis Tool; http://pedant.gsf.de) (Riley et al. 2005), the Neurospora crassa e-Compendium (http://bioinformatics.leeds.ac.uk/~gen6ar/newgenelist/genes/), and the Broad Institute of MIT and Harvard (http://www.broadinstitute.org/annotation/genome/neurospora/). Annotation data for Aspergillus fumigatus, A. nidulans, Coccidioides immitis, Coprinus cinereus, Fusarium graminearum, F. verticillioides, Histoplasma capsulatum, and Magnaporthe grisea were obtained from the Broad Institute. Additionally, pathway information for several species was extracted from KEGG (Kyoto Encyclopedia of Genes and Genomes) (Kanehisa and Goto 2000).
2.3. Normalization
To ease data analysis, FFGED features an online normalization tool, providing normalization for two-color arrays as in (Townsend et al. 2003). For evaluation of hybridization quality a helpful illustrative plot of the normalization regression, can be constructed and exported as an image in SVG or PNG formats.
2.4. Identification of related experiments
To link related experiments together, FFGED calculates the cosine similarity coefficient for two experiments. The cosine similarity coefficient is widely used in information retrieval and data mining (Salton and McGill 1983; Baeza-Yates and Ribeiro-Neto 1999; Dhillon and Modha 2001). It is the cosine of the angle (θ) between two vectors of n dimensions when each experiment is represented by its experimental design, which can include multiple developmental, genetic, and/or environmental states. Therefore, given two vectors of experiments x(x1, x2, …, xn) and y(y1, y2, …, yn), the cosine similarity coefficient between x and y is obtained by
| (1) |
where and .
3. Results
3.1. Data submission and management
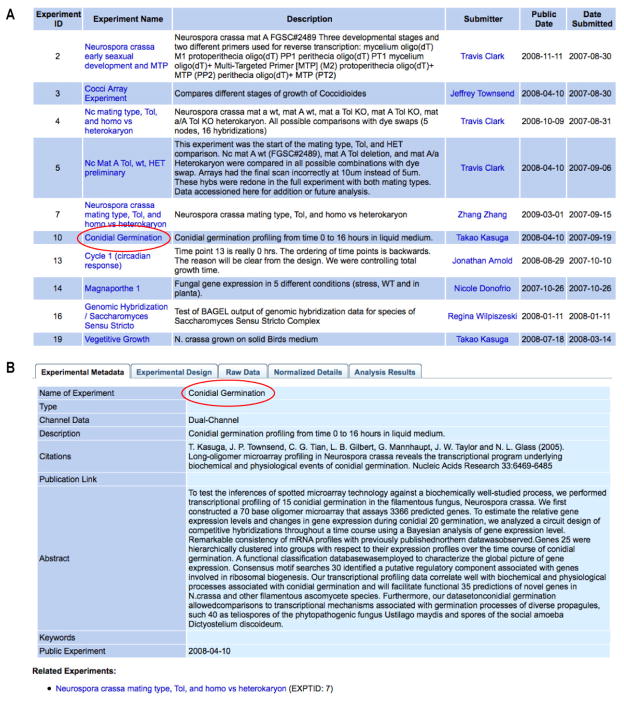
In FFGED, any data can be submitted in the process of an experiment or after its completion. Entry of an experiment is divided into five sections: experimental Metadata (e.g., experimental name), experimental Design (e.g., cyanine-3 and cyanine-5 treatments), Raw data (e.g., GenePix Results), Normalized details (e.g., normalization methods), and analysis Results (e.g., gene expression levels). This division is hereafter denoted as MDRNR (see Figure 2B). MDRNR is also compatible with MIAME (Minimum Information about Microarray Experiment; http://www.mged.org/Workgroups/MIAME/miame.html) (Brazma et al. 2001), further facilitating data exchange with other institutions. By providing resources for archiving raw data through processed results at each of these stages, FFGED facilitates use as an “active experiment” database, increasing the immediate utility to the user and decreasing the burden of post-experimental submission and annotation.
Figure 2.
Screenshots from public experiments, shown in a tab format. All information in public experiments can be accessed by any user. (A) Some public experiments submitted by users. (B) Detailed information for a given experiment, including experimental metadata, experimental design, raw data, normalized details, and analysis results (only “experimental metadata” shown), and listing related experiments at the bottom.
