Summary
Post-translational modifications of viral replication proteins could be widespread phenomena during the replication of plus-stranded RNA viruses. In this paper, we identify two lysines in the tombusvirus p33 replication co-factor involved in ubiquitination and show that the same lysines are also important for the p33 to interact with the host Vps23p ESCRT-I factor. We find that the interaction of p33 with Vps23p is also affected by a "late-domain"-like sequence in p33. The combined mutations of the two lysines and the late-domain-like sequences in p33 reduced replication of a replicon RNA of Tomato bushy stunt virus in yeast model host, in plant protoplasts and plant leaves, suggesting that p33-Vps23p ESCRT protein interaction affects tombusvirus replication. Using ubiquitin-mimicking p33 chimeras, we demonstrate that high level of p33 ubiquitination is inhibitory for TBSV replication. These findings argue that optimal level of p33 ubiquitination plays a regulatory role during tombusvirus infections.
INTRODUCTION
Most plus-stranded (+)RNA viruses have small genomes with limited coding capacity. However, the viral-coded proteins have multiple functions, thus extending the functional repertoire of these proteins. In addition, post-translational modifications of the viral proteins, such as phosphorylation, ubiquitination, acetylation, and glycosylation, can further increase the variety of functions performed by these proteins. Post-translational modifications may also affect the localization and stability of the viral proteins or serve as molecular switches between different functions (Brigati et al., 2003; Cereseto et al., 2005; Jakubiec and Jupin, 2007; Vigerust and Shepherd, 2007). Also, post-translational modifications of the viral proteins could affect their abilities to interact with selected host proteins. In spite of the possible significance of post-translational modifications, our understanding of the roles of protein modifications in virus replication is limited. Accumulating data show that phosphorylation of the viral replication proteins could affect interactions between 1a and 2a replication proteins of Cucumber mosaic virus (Kim, Palukaitis, and Park, 2002) or the NS3 and NS5 replication proteins of Dengue virus (Kapoor et al., 1995), the ability of the p33 tombusvirus replication proteins to bind to the viral RNA (Shapka, Stork, and Nagy, 2005; Stork, Panaviene, and Nagy, 2005), or interaction between the NS5A replication protein of hepatitis C virus (HCV) and hVAP-A (human vesicle-associated membrane protein-associated protein A), which is proposed to facilitate the assembly of the viral replicase complex by acting as the membrane docking site (Evans, Rice, and Goff, 2004; Gao et al., 2004).
Protein ubiquitination is a common post-translational modification in eukaryotic cells (Pickart, 2001; Pickart and Eddins, 2004). Poly-ubiquitination of proteins frequently lead to their degradation by the 26S proteosome, while mono-ubiquitination of proteins could affect their functions and localizations. For many membrane proteins, mono-ubiquitination is important for targeting to the endosomal, vacuolar or plasma membranes or for their degradation in the vacuoles (lysosomes in mammalian cells). Ubiquitination of client proteins is performed by a chain of enzymes, including E1 ubiquitin activating protein, which binds to the 76 amino-acid-long ubiquitin (Ub); E2 Ub-conjugating enzyme and E3 Ub-ligases. The selection of a given client protein is usually done by specific E3 Ub-ligases coded by several hundred genes in the mammalian genomes, though E2 Ub- conjugating enzymes could occasionally also select client proteins for ubiquitination (Pickart, 2001; Pickart and Eddins, 2004; Roos-Mattjus and Sistonen, 2004). As expected, ubiquitination of viral proteins has been documented previously. The HCV NS5B RdRp protein has been shown to interact with a ubiquitin-like protein hPLIC1, which is associated with E3 Ub-ligases and the proteasome (Gao et al., 2003). Over-expression of hPLIC1 led to ubiquitination and degradation of NS5B, suggesting that this host protein is a regulator of HCV replication. Replication of coxsackievirus B3 (CVB3) is affected by inhibition of ubiquitination and by proteosome inhibitors, probably due to their effect on the proteosome-based protein degradation (Si et al., 2008). Ubiquitination of the 3D polymerase of CVB3 has been demonstrated, implicating that Ub might affect the functions of the 3D polymerase (Si et al., 2008). Polyubiquitination of the movement protein of Turnip yellow mosaic virus (TYMV) is proposed to play a role in its degradation and regulation of transient cell-to-cell movement process (Drugeon and Jupin, 2002). Also, the amount of RNA-dependent RNA polymerase of TYMV might be regulated by ubiquitination (Hericourt et al., 2000). These examples show the importance of ubiquitination during viral infections and the transient nature of the ubiquitination process.
