Abstract
Ube2g2 is an E2 enzyme which functions as part of the endoplasmic reticulum-associated degradation (ERAD) pathway responsible for identification and degradation of misfolded proteins in the endoplasmic reticulum. In tandem with a cognate E3 ligase, Ube2g2 assembles K48-linked polyubiquitin chains and then transfers them to substrate, leading ultimately to proteasomal degradation of the polyubiquitin-tagged substrate. We report here the solution structure and backbone dynamics of Ube2g2 solved by nuclear magnetic resonance spectroscopy. Although the solution structure agrees well with crystallographic structures for the E2 core, catalytically important loops (encompassing residues 95-107 and 130-135) flanking the active site cysteine are poorly defined. 15N spin relaxation and residual dipolar coupling analysis directly demonstrates that these two loops are highly dynamic in solution. These results suggest that Ube2g2 requires one or more of its protein partners, such as cognate E3, acceptor ubiquitin substrate or thiolester-linked donor ubiquitin, in order to assume its catalytically relevant conformation. Within the NMR structural ensemble, interactions were observed between His94 and the highly mobile loop residues Asp98 and Asp99, supporting a possible role for His94 as a general base activated by the carboxylate side-chains of Asp98 or Asp99.
Keywords: UBC7, Ube2g2, ubiquitin conjugating enzyme, ERAD, NMR spectroscopy, spin relaxation
Introduction
K48 polyubiquitination of a substrate protein destines that protein for proteasomal degradation. The pathway leading to polyubiquitination of a substrate protein involves a cascade of enzymatic activity. Initially, ubiquitin is activated by the ATP-dependent formation of a thiolester bond between the C-terminal carboxylate of ubiquitin and a cysteine residue on E1 (ubiquitin activating) enzyme. The activated ubiquitin is then transferred to the active site cysteine residue of a specific E2 (ubiquitin conjugating) enzyme. Finally, an E3 (ubiquitin ligase) enzyme recruits both substrate protein and the appropriate E2 in order to mediate the assembly of the polyubiquitin chain and its covalent attachment to substrate.
Human Ube2g2, or UBC7, is an E2 protein expressed endogenously in all human tissues1 and which constitutes an essential component of the endoplasmic reticulum-associated degradation (ERAD) pathway. The ERAD pathway targets misfolded, unassembled or tightly regulated proteins of the endoplasmic reticulum (ER) for K48-linked polyubiquitination and ultimately proteasomal degradation2. Proper function of the ERAD pathway is critical for many cellular processes such as signal transduction, cell cycle progress, cellular proliferation and apoptosis. Malfunction of the ERAD is associated with human diseases such as Parkinson's disease and cystic fibrosis3. As ubiquitin is not found in the ER lumen, target proteins identified by the ER quality control system4 must be retrotranslocated to cytoplasm for tagging with a K48 polyubiquitin chain and ultimately degradation by the proteasome. It has been proposed that the role of Ube2g2 is to preassemble K48-linked polyubiquitin chains on its active site cysteine and then transfer the preassembled chain to substrate in a fashion which is dependent upon interaction with one of its cognate E3 ligases5-8.
Several ubiquitin ligases (E3s) are known to interact with Ube2g2 including gp78 (or autocrine motility factor receptor)9, parkin10, HRD111 and TEB412. Together with its E3 partners, Ube2g2 mediates the K48-polyubiquitination and degradation of substrates such as the T-Cell Receptor α subunit (TCR-α), the thymocyte maturation protein CD3 δ subunit (CD3-δ)13 and human liver cytochrome P450 CYP3A414. It has also been implicated in the down regulation of the Pael 15, and inositol 1,4,5-trisphosphate 16 receptors.
Of the more than 30 human E2's identified17, all contain an active site cysteine residue to which ubiquitin molecules are linked by means of a thiolester bond. The active site cysteine residue of Ube2g2 is situated within a conserved ∼150 residue structural core which adopts an α/β fold18-25. Many E2 proteins contain sequence insertions or extensions of its 150 residue core sequence. Ube2g2 is a class I E2 enzyme (due to the absence of either N- or C-terminal extensions) which contains a 13-residue sequence insertion into its sequence just downstream from the active site cysteine. In addition to Ube2g2, this insertion occurs in human UBC3 and their yeast orthologs Ubc7p and Cdc34. A sequence alignment of the active site region is shown in Table I for these four E2s along with several other well studied E2 proteins. All four of the E2s that contain this 13-residue insertion as well as E2-25K (which lacks the insertion) have been shown to specifically catalyze the formation of K48-linked polyubiquitin chains12,26-28.
Table I.
Primary sequence alignment of selected ubiquitin conjugating enzymes
| Ube2g2 | 73 | FTCEMFHPNIYPD-GRVCISILHAPGDDPMGYESSAERWSPVQSVEKILLSVVSMLAEPNDESGANVD | 139 |
| Ubc7p | 73 | FTPSILHPNIYPN-GEVCISILHSPGDDPNMYELAEERWSPVQSVEKILLSVMSMLSEPNIESGANID | 139 |
| UBC3 | 77 | FLTKMWHPNIYET-GDVCISILHPPVDDPQSGELPSERWNPTQNVRTILLSVISLLNEPNTFSPANVD | 143 |
| Cdc34 | 79 | FTPAIYHPNVYRD-GRLCISILHQSGD-PMTDEPDAETWSPVQTVESVLISIVSLLEDPNINSPANVD | 144 |
| UBC9 | 77 | FEPPLFHPNVYPS-GTVCLSILE-----------EDKDWRPAITIKQILLGIQELLNEPNIQDPAQAE | 132 |
| UBCH5B | 69 | FTTRIYHPNINSN-GSICLDILR-------------SQWSPALTISKVLLSICSLLCDPNPDDPLVPE | 122 |
| Ubc4 | 70 | FTTKIYHPNINAN-GNICLDILK-------------DQWSPALTLSKVLLSICSLLTDANPDDPLVPE | 123 |
| Ubc13 | 71 | FLTKIYHPNIDRL-GRICLDVLK-------------TNWSPALQIRTVLLSIQALLASPNPNDPLAND | 124 |
| E2-25K | 75 | FITKIWHPNISSVTGAICLDILK-------------DQWAAAMTLRTVLLSLQALLAAAEPDDPQDAV | 129 |
H. sapien Ube2g2 (GI:29893557); S. cerevisiae Ubc7p (GI:6323664); H. sapien UBC3 (GI:16357477); S. cerevisiae Cdc34 (GI:1431497); H. sapien UBC9 (GI:54039791); H. sapien UBCH5B (GI:4507775); S. cerevisiae Ubc4 (GI:536344); S. cerevisiae Ubc13 (GI:1717864); H. sapien E2-25K (GI:4885417)
Ube2g2, UBC3 and their yeast orthologs all contain four conserved acidic residues proximate or within their 13-residue sequence insertions which are required for proper catalytic function. Li et al5 demonstrated that substitution of all four acidic residues in the loop with alanine abolished polyubiquitin chain assembly on either Ube2g2 or substrate. In Cdc34, mutation of the conserved acidic residues compromises the processivity and linkage specificity of polyubiquitin chain synthesis27. Gazdoiu et al. showed that mutation of the acidic residues D102/D103 of UBC3 led to accumulation of monoubiquitinated IĸBα substrate, while mutations of residues E108/E112 abolished both poly- and mono-ubiquitination of IĸBα29. Beyond the essential role of these acidic residues in K48-polyubiquitin chain formation, few details are known about how the 13-residue sequence insertion promotes catalytic function. Few insights are provided by available crystal structures of Ubc7p and Ube2g2, which indicate that the 13-residue insertion forms an extended loop that can assume different conformations depending on the local packing forces7,8,18,30.