Unlike existing related databases (Killion et al. 2003; Demeter et al. 2007; Hong et al. 2008; Barrett et al. 2009; Parkinson et al. 2009), FFGED not only functions as a storage database but also as an analysis tool that facilitates progression and processing of data through the MDRNR classifications. Several online programs, for example, normalization (Townsend et al. 2003), generation of an input file for BAGEL (Bayesian Analysis of Gene Expression Levels; (Townsend and Hartl 2002), annotation of results, and figure plotting (implemented with JfreeChart, an open-source framework for creation of professional charts; http://www.jfree.org), have been integrated into FFGED. These tools greatly ease data analysis and visualization. In addition, user-friendly web interfaces with clear progression monitoring of experiment submission are provided for diverse competencies of submitters, lowering technological entrance barriers (Ball et al. 2004).
3.2. Public experiments
Any user can submit multiple experiments to FFGED. Each experiment can be set to be private or public. Public experiments are freely accessible by users. MDRNR classifies gene expression data into five different sections, with “experimental design” as the core section that shapes the entries for other sections. Although experiments may differ greatly, similar experimental states submitted in experimental design indicate that experiments may be compared very productively. Therefore, FFGED employs experimental states to link each experiment with similar experiments made public by other users.
For each public experiment in FFGED, detailed information is shown in a tab format according to MDRNR (Figure 2), and related experiments are also displayed according to their similarity coefficients (see Materials and Methods). As a result, related experiments are identified, which can lead to synthetic knowledge discovery. Furthermore, users with similar experiments are connected to each other, enhancing communication and collaboration within the scientific community.
3.3. Public results
Public results are gene expression levels under a variety of experimental states, extracted from analysis results within public experiments. Such public results provide a forum for the study of the expression of individual genes and their correlations under different experimental states. For each gene, FFGED quantifies expression levels under diverse experimental variables within different public experiments (Figure 3), enabling comparative gene expression analysis.
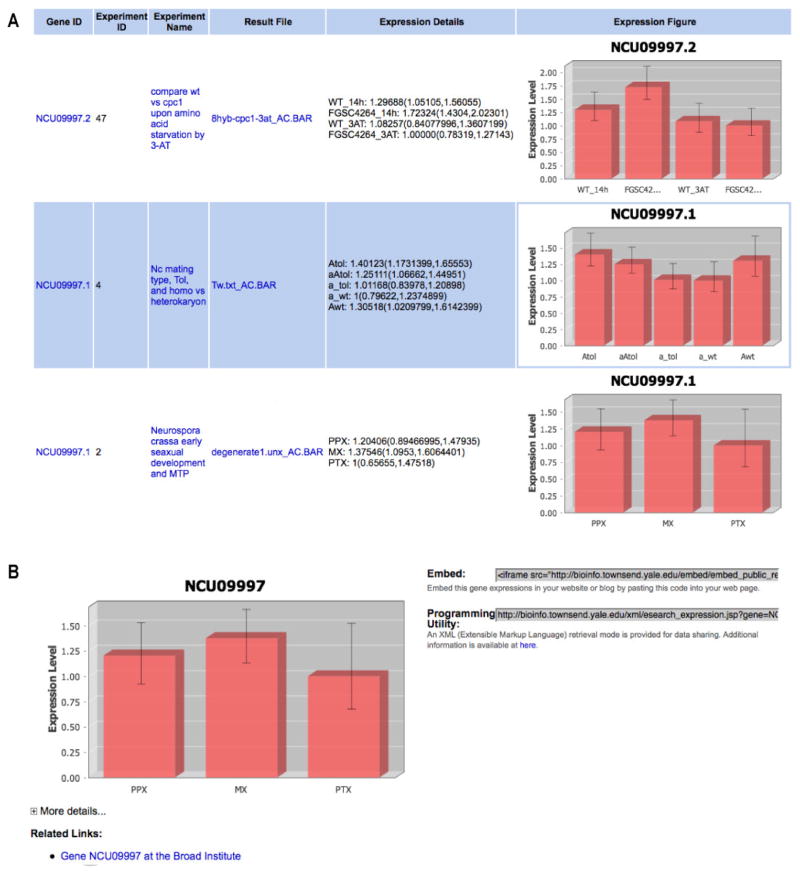
Figure 3.
Screenshots from public results, taking Neurospora crassa Gene NCU09997 as an example. All information in public results can be accessed by any user. (A) Results for expression of gene NCU09997 are included in three public experiments. (B) Gallery show of expression information for gene NCU09997. The gallery show for expression of a gene, for instance, can be embedded into any web site or exchanged by an XML retrieval mode. Expression information displayed in this manner is embedded into gene descriptions for Neurospora at the Broad Institute website (e.g. http://www.broadinstitute.org/annotation/genome/neurospora/GeneLocus.html?sp=SNCU09997). The 3D figure for gene expression level can be freely downloaded in PNG and SVG formats.