Tomato bushy stunt virus (TBSV) is a small (+)RNA virus of plants, which has recently emerged as a model virus to study virus replication, recombination, and virus - host interactions. Progress in these areas is facilitated by the development of yeast (Saccharomyces cerevisiae) as a model host (Nagy and Pogany, 2006; Panavas and Nagy, 2003; Panaviene et al., 2004; Pogany and Nagy, 2008; White and Nagy, 2004). The auxiliary p33 replication protein is involved in the recruitment of the TBSV (+)RNA to the site of replication, which is the cytosolic surface of peroxisomal membranes (Jonczyk et al., 2007; Panavas et al., 2005a; Pogany, White, and Nagy, 2005). The p92pol RNA-dependent RNA polymerase (RdRp) protein, which is the translational readthrough product of the p33 stop codon, binds to p33 replication protein leading to the assembly of the functional membrane-bound replicase complex (Panavas et al., 2005a; Panaviene, Panavas, and Nagy, 2005; Panaviene et al., 2004; Pogany and Nagy, 2008). These viral replication proteins bind to over 50 host proteins in vitro (Li et al., 2008), suggesting the existence of protein networks subverted by tombusviruses for their replication.
The complexicity of host - virus interactions is revealed by recent genome-wide screens covering 95% of yeast genes that have identified more than 150 host genes affecting TBSV replication or recombination (Jiang et al., 2006; Panavas et al., 2005b; Serviene et al., 2006; Serviene et al., 2005). Also, proteomics analysis of the highly purified tombusvirus replicase complex identified the presence of 6–10 host proteins in the replicase in addition to the viral p33 and p92pol replication proteins (Li et al., 2008; Li et al., 2009; Serva and Nagy, 2006). These host proteins, such as heat shock protein 70 (hsp70) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) affect the assembly of the viral replicase complex (Pogany et al., 2008; Wang, Stork, and Nagy, 2009) (Serva and Nagy, 2006; Wang et al., 2009), or regulate viral RNA replication by selectively binding to the minus-stranded TBSV RNA and affecting plus-strand synthesis (Wang and Nagy, 2008). Additional defined components in the viral replicase complex are Cdc34p Ub-conjugating enzyme, which is involved in ubiquitination of the p33 replication co-factor (Li et al., 2008) and eEF1a translation elongation factor (Li et al., 2009). Moreover, Pex19p cytosolic transport protein is involved in the transport of the replication proteins to the peroxisomal membranes (Pathak, Sasvari, and Nagy, 2008).
The possible involvement of ubiquitination of p33 replication co-factor in tombusvirus replication is raised by several independent observations. First, ubiquitination of the tombusvirus p33 replication protein has been demonstrated in yeast (Li et al., 2008). Second, genome-wide screens to identify host factors affecting TBSV RNA replication or RNA recombination in yeast led to the identification of BRE1, DOA4, RAD6, LGE1, UBP3 genes known to be involved in various aspects of protein ubiquitination (Jiang et al., 2006; Panavas et al., 2005b; Serviene et al., 2006; Serviene et al., 2005). Third, a proteomics screen based on a yeast protein array carrying 4,100 purified yeast proteins revealed interaction of p33 replication protein with Uba1p Ub-activating enzyme, Cdc34p E2 Ub-conjugating enzyme, Rsp5p E3 Ub-ligase, Ubp10p and Ubp15p Ub-specific proteases (Li et al., 2008). Moreover, subsequent work has demonstrated that Cdc34p E2 Ub-conjugating enzyme is present in the tombusvirus replicase complex and it can mono- and bi-ubiquitinate p33 in vitro in the absence of an E3 Ub-ligase (Li et al., 2008). The Nedd4-type Rsp5p E3 Ub-ligase has also been shown to bind to and ubiquitinate p33 replication protein in vitro (Barajas, Li, and Nagy, 2009). The identification of many host proteins involved in ubiquitination/deubiquitination of TBSV replication proteins suggests a complex role for this post-translational modification in p33 and p92pol functions.