In order to obtain further insights into the role of this 13-residue insertion loop, we have determined the NMR solution structure and characterized the backbone dynamics of Ube2g2. Our calculated solution structure is in good agreement with the conserved E2 core, however the 13-residue insertion as well as a loop stretching from residues 130-135 are poorly defined by the NMR restraints. Subsequent 15N spin relaxation studies and a residual dipolar coupling analyses demonstrate that both of these aforementioned loops, which flank the active site of Ube2g2, undergo extensive motions and sample a much broader range of conformations than reflected by the variation among solid state structures7,8,30.
Materials and Methods
Expression and purification of the human Ube2g2
The codon optimized31 human Ube2g2 gene (geneID 7327) was obtained from Integrated DNA Technologies (Coralville, IA) and then cloned into the His-tagged bacterial expression vector pET-28b in which the thrombin cleavage site had been replaced by a TEV cleavage site, a sequence which was also prepared by Integrated DNA Technologies. The resulting construct was transformed into the Escherichia coli strain BL21(DE3) and a single colony selected to inoculate a 5 ml overnight culture of LB media containing kanamycin (50 ug/mL). This overnight culture was subsequently diluted into 1L of identical media and grown at 37 °C. Upon reaching an OD600 of 0.8-1, 0.5 mM isopropyl β-D-1-thiogalactopyranoside was added to induce protein expression and the culture incubated at 18°C overnight (∼ 18 h). Cells were harvested by centrifugation and pellets stored at -80°C until purification. Uniform 13C and/or 15N-labeling of the protein was achieved by replacement of the LB media prior to induction with M9 minimal medium containing 4 g/L [13C]-D-glucose and/or 1g/L 15NH4Cl as the sole carbon and nitrogen sources, respectively32.
For purification of Ube2g2, the cell pellets were resuspended and lysed using BugBuster Protein Extraction Reagent (Novagen, Darmstadt, Germany) containing 0.5 uL/mL benzonase nuclease and one Complete EDTA-free Protease Inhibitor Cocktail Tablet (Roche, Basel, Switzerland). After removing cell debris by centrifugation (16,000 × g, 40 min), the clarified lysate was loaded onto a gravity flow nickel column (Sigma) pre-equilibrated with buffer A (25 mM Tris, 50 mM NaCl, pH 7.5). After wash with buffer A, the protein was eluted with buffer B (25 mM Tris, 50 mM NaCl, 200 mM imidazole, pH 7.5). The fractions containing Ube2g2 were identified by SDS-PAGE, pooled and then dialyzed against buffer A. Histidine-tagged tobacco etch virus (TEV) NIa protease was then added to the Ube2g2 solution and held at 4°C overnight in order to cleave off the poly-His tag. The mixture was then passed again over the gravity flow nickel column pre-equilibrated with buffer A for a second time to remove histidine-tagged TEV protease and uncleaved Ube2g2. The now tag-less Ube2g2 (two residues (GlyHis) remain N-terminal to the native Ube2g2 sequence) was finally purified by size-exclusion chromatography (superdex 75, 16/60, GE healthcare) equilibrated and eluted with the NMR buffer (20 mM NaPi, 50 mM NaCl, 0.5 mM EDTA, 1 mM TCEP, pH 7.0).
NMR data acquisition and analysis
All NMR measurements were performed at 295 K on Varian Inova 800 MHz or Bruker Avance and Avance II 600 MHz spectrometers equipped with cryogenically cooled probeheads. Although the backbone and side-chain chemical shift assignments have previously been published33, the completeness of the side-chain assignments were improved further by utilizing a 3D HCCH-COSY experiment for assignment of prochiral methyls and based on NOE assignments made during structure calculations. The NOE restraints were obtained from 3D 15N-edited NOESY-HSQC, aliphatic 3D 13C-edited NOESY-HSQC, and 2D 1H-1H NOESY spectra acquired with a mixing time of 60 msec on a 1 mM solution of Ube2g2 in NMR buffer. Proton chemical shifts were referenced to 3-(trimethyl-silyl)-1-propanesulfonate (DSS) resonance, while 13C and 15N chemical shifts were referenced indirectly to proton using the absolute frequency ratios. NMRPipe software34 was used to process all NOESY datasets. Prior to Fourier Transformation, time domain data were apodized using squared sine bell functions (shifted by 74° in 1H and 90° in 15N/13C dimensions) and zero-filled to achieve a final data matrix size of at least 2048×512×128 points. Two sets of residual dipolar couplings (RDCs) were measured in NMR buffer for 1 mM Ube2g2 in a 4.5% stretched polyacrylamide gel sample and 0.7 mM Ube2g2 in a 8 mg/ml bacteriophage Pf1 sample35,36. All 1JNH coupling measurements were performed using the IPAP-HSQC experiment37 with one bond 15N-1H RDCs taken to be the difference between 1JNH measurements in isotropic and aligned samples. 3JHNHα coupling constants were measured using the 3D HNHA38 experiment.
