In 1993, an enzyme with an ATP-dependent microtubule severing activity was purified from sea urchin eggs and named katanin, after the Japanese for sword. Now we know that katanin, spastin and fidgetin form a family of closely related microtubule severing enzymes that is widely distributed in eukaryotes ranging from Tetrahymena and Chlamydomonas to humans (Table 1, Figure 1A). Here we review the diverse in vivo functions of these proteins and the recent significant advances in deciphering the biophysical mechanism of microtubule severing.
Table 1.
Katanin-related microtubule-severing proteins in 6 representative species, Homo sapiens, Drosophila melanogaster, Caenorhabditis elegans, Arabidopsis thaliana, Tetrahymena thermophila [25] and Trypanosoma brucei [55]. Corresponding sequences can be retrieved at http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene.
| Species | H. sapiens | D. melanogaster | C. elegans | A. thaliana | T. thermophila | T. brucei | |
|---|---|---|---|---|---|---|---|
| P80 katanin | canonical | KATNB1 | CG13956 | F47G4.4 | AT5G08390 AT5G23430 AT1G11160 AT1G61210 |
TTHERM_00497660 | Tb09.211.1500 |
| WD40less | C15orf29 | MEI-2 F47G4.5 |
|||||
| P60 katanin | canonical | KATNA1 KATNAL1 |
CG10229 | MEI-1 | AT1G80350 | TTHERM_00322760 | Tb11.01.0200 Tb10.70.6880 |
| lisH | KATNAL2 | CG10793 | TTHERM_00414230 | Tb11.02.1370 | |||
| orphan | CG1193 | ||||||
| spastin | SPAST | CG5977 | SPAS-1 | AT2G45500 | TTHERM_01128570 | Tb927.3.1440 | |
| fidgetin | FIGN FIGNL1 FIGNL2 |
CG3326 | FIGL-1 | AT3G27120 | TTHERM_00088010 | Tb927.5.1870 | |
| VPS4 | VPS4A VPS4B |
CG6842 | VPS-4 | AT2G27600 | TTHERM_00036960 TTHERM_00989490 |
Tb927.3.3280 | |
Figure 1.
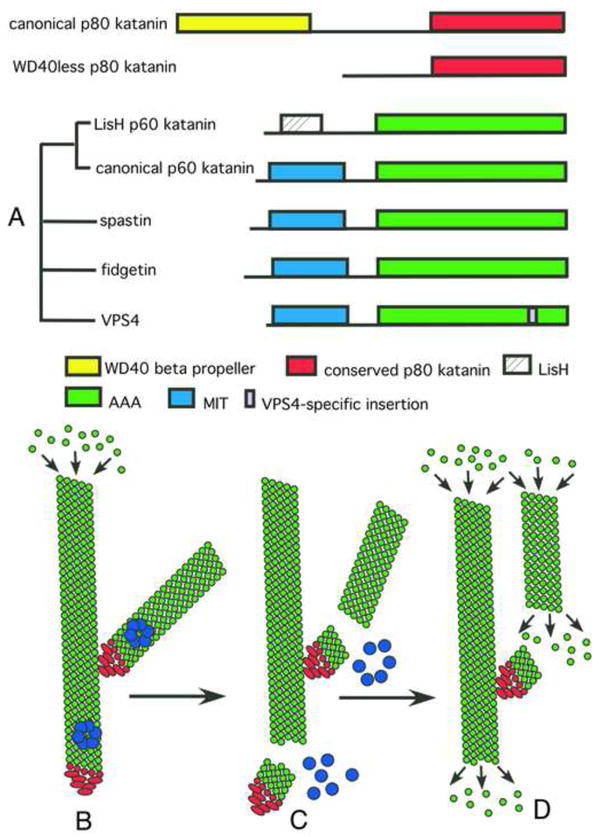
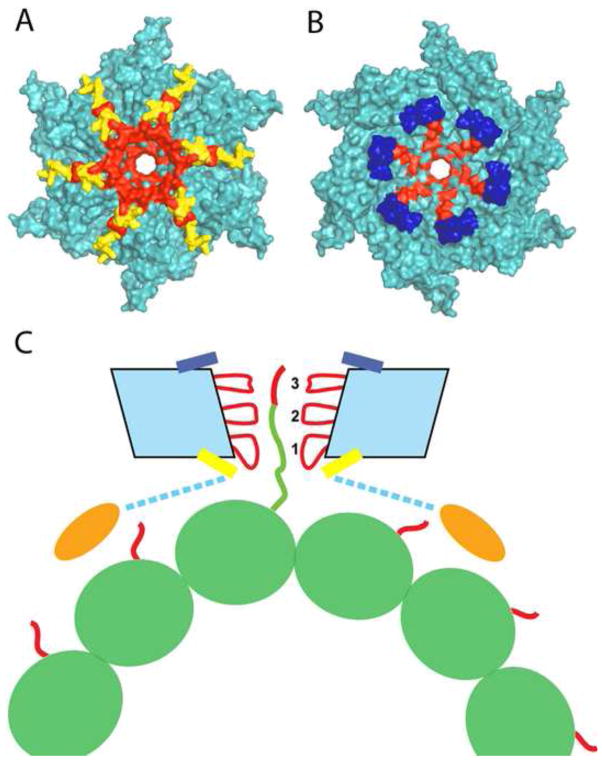
(A) Schematic representation of the domain architecture of microtubule-severing enzymes. Katanin purified from sea urchin eggs is a heterodimer with a regulatory, non-AAA subunit, p80 katanin, and a AAA catalytic subunit, p60 katanin. The phylogenetic relationship of proteins related to p60 katanin is indicated on the left. Based on their representation in sequenced genomes, we speculate that the LisH family of p60 katanins diverged early in eukaryotic evolution whereas the WD40less family of p80 katanins evolved independently in chordates and nematodes. (B–D) Schematic representation of branched nucleation and severing to generate parallel, treadmilling microtubule arrays in plant cells. (B) γ–tubulin ring complex (γTuRC), shown in red, binds to the wall of a pre-existing microtubule and nucleates polymerization of a new microtubule at a 40° angle. (C) Assembly of katanin rings, shown in blue, results in severing of these branched structures. (D) Severing by katanin frees microtubule minus ends to allow depolymerization of minus ends and allows branched arrays to re-arrange into parallel arrays.