To determine the functional significance of p33 ubiquitination in tombusvirus replication, in this work, we identified lysines in p33, which are targeted for mono-ubiquitination. We also show that these lysines, together with the so-called “late-domain-like” sequence in p33, are important for binding to Vps23p ESCRT (endosomal complexes required for transport) protein. Combined mutation of these sequences in p33 affected replication of TBSV in yeast and in plant hosts. These observations open the possibility that ubiquitination of p33 is involved in recruitment of Vps23p and likely other ESCRT proteins for TBSV replication.
RESULTS
Ubiquitination of K70 and K76 lysines in the TBSV p33 replication protein
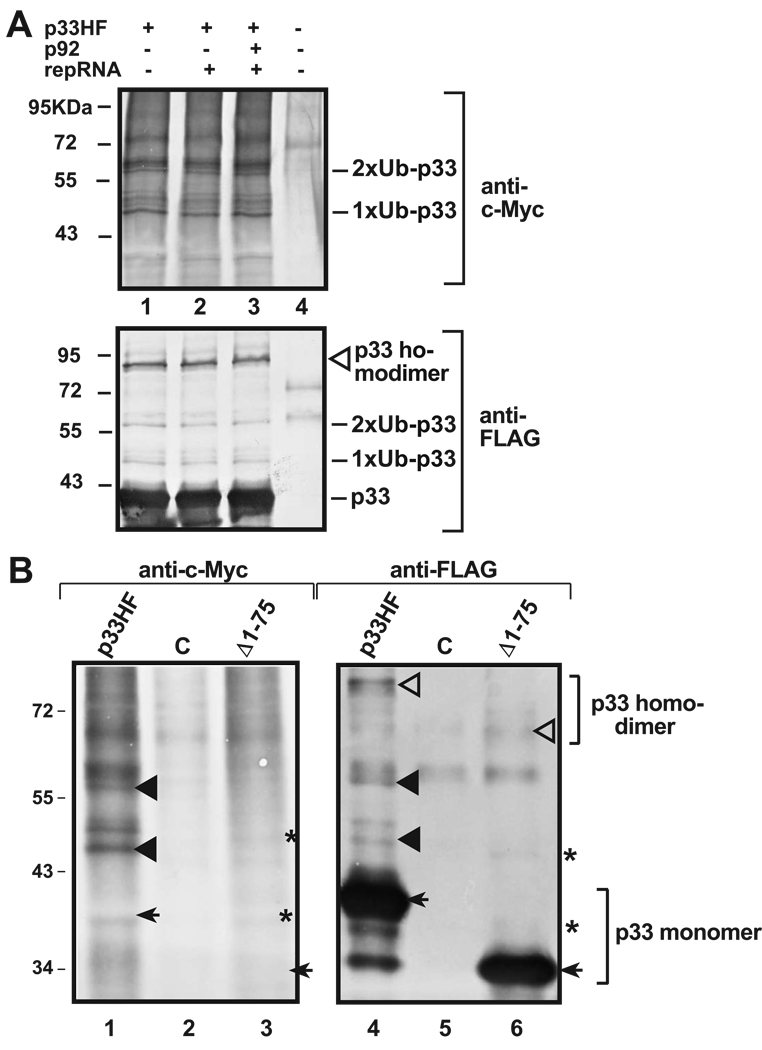
We have shown previously that a small fraction of TBSV p33 molecules becomes mono- and biubiquitinated in yeast cells (Li et al., 2008). To determine if p33 must be part of the viral replicase complex in order to become ubiquitinated, we expressed p33HF (which is 6×His and FLAG-tagged) alone in yeast cells or together with p92pol replication protein and/or the viral RNA as well as Ub tagged with c-Myc from separate expression plasmids. Then, the ubiquitination status of the FLAG-affinity purified p33HF was examined by Western blot analysis based on anti-c-Myc antibody as well as a shift in MW of p33HF (mono-and bi-ubiquitination causes ~ 8 and 16KDa increase, respectively). Similar levels of mono- and bi-ubiquitinated p33HF were detected with anti-cMyc antibody when p33HF was expressed alone or in combination with p92pol and/or replicon (rep)RNA (Fig. 1A, lanes 1–3, upper panel), suggesting that ubiquitination of p33HF is not affected whether it is part of the replicase complex or not.