Structural calculations
The amino acid sequence, chemical shift assignment list and the three aforementioned NOESY spectra were supplied as input to the automated NOE assignment/structure determination software suite ATNOS/CANDID/CYANA. After manual inspection and correction of automatically generated NOE crosspeak lists, ATNOS/CANDID/CYANA successfully generated unambiguous NOE assignments and an ensemble of structural models. The CYANA-generated structural models were then ported to the software suite ARIA2.2/CNS for refinement with additional non-NOE experimental restraints. These additional restraints included two independent residual dipolar coupling (RDC) datasets, 3JHNHα coupling constants, and hydrogen bonding restraints based on NOE patterns in regions of regular secondary structure39. The force constants of the two RDC datasets were adjusted by trial and error such that the calculated structures agreed well with both datasets without introducing local geometric distortions (Stretched gel: kcool1/kcool2 = 0.02/2.0, phage Pf1 kcool1/kcool2 = 0.8/12). All other force constants were set to the default values (unambiguous distance restraints: 10/50, scalar coupling restraints: 0.2/1.0, H-bonding restraints: 10/50). Initial ARIA calculations used the CYANA-generated structural model in order to assign additional unambiguous NOE restraints, which were combined with those already identified by CYANA after manual validation. In the final ARIA run, 100 structures were calculated followed by water refinement of the 10 lowest energy structures. The quality of the final calculated structural ensemble was assessed by the programs NMR-PROCHECK, WHAT-CHECK and MolProbity. The updated chemical shift assignments, experimental constraints and solution structural coordinates of Ube2g2 have been submitted to the BMRB (accession code 16404) and the PDB (ID code 2kly)
15N spin relaxation analysis of Ube2g2
Measurement of 15N T1, T2 and steady state NOE values were carried out using established pulse sequences40,41 with WATERGATE modifications42. Complete relaxation datasets were collected at 295 K on both Varian Inova 800 MHz and Bruker Avance 600 MHz spectrometers equipped with cryogenic probes. Sample conditions were 200 μM 15N Ube2g2 in 10 mM Tris pH 7.5, 1 mM TCEP, 0.05% sodium azide, 0.1mM EDTA, and 10% D2O. For T1 measurements, the recycle delay between transients was set to 2.5 seconds and relaxation delays were set to 42.03, 287.17, 574.34, 707.41, 847.49, 1127.65, 1547.89, 1968.13, and 2388.37 msec for the 600 MHz dataset and 162.65, 325.30, 487.95, 650.60, 813.25, 975.90, 1301.20, 1463.85, and 1626.50 ms for the 800 MHz dataset. For T2 measurements, the recycle delay between transients was set to 3.0 seconds and relaxation delays were set to 16.80, 33.60, 50.40, 67.20, 84.00 and 100.80 msec for the 600 MHz data and 16.67, 33.34, 50.02, 66.69, and 83.36 msec for the 800 MHz data. After spectral processing using NMRPipe34, the program SPARKY43 was used to assign peaks and extract T1 and T2 relaxation times by non-linear least squares fitting. Heteronuclear NOE values were taken as the ratio of peak intensities observed for experiments with and without 3.0 seconds of 1H-presaturation during the recycle delay. Experimental errors for the relaxation time measurements were estimated based on the observed RMSDs between peak intensities measured for replicate experiments acquired for a single relaxation time point in each series. Utilizing the relaxation rates measured at both 600 and 800 MHz, a Lipari-Szabo model free analysis44 was carried out using FAST-Modelfree45 assuming a 1.02 Å 1H -15N bond distance, and a 15N CSA of -172. The rotational diffusion tensor was assumed to be axially symmetric, with a complete Modelfree optimization carried out for each set of systematically varied diffusion tensor parameters.
Determination of histidine pKa values
NMR samples spanning pH values ranging from 5.7-10 were prepared by buffer exchange into the following freshly prepared buffers: Phosphate-citrate(5.7-6.6), sodium phosphate(7-8), TRIS(8.5-8.8) and borax/sodium hydroxide(9.2-10). All samples contained 20 mM buffering agent as well as 50 mM NaCl, 0.5 mM EDTA, 1 mM TCEP and 1 mM Ube2g2. pH titration experiments were performed at 295K on a Varian Inova 800 MHz spectrometer. Histidine side-chain resonances were monitored by long range HMQC experiments, which correlated the non-labile ε1 and δ2 ring protons with the ε2 and δ1 ring nitrogen resonances46. pKa values were estimated by fitting the observed 1H-ε1 chemical shifts to a modified Henderson-Hasselbalch equation47 in which the pKa, Hill coefficient, and the acid and base limiting chemical shift values were all allowed to float.
Results and Discussion
Solution structure of Ube2g2
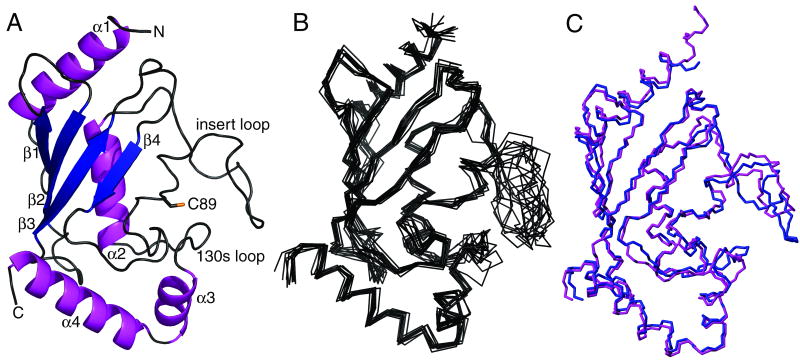
The solution structure of Ube2g2 was determined on the basis of 2501 experimental NOE distance restraints and refined with 3JHNHα couplings, H-bonding restraints and 2 sets of RDCs. Fig. 1A shows the closest to mean structure in ribbon form, and Fig. 1B shows an overlay of the 10 lowest energy structural models of Ube2g2 in solution. For the final ensemble of 10 structures, the backbone and heavy atom RMSDs for the E2 core (ie. excluding residues 96-109) are 0.77 and 1.18 Å, respectively. Statistics describing the structural ensemble are summarized in Table II. As anticipated, the solution structure exhibits a canonical E2 fold in good agreement with the previously reported crystal structure of Ube2g2 for the E2 core 30. For core residues 3-95,110-129, and 136-163, the closest to mean NMR structural model agrees with the crystal structure with a backbone (C′, CA and N) RMSD of 0.95 Å (Fig. 1C). The closest to mean solution structure of Ube2g2 differs more substantially from the crystal structure in the two loop regions which flank the active site cysteine, and which correspond approximately to the 13-residue insertion (residues 95-107) and the loop spanning residues 130-135. These loops are in fact very poorly defined in the NMR structural ensemble, with an average backbone RMSD (from the closest to mean NMR model) of 1.8 Å for residues 96-109 and 1.07 Å for residues 130-135. The poor definition of the insert loop and the 130s loop in the NMR ensemble clearly arises from the near absence of long range NOE restraints for these regions as shown in Fig. 2. Interestingly, the majority (7/9) of the long range NOEs recorded for these two loops were inter-loop NOEs observed between residues M101 and A136 and between residues Y103 and E133/S134/A136.
Figure 1.
A) Ribbon diagram of the closest-to-mean solution structure of Ube2g2 determined by NMR with the side-chain of catalytic residue C89 depicted as sticks. B) Backbone diagram of 10 lowest energy NMR structures of Ube2g2. C) Overlay of the closest-to-mean solution structure (blue) and the closest-to-mean crystal structure (PDB ID: 2cyx) 30 (magenta) of Ube2g2.
Table II.