Introduction
Microtubule severing is a reaction that generates an internal break in a microtubule. Unlike depolymerization from the microtubule ends, severing does not occur spontaneously in solutions of pure tubulin. Purified preparations of katanin [1–3] and spastin [4–6] from several species catalyze ATP-dependent microtubule severing in vitro, and fidgetin causes microtubule disassembly when over-expressed in vivo [7]. First we highlight new information about the in vivo functions of these enzymes, and then discuss recent structural work on spastin and the mechanism of microtubule severing.
Microtubule severing in vivo
The first insight into the in vivo function of a microtubule severing enzyme came from studies in C. elegans that identified two genes, MEI-1 and MEI-2 (the names for the C. elegans katanin catalytic and regulatory subunits, respectively) as essential for the assembly of the acentriolar female meiotic spindle [8,9]. Recent studies have brought fresh mechanistic insights into katanin’s role in building the C. elegans meiotic spindle and established the importance of the microtubule severing enzymes katanin, spastin and fidgetin in additional fundamental cellular processes such as mitosis, cilia biogenesis, deflagellation and neurogenesis.
In C. elegans oocytes, a katanin loss of function mutation results in failure to form a bipolar meiotic spindle [9]. EM tomography of these mutant spindles revealed that they have fewer microtubules and that microtubules are longer than in wild-type embryos [10]. Long microtubules are consistent with the long meiotic spindles and spindle shortening defects observed in a katanin partial loss of function mutant [3], however it is still unclear why long microtubules would result in complete spindle assembly failure. Interestingly, the EM tomographic analysis showed a large decrease in the total microtubule mass in the katanin mutant meiotic spindle relative to wild-type, but only a small decrease in polymer mass was observed by light microscopy [3,10]. Consistent with a role in regulating microtubule density, a synergistic loss of polymer mass was observed in a katanin γ-tubulin double mutant [3]. These results suggest that microtubule severing during C. elegans meiotic spindle assembly may increase polymer mass by generating shorter microtubules that can serve as seeds for nucleating new microtubules [11,12]. By regulating the stability of the seeds (whether they depolymerize or not), microtubule density can be controlled. The stability of the seeds is most likely influenced by microtubule-associated factors and this will likely be an interesting area of future investigations.
In C. elegans, persistence of katanin beyond meiosis results in damage to the mitotic spindle and chromosome segregation failure [13], underscoring that cellular context is critical for the phenotypic outcomes of katanin severing. Thus, katanin levels are stringently controlled upon the transition from meiosis to mitosis via two parallel proteolytic degradation pathways, the CUL-3 and MBK-2 pathways [14]. This control mechanism is conserved in mammals [15,16], however with a less dramatic outcome than in C. elegans since katanin is still present in the mitotic spindles of vertebrates [17], while absent from C. elegans mitotic spindles. The significance of this difference is presently unclear.
Direct evidence for the role of severing in microtubule nucleation was uncovered in plants. Cortical microtubules in plants are arranged in a parallel array and are responsible for controlling the direction of cellulose deposition, and thereby cell shape. In vivo imaging of GFP-tubulin revealed that many cortical microtubules nucleate off the wall of pre-existing microtubules at discrete 40° angles [18]–[19]. The new microtubules are released from the branch point and then “move” predominantly by treadmilling [18], which presumably allows them to arrange into a parallel array. In a katanin mutant (AT1G80350 in Table 1), release of nascent microtubules from branch points is blocked [19], explaining the lack of parallel microtubule organization, defective cellulose deposition and abnormal cell shape [20]. Branch points contain γ-tubulin [21] and interestingly, a point mutation in a γ-tubulin ring complex (γ-TuRC) subunit changes the angle at which the microtubule branches [19], strongly indicating that its minus end is capped by a γ–TuRC. Release of microtubules from branch points by katanin likely removes the γ-TuRC from the minus end, a requirement for treadmilling-based movement of the freed microtubule (Figure 1B–D).
