Summary
The basic helix-loop-helix protein Myc is a renowned transcription factor controlling disparate aspects of cell physiology that, together, allow efficient proliferation of somatic cells. This ability, together with the observation that its deregulated expression occurs in the majority of human cancers, suggests that Myc could be a good therapeutic target. However, several aspects of Myc biology remain elusive: what is the major difference between oncogenic and physiological Myc? How does oncogenic Myc evade the intrinsic tumor surveillance pathways provided by evolution? If Myc inhibition were even possible, what would be the consequences for the homeostasis of normal proliferating tissues versus the fate of cancer cells? Here we summarize the latest works addressing these issues.
The Myc family comprises three evolutionary conserved bHLHZip transcription factors – c-, N- and L-Myc - that coordinate diverse aspects of somatic and germ cell proliferation, including intracellular functions like cell growth, cell cycle progression, biosynthetic metabolism and apoptosis, as well as extracellular processes that coordinate cell proliferation with its adjacent somatic microenvironment, such as angiogenesis, invasion, stromal remodeling and inflammation [1]. The Myc proteins remain enigmatic for several reasons. Most notably, the repertoire of Myc target genes is very large and extremely diverse. It comprises RNA polymerase II protein coding target genes [2], RNA polymerase I and RNA polymerase III RNAs involved in translation and growth [3,4], and miRNAs likely to have key roles in cell proliferation, cancer and stem cell maintenance [5,6]. Second, the effect of Myc on individual genes is typically very modest and there appear to be few if any genes for which Myc is the sole, or even the principal, transcriptional regulator. More likely, Myc seems to generate global alterations in chromatin structure, which in turn modulate transcription [7]. Nonetheless, all studies concur that Myc function is essential for the efficient and orderly proliferation of somatic cells. Germ line deletion of either c-myc or N-myc leads to embryonic death around E11 due to widespread failures in organ and tissue growth, while fibroblasts lacking c-Myc proliferate very slowly and inefficiently. Third, the net consequences of Myc activity are highly context dependent, varying greatly with cell type and circumstance. Such extreme and contingent pleiotropy makes it meaningless to talk of any unitary Myc “function.” It also implies that Myc’s essentiality derives not from any specific sub-set of its functions but through its unique capacity to coordinate and integrate the diverse gamut of processes that, operating together, underpin somatic cell expansion.
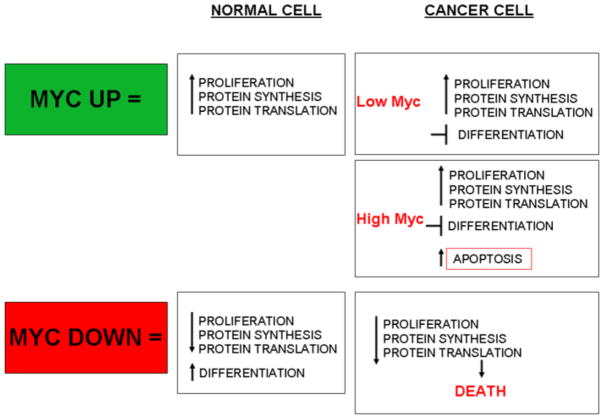
In normal cells, Myc function is tightly regulated by developmental or mitogenic signals. Myc mRNAs and proteins are very short-lived and, in the absence of such sustained proactive signals, Myc transcription is curtailed and Myc protein levels rapidly fall, triggering growth arrest. Such exquisite and continuous dependence of Myc activity on mitogenic signals safeguards against untoward somatic cell expansion. Indeed, Myc activity is almost always deregulated in tumor cells, sometimes through mutations within Myc genes themselves but, more often, through their incessant induction by upstream oncogenic signals in pathways such as RTK-Ras, Wnt-β-catenin or Notch. Interest in Myc is focused on these two related but discrete aspects of Myc biology: its role in normal cells and in tissue homeostasis, and the role played by deregulated Myc in driving and maintaining neoplasia (Fig 1).