Structural and CNS Refinement Statistics for Ube2g2
| Total number of experimental restraints | 2867 |
| Unambiguous distance restraints | 2501 |
| Intra residue |i-j| = 0 | 778 |
| Sequential |i-j| = 1 | 572 |
| Short range |i-j| = 2 - 4 | 526 |
| Long range |i-j| = 5 + | 625 |
| Residual dipolar couplings (Stretched polyacrylamide gel) | 90 |
| Residual dipolar couplings (Bacteriophage pf1) | 101 |
| J-coupling restraints | 69 |
| H-bonding restraints | 106 |
| Number of experimental restraints per residue | 17.4 |
| Structure Quality Analysis | |
| Ramachandran analysis of all residuesa | |
| residues in most favored regions | 79.5% |
| residues in additional allowed regions | 15.9% |
| residues in generously allowed regions | 3.2% |
| residues in disallowed regions | 1.4% |
| Deviations from idealized covalent geometrya | |
| Bonds (Å) | 0.0040±0.0002 |
| Angles (°) | 0.62±0.02 |
| Impropers (°) | 1.67±0.09 |
| RMSD within the bundle in the core region (3-95,110-163) (Å) | |
| Backbone (C′, CA, N) | 0.77 |
| Heavy atoms | 1.18 |
| Goodness of fit of RDC restraints in the core region (3-95,110-163)a | |
| Stretched polyacrylamide gel | Q = 0.04 |
| Bacteria phage pf1 | Q = 0.11 |
| Goodness of fit of all RDC restraints | |
| Stretched polyacrylamide gel | Q = 0.32 |
| Bacteriophage pf1 | Q = 0.33 |
| The average RDC fit to all 10 structuresb | |
| Stretched polyacrylamide gel | Q = 0.16 |
| Bacteriophage pf1 | Q = 0.18 |
Average of the 10 lowest energy NMR structures.
Q-value calculated by in-house software multi_fitalign.
Figure 2.
Distribution of observed NOE restraints utilized for the solution structure determination of Ube2g2. gray: Short/medium range NOEs (i-j <= 4); black: long range NOE's (i-j >= 5). Relatively few NOE restraints were observed for the 13-residue insertion loop (residues 96-107) and the loop encompassing residues 130-135.
The lack of a well defined conformation for the 13-residue insertion loop and the 130s loop is mirrored by several crystallographic studies of E2 proteins. For example, in the Ube2g2 crystal structure30, residues 96-109 exhibit an average backbone RMSD among the three molecules in the asymmetric unit of 0.8 Å compared to a backbone RMSD of 0.046 Å for the E2 core (residues 3-95, 110-163). In a pair of crystallographic studies of Ube2g2 in complex with the gp78 G2BR peptide, substantially different conformations are observed for both the 130s loop and the 13-residue insertion loop7,8. These differences are accompanied by the observation of elevated B-factors8 or poor electron density7. In the structure of the ubiquitin conjugating enzyme Rad648, the 130s loop exhibits different conformations among the different molecules in the asymmetric unit. These results together suggest that these functionally important loops may be dynamic when Ube2g2 is free in solution. As such, we set out to directly establish the extent of Ube2g2 motions in solution using NMR techniques.
Characterization of Ube2g2 dynamics
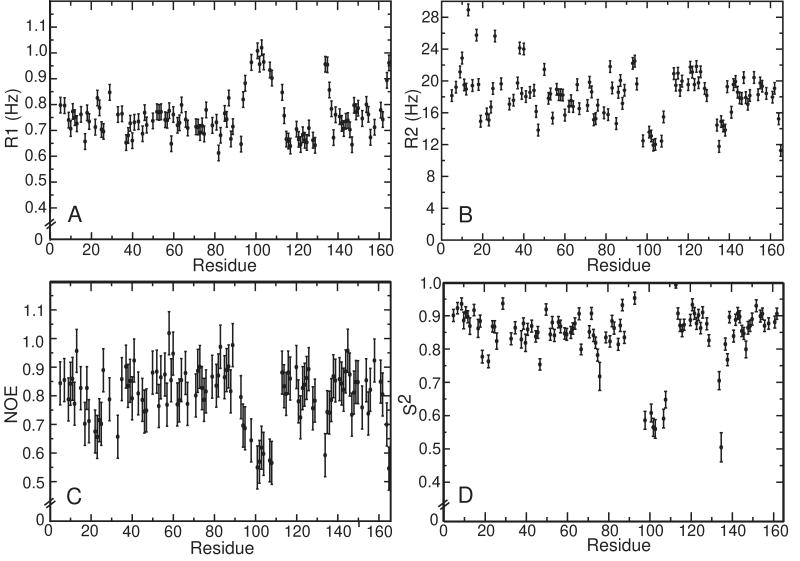
In order to probe fast, picosecond timescale motions, 15N spin relaxation measurements were made at both 14 T and 18.6 T field strengths and subjected to a Lipari-Szabo model free analysis44. In the course of carrying out the 15N spin relaxation study it was found that at a Ube2g2 concentration of 800 μM, Ube2g2 tumbles with a correlation time τc of ca. 14 nsec, which suggests self-association. Yet amide 15N and 1H chemical shifts remain invariant over a wide concentration range, indicating that the association is non-specific in nature and that individual Ube2g2 molecules remain fully solvated. Measured T2 relaxation times for Ube2g2 remain constant below concentrations of ca. 350 μM, and correspond to a rotational correlation time of ca. 11 nsec, which is close to expectations for a protein of Ube2g2's molecular weight. As such, the 15N spin relaxation study was carried out at a Ube2g2 concentration of 200 μM. At both fields, strikingly increased R1, decreased R2, and decreased heteronuclear NOE values were observed for residues in the 13-residue insertion loop and the 130s loop relative to the rest of the protein, which is a signature of substantial internal mobility (Fig. 3A-C, S1). A Lipari-Szabo model free analysis of the relaxation data indicated that overall molecular tumbling of Ube2g2 was fit best using an axially symmetric rotational diffusion tensor with correlation time τm = 11.9 nsec and anisotropy Dratio = 1.45. The resulting orientation and anisotropy of the rotational diffusion tensor agree very well with hydrodynamic calculations performed using the program HydroNMR49, as summarized in Table III. The hydrodynamic calculations better replicate the observed anisotropy when the 13-residue loop is excised, but in all cases the principal axes of rotational diffusion agree very closely and are nearly coincident with the direction of the α2 helix, which spans residues 115 to 128. The squared generalized order parameters (S2) resulting from the model free analysis are shown in Fig. 3D. Although resonance overlap reduces the number of residues to which the analysis can be applied, it is clear that the two loop regions flanking the active site (ie. 96-109 and 130-135) are undergoing extensive dynamic fluctuations with an average S2 of 0.61. By comparison, the average S2 for the rest of the protein, excluding both termini, is 0.87. Dynamics of this magnitude are consistent with the relatively few long range NOE's observed for residue 96-109 and 130-135 (Fig. 2).