Our understanding of the cellular mechanism of katanin is more advanced in plants than in other systems because loss of function mutants are viable and katanin acts on interphase microtubules that are adequately spaced to allow direct observation of individual microtubules by light microscopy. In contrast, other subcellular structures where microtubule severing enzyme function is important, such as spindles, axons, dendrites and cilia, contain bundled microtubules with less than 200 nm spacing, thus currently precluding direct observation of individual microtubules by light microscopy.
The most complete functional analysis of microtubule severing enzymes has been in mitosis. In Drosophila S2 spindles, spastin and fidgetin both localize to the centrosome and are required for microtubule minus end depolymerization, while katanin is required for plus end depolymerization [7]. Spastin and fidgetin in mitotic S2 cells may uncap minus ends from γ-tubulin ring complexes as in the plant cortical cytoskeleton, but these events have not been directly observed at spindle poles due to the high microtubule density. It is worth noting that while fidgetin over-expression does cause microtubule disassembly in Drosophila S2 cells [7], purified fidgetin, unlike katanin and spastin, has not yet been shown to sever microtubules in vitro. The C. elegans fidgetin mutant shows defects consistent with a role during mitosis in the germline [22] and mouse fidgetin mutants show an array of phenotypes [23]; however, the specific cellular function of fidgetin has not been elucidated in these systems.
One of the most conserved roles for katanin is likely in the assembly and disassembly of cilia and flagella. Loss of function mutations in katanin subunit genes result in the assembly of flagella or cilia without a central microtubule pair in both Chlamydomonas [24] and Tetrahymena [25], two very distantly related organisms. Coincidentally, Arabidopsis and C. elegans, organisms with well-characterized loss of function katanin mutants, do not have cilia with a central pair at any time during their wild-type development. It is likely that a role for katanin in ciliogenesis in Drosophila and mouse will be revealed when appropriate mutants are isolated. Recently, katanin has also been implicated in severing of basal bodies from the transition zone at the ends of resorbing flagella in Chlamydomonas, a process that appears essential for the use of the basal bodies as mitotic spindle poles. Depletion of katanin is lethal but dividing cells with intact flagella accumulate in the population of dying cells [26]. This is definitely an area that requires further investigation since resorption of cilia during mitosis likely occurs in most cells of the human body, but it is not known whether severing of the basal body is a required step in this process.
Consistent with the role of microtubule severing enzymes in the formation of complex, dense microtubule arrays such as those found in spindles or cilia, both spastin and katanin have functions in axonal elongation as well as branch formation in neurons. Spastin was originally identified as one of the most commonly mutated genes in hereditary spastic paraplegia [27], a human neurodegenerative disease characterized by lower extremity weakness due to axonopathy. Disease mutations in humans either inactivate or downregulate spastin severing activity [4]. In neurons, spastin localizes to the centrosome, synaptic boutons, points of new branch formation and its overexpression induces excessive branching [28,29]. Loss of spastin function leads to sparse and disorganized microtubule arrays at the synaptic terminals of the neuromuscular junction in Drosophila [28,30], as well as defects in dendritic arbor outgrowth and branching of a subclass of neurons characterized by their elaborate dendritic arbors [31]. In zebra fish, spastin loss leads to a disorganized and sparse axonal microtubule array and impaired axonal outgrowth [32]. Earlier work in hippocampal neurons also established a role for katanin in axonal elongation, presumably through the release of microtubules from the centrosome [33].
Recently, a katanin-like protein (CG1193 in Table 1) has been implicated in dendritic pruning in the Drosophila nervous system [34]. Specific sensory neurons initially have numerous dendrites that are eliminated during the transition from larvae to adult, reflecting a type of neural plasticity. In wild-type larvae, discontinuities or “breaks” in GFP-tubulin fluorescence within these dendrites precede visible breaks in a membrane marker, indicating that localized microtubule disassembly precedes severing of these dendrites. Both tissue specific RNAi and insertion mutations in a p60 katanin like protein cause a pronounced delay in the appearance of breaks in the tubulin fluorescence and a delay in dendrite severing [34]. These results show a requirement for a katanin-like protein in localized microtubule disassembly during dendritic pruning; however, dendrites contain dense microtubule bundles [35] making it very challenging to directly observe microtubule severing during dendritic pruning in wild-type neurons.
Phylogeny and architecture: one ring to rule them all
Katanin, spastin and fidgetin form together with VPS4 a subgroup (Figure 1A) of the large family of AAA ATPases. Members of this family participate in every major biochemical pathway in the human body and act on a dizzying array of substrates [36]. Despite their diversity of function, AAA ATPases have one common feature: they use the energy of ATP hydrolysis to take apart or remodel large molecular assemblies in the cell.
Katanin and spastin are microtubule-stimulated ATPases, and ATP hydrolysis is required for them to sever and disassemble stable microtubules [4–6,37]. VPS4 does not sever microtubules but instead disassembles protein complexes involved in membrane trafficking [38]. The ATP is hydrolyzed in their C-terminal AAA ATPase domains that are highly conserved between these enzymes. X-ray crystallographic analysis of spastin revealed that the AAA ATPase domain contains a canonical/nucleotide binding domain embraced by two helices, and a smaller four-helix bundle domain. This AAA motor module is connected via a poorly conserved linker to an N-terminal domain that binds microtubules with low affinity [39,40]. Recent structural studies have shown that the N-terminal domain in spastin and katanin contains a Microtubule Interacting and Trafficking (MIT) domain consisting of a three-helix bundle (Figure 1,[41,42]; PDB ID 2rpa). The MIT and AAA domains are sufficient for ATP-dependent microtubule severing for both katanin and spastin [5,39,40].