Fig. 1.
Ubiquitination of p33 replication protein in yeast. (A) The effect of co-expression of p92pol and repRNA on p33 ubiquitination. The membrane-bound p33HF (tagged with 6×His and FLAG) was purified via FLAG-affinity chromatography after membrane solubilization from yeast co-expressing c-Myc-tagged ubiquitin from a plasmid. The ubiquitinated p33 was detected with Western blot using anti-c-Myc (top panel) or anti-FLAG antibodies (bottom panel), based on the 8 and 16 KDa increase of MW for mono (1×Ub)- and bi (2×Ub)-ubiquitination products of p33. Note that the additional p33-specific bands near the mono- and bi-ubiquitinated bands could be phosphorylated forms (Shapka, Stork, and Nagy, 2005) of the ubiquitinated p33HF. The origin of smear detected with anti-c-Myc antibody is unknown, though they are probably co-purified ubiquitinated host proteins, since the p33-derived products would also be detected by the anti-FLAG antibody (bottom panel). Also, these products are missing in the control sample from yeast expressing 6× His-FLAG peptide from pYC-HF (lane 4). The p33 homodimer is marked with an empty arrowhead. Note that co-expression of TBSV repRNA and/or p92 RdRp protein did not affect the ubiquitination status of p33. (B) Testing the ubiquitination status of an N-terminal deletion mutant of p33 replication protein in yeast. The positions of the deleted amino acids are indicated and the expected FLAG-purified p33HF products are marked with arrows. The ubiquitinated p33 was detected with Western blot using anti-c-Myc (left panel) or anti-FLAG antibodies (right panel). The mono- and biubiquitinated p33HF products are shown with black arrowheads, while the p33HF-homodimer is marked with an open arrowhead. The calculated sizes of the expected, but not detected, mono (1×Ub)- and bi (2×Ub)-ubiquitination products of the p33 mutant are marked with asterisks. Note that the N-terminal deletion in p33 eliminated p33 mono- and biubiquitination in yeast. See further details in panel A.
We also analyzed the same FLAG-purified p33HF by using anti-FLAG antibody (Fig. 1A, bottom panel) (Serva and Nagy, 2006). Accordingly, we have detected p33HF-specific bands with ~8 and 16 KDa increase in the MW (Fig. 1A, bottom panel, lanes 1–3), which is consistent with mono- and bi-ubiquitinated p33HF. The mono- and bi-ubiquitinated p33HF represented a small fraction (~5%) of the total p33HF, suggesting that not all p33HF is ubiquitinated or Ub-P33HF can be rapidly deubiquitinated in yeast cells. The estimated amount, however, might be lower than the actual percentage of ubiquitinated p33HF, since yeast expressed not only the cMyc-tagged Ub, but the untagged wt Ub from its natural chromosomal location as well. We used inhibitors of de-ubiquitination during the isolation to minimize the chance of reducing Ub-p33HF in the samples (see M&M). Altogether, these experiments revealed that mono-ubiquitination of p33 was not affected by the presence of p92pol replication protein or the viral RNA (Fig. 1A).