Figure 1.
A consequence of Myc’s pleiotropy is its widespread impact on many, diverse cell and tissue processes, both directly and indirectly through its modulation of downstream transcriptional programs. Such secondary programs not only ramify all aspects of cell and tissue biology but they also feed back in a context and cell type-specific way to modulate how Myc acts. Deconvoluting any recognizable causal Myc signature from such networks has proved vexing, and the many independent attempts at gene expression profiling Myc target genes have yielded data sets with little overlap [8]. Nonetheless, a recent meta analysis of Myc-dependent expression culled from many different cell and tissue types has successfully revealed a provocative concordance between Myc targets and an embryonic stem cell signature [9]. Furthermore, Myc is one of four transcription factors that are capable of reprogramming differentiated adult cells back to a pluripotent state [10], and it can also regulate miRNAs involved in self-renewal and repression of differentiation [6], suggesting a potential role for Myc in the genesis of the infamous cancer stem cells.
Switching Myc on
A complementary strategy for unpicking the direct and indirect consequences of Myc function is to use switchable technologies that allow for acute activation or inactivation of Myc. Cause and consequence relationships can then be deciphered by following how the response evolves over time in the affected cell or tissue. Several mature techniques exist for regulating protein or gene function in vitro and in vivo, including various tetracycline regulatable expression systems (e.g. tet off and tet on) and conditional excision of genes (e.g. cre-lox and flp-frt). Each has its own advantages with respect to reversibility and rapidity, as well as disadvantages in terms of efficiency and leakiness.
Fusing Myc to the hormone-binding domain of the estrogen receptor (ER) renders all known Myc functions dependent upon the presence of the ER ligand β-estradiol, which displaces ER from inactive association with Hsp90 and promotes translocation of Myc to the nucleus. Later enhancements make use of mutant ERs (e.g. ERTAM [11] and ERT2 [12]) that are unresponsive to physiological estrogens yet activatable by the synthetic ligand 4-hydroxytamoxifen (4-OHT) and can therefore be used in vivo. ERTAM-based switchable systems are somewhat limited by their continuous dependence on 4-OHT and by the known estrogenic and anti-estrogenic effects of 4-OHT. In practice, this necessitates daily bolus administration of 4-OHT or Tamoxifen base (which is metabolized by the liver to 4-OHT) and avoidance of estrogenic tissues. Nonetheless, they enjoy certain advantages over other regulatable systems: ERTAM fusion proteins in tissues are synchronously activated within minutes of systemic administration of 4-OHT and such activation is fully and rapidly reversible. Expression of MycERTAM can be directed to any target tissue using a tissue specific promoter/enhancer and, because 4-OHT regulates activity of the MycERTAM fusion protein rather than its transcription, levels of the activated gene product in the target cell type are both predictable and consistent. This latter is an underappreciated advantage that is especially important in the case of Myc, where different levels of the oncoprotein elicit dramatically different biological outputs. Importantly, the rapid switchability of MycERTAM overcomes a profound limitation of classical transgenic technology wherein the active gene product of interest is ectopically expressed throughout the ontogeny of the target tissue, eliciting varying degrees of compensation and adaptation.