The N-terminus of VPS4 also contains an MIT domain that is responsible for binding multiple chromatin modifying proteins (CHMPs) that are part of ESCRTIII complexes and recent structural studies have revealed different modes of binding to these various partners [43–45]. The MIT domain of spastin is also known to interact with the ESCRTIII protein CHM1B and this interaction is important for targeting spastin to the midbody [41,46]. While spastin depletion does not affect endosomal degradation of model substrates, it does result in longer times to complete cytokinesis and an inability to disrupt the microtubule intercellular bridge during the final abscission process after membrane invagination has occurred, suggesting that spastin might be responsible for severing the stable microtubule bundles in the midbody [46]. It is not yet clear whether binding to the microtubule and CHMP1B are mutually exclusive, but the convergence of interacting partners from the microtubule cytoskeleton and membrane compartments is an intriguing area of future exploration, as has been recently highlighted by the double lives of dynamin as a membrane and microtubule dynamics regulator or the role of Golgi residing proteins as microtubule nucleation factors [47,48].
Members of the AAA family tend to assemble into ring-shaped oligomers, and only in this oligomeric state can they bind their substrate with high affinity. Unlike most AAA ATPases, katanin and spastin are monomeric when bound to ADP, and form hexamers only in the presence of ATP [39,40]. This ATP dependent hexamerization is also accompanied by a marked increase in binding affinity for microtubules (more than 20 fold) [40]. The hexamer is a labile structure, as both spastin and katanin exist mostly as monomers at submicromolar concentrations, even in the presence of ATP [39]. Based on the observation that the critical concentration for oligomerization of katanin can be lowered by the presence of microtubules, Vale and colleagues have advanced the model that katanin hexamers are assembled in a templated fashion on the microtubule lattice by multivalent interaction with the tubulin dimer [40]. This is a very attractive model that also suggests potential models of katanin regulation at the assembly step. The closely related subfamily member Vps4 also displays ATP dependent oligomerization; however, unlike katanin and spastin, Vps4 assembles into a dodecameric double ringed structure [49]. No evidence for higher order assemblies was found for either spastin or katanin [39,40,50] and the reasons for this difference in stoichiometry of assembly are presently unclear.
Recent structural work using a combination of X-ray crystallography and solution light and X-ray scattering coupled with atomic docking revealed that the active spastin hexamer displays the ring structure common to AAA AATPases, with a prominent conserved central pore and six radiating arms that may be used by the enzyme to dock onto the microtubule [39]. Both sides of the pore are lined by a corona of helices conserved in spastin and katanin (Figure 2A, B). Mutagenesis has shown these helices to be important for ATPase and microtubule severing activity [39].
Figure 2.
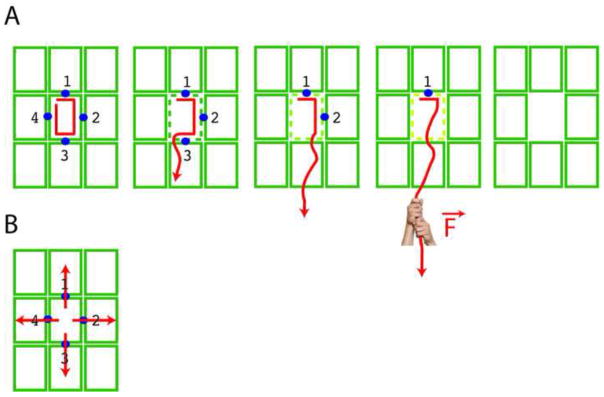
(A, B) Molecular surface of the spastin hexamer (after [39]). The N- and C-terminal helices that surround the entry and exit of the pore are shown in yellow and blue, respectively. Residues that line the entrance and the interior of the pore and are important for microtubule severing are shown in red. (C) Proposed mechanism for microtubule severing by spastin. The spastin AAA core is shown in cyan with Pore Loops 1,2 and 3 highlighted in red. The MIT domains are shown as gold ovals. The valency of the interaction of the MIT domains with the microtubule is unknown. Tubulin is shown in green, while the C-terminal tubulin tails are shown in red. Spastin uses its pore loops to exert “tugs” on the C-terminal tails and dislodge tubulin out of the microtubule lattice.
Microtubule severing mechanism: grab that tail?
Severing is a demanding molecular task, as the enzyme needs to break both longitudinal and lateral contacts in the microtubule lattice. At first glance, severing would require huge energy expenditure, since the microtubule is a very stiff polymer with a persistence length of millimeters and a lateral rigidity comparable to that of Plexiglas [51]. How then do severing enzymes perform the remarkable feat of severing the armor of the microtubule?