Figure 3.
Residue specific 15N spin relaxation rates measured for Ube2g2 at 800 MHz: A) R1 values. B) R2 values and C) Steady state heteronuclear NOE values. Shown in D) are the squared generalized order parameters (S2) as a function of residue resulting from a model free 15N spin relaxation analysis of Ube2g2.
Table III.
Rotational diffusion tensor parameters
| Orientationa | Dratiob | ||
|---|---|---|---|
| θ | ϕ | ||
| ModelFree | 83.9 | 171 | 1.45 |
| HydroNMR | 87.2 | -178 | 1.34 |
| HydroNMR (no loop)c | 84.9 | 177.2 | 1.49 |
Orientation of the principal axis of the rotational diffusion tensor relative to the Ube2g2 structural coordinates (PDB ID: 2kly model 6).
Dratio = Dzz / (0.5(Dxx + Dyy)).
Results with loop residues 96-109 excised.
In addition to their exquisite sensitivity to bond orientations, RDCs are sensitive to motions occurring on timescales ranging from picoseconds to milliseconds, although they depend on the generalized order parameter S rather than on its square50. In the case of Ube2g2, two independent sets of RDCs were employed for refinement of the structure and the RDCs corresponding to residues 96-109 were excluded from the structural refinement due to very poor agreement with the refined structural models. Notably, the measured RDCs corresponding to residues 96-107 are uniformly < 20% of the maximum RDC observed in each of the two datasets (Fig. 4A). This is indicative of dynamic averaging which is much more extensive than that occurring on the picosecond timescale, as reported by the 15N spin relaxation results. Indeed, the agreement between measured RDCs and those calculated from any member of the structural ensemble is invariably very poor for some subset of insertion loop residues (eg. Fig. 4B). Yet if RDCs are calculated assuming equiprobable population of all ten of the NMR structures in the ensemble, the agreement with the measured RDCs is improved considerably although the magnitudes of some of the calculated RDCs still remain larger than observation (Fig. 4C). Together these observations indicate that very extensive motions of the 13-residue insertion loop are occurring which are if anything underestimated by the observed conformational breadth of the NMR structural ensemble. Moreover, these motions are occurring over timescales which stretch beyond the picosecond timescale to which 15N spin relaxation derived order parameters are sensitive. In contrast, the RDC measurements corresponding to the 130s loop do not support the existence of significant motions beyond what is reflected by the 15N spin relaxation order parameters.
Figure 4.
Residual dipolar coupling (RDC) analysis of Ube2g2. A) Observed RDCs for Ube2g2 aligned in 8 mg/ml bacteriophage Pf1 (top) and in 4.5% stretched acrylamide gel (bottom). B) Best fit of the amide 15N-1H RDC data (using the 4.5% stretched polyacrylamide gel medium) to a typical member of the NMR structural ensemble. Annotations refer to specific residue numbers. C) Same as B except the best fit is to the average internuclear vector coordinates of the 10 member NMR structural ensemble assuming that each structural member is equally populated.
Insights into Ube2g2 function
In addition to the active site cysteine residue, all E2s contain a strictly conserved asparagine residue (N81 in Ube2g2) upstream from the active site. This conserved asparagine residue has been shown to be essential for the catalytic function of E2s, presumably by stabilizing the oxyanion intermediate51. It has been noted that in many structures the side-chain of this conserved Asn points away from the active site such that it cannot interact with the thiolester moiety25,30,48. As shown in Fig. 5A (in blue), the N81 side-chains in the solution structural ensemble are all oriented towards the active site cysteine residue, with one structural model exhibiting a distance of only 2.8 Å between the backbone oxygen of C89 and the side-chain amide nitrogen of N81. This contrasts with the orientation of the N81 side-chain (shown in magenta in Fig. 5A) observed in the crystal structure of Ube2g2. Closer examination reveals that this observation is primarily due to differences in χ1 rotameric state between the crystal (gauche-) and the solution (trans) structures. The solution state χ1 conformation is directly supported by NOE contacts observed between the N81/Hβ protons and the N81/HN and H79/Hε1 protons. N81 side-chain orientations consistent with its proposed role have also been observed in the solution state structures of UbcH5B21 and Ubc152, suggesting that the observed solid state side-chain conformation of N81 may result from crystal packing forces due to its proximity to the flexible 130s loop, with which N81 makes a number of H-bonding contacts.
Figure 5.
Structural features observed for the active site of the Ube2g2 solution structure. A) Overlay of the crystal structures (PDB ID: 2cyx) 30 (in magenta) and the 10 member NMR structural ensemble (in black) of Ube2g2. The side-chains of residue N81, Y83, C89 and S134 are shown as sticks. Compared to the crystal structure, the side-chains of N81 and S134 in the NMR structures are in closer proximity to the active site cysteine residue, C89. B) Selected member of Ube2g2 solution structural ensemble in which the side-chains of residues H94 and D98 are within hydrogen bonding distance. C) Selected member of Ube2g2 solution structural ensemble in which the side-chain of residue H94 is within hydrogen bonding distance of both C89 and D99. D) Selected member of the Ube2g2 NMR structural ensemble in which N81, Y83 and S134 are all in close proximity to the active site cysteine, C89.
As shown in Table I, all four of the E2s containing 13-residue sequence insertions have a histidine residue (H94 in Ube2g2) just prior to the insertion point. This histidine residue is in very close spatial proximity to the active site cysteine residue. In two members of the NMR structural ensemble, the side-chain carboxylate groups of either D98 or D99 were found to be within hydrogen bonding distance of the side-chain of H94 as shown in Figures 5B and 5C. These interactions are not observed in any of the available crystal structures. While E2s have been recognized to lack residues which might participate in catalysis51, the proximity of H94 and D98/D99 residues suggests that 13-residue insert containing E2s may be an exception. An obvious role for H94 would be to serve as a general base in catalysis either by promoting deprotonation of the attacking lysine or deprotonation of the active site cysteine. Aside from the common occurrence in cysteine or serine proteases53, such a histidine:aspartate pair occurs in the deubiquitinating enzyme HAUSP, with the histidine observed to be only 3.6 Å away from the catalytic cysteine residue when ubiquitin is bound54.
The protonation state of the two histidine residues of Ube2g2 was monitored by recording long range 15N-1H chemical shift correlation experiments as a function of pH46,55. The observed pH dependence of histidine imidazole ring chemical shifts are displayed in Figure 6. In the case of H79, the Nε2 and Nδ1 chemical shifts are characteristic of the exclusive population of the ε tautomer. No changes in intensities or chemical shifts are observed for H79 until pH < 6.5 when the resonances weaken and disappear. Ube2g2 starts to precipitate out of solution when below a pH of 5.7. H94 exhibits strikingly different behavior in that it titrates over the entire the entire range of pHs between 5.7 and 8.0, above which its resonances are broadened beyond detection. The relative similarity of Nε2 and Nδ1 chemical shifts is indicative of rapid exchange between δ and ε tautomers, with the ε tautomer being the majority species in the case of H94. A fit of the H94 data to the modified Henderson-Hasselbalch equation with a floating Hill coefficient, n, yielded the best fit to the data with n = 0.55 and a pKa = 6.25. Although H94 exhibits a highly typical pKa, this is likely just a reflection of the extensive mobility of the insertion loop when Ube2g2 is free in solution. The question that remains is whether the pKa of H94 becomes elevated when the Ube2g2 assumes a catalytically viable conformation.
Figure 6.
pH dependence of imidazole ring chemical shifts for His94 (circles) and His79 (diamonds). A) pH dependence of 15Nδ1 and 15Nε2 ring chemical shifts. B) pH dependence of 1Hε1 ring chemical shifts. The solid line denotes the resulting best fit of the corresponding data points to the modified Henderson-Hasselbalch equation.
Although the actual role of H94 has not been established, D98 and D99 have been shown to be functionally important. A quadruple mutant of Ube2g2 in which all acidic residues in the insertion loop are mutated to alanine (D98A/D99A/E104A/E108A) was defective in both ubiquitin chain elongation and substrate polyubiquitination5. Furthermore, the two aspartate residues corresponding to D98 and D99 in UBC3 are required for proper formation of polyubiquitin chains29. Although polarization of the H94 side-chain is one possible role of D98/D99, other possible roles cannot be ruled out at this time, such as involvement of D98/D99 in properly orienting the lysine nucleophile. It should also be noted that D98 and D99 are in a loop shown by the present study to populate a very wide range of conformations in solution. The catalytic importance of these residues implies that this loop must rigidify into a catalytically viable conformation when in the presence of one or more of the required elements for catalysis: a thiolester-linked donor ubiquitin, bound E3 or bound acceptor ubiquitin.
Residues corresponding to Y83 and S134 in Ube2g2 are partially conserved and observed to make important contacts with the substrate lysine in crystal structures of Ubc9 bound to the substrate RanGAP156,57. A mutagenesis study of UBC3 has shown that mutation of the residues corresponding to Y83 and S134 are to Ala are both defective (with Y83 less so) in poly-ubiquitination of substrate29. The solution structure of Ube2g2 is consistent with a role of Y83 and S134 in catalysis as both are observed to be in close proximity to the active site cysteine (Fig. 5D). However, S134 assumes many different conformations across the NMR ensemble (Fig. 5A) and has a low squared generalized order parameter (S2 = 0.61) resulting from 15N spin relaxation analysis (Fig. 3D). This suggests that, as proposed for the 13-residue insertion loop, the loop containing S134 may also rigidify upon binding of substrate molecules and/or E3.
We have reported here the solution structure of Ube2g2 and demonstrated that the two loops (residues 130-135 and 95-107) flanking the active site cysteine undergo extensive dynamics. Due to the catalytic importance of many residues in these loops, it appears that Ube2g2 assumes a catalytically viable conformation only when one or more of the other proteins required for catalysis are present. This likely involves a rigidification of the 13-residue insertion loop upon binding of E3, donor or acceptor ubiquitin molecules, or upon self-association of Ube2g27. The observed flexibility of Ube2g2 could play an important functional role by enabling the adaptive recognition of different binding partners8. The backbone dynamics results are consistent with motions which sample conformations at least as broadly as observed within the NMR structural ensemble. It is therefore feasible that the observation of contacts in some members of the NMR structural ensemble between residues H94 and D98 or D99 corresponds to a genuine catalytic role for H94 as a general base. Further clarification of these questions awaits additional structural and biochemical studies.
Supplementary Material
Acknowledgments
We are grateful for support from the NIH biophysics training grant T32GM008403 (WB) and helpful discussions with Prof. Juliette Lecomte.
Footnotes
Institution at which the work was performed: Johns Hopkins University
References
- 1.Katsanis N, Fisher EM. Identification, expression, and chromosomal localization of ubiquitin conjugating enzyme 7 (UBE2G2), a human homologue of the Saccharomyces cerevisiae ubc7 gene. Genomics. 1998;51(1):128–131. doi: 10.1006/geno.1998.5263. [DOI] [PubMed] [Google Scholar]
- 2.Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9(12):944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim I, Xu W, Reed JC. Cell death and endoplasmic reticulum stress: disease relevance and therapeutic opportunities. Nat Rev Drug Discov. 2008;7(12):1013–1030. doi: 10.1038/nrd2755. [DOI] [PubMed] [Google Scholar]
- 4.Simoes-Correia J, Figueiredo J, Oliveira C, van Hengel J, Seruca R, van Roy F, Suriano G. Endoplasmic reticulum quality control: a new mechanism of E-cadherin regulation and its implication in cancer. Hum Mol Genet. 2008;17(22):3566–3576. doi: 10.1093/hmg/ddn249. [DOI] [PubMed] [Google Scholar]
- 5.Li W, Tu D, Brunger AT, Ye Y. A ubiquitin ligase transfers preformed polyubiquitin chains from a conjugating enzyme to a substrate. Nature. 2007;446(7133):333–337. doi: 10.1038/nature05542. [DOI] [PubMed] [Google Scholar]
- 6.Ravid T, Hochstrasser M. Autoregulation of an E2 enzyme by ubiquitin-chain assembly on its catalytic residue. Nat Cell Biol. 2007;9(4):422–427. doi: 10.1038/ncb1558. FIELD Full Journal Title:Nature Cell Biology. [DOI] [PubMed] [Google Scholar]
- 7.Li W, Tu D, Li L, Wollert T, Ghirlando R, Brunger AT, Ye Y. Mechanistic insights into active site-associated polyubiquitination by the ubiquitin-conjugating enzyme Ube2g2. Proc Natl Acad Sci U S A. 2009;106(10):3722–3727. doi: 10.1073/pnas.0808564106. FIELD Full Journal Title:Proceedings of the National Academy of Sciences of the United States of America. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das R, Mariano J, Tsai YC, Kalathur RC, Kostova Z, Li J, Tarasov SG, McFeeters RL, Altieri AS, Ji X, Byrd RA, Weissman AM. Allosteric activation of E2-RING finger-mediated ubiquitylation by a structurally defined specific E2-binding region of gp78. Molecular Cell. 2009;34(6):674–685. doi: 10.1016/j.molcel.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen B, Mariano J, Tsai YC, Chan AH, Cohen M, Weissman AM. The activity of a human endoplasmic reticulum-associated degradation E3, gp78, requires its Cue domain, RING finger, and an E2-binding site. Proc Natl Acad Sci U S A. 2006;103(2):341–346. doi: 10.1073/pnas.0506618103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imai Y, Soda M, Inoue H, Hattori N, Mizuno Y, Takahashi R. An unfolded putative transmembrane polypeptide, which can lead to endoplasmic reticulum stress, is a substrate of Parkin. Cell. 2001;105(7):891–902. doi: 10.1016/s0092-8674(01)00407-x. [DOI] [PubMed] [Google Scholar]
- 11.Kikkert M, Doolman R, Dai M, Avner R, Hassink G, van Voorden S, Thanedar S, Roitelman J, Chau V, Wiertz E. Human HRD1 is an E3 ubiquitin ligase involved in degradation of proteins from the endoplasmic reticulum. J Biol Chem. 2004;279(5):3525–3534. doi: 10.1074/jbc.M307453200. [DOI] [PubMed] [Google Scholar]
- 12.Hassink G, Kikkert M, van Voorden S, Lee SJ, Spaapen R, van Laar T, Coleman CS, Bartee E, Fruh K, Chau V, Wiertz E. TEB4 is a C4HC3 RING finger-containing ubiquitin ligase of the endoplasmic reticulum. Biochem J. 2005;388(Pt 2):647–655. doi: 10.1042/BJ20041241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tiwari S, Weissman AM. Endoplasmic reticulum (ER)-associated degradation of T cell receptor subunits. Involvement of ER-associated ubiquitin-conjugating enzymes (E2s) J Biol Chem. 2001;276(19):16193–16200. doi: 10.1074/jbc.M007640200. [DOI] [PubMed] [Google Scholar]
- 14.Pabarcus MK, Hoe N, Sadeghi S, Patterson C, Wiertz E, Correia MA. CYP3A4 ubiquitination by gp78 (the tumor autocrine motility factor receptor, AMFR) and CHIP E3 ligases. Arch Biochem Biophys. 2009;483(1):66–74. doi: 10.1016/j.abb.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi R, Imai Y. Pael receptor, endoplasmic reticulum stress, and Parkinson's disease. J Neurol. 2003;250 3:III25–29. doi: 10.1007/s00415-003-1305-8. [DOI] [PubMed] [Google Scholar]
- 16.Webster JM, Tiwari S, Weissman AM, Wojcikiewicz RJ. Inositol 1,4,5-trisphosphate receptor ubiquitination is mediated by mammalian Ubc7, a component of the endoplasmic reticulum-associated degradation pathway, and is inhibited by chelation of intracellular Zn2+ J Biol Chem. 2003;278(40):38238–38246. doi: 10.1074/jbc.M305600200. [DOI] [PubMed] [Google Scholar]
- 17.Kostova Z, Tsai YC, Weissman AM. Ubiquitin ligases, critical mediators of endoplasmic reticulum-associated degradation. Semin Cell Dev Biol. 2007;18(6):770–779. doi: 10.1016/j.semcdb.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cook WJ, Martin PD, Edwards BF, Yamazaki RK, Chau V. Crystal structure of a class I ubiquitin conjugating enzyme (Ubc7) from Saccharomyces cerevisiae at 2.9 angstroms resolution. Biochemistry. 1997;36(7):1621–1627. doi: 10.1021/bi962639e. [DOI] [PubMed] [Google Scholar]
- 19.Giraud MF, Desterro JM, Naismith JH. Structure of ubiquitin-conjugating enzyme 9 displays significant differences with other ubiquitin-conjugating enzymes which may reflect its specificity for sumo rather than ubiquitin. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):891–898. doi: 10.1107/s0907444998002480. [DOI] [PubMed] [Google Scholar]
- 20.Haldeman MT, Xia G, Kasperek EM, Pickart CM. Structure and function of ubiquitin conjugating enzyme E2-25K: the tail is a core-dependent activity element. Biochemistry. 1997;36(34):10526–10537. doi: 10.1021/bi970750u. [DOI] [PubMed] [Google Scholar]
- 21.Houben K, Dominguez C, van Schaik FM, Timmers HT, Bonvin AM, Boelens R. Solution structure of the ubiquitin-conjugating enzyme UbcH5B. J Mol Biol. 2004;344(2):513–526. doi: 10.1016/j.jmb.2004.09.054. [DOI] [PubMed] [Google Scholar]
- 22.Merkley N, Shaw GS. Solution structure of the flexible class II ubiquitin-conjugating enzyme Ubc1 provides insights for polyubiquitin chain assembly. J Biol Chem. 2004;279(45):47139–47147. doi: 10.1074/jbc.M409576200. [DOI] [PubMed] [Google Scholar]
- 23.Pichler A, Knipscheer P, Oberhofer E, van Dijk WJ, Korner R, Olsen JV, Jentsch S, Melchior F, Sixma TK. SUMO modification of the ubiquitin-conjugating enzyme E2-25K. Nat Struct Mol Biol. 2005;12(3):264–269. doi: 10.1038/nsmb903. [DOI] [PubMed] [Google Scholar]
- 24.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 25.VanDemark AP, Hofmann RM, Tsui C, Pickart CM, Wolberger C. Molecular insights into polyubiquitin chain assembly: crystal structure of the Mms2/Ubc13 heterodimer. Cell. 2001;105(6):711–720. doi: 10.1016/s0092-8674(01)00387-7. [DOI] [PubMed] [Google Scholar]
- 26.Plon SE, Leppig KA, Do HN, Groudine M. Cloning of the human homolog of the CDC34 cell cycle gene by complementation in yeast. Proc Natl Acad Sci U S A. 1993;90(22):10484–10488. doi: 10.1073/pnas.90.22.10484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petroski MD, Deshaies RJ. Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell. 2005;123(6):1107–1120. doi: 10.1016/j.cell.2005.09.033. [DOI] [PubMed] [Google Scholar]
- 28.Chen Z, Pickart CM. A 25-kilodalton ubiquitin carrier protein (E2) catalyzes multi-ubiquitin chain synthesis via lysine 48 of ubiquitin. J Biol Chem. 1990;265(35):21835–21842. [PubMed] [Google Scholar]