Activation of MycERTAM is, alone, sufficient to drive proliferation in multiple adult tissues in vivo. For example, activation of high levels of MycERTAM expressed from the involucrin promoter in suprabasal skin drives proliferation and disrupts differentiation of postmitotic keratinocytes, the consequent progressive accumulation of cells resulting in dramatic papillomatosis that rapidly regresses upon subsequent deactivation of MycERTAM [13]. When targeted to the basal keratinocyte compartment, activated MycERTAM forces cells out of the stem cell niche, generating a grossly expanded transit amplifying population that persists even after MycERTAM has been deactivated [14]. Unexpectedly, the epidermal hyperplasia elicited by acute Myc activation involved not only expansion of keratinocytes but also of the adjacent dermal vasculature hyperplasia, providing the first inkling that activation of Myc in one cell elicits cooperative changes in that cell’s adjacent niche. Similarly, activation of high-level MycERTAM in pancreatic β cells driven from the insulin promoter is also alone sufficient to drive β cells into cycle. Unlike keratinocytes, however, such acute activation of Myc does not trigger net β cell expansion because of widespread apoptosis that rapidly overwhelms β cell proliferation and results in islet attrition and consequent diabetes. Myc-induced apoptosis is a prototypical example of intrinsic tumor suppression – an evolved tumor defense mechanism that, like senescence, serves to quell the oncogenic potential of most, perhaps all, dominant oncogenes [15–17]. Oncogenic Myc will also induce apoptosis in keratinocytes, but execution of the apoptotic program is suppressed in intact skin by local survival factors. In β cells, Myc induced apoptosis can be suppressed by transgenic co-expression of the anti-apoptotic protein Bcl-xL whereupon Myc activation triggers rapid, progressive and uniform expansion of β cells turning all pancreatic islets into angiogenic and often invasive tumors [17]. Such Myc-driven β cell expansion is accompanied by rapid and synchronous β cell de-differentiation, down regulation of E-cadherin and loss of cell-cell adhesion, local invasion of β cells into adjacent exocrine and vascular compartments and (just as in skin) concomitant expansion and elaboration of the entire islet microenvironment, with proliferation of adjoining islet vasculature, influx of inflammatory cells and activation of stromal fibroblasts, and formation of angiogenic and invasive islet tumors [17]. Furthermore, subsequent Myc de-activation triggers regression of these complex neoplastic lesions. The rapidity and synchrony with which such multifarious traits appear in each islet mass, as well as their continuous dependence on Myc activity, clearly indicate that they are all instructed by Myc – either directly within each β cell, or indirectly through signals produced by β cells as a consequence of Myc activation within them. This, together with the reversible switchability of MycERTAM in vivo provides an unparalleled opportunity for constructing the cause and effect sequence of processes that link Myc activation with the complex phenotypes it elicits.
Kinetic expression array analysis following acute activation, and subsequent deactivation, of MycERTAM in β cells confirmed that Myc regulates multiple genes involved in cell cycle regulation, growth and metabolism. Unexpectedly, however, a significant proportion of the genes Myc was found to regulate are cell type specific [18]. In addition, a limited repertoire of candidate tumor maintenance genes could be identified on the basis that they are inversely regulated during Myc activation-induced tumor progression versus Myc deactivation-induced tumor regression [18]. The same kinetic analysis also demonstrated a direct instructive role for Myc in driving angiogenesis and the tumor microenvironment: acute Myc activation in β-cells of pancreatic islets rapidly induces expression of the pro-inflammatory cytokine interleukin 1β beta, which triggers release of sequestered VEGF from the islet extracellular matrix whereupon it homes to the endothelial compartment, and induces endothelial cell proliferation and islet angiogenesis [19]. Myc activation also induces expression of a cluster of chemokines implicated in the recruitment of mast cells, macrophages and neutrophils. Subsequently, mast cells were shown to have an essential causal role in the expansion and maintenance of the islet tumor vasculature [20]. Both these studies proved the crucial role of Myc in governing intracellular and extracellular aspects of tumorigenesis.