Several AAA proteins (e.g. ClpX, ClpA, and ClpB) remodel their protein substrates by threading the end of their polypeptide chains through a central pore in their AAA ATPase rings [52]. The microtubule severing activities of spastin and katanin depend on the disordered and negatively charged C-terminal tails of tubulin [5,37] and genetic data show that mutations in this region of tubulin suppress the lethality of ectopic katanin activity in C. elegans [53] and phenocopy a katanin loss of function mutation in Tetrahymena [25]. Moreover, C-terminal tubulin peptides as well as antibodies against these C-terminal tails inhibit spastin activity [39,50]. Structural analysis also revealed that the entry on one side of the central pore of the spastin AAA ATPase ring is positively charged and the lumen of the channel is lined with three highly conserved loops that are essential for microtubule severing (Figure 2, [39]). These results suggest that spastin and katanin are using their pore loops to tug on the C-terminal tail of tubulin, generating a mechanical force that may partially unfold tubulin or locally destabilize tubulin-tubulin interactions within the microtubule lattice, leading to catastrophic breakdown of the microtubule (Figure 2C, [39,50]). It is possible that spastin and katanin may not need to completely translocate the tubulin polypeptide substrate, but just exert multiple tugs on it to pull it from the lattice. Consistent with this idea, tubulin released from the microtubule via severing by katanin is competent to repolymerize [37].
Spastin and katanin seem to exploit the fact that the microtubule, despite being a very long and stiff cellular structure, is a polymer built from individual polypeptide chains (the tubulins) that can unfold or be unraveled mechanically. By pulling on the C-terminal tails of tubulin, the enzymes achieve stepwise destabilization of local microtubule lattice contacts, ultimately leading to breakdown of the cylindrical polymer (Figure 3). The activation energy required for this mechanism of severing is considerably less than that required to simultaneously break the lateral and longitudinal contacts in the lattice of the cylindrical polymer. This is because disruption of lateral and longitudinal lattice contacts would need to be simultaneous, as the low-affinity, highly cooperative nature of the microtubule lattice would otherwise easily drive it to re-anneal.
Figure 3.
Schematic representation of the proposed mechanisms of microtubule severing for spastin and katanin (A) by sequential unfolding of the tubulin peptide chain, leading to gradual loss of the lattice contact points (blue dots) and complete removal of a tubulin subunit (green rectangle); (B) by prying apart the tubulin subunits in the microtubule lattice. The green rectangles represent the tubulin subunits within a small region of a microtubule.
Future directions
The cylindrical structure of the microtubule and the highly cooperative nature of its lattice raise the interesting question of how many tubulin dimers need to be removed to break the microtubule? Do multiple hexamers cooperate at a severing site? Is the severing activity of spastin and katanin affected by microtubule curvature? Waterman and Salmon observed that in newt lung cells microtubule breakage occurred on buckled microtubules with similar curvatures [54]. Microtubules in vitro can bend 180 without breaking, however can microtubule severing enzymes bind preferentially or sever more efficiently bent microtubules with a stressed lattice?
While we now have an attractive model for microtubule severing, there are many salient questions that remain unanswered. What is the nature of the conformational changes driven by ATP hydrolysis? How do spastin and katanin recognize the microtubule polymer and the tubulin tails? Are the MIT domains involved in recognizing the tubulin tails? The C-terminal tails of tubulin are subject to complex posttranslational modifications such as polyglutamylation and polyglycylation. Both spastin and katanin localize to subcellular structures that are rich in these modifications. Do posttranslational modifications on the tubulin tails regulate spastin and katanin function? Interestingly, spastin in vitro severing activity is inhibited by antibodies that recognize the tubulin tails of Glu-tubulin, but not Tyr-tubulin [39] and in Tetrahymena, katanin severs preferentially the axonemal B-tubule [25] that has higher levels of polyglutamylation than the A-tubule or the central microtubule pair.
In vivo, one of the most interesting questions is the fate of the two ends generated by a microtubule severing event. In spindles, axons, dendrites and cilia, the density of microtubules makes light microscopic observation of the ends generated by severing very challenging. Katanin-dependent breaks have been observed in C. elegans meiotic spindles [10], but the fate of these ends could not be followed. The rapid advance in high-resolution light microscopy might finally allow us to “glimpse” the severing action of these enzymes in the dense microtubule structures where they function and provide us with some answers. Future work may also reveal how other proteins determine whether microtubule plus and minus ends generated by a severing protein will polymerize, depolymerize or remain stable. Almost two decades after the discovery of the first microtubule severing enzyme, we are finally starting to get a glimpse into the biophysical mechanism and multifaceted in vivo functions of these proteins.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Antonina Roll-Mecak, Cell Biology and Biophysics Unit, NINDS Porter Neuroscience Research Center, Building 35, Room 3B-203, 35 Convent Drive, MSC 3701, Bethesda, MD 20892-3701.
Francis J. McNally, Dept. of Molecular and Cellular Biology, University of California, Davis, Davis, CA 95616.