To share this information with the scientific community at large and to provide content for other institutions, we provide an XML retrieval mode via Internet for structured data exchange. In addition, FFGED offers a dynamic interface to embed up-to-the-minute experimental results for the expression of a relevant gene into any web site. For instance, the N. crassa database at the Broad Institute has implemented this tool, embedding experimental results for each gene’s expression into its gene-specific web page (See the FAQ page at http://bioinfo.townsend.yale.edu/faq.jsp#sharing).
4. Discussion and Future Development
Our newly developed database, FFGED, is a collective and collaborative platform that provides a useful tool for filamentous fungal gene expression studies. The AJAX-based database has several unique advantages over other currently available databases; the database provides users with friendly and highly interactive web interfaces, lowers technological barriers and emphasizes ease of data submission, retrieval, visualization, and sharing. Furthermore, unlike existing generalized databases, FFGED provides a specific platform for filamentous fungal gene expression studies, and most importantly, facilitates direct comparisons of diverse experimental designs and correlations of different experimental results for filamentous fungi. Additionally, in FFGED, gene expression data can be shared rapidly within the scientific community, related gene expression experiments can be identified easily, and gene expression systems can be compared virtually under multiple experimental designs across diverse fungi.
Regarding gene expression data storage and management, FFGED provides a flexible framework, MDRNR, which clearly classifies gene expression data into five different sections and can be used with diverse designs and platforms (Burgoon 2006). This classification scheme accordingly makes FFGED completely different from other databases. Compared with extant unclassified databases, FFGED also promises to provide correlations among different experiments that may be easily retrieved and interpreted. Moreover, MDRNR separates wet-bench work conducted by biologists (experimental metadata and experimental design) from dry-bench work done by bioinformaticians (raw data, normalized details and analysis results).
In contrast with some databases which are not free of charge for all users (e.g., SMD), FFGED features free private submission and free public access to public data for any user, with the aim of increasing collaboration by maximizing data sharing and facilitating knowledge discovery from collective data. This feature of knowledge discovery through collective public data is a key element for data-intensive bioinformatics studies (Stajich and Lapp 2006; Zhang et al. 2009).
FFGED has served as an in-house database for gene expression data since October 2007, and has been regularly updated and improved. Potential expansions of this database would be the addition of other fungal genome browsers (http://fungal.genome.duke.edu), facilitating genome annotations and comparative sequence analysis. In addition, providing specific formats for submission of other types of gene expression data, such as MPSS (Massively Parallel Signature Sequencing, Reinartz et al. 2002; Torres et al. 2008) and DGE (Digital Gene Expression) profiling (Linsen et al. 2009), into the expression level tables would facilitate further gene expression comparisons across multiple samples, as with DNA microarrays and other gene expression platforms (Nakano et al. 2006). Moreover, integrating more online tools for gene expression analysis, for example, tools for detecting co-expressed genes within and among experiments, would greatly facilitate expression analysis. Lastly, expanding comparative expression analysis across fungi would be essential for studying their vast and diverse biology and morphology (Galagan et al. 2005b).
In summary, we have constructed the Filamentous Fungal Gene Expression Database and it features user-friendly web interfaces, free access, clear classification of expression data (MDRNR), novel identification of similar related experiments and correlation of diverse gene expression levels within multiple experiments. It will facilitate in-depth comparative expression studies on diverse filamentous fungi.
Acknowledgments
We thank Dr. Jay Dunlap, Dr. Louise Glass, Dr. Matthew Sachs, our colleagues at the Broad Institute of MIT and Harvard, and everyone involved in the Neurospora PO1 functional analysis of a model filamentous fungus project for providing feedback and a highly productive collaborative community. We also thank Dr. Takao Kasuga for valuable suggestions, Dr. Namboori B. Raju for providing an image for the database logo, and many users as well as members of the Townsend lab for reporting bugs and sending comments.
Funding
This work was supported by program project grant GM068087 from the National Institute of General Medical Sciences at the US National Institutes of Health.
Footnotes
Author Contributions
ZZ designed, programmed, and implemented this database and drafted the manuscript. JPT contributed core design principles, supervised the development, and revised the manuscript. Both authors have read and approved the final manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Zhang Zhang, Email: Zhang.Zhang@Yale.edu.
Jeffrey P. Townsend, Email: Jeffrey.Townsend@Yale.edu.