These observations in the pancreatic islet MycERTAM model confirm the important notion that Myc coordinates both the intracellular programs required for cell proliferation and also the many, and tissue type-specific, extracellular processes that proliferating cells require for their expansion within the somatic milieu. However, the requirement for co-expression of Bcl-xL in the same model also underscores the potency of Myc-induced apoptosis as a mechanism to suppress untoward proliferation. This raises an important problem: since Myc is required to integrate the proliferative programs of all normal cells, how is Myc-induced apoptosis confined only to tumor, and not normal, cells? Myc is deregulated and/or elevated in most human cancers (reviewed in [21]) but until recently it has been unclear whether it is deregulation or over expression that is required for Myc oncogenic activity. However, in a variant MycERTAM model in which MycERTAM is driven by the very weak but constitutively Rosa26 promoter, expression of Myc is deregulated (in the presence of 4-OHT) but expressed at low, physiological levels [22]. Activation of MycERTAM in tissues of such animals drives proliferation without any attendant apoptosis, demonstrating that distinct threshold levels of Myc govern its biological output in vivo: low levels of “deregulated” Myc induce somatic cell proliferation and, when deregulated, tumorigenesis in multiple tissues, but substantially elevated levels of Myc are required to engage tumor suppressor mechanisms – principally apoptosis and the ARF/p53 pathway [22] (Fig 1). Such observations have important and surprising implications for tumor evolution: they indicate that low-level Myc deregulation can drive indolent oncogenesis covertly in somatic tissues without alerting our evolved mechanisms for tumor surveillance and suppression. They also offer an explanation for why Myc is so frequently activated indirectly rather than through direct mutation: while indirect activation of Myc by upstream oncogenic signals deregulates Myc activity, it does not lead to significant over-expression and so fails to engage tumor suppression. This phenomenon is starkly demonstrated by the differences between indirect activation of Myc in intestinal epithelium through activation of the Wnt-β-catenin pathway, which engages only Myc’s proliferative programs [23,24] versus direct transgenic activation and concomitant over expression of Myc, which triggers activation of p53-mediated tumor suppression [25].
Switching Myc off
Taken together, the data supporting the pivotal role that Myc plays as a necessary and non-redundant coordinator of the many intra and extracellular programs required for somatic cell proliferation suggest that Myc would make an excellent target for cancer therapy. Unfortunately, there are several caveats. Most notably, Myc is widely considered “undruggable” since it has no “active site” amenable to binding by conventional small drug-like molecules. Moreover, Myc is required for the proliferation of all normal cells, so the side effects of its inhibition could be as severe as those elicited by conventional chemo and radiotherapy. Myc also plays a critical role as gatekeeper of the stem cell compartment [6,26], so even transient inhibition of Myc might cause permanent disruption of renewing tissues. Unfortunately, the embryonic lethality associated with germ line deletion of the two principal Myc family members implicated in cancer, c-Myc and N-Myc, precludes any meaningful analysis of their roles in adult tissues, so most published data regarding the impact of Myc inhibition in vivo has come from conditional knockouts of the c-myc gene using Cre mediated recombination [26–28]. While such data are useful guides as to the essentiality of c-Myc in adult tissues, the approach suffers from several confounding problems. First, they are limited to c-myc, but one member of the largely isofunctional myc gene family [26], members of which are frequently co-expressed in tissues. Second, the extents of c-myc deletion within the target cells of any tissue achievable by cre-lox technology are highly variable. The unpredictable extents to which c-Myc-deleted versus c-Myc-competent cells then contribute to tissue function have led to wildly disparate conclusions concerning the necessity of c-Myc for tissue maintenance (e.g. [27] vs. [29]) Third, conditional knockouts are irreversible, making it impossible to ascertain the impact of the type of transient Myc inhibition that would be achieved with a Myc-inhibitor drug. Recently, an alternative genetic approach to modeling Myc inhibition has made use of a reversibly inducible dominant negative Myc mutant, Omomyc [20,30–32], which competitively inhibits Myc-dependent gene transactivation by blocking the obligate dimerization of all three Myc proteins with their obligate partner Max. Omomyc inhibits binding of Myc to its consensus E-box CACGTG DNA elements and so blocks Myc-dependent transactivation of its target genes. Of note, Omomyc does not inhibit, and may even augment, Myc-dependent transrepression [30,31]. By directing transgenic Omomyc expression ubiquitously under the control of a tetracycline-responsive promoter element, Soucek et al. could systemically and reversibly shut down Myc trans-activation activity in mice by administration of doxycyclin [20]. Such mice were then crossed with the well-established LSL–KrasG12D murine model of non-small cell lung cancer [33] to study both the therapeutic impact and the side effects of systemic Myc inhibition. In LSL–KrasG12D mice, irreversible activation of oncogenic KRasG12D driven from the endogenous kras promoter is initiated in mice by inhalation of adenovirus expressing Cre recombinase. Multifocal lung tumorigenesis then ensues and by 18 weeks each lung harbors multiple independent tumors at all stages of evolution through to adenocarcinoma [33]. Remarkably, induction of Omomyc expression for as little as 3 days triggers profound tumor shrinkage and, after 28 days of sustained Omomyc expression, animals are overtly tumor free [20]. Surprisingly, such extended systemic inhibition of Myc elicits only mild and well tolerated side effects in normal proliferating tissues. Mice exhibit no signs of distress, maintain their weight, hydration and normal blood chemistry and, while proliferating tissues such as intestine, bone marrow, skin and testis exhibit varying degrees of attrition, cell death does not occur in any adult tissue, all of which maintain structural integrity [20]. Moreover, all effects of Myc inhibition on normal tissue are fully and rapidly reversed upon restoration of endogenous Myc function.
These surprising observations lead to two unexpected conclusions. First, endogenous Myc is required not only for the proliferation of Ras-driven lung tumors but also for their survival. Second, such dependency on endogenous Myc for cell survival is specific to tumor cells and absent from all normal proliferating somatic cells (Fig 1). Together, these two observations explain the remarkable therapeutic index enjoyed by systemic Myc inhibition and strongly support rekindling interest in Myc as a therapeutic cancer target.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Laura Soucek, Email: laura.soucek@ucsf.edu.
Gerard I. Evan, Email: gerard.evan@ucsf.edu.
References
- 1.Eisenman RN. Deconstructing myc. Genes Dev. 2001;15:2023–2030. doi: 10.1101/gad928101. [DOI] [PubMed] [Google Scholar]
- 2.Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN, White RJ. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol. 2005;7:311–318. doi: 10.1038/ncb1224. [DOI] [PubMed] [Google Scholar]
- 3.Arabi A, Wu S, Ridderstrale K, Bierhoff H, Shiue C, Fatyol K, Fahlen S, Hydbring P, Soderberg O, Grummt I, et al. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol. 2005;7:303–310. doi: 10.1038/ncb1225. [DOI] [PubMed] [Google Scholar]
- 4.Gomez-Roman N, Grandori C, Eisenman RN, White RJ. Direct activation of RNA polymerase III transcription by c-Myc. Nature. 2003;421:290–294. doi: 10.1038/nature01327. [DOI] [PubMed] [Google Scholar]