References
- 1.Hartman JJ, Mahr J, McNally K, Okawa K, Iwamatsu A, Thomas S, Cheesman S, Heuser J, Vale RD, McNally FJ. Katanin, a microtubule-severing protein, is a novel AAA ATPase that targets to the centrosome using a WD40-containing subunit. Cell. 1998;93:277–287. doi: 10.1016/s0092-8674(00)81578-0. [DOI] [PubMed] [Google Scholar]
- 2.Stoppin-Mellet V, Gaillard J, Vantard M. Functional evidence for in vitro microtubule severing by the plant katanin homologue. Biochem J. 2002;365:337–342. doi: 10.1042/BJ20020689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McNally K, Audhya A, Oegema K, McNally FJ. Katanin controls mitotic and meiotic spindle length. J Cell Biol. 2006;175:881–891. doi: 10.1083/jcb.200608117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Evans KJ, Gomes ER, Reisenweber SM, Gundersen GG, Lauring BP. Linking axonal degeneration to microtubule remodeling by Spastin-mediated microtubule severing. J Cell Biol. 2005;168:599–606. doi: 10.1083/jcb.200409058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roll-Mecak A, Vale RD. The Drosophila homologue of the hereditary spastic paraplegia protein, spastin, severs and disassembles microtubules. Curr Biol. 2005;15:650–655. doi: 10.1016/j.cub.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 6.Salinas S, Carazo-Salas RE, Proukakis C, Cooper J, Weston A, Schiavo G, Warner TT. Human spastin has multiple microtubule-related functions. J Neurochem. 2005;95:1411–1420. doi: 10.1111/j.1471-4159.2005.03472.x. [DOI] [PubMed] [Google Scholar]
- 7*.Zhang D, Rogers GC, Buster DW, Sharp DJ. Three microtubule severing enzymes contribute to the “Pacman-flux” machinery that moves chromosomes. J Cell Biol. 2007;177:231–242. doi: 10.1083/jcb.200612011. A complete analysis of the function of the microtubule-severing proteins, katanin, spastin and fidgetin, during Drosophila S2 cell mitosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark-Maguire S, Mains PE. mei-1, a gene required for meiotic spindle formation in Caenorhabditis elegans, is a member of a family of ATPases. Genetics. 1994;136:533–546. doi: 10.1093/genetics/136.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mains PE, Kemphues KJ, Sprunger SA, Sulston IA, Wood WB. Mutations affecting the meiotic and mitotic divisions of the early Caenorhabditis elegans embryo. Genetics. 1990;126:593–605. doi: 10.1093/genetics/126.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Srayko M, O’toole ET, Hyman AA, Müller-Reichert T. Katanin disrupts the microtubule lattice and increases polymer number in C. elegans meiosis. Curr Biol. 2006;16:1944–1949. doi: 10.1016/j.cub.2006.08.029. [DOI] [PubMed] [Google Scholar]
- 11.Ribbeck K, Mitchison TJ. Meiotic spindle: sculpted by severing. Curr Biol. 2006;16:R923–925. doi: 10.1016/j.cub.2006.09.048. [DOI] [PubMed] [Google Scholar]
- 12.Roll-Mecak A, Vale RD. Making more microtubules by severing: a common theme of noncentrosomal microtubule arrays? J Cell Biol. 2006;175:849–851. doi: 10.1083/jcb.200611149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dow MR, Mains PE. Genetic and molecular characterization of the caenorhabditis elegans gene, mel-26, a postmeiotic negative regulator of mei-1, a meiotic-specific spindle component. Genetics. 1998;150:119–128. doi: 10.1093/genetics/150.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu C, Mains PE. The C. elegans anaphase promoting complex and MBK-2/DYRK kinase act redundantly with CUL-3/MEL-26 ubiquitin ligase to degrade MEI-1 microtubule-severing activity after meiosis. Dev Biol. 2007;302:438–447. doi: 10.1016/j.ydbio.2006.09.053. [DOI] [PubMed] [Google Scholar]
- 15.Cummings CM, Bentley CA, Perdue SA, Baas PW, Singer JD. The Cul3/Klhdc5 E3 ligase regulates p60/katanin and is required for normal mitosis in mammalian cells. J Biol Chem. 2009;284:11663–11675. doi: 10.1074/jbc.M809374200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maddika S, Chen J. Protein kinase DYRK2 is a scaffold that facilitates assembly of an E3 ligase. Nat Cell Biol. 2009;11:409–419. doi: 10.1038/ncb1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McNally FJ, Thomas S. Katanin is responsible for the M-phase microtubule-severing activity in Xenopus eggs. Mol Biol Cell. 1998;9:1847–1861. doi: 10.1091/mbc.9.7.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shaw SL, Kamyar R, Ehrhardt DW. Sustained microtubule treadmilling in Arabidopsis cortical arrays. Science. 2003;300:1715–1718. doi: 10.1126/science.1083529. [DOI] [PubMed] [Google Scholar]
- 19.Murata T, Sonobe S, Baskin TI, Hyodo S, Hasezawa S, Nagata T, Horio T, Hasebe M. Microtubule-dependent microtubule nucleation based on recruitment of gamma-tubulin in higher plants. Nat Cell Biol. 2005;7:961–968. doi: 10.1038/ncb1306. [DOI] [PubMed] [Google Scholar]
- 20**.Nakamura M, Hashimoto T. A mutation in the Arabidopsis {gamma}-tubulin-containing complex causes helical growth and abnormal microtubule branching. J Cell Sci. 2009;122:2208–2217. doi: 10.1242/jcs.044131. This paper provides direct evidence that γ-tubulin ring complexes form the branch points in plant cortical microtubules and that katanin is responsible for debranching. [DOI] [PubMed] [Google Scholar]