- 29.Gazdoiu S, Yamoah K, Wu K, Pan ZQ. Human Cdc34 employs distinct sites to coordinate attachment of ubiquitin to a substrate and assembly of polyubiquitin chains. Mol Cell Biol. 2007;27(20):7041–7052. doi: 10.1128/MCB.00812-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arai R, Yoshikawa S, Murayama K, Imai Y, Takahashi R, Shirouzu M, Yokoyama S. Structure of human ubiquitin-conjugating enzyme E2 G2 (UBE2G2/UBC7) Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006;62(Pt 4):330–334. doi: 10.1107/S1744309106009006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharp PM, Cowe E, Higgins DG, Shields DC, Wolfe KH, Wright F. Codon usage patterns in Escherichia coli, Bacillus subtilis, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster and Homo sapiens; a review of the considerable within-species diversity. Nucleic Acids Res. 1988;16(17):8207–8211. doi: 10.1093/nar/16.17.8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marley J, Lu M, Bracken C. A method for efficient isotopic labeling of recombinant proteins. J Biomol NMR. 2001;20(1):71–75. doi: 10.1023/a:1011254402785. [DOI] [PubMed] [Google Scholar]
- 33.Briggman KB, Majumdar A, Coleman CS, Chau V, Tolman JR. NMR assignment of human ubiquitin conjugating enzyme Ubc7. J Biomol NMR. 2005;32(4):340. doi: 10.1007/s10858-005-1257-7. [DOI] [PubMed] [Google Scholar]
- 34.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 35.Hansen MR, Mueller L, Pardi A. Tunable alignment of macromolecules by filamentous phage yields dipolar coupling interactions. Nat Struct Biol. 1998;5(12):1065–1074. doi: 10.1038/4176. [DOI] [PubMed] [Google Scholar]
- 36.Tycko R, Blanco FJ, Ishii Y. Alignment of biopolymers in strained gels: A new way to create detectable dipole-dipole couplings in high resolution biomolecular NMR. J Am Chem Soc. 2000;122:9340–9341. [Google Scholar]
- 37.Ottiger M, Delaglio F, Bax A. Measurement of J and dipolar couplings from simplified two- dimensional NMR spectra. J Magn Reson. 1998;131(2):373–378. doi: 10.1006/jmre.1998.1361. [DOI] [PubMed] [Google Scholar]
- 38.Vuister GW, Bax A. Quantitative J Correlation: A New Approach for Measuring Homonuclear Three-Bond J(HNHα) Coupling Constants in 15N-Enriched Proteins. Journal of the American Chemical Society. 1993;115:7772–7777. [Google Scholar]
- 39.Wüthrich K. NMR of Proteins and Nucleic Acids. New York: Wiley; 1986. [Google Scholar]
- 40.Barbato G, Ikura M, Kay LE, Pastor RW, Bax A. Backbone dynamics of calmodulin studied by 15N relaxation using inverse detected two-dimensional NMR spectroscopy: the central helix is flexible. Biochemistry. 1992;31(23):5269–5278. doi: 10.1021/bi00138a005. [DOI] [PubMed] [Google Scholar]
- 41.Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry. 1989;28(23):8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 42.Piotto M, Saudek V, Sklenar V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J Biomol NMR. 1992;2(6):661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 43.Goddard TD, Kneller DG. SPARKY 3. San Francisco: University of California; [Google Scholar]
- 44.Lipari G, Szabo A. Model-Free Approach to the Interpretation of Nuclear Magnetic Resonance Relaxation in Macromolecules. 1. Theory and Range of Validity. Journal of the American Chemical Society. 1982;104:4546–4559. [Google Scholar]
- 45.Cole R, Loria JP. FAST-Modelfree: a program for rapid automated analysis of solution NMR spin-relaxation data. J Biomol NMR. 2003;26(3):203–213. doi: 10.1023/a:1023808801134. [DOI] [PubMed] [Google Scholar]
- 46.Pelton JG, Torchia DA, Meadon ND, Roseman S. Tautomeric states of the active-site histidines of phosphorylated and unphosphorylated IIIGlc, a signal-transducing protein from Escherichia coli using two-dimensional heteronuclear NMR techniques. Protein Science. 1993;2(4):543–558. doi: 10.1002/pro.5560020406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee KK, Fitch CA, Lecomte JTJ, Garcia-Moreno BE. Electrostatic Effects in Highly Charged Proteins: Salt Sensitivity of pKa Values of Histidines in Staphylococcal Nuclease. Biochemistry. 2002;41(17):5656–5667. doi: 10.1021/bi0119417. [DOI] [PubMed] [Google Scholar]
- 48.Worthylake DK, Prakash S, Prakash L, Hill CP. Crystal structure of the Saccharomyces cerevisiae ubiquitin-conjugating enzyme Rad6 at 2.6 A resolution. J Biol Chem. 1998;273(11):6271–6276. doi: 10.1074/jbc.273.11.6271. [DOI] [PubMed] [Google Scholar]
- 49.Garcia de la Torre J, Huertas ML, Carrasco B. HYDRONMR: Prediction of NMR Relaxation of Globular Proteins from Atomic-Level Structures and Hydrodynamic Calculations. J Magn Reson. 2000;147(1):138–146. doi: 10.1006/jmre.2000.2170. [DOI] [PubMed] [Google Scholar]
- 50.Tolman JR, Ruan K. NMR residual dipolar couplings as probes of biomolecular dynamics. Chem Rev. 2006;106(5):1720–1736. doi: 10.1021/cr040429z. [DOI] [PubMed] [Google Scholar]
- 51.Wu PY, Hanlon M, Eddins M, Tsui C, Rogers RS, Jensen JP, Matunis MJ, Weissman AM, Wolberger C, Pickart CM. A conserved catalytic residue in the ubiquitin-conjugating enzyme family. EMBO J. 2003;22(19):5241–5250. doi: 10.1093/emboj/cdg501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hamilton KS, Ellison MJ, Barber KR, Williams RS, Huzil JT, McKenna S, Ptak C, Glover M, Shaw GS. Structure of a conjugating enzyme-ubiquitin thiolester intermediate reveals a novel role for the ubiquitin tail. Structure. 2001;9(10):897–904. doi: 10.1016/s0969-2126(01)00657-8. [DOI] [PubMed] [Google Scholar]
- 53.Rawlings ND, Barrett AJ. Families of serine peptidases. Methods Enzymol. 1994;244:19–61. doi: 10.1016/0076-6879(94)44004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu M, Li P, Li M, Li W, Yao T, Wu JW, Gu W, Cohen RE, Shi Y. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell. 2002;111(7):1041–1054. doi: 10.1016/s0092-8674(02)01199-6. [DOI] [PubMed] [Google Scholar]
- 55.Shimahara H, Yoshida T, Shibata Y, Shimizu M, Kyogoku Y, Sakiyama F, Nakazawa T, Tate Si, Ohki Sy, Kato T, Moriyama H, Kishida Ki, Tano Y, Ohkubo T, Kobayashi Y. Tautomerism of Histidine 64 Associated with Proton Transfer in Catalysis of Carbonic Anhydrase. Journal of Biological Chemistry. 2007;282(13):9646–9656. doi: 10.1074/jbc.M609679200. [DOI] [PubMed] [Google Scholar]
- 56.Yunus AA, Lima CD. Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway. Nat Struct Mol Biol. 2006;13(6):491–499. doi: 10.1038/nsmb1104. [DOI] [PubMed] [Google Scholar]
- 57.Capili AD, Lima CD. Taking it step by step: mechanistic insights from structural studies of ubiquitin/ubiquitin-like protein modification pathways. Curr Opin Struct Biol. 2007;17(6):726–735. doi: 10.1016/j.sbi.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.