- 5.Lotterman CD, Kent OA, Mendell JT. Functional integration of microRNAs into oncogenic and tumor suppressor pathways. Cell Cycle. 2008;7:2493–2499. doi: 10.4161/cc.7.16.6452. ? Summary of evidences that miRNAs function as critical effectors of several canonical oncogenic and tumor suppressor pathways, including those controlled by Myc and p53, directly influencing neoplastic phenotypes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin CH, Jackson AL, Guo J, Linsley PS, Eisenman RN. Myc-regulated microRNAs attenuate embryonic stem cell differentiation. EMBO J. 2009 doi: 10.1038/emboj.2009.254. ? Using a quantitative primer-extension PCR assay, the authors identified miRNAs, whose expression is regulated by c-Myc in ES cells and showed that in ES cells c-Myc acts, in part, through a subset of miRNAs to attenuate differentiation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y, Blackwell T, Chen J, Gao J, Lee A, States D. Integration of genome and chromatin structure with gene expression profiles to predict c-MYC recognition site binding and function. PLoS Comput Biol. 2007;3:e63. doi: 10.1371/journal.pcbi.0030063. ? Making use of a computational model that integrates multiple sources of evidence to predict which genes will bind and be regulated by MYC in vivo, the authors predict at least 460 likely c-MYC target genes in the human genome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong DJ, Liu H, Ridky TW, Cassarino D, Segal E, Chang HY. Module Map of Stem Cell Genes Guides Creation of Epithelial Cancer Stem Cells. Cell Stem Cell. 2008;2:333–344. doi: 10.1016/j.stem.2008.02.009. ? Using a gene module map to systematically relate transcriptional programs in embryonic stem cells (ESCs), adult tissue stem cells, and human cancers, the authors show that c-Myc, but not other oncogenes, is sufficient to reactivate the ESC-like program in normal and cancer cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 11.Littlewood TD, Hancock DC, Danielian PS, Parker MG, Evan GI. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 1995;23:1686–1690. doi: 10.1093/nar/23.10.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Indra AK, Warot X, Brocard J, Bornert JM, Xiao JH, Chambon P, Metzger D. Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: comparison of the recombinase activity of the tamoxifen- inducible Cre-ER(T) and Cre-ER(T2) recombinases. Nucleic Acids Res. 1999;27:4324–4327. doi: 10.1093/nar/27.22.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pelengaris S, Littlewood T, Khan M, Elia G, Evan G. Reversible activation of c-Myc in skin: induction of a complex neoplastic phenotype by a single oncogenic lesion. Mol Cell. 1999;3:565–577. doi: 10.1016/s1097-2765(00)80350-0. [DOI] [PubMed] [Google Scholar]
- 14.Arnold I, Watt FM. c-Myc activation in transgenic mouse epidermis results in mobilization of stem cells and differentiation of their progeny. Curr Biol. 2001;11:558–568. doi: 10.1016/s0960-9822(01)00154-3. [DOI] [PubMed] [Google Scholar]
- 15.Evan G, Harrington E, Fanidi A, Land H, Amati B, Bennett M. Integrated control of cell proliferation and cell death by the c-myc oncogene. Philos Trans R Soc Lond B Biol Sci. 1994;345:269–275. doi: 10.1098/rstb.1994.0105. [DOI] [PubMed] [Google Scholar]
- 16.Evan G, Littlewood T. A matter of life and cell death. Science. 1998;281:1317–1322. doi: 10.1126/science.281.5381.1317. [DOI] [PubMed] [Google Scholar]
- 17.Pelengaris S, Khan M, Evan GI. Suppression of Myc-induced apoptosis in beta cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression. Cell. 2002;109:321–334. doi: 10.1016/s0092-8674(02)00738-9. [DOI] [PubMed] [Google Scholar]
- 18.Lawlor ER, Soucek L, Brown-Swigart L, Shchors K, Bialucha CU, Evan GI. Reversible Kinetic Analysis of Myc Targets In vivo Provides Novel Insights into Myc-Mediated Tumorigenesis. Cancer Res. 2006;66:4591–4601. doi: 10.1158/0008-5472.CAN-05-3826. [DOI] [PubMed] [Google Scholar]