To define which Lysine becomes ubiquitinated in p33HF, first, we have used a set of p33HF mutants truncated at the N-terminus (Fig. 1B). These experiments defined that mono-and bi-ubiquitination did not occur when the N-terminal 75 (Fig. 1B, lanes 3 and 6) or 150 aa (not shown) segments of p33 were deleted. This suggests that the N-terminal 75 aa in p33 includes the lysines that are targeted for mono-and bi-ubiquitination in yeast.
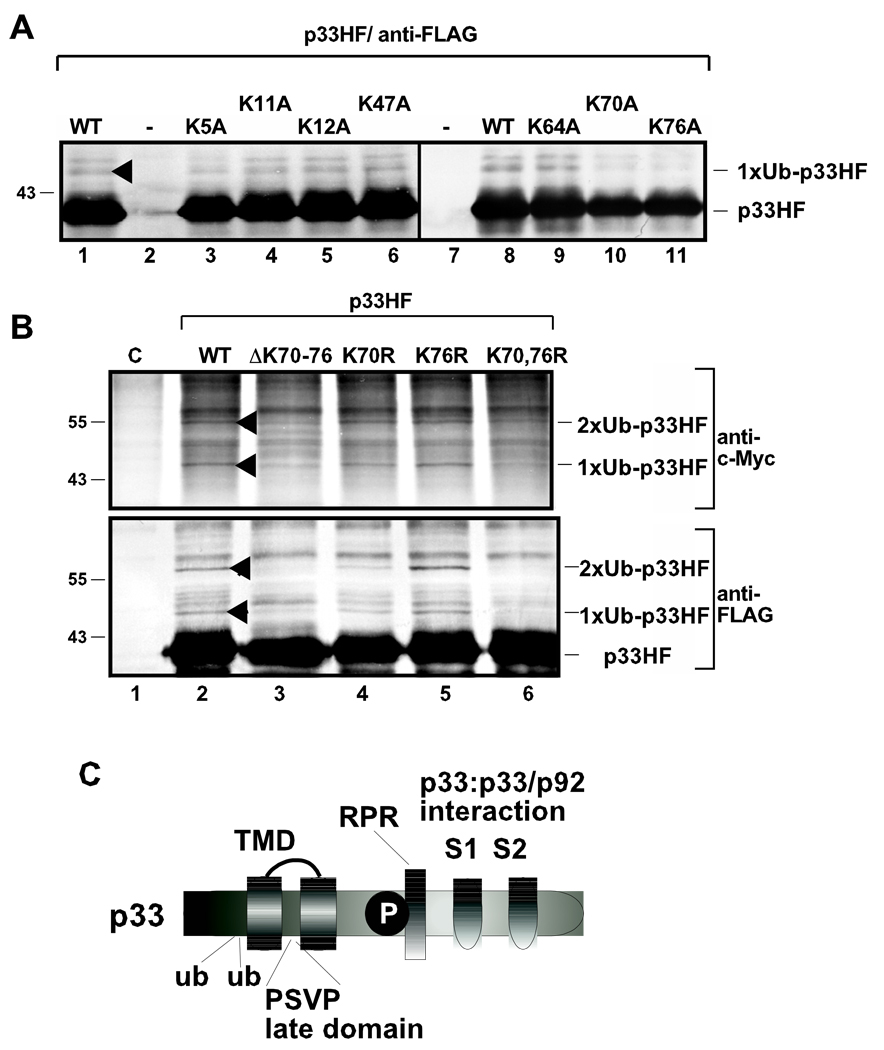
Individual mutagenesis of each of the seven conserved lysines to alanines (incompatible with ubiquitination) within the N-terminal 75aa segment of p33HF revealed that K70 and K76 were the most important targets for ubiquitination (Fig. 2A). Moreover, individual or combined mutagenesis of K70 and K76 to arginines (the most conservative change preserving the positive charge of the position and likely causing the least local or global structural changes in the structure of the protein) revealed that K70 was the primary target for mono- and bi-ubiquitination (Fig. 2B, lane 4), while K76 served as the secondary target when K70 was mutated. Accordingly, K70,76R double-mutant was the most deficient in both mono- and bi-ubiquitination (Fig. 2B, lane 6). The importance of these lysines in ubiquitination is also confirmed by deletion of K70-K76-stretch in p33HF that greatly reduced mono- and bi-ubiquitinated forms of p33HF in yeast cells (Fig. 2B, lane 3). Overall, these studies supported a major role for K70 in mono- and bi-ubiquitination of p33 (Fig. 2C).