- 21.Burk DH, Ye ZH. Alteration of oriented deposition of cellulose microfibrils by mutation of a katanin-like microtubule-severing protein. Plant Cell. 2002;14:2145–2160. doi: 10.1105/tpc.003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luke-Glaser S, Pintard L, Tyers M, Peter M. The AAA-ATPase FIGL-1 controls mitotic progression, and its levels are regulated by the CUL-3MEL-26 E3 ligase in the C. elegans germ line. J Cell Sci. 2007;120:3179–3187. doi: 10.1242/jcs.015883. [DOI] [PubMed] [Google Scholar]
- 23.Yang Y, Mahaffey CL, Bérubé N, Frankel WN. Interaction between fidgetin and protein kinase A-anchoring protein AKAP95 is critical for palatogenesis in the mouse. J Biol Chem. 2006;281:22352–22359. doi: 10.1074/jbc.M603626200. [DOI] [PubMed] [Google Scholar]
- 24.Dymek EE, Lefebvre PA, Smith EF. PF15p is the chlamydomonas homologue of the Katanin p80 subunit and is required for assembly of flagellar central microtubules. Eukaryotic Cell. 2004;3:870–879. doi: 10.1128/EC.3.4.870-879.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25*.Sharma N, Bryant J, Wloga D, Donaldson R, Davis R, Jerka-Dziadosz M, Gaertig J. Katanin regulates dynamics of microtubules and biogenesis of motile cilia. J Cell Biol. 2007;178:1065–1079. doi: 10.1083/jcb.200704021. An analysis of the roles of different microtubule-severing proteins in ciliagenesis and cell division in Tetrahymena. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26*.Rasi MQ, Parker JD, Feldman JL, Marshall WF, Quarmby LM. Katanin knockdown supports a role for microtubule severing in release of basal bodies before mitosis in Chlamydomonas. Mol Biol Cell. 2009;20:379–388. doi: 10.1091/mbc.E07-10-1007. This paper reveals a role for katanin in releasing flagellar basal bodies from flagella during mitosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hazan J, Fonknechten N, Mavel D, Paternotte C, Samson D, Artiguenave F, Davoine CS, Cruaud C, Dürr A, Wincker P, et al. Spastin, a new AAA protein, is altered in the most frequent form of autosomal dominant spastic paraplegia. Nat Genet. 1999;23:296–303. doi: 10.1038/15472. [DOI] [PubMed] [Google Scholar]
- 28.Sherwood N, Sun Q, Xue M, Zhang B, Zinn K. Drosophila Spastin Regulates Synaptic Microtubule Networks and Is Required for Normal Motor Function. Plos Biol. 2004;2:e429. doi: 10.1371/journal.pbio.0020429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu W, Qiang L, Solowska JM, Karabay A, Korulu S, Baas PW. The microtubule-severing proteins spastin and katanin participate differently in the formation of axonal branches. Mol Biol Cell. 2008;19:1485–1498. doi: 10.1091/mbc.E07-09-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trotta N, Orso G, Rossetto MG, Daga A, Broadie K. The hereditary spastic paraplegia gene, spastin, regulates microtubule stability to modulate synaptic structure and function. Curr Biol. 2004;14:1135–1147. doi: 10.1016/j.cub.2004.06.058. [DOI] [PubMed] [Google Scholar]
- 31*.Jinushi-Nakao S, Arvind R, Amikura R, Kinameri E, Liu AW, Moore AW. Knot/Collier and cut control different aspects of dendrite cytoskeleton and synergize to define final arbor shape. Neuron. 2007;56:963–978. doi: 10.1016/j.neuron.2007.10.031. This study shows that the transcription factor Knot activates a microtubule based cellular program for dentritic arbor outgrowth and branching by upregulating the expression levels of the microtubule severing protein spastin. [DOI] [PubMed] [Google Scholar]
- 32.Wood JD, Landers JA, Bingley M, McDermott CJ, Thomas-McArthur V, Gleadall LJ, Shaw PJ, Cunliffe VT. The microtubule-severing protein Spastin is essential for axon outgrowth in the zebrafish embryo. Hum Mol Genet. 2006;15:2763–2771. doi: 10.1093/hmg/ddl212. [DOI] [PubMed] [Google Scholar]
- 33.Ahmad FJ, Yu W, McNally FJ, Baas PW. An essential role for katanin in severing microtubules in the neuron. J Cell Biol. 1999;145:305–315. doi: 10.1083/jcb.145.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee HH, Jan LY, Jan YN. Drosophila IKK-related kinase Ik2 and Katanin p60-like 1 regulate dendrite pruning of sensory neuron during metamorphosis. Proc Natl Acad Sci USA. 2009;106:6363–6368. doi: 10.1073/pnas.0902051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baas PW, Deitch JS, Black MM, Banker GA. Polarity orientation of microtubules in hippocampal neurons: uniformity in the axon and nonuniformity in the dendrite. Proc Natl Acad Sci USA. 1988;85:8335–8339. doi: 10.1073/pnas.85.21.8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frickey T, Lupas AN. Phylogenetic analysis of AAA proteins. J Struct Biol. 2004;146:2–10. doi: 10.1016/j.jsb.2003.11.020. [DOI] [PubMed] [Google Scholar]
- 37.McNally FJ, Vale RD. Identification of katanin, an ATPase that severs and disassembles stable microtubules. Cell. 1993;75:419–429. doi: 10.1016/0092-8674(93)90377-3. [DOI] [PubMed] [Google Scholar]