References
- Baeza-Yates R, Ribeiro-Neto B. Modern information retrieval. ACM Press; New York: 1999. [Google Scholar]
- Ball CA, Brazma A, Causton H, Chervitz S, Edgar R, Hingamp P, et al. Submission of microarray data to public repositories. PLoS Biol. 2004;2:E317. doi: 10.1371/journal.pbio.0020317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball CA, Jin H, Sherlock G, Weng S, Matese JC, Andrada R, et al. Saccharomyces Genome Database provides tools to survey gene expression and functional analysis data. Nucleic Acids Res. 2001;29:80–81. doi: 10.1093/nar/29.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballance DJ. Sequences important for gene expression in filamentous fungi. Yeast. 1986;2:229–236. doi: 10.1002/yea.320020404. [DOI] [PubMed] [Google Scholar]
- Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadle GW, Tatum EL. Genetic Control of Biochemical Reactions in Neurospora. Proc Natl Acad Sci U S A. 1941;27:499–506. doi: 10.1073/pnas.27.11.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkovich KA, Alex LA, Yarden O, Freitag M, Turner GE, Read ND, et al. Lessons from the genome sequence of Neurospora crassa: Tracing the path from genomic blueprint to multicellular organism. Microbiology and Molecular Biology Reviews. 2004;68:1–108. doi: 10.1128/MMBR.68.1.1-108.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- Breakspear A, Momany M. The first fifty microarray studies in filamentous fungi. Microbiology. 2007;153:7–15. doi: 10.1099/mic.0.2006/002592-0. [DOI] [PubMed] [Google Scholar]
- Bruns TD, White TJ, Taylor JW. Fungal Molecular Systematics. Annual Review of Ecology and Systematics. 1991;22:525–564. [Google Scholar]
- Burgoon LD. The need for standards, not guidelines, in biological data reporting and sharing. Nat Biotechnol. 2006;24:1369–1373. doi: 10.1038/nbt1106-1369. [DOI] [PubMed] [Google Scholar]
- Cuomo CA, Guldener U, Xu JR, Trail F, Turgeon BG, Di Pietro A, et al. The Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science. 2007;317:1400–1402. doi: 10.1126/science.1143708. [DOI] [PubMed] [Google Scholar]
- Davis RH. Neurospora. Science. 1966;153:1553–1556. doi: 10.1126/science.153.3743.1553. [DOI] [PubMed] [Google Scholar]
- Davis RH, Perkins DD. Timeline: Neurospora: a model of model microbes. Nat Rev Genet. 2002;3:397–403. doi: 10.1038/nrg797. [DOI] [PubMed] [Google Scholar]
- Demeter J, Beauheim C, Gollub J, Hernandez-Boussard T, Jin H, Maier D, et al. The Stanford Microarray Database: implementation of new analysis tools and open source release of software. Nucleic Acids Res. 2007;35:D766–770. doi: 10.1093/nar/gkl1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon IS, Modha DS. Concept decompositions for large sparse text data using clustering. Machine Learning. 2001;42:143–175. [Google Scholar]
- Dong W, Tang X, Yu Y, Nilsen R, Kim R, Griffith J, et al. Systems Biology of the Clock in Neurospora crassa. PLoS ONE. 2008;3:e3105. doi: 10.1371/journal.pone.0003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap JC, Borkovich KA, Henn MR, Turner GE, Sachs MS, Glass NL, et al. Enabling a community to dissect an organism: overview of the Neurospora functional genomics project. Adv Genet. 2007;57:49–96. doi: 10.1016/S0065-2660(06)57002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan JE, Calvo SE, Borkovich KA, Selker EU, Read ND, Jaffe D, et al. The genome sequence of the filamentous fungus Neurospora crassa. Nature. 2003;422:859–868. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005a;438:1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- Galagan JE, Henn MR, Ma LJ, Cuomo CA, Birren B. Genomics of the fungal kingdom: insights into eukaryotic biology. Genome Res. 2005b;15:1620–1631. doi: 10.1101/gr.3767105. [DOI] [PubMed] [Google Scholar]
- Gow NA, Brown AJ, Odds FC. Fungal morphogenesis and host invasion. Curr Opin Microbiol. 2002;5:366–371. doi: 10.1016/s1369-5274(02)00338-7. [DOI] [PubMed] [Google Scholar]
- Hong EL, Balakrishnan R, Dong Q, Christie KR, Park J, Binkley G, et al. Gene Ontology annotations at SGD: new data sources and annotation methods. Nucleic Acids Res. 2008;36:D577–D581. doi: 10.1093/nar/gkm909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killion PJ, Sherlock G, Iyer VR. The Longhorn Array Database (LAD): an open-source, MIAME compliant implementation of the Stanford Microarray Database (SMD) BMC Bioinformatics. 2003;4:32. doi: 10.1186/1471-2105-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn LM. Mechanisms of fungal speciation. Annu Rev Phytopathol. 2005;43:279–308. doi: 10.1146/annurev.phyto.43.040204.135958. [DOI] [PubMed] [Google Scholar]
- Linsen SE, de Wit E, Janssens G, Heater S, Chapman L, Parkin RK, et al. Limitations and possibilities of small RNA digital gene expression profiling. Nat Methods. 2009;6:474–476. doi: 10.1038/nmeth0709-474. [DOI] [PubMed] [Google Scholar]
- Merrow M, Roenneberg T. Circadian entrainment of Neurospora crassa. Cold Spring Harb Symp Quant Biol. 2007;72:279–285. doi: 10.1101/sqb.2007.72.032. [DOI] [PubMed] [Google Scholar]
- Nakano M, Nobuta K, Vemaraju K, Tej SS, Skogen JW, Meyers BC. Plant MPSS databases: signature-based transcriptional resources for analyses of mRNA and small RNA. Nucleic Acids Res. 2006;34:D731–735. doi: 10.1093/nar/gkj077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson H, Kapushesky M, Kolesnikov N, Rustici G, Shojatalab M, Abeygunawardena N, et al. ArrayExpress update--from an archive of functional genomics experiments to the atlas of gene expression. Nucleic Acids Res. 2009;37:D868–872. doi: 10.1093/nar/gkn889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinartz J, Bruyns E, Lin JZ, Burcham T, Brenner S, Bowen B, et al. Massively parallel signature sequencing (MPSS) as a tool for in-depth quantitative gene expression profiling in all organisms. Brief Funct Genomic Proteomic. 2002;1:95–104. doi: 10.1093/bfgp/1.1.95. [DOI] [PubMed] [Google Scholar]
- Riley ML, Schmidt T, Wagner C, Mewes HW, Frishman D. The PEDANT genome database in 2005. Nucleic Acids Res. 2005;33:D308–310. doi: 10.1093/nar/gki019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salton G, McGill MJ. Introduction to modern information retrieval. McGraw-Hill, Inc; New York: 1983. [Google Scholar]
- Schoberle T, May GS. Fungal genomics: a tool to explore central metabolism of Aspergillus fumigatus and its role in virulence. Adv Genet. 2007;57:263–283. doi: 10.1016/S0065-2660(06)57007-5. [DOI] [PubMed] [Google Scholar]
- Soderlund C, Haller K, Pampanwar V, Ebbole D, Farman M, Orbach MJ, et al. MGOS: A resource for studying Magnaporthe grisea and Oryza sativa interactions. Mol Plant Microbe Interact. 2006;19:1055–1061. doi: 10.1094/MPMI-19-1055. [DOI] [PubMed] [Google Scholar]
- Stajich JE, Lapp H. Open source tools and toolkits for bioinformatics: significance, and where are we? Brief Bioinform. 2006;7:287–296. doi: 10.1093/bib/bbl026. [DOI] [PubMed] [Google Scholar]
- Torres TT, Metta M, Ottenwalder B, Schlotterer C. Gene expression profiling by massively parallel sequencing. Genome Res. 2008;18:172–177. doi: 10.1101/gr.6984908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend JP, Cavalieri D, Hartl DL. Population genetic variation in genome-wide gene expression. Mol Biol Evol. 2003;20:955–963. doi: 10.1093/molbev/msg106. [DOI] [PubMed] [Google Scholar]
- Townsend JP, Hartl DL. Bayesian analysis of gene expression levels: statistical quantification of relative mRNA level across multiple strains or treatments. Genome Biol. 2002;3:RESEARCH0071. doi: 10.1186/gb-2002-3-12-research0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunlid A, Talbot NJ. Genomics of parasitic and symbiotic fungi. Curr Opin Microbiol. 2002;5:513–519. doi: 10.1016/s1369-5274(02)00355-7. [DOI] [PubMed] [Google Scholar]
- Wu C, Amrani N, Jacobson A, Sachs MS. The use of fungal in vitro systems for studying translational regulation. Methods Enzymol. 2007;429:203–225. doi: 10.1016/S0076-6879(07)29010-X. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Cheung KH, Townsend JP. Bringing Web 2.0 to bioinformatics. Brief Bioinform. 2009;10:1–10. doi: 10.1093/bib/bbn041. [DOI] [PMC free article] [PubMed] [Google Scholar]