Fig. 2.
Identification of the ubiquitinated lysines in p33 replication protein in yeast. (A) Testing mono-ubiquitination of the first seven N-terminal lysines in the p33 replication protein. The positions of the lysines mutated to alanines are indicated and the expected FLAG-purified p33HF products are marked. The purified p33HF was detected with Western blot using anti-FLAG antibody. See further details in Fig. 1A. (B) The positions of lysine to arginine mutations or deletion are indicated. The membrane-bound p33HF was purified via FLAG-affinity chromatography. The ubiquitinated p33 was identified via anti-c-Myc antibody (top panel) or via anti-FLAG antibody (bottom panel). Note that single K76R mutation in p33HF did not have effect on ubiquitination of p33, likely due to ubiquitination of an alternative lysine in this mutant. See further details in Fig. 1A. (C) Schematic representation of the ubiquitinated lysines and known functional motifs/domains in p33 replication protein. TMD, transmembrane domain; ub, ubiquitinated lysines; P, phosphorylated serine/threonine; RPR, proline/arginine-rich RNA binding region; S1 and S2 subdomains involved in p33:p33/p92 interaction.
Binding of the p33 replication protein to the host Vps23p ESCRT-I protein is affected by the ubiquitination region in p33
To identify the possible function(s) of mono-ubiquitination of the tombusvirus p33 replication co-factor, we assumed that ubiquitination could affect the ability of p33 to bind to host proteins. To select host protein candidates binding to Ub-p33, we searched our database of ~200 host factors identified via genome-wide screens and proteomics approaches for affecting TBSV replication/ recombination or for binding of p33 to host proteins in vitro (Jiang et al., 2006; Li et al., 2008; Panavas et al., 2005a; Serva and Nagy, 2006; Serviene et al., 2006; Serviene et al., 2005). An interesting candidate was Vps23p ESCRT-I protein, which binds to ubiquitinated membrane proteins (Morita and Sundquist, 2004; Usami et al., 2009) as well as to the tombusvirus p33 replication protein (Barajas, Jiang and Nagy, submitted). In addition, Vps23p is known to bind to additional ESCRT proteins (Morita and Sundquist, 2004; Usami et al., 2009) and is one of the seven members of the endosome sorting pathway affecting TBSV replication (Panavas et al., 2005b). We hypothesized that Ub-p33 might be involved in assisting the recruitment of Vps23p, which then mediates additional recruitment of selected ESCRT proteins for TBSV replication, possibly for facilitating the formation of the viral replicase.