- 38.Lata S, Schoehn G, Jain A, Pires R, Piehler J, Gottlinger HG, Weissenhorn W. Helical structures of ESCRT-III are disassembled by VPS4. Science. 2008;321:1354–1357. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39**.Roll-Mecak A, Vale RD. Structural basis of microtubule severing by the hereditary spastic paraplegia protein spastin. Nature. 2008;451:363–367. doi: 10.1038/nature06482. This study provides the first structural information on a microtubule severing enzyme and proposes a mechanism for microtubule severing via local unfolding of the tubulin monomer in the microtubule lattice. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hartman JJ, Vale RD. Microtubule disassembly by ATP-dependent oligomerization of the AAA enzyme katanin. Science. 1999;286:782–785. doi: 10.1126/science.286.5440.782. [DOI] [PubMed] [Google Scholar]
- 41.Yang D, Rismanchi N, Renvoisé B, Lippincott-Schwartz J, Blackstone C, Hurley JH. Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B. Nat Struct Mol Biol. 2008;15:1278–1286. doi: 10.1038/nsmb.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rigden DJ, Liu H, Hayes SD, Urbé S, Clague MJ. Ab initio protein modelling reveals novel human MIT domains. FEBS Lett. 2009;583:872–878. doi: 10.1016/j.febslet.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 43.Stuchell-Brereton MD, Skalicky JJ, Kieffer C, Karren MA, Ghaffarian S, Sundquist WI. ESCRT-III recognition by VPS4 ATPases. Nature. 2007;449:740–744. doi: 10.1038/nature06172. [DOI] [PubMed] [Google Scholar]
- 44.Obita T, Saksena S, Ghazi-Tabatabai S, Gill DJ, Perisic O, Emr SD, Williams RL. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature. 2007;449:735–739. doi: 10.1038/nature06171. [DOI] [PubMed] [Google Scholar]
- 45.Kieffer C, Skalicky JJ, Morita E, De Domenico I, Ward DM, Kaplan J, Sundquist WI. Two distinct modes of ESCRT-III recognition are required for VPS4 functions in lysosomal protein targeting and HIV-1 budding. Dev Cell. 2008;15:62–73. doi: 10.1016/j.devcel.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 46.Connell JW, Lindon C, Luzio JP, Reid E. Spastin couples microtubule severing to membrane traffic in completion of cytokinesis and secretion. Traffic. 2009;10:42–56. doi: 10.1111/j.1600-0854.2008.00847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanabe K, Takei K. Dynamic instability of microtubules requires dynamin 2 and is impaired in a Charcot-Marie-Tooth mutant. J Cell Biol. 2009;185:939–948. doi: 10.1083/jcb.200803153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rivero S, Cardenas J, Bornens M, Rios RM. Microtubule nucleation at the cis-side of the Golgi apparatus requires AKAP450 and GM130. EMBO J. 2009;28:1016–1028. doi: 10.1038/emboj.2009.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scott A, Chung H, Gonciarz-Swiatek M, Hill G, Whitby FG, Gaspar J, Holton J, Viswanathan R, Ghaffarian S, Hill CP, et al. Structural and mechanistic studies of VPS4 proteins. EMBO J. 2005;24:3658–3669. doi: 10.1038/sj.emboj.7600818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50*.White SR, Evans KJ, Lary J, Cole JL, Lauring B. Recognition of C-terminal amino acids in tubulin by pore loops in Spastin is important for microtubule severing. J Cell Biol. 2007;176:995–1005. doi: 10.1083/jcb.200610072. This study provides evidence for the interaction of the C-terminal tails of tubulin with spastin and proposes a mechanism for spastin mediated microtubule severing based on threading of the tubulin polypeptide chain through the spastin central pore. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gittes F, Mickey B, Nettleton J, Howard J. Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. J Cell Biol. 1993;120:923–934. doi: 10.1083/jcb.120.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sauer RT, Bolon DN, Burton B, Burton RE, Flynn JM, Grant RA, Hersch GL, Joshi SA, Kenniston JA, Levchenko I, et al. Sculpting the proteome with AAA(+) proteases and disassembly machines. Cell. 2004;119:9–18. doi: 10.1016/j.cell.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu C, Srayko M, Mains PE. The Caenorhabditis elegans microtubule-severing complex MEI-1/MEI-2 katanin interacts differently with two superficially redundant beta-tubulin isotypes. Mol Biol Cell. 2004;15:142–150. doi: 10.1091/mbc.E03-06-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Waterman-Storer CM, Salmon ED. Actomyosin-based retrograde flow of microtubules in the lamella of migrating epithelial cells influences microtubule dynamic instability and turnover and is associated with microtubule breakage and treadmilling. J Cell Biol. 1997;139:417–434. doi: 10.1083/jcb.139.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Casanova M, Crobu L, Blaineau C, Bourgeois N, Bastien P, Pages M. Microtubule-severing proteins are involved in flagellar length control and mitosis in Trypanosomatids. Mol Microbiol. 2009;71:1353–1370. doi: 10.1111/j.1365-2958.2009.06594.x. [DOI] [PubMed] [Google Scholar]