- 19.Shchors K, Shchors E, Rostker F, Lawlor ER, Brown-Swigart L, Evan GI. The Myc-dependent angiogenic switch in tumors is mediated by interleukin 1beta. Genes Dev. 2006;20:2527–2538. doi: 10.1101/gad.1455706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, Karnezis AN, Swigart LB, Nasi S, Evan GI. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–683. doi: 10.1038/nature07260. ?? Making use of Omomyc, a dominant negative of Myc transactivation function, the authors show that Myc inhibition triggers rapid regression of incipient and established lung tumours, while having relatively mild side effects in normal tissues. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nesbit CE, Tersak JM, Prochownik EV. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18:3004–3016. doi: 10.1038/sj.onc.1202746. [DOI] [PubMed] [Google Scholar]
- 22.Murphy DJ, Junttila MR, Pouyet L, Karnezis A, Shchors K, Bui DA, Brown-Swigart L, Johnson L, Evan GI. Distinct Thresholds Govern Myc’s Biological Output In Vivo. Cancer Cell. 2008;14:447–457. doi: 10.1016/j.ccr.2008.10.018. ?? Using a deregulated, but not overexpressed switchable Myc, the authors show that distinct threshold levels of Myc govern its output in vivo: low levels of deregulated Myc are competent to drive proliferation of somatic cells and oncogenesis, but activation of the apoptotic and ARF/p53 intrinsic tumor surveillance pathways requires Myc overexpression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sansom OJ, Meniel VS, Muncan V, Phesse TJ, Wilkins JA, Reed KR, Vass JK, Athineos D, Clevers H, Clarke AR. Myc deletion rescues Apc deficiency in the small intestine. Nature. 2007;446:676–679. doi: 10.1038/nature05674. ? By simultaneously deleting both Apc and Myc in the adult murine small intestine, the authors show that loss of Myc rescued the phenotypes of perturbed differentiation, migration, proliferation and apoptosis, which occur on deletion of Apc. [DOI] [PubMed] [Google Scholar]
- 24.Wilkins J, Sansom O. C-Myc Is a Critical Mediator of the Phenotypes of Apc Loss in the Intestine. Cancer Research. 2008;68:4963–4966. doi: 10.1158/0008-5472.CAN-07-5558. [DOI] [PubMed] [Google Scholar]
- 25.Finch AJ, Soucek L, Junttila MR, Swigart LB, Evan GI. Acute overexpression of Myc in intestinal epithelium recapitulates some but not all the changes elicited by Wnt/beta-catenin pathway activation. Mol Cell Biol. 2009;29:5306–5315. doi: 10.1128/MCB.01745-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson A, Murphy MJ, Oskarsson T, Kaloulis K, Bettess MD, Oser GM, Pasche AC, Knabenhans C, Macdonald HR, Trumpp A. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev. 2004;18:2747–2763. doi: 10.1101/gad.313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bettess MD, Dubois N, Murphy MJ, Dubey C, Roger C, Robine S, Trumpp A. c-Myc is required for the formation of intestinal crypts but dispensable for homeostasis of the adult intestinal epithelium. Mol Cell Biol. 2005;25:7868–7878. doi: 10.1128/MCB.25.17.7868-7878.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oskarsson T, Essers MA, Dubois N, Offner S, Dubey C, Roger C, Metzger D, Chambon P, Hummler E, Beard P, et al. Skin epidermis lacking the c-Myc gene is resistant to Ras-driven tumorigenesis but can reacquire sensitivity upon additional loss of the p21Cip1 gene. Genes Dev. 2006;20:2024–2029. doi: 10.1101/gad.381206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muncan V, Sansom OJ, Tertoolen L, Phesse TJ, Begthel H, Sancho E, Cole AM, Gregorieff A, de Alboran IM, Clevers H, et al. Rapid loss of intestinal crypts upon conditional deletion of the Wnt/Tcf-4 target gene c-Myc. Mol Cell Biol. 2006;26:8418–8426. doi: 10.1128/MCB.00821-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soucek L, Helmer-Citterich M, Sacco A, Jucker R, Cesareni G, Nasi S. Design and properties of a Myc derivative that efficiently homodimerizes. Oncogene. 1998;17:2463–2472. doi: 10.1038/sj.onc.1202199. [DOI] [PubMed] [Google Scholar]
- 31.Soucek L, Jucker R, Panacchia L, Ricordy R, Tato F, Nasi S. Omomyc, a potential Myc dominant negative, enhances Myc-induced apoptosis. Cancer Res. 2002;62:3507–3510. [PubMed] [Google Scholar]
- 32.Soucek L, Evan G. Myc-Is this the oncogene from Hell? Cancer Cell. 2002;1:406–408. doi: 10.1016/s1535-6108(02)00077-6. [DOI] [PubMed] [Google Scholar]
- 33.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]