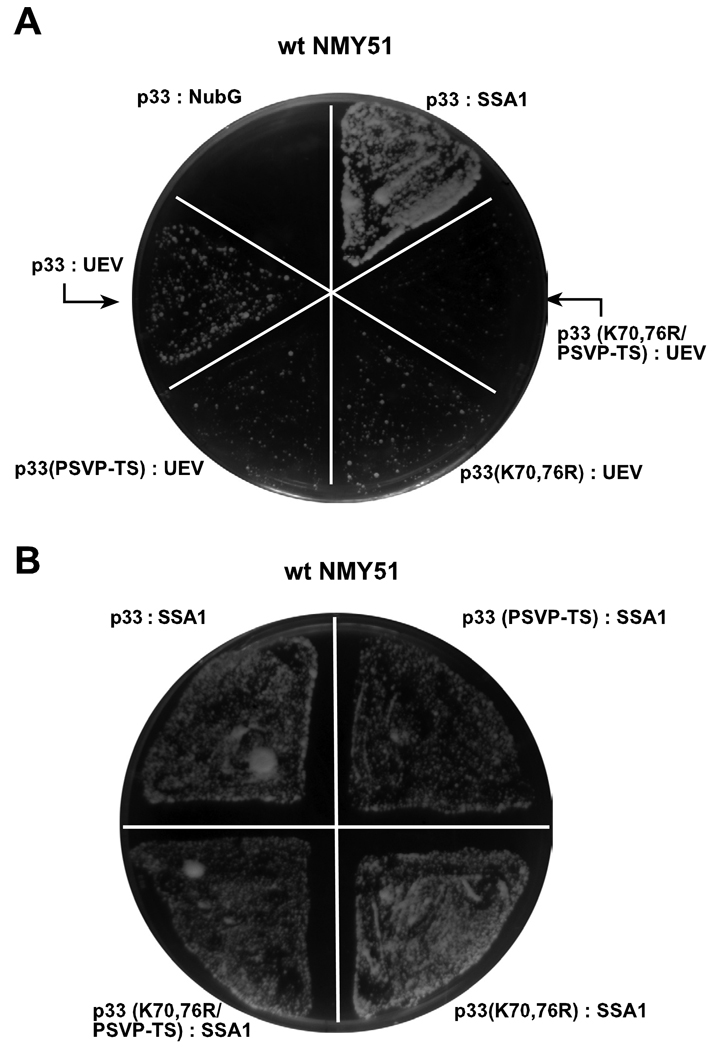
To test if the interaction between p33 and Vps23p is affected by either the ubiquitinated lysines, we used the split-ubiquitin assay, a variant of the yeast two-hybrid approach (Thaminy, Miller, and Stagljar, 2004). Since it is known that Vps23p interacts weakly/temporarily with client proteins and with p33 (Barajas, Jiang and Nagy, submitted), we used the protein-binding domain, called the UEV (ubiquitin E2 variant) domain of Vps23p in this assay (Fig. 3A).
Fig. 3.
Interaction between Vps23p ESCRT protein and p33 replication protein. (A) Split ubiquitin assay was used to test binding between p33 or its mutants and the N-terminal UEV domain of Vps23p in yeast. The bait p33 or its mutant derivatives were co-expressed with the shown prey proteins. Ssa1p (HSP70 chaperone), and the empty prey vector (NubG) were used as positive and negative controls, respectively. (B) A control split ubiquitin assay was based on using the documented interaction between p33 and Ssa1p. Note that the p33 mutants showed comparable level of interaction with Ssa1p to that of wt p33, suggesting that the p33 mutants were expressed and maintained stably in yeast.
Using K70,76R p33 ubiquitination-deficient mutant in the split-ubiquitination assay, we found that K70,76R p33 had weakened interaction with the UEV domain of Vps23p when compared with wt p33 (Fig. 3A), suggesting that ubiquitination of p33 could contribute to interaction with Vps23p. Importantly, the weaker interactions between the p33 mutant and UEV is not due to instability of the p33 K70,76R mutant in yeast, because this p33 mutant and the wt p33 showed comparable level of interaction with Ssa1p HSP70 (Fig. 3B), one of the host proteins present as a permanent resident in the tombusvirus replicase complex (Serva and Nagy, 2006; Wang, Stork, and Nagy, 2009).
The 'late-domain'-like motif in p33 replication protein affects p33-Vps23p interaction
Another interesting feature of Vps23p is that it recognizes the 'late domain' [P(T/S)XP sequence] in Vps27p, the initiator protein of the endosome pathway (Hurley and Emr, 2006; Morita and Sundquist, 2004). Interestingly, p33 replication protein contains one PSVP sequence (Fig. 2C), which is similar to the consensus late-domain P(T/S)XP motif recognized by Vps23p.
To test if the PSVP sequence in p33 (Fig. 2C) could affect its ability to interact with Vps23p, we used the split-ubiquitin assay. Indeed, we found that changing the PSVP sequence to TS reduced the number of yeast colonies (Fig. 3A), indicating a weaker interaction between p33PSVP-TS and UEV. Interestingly when mutations in the PSVP sequence and in lysines 70 and 76 were combined, the resulting p33 mutant (K70, 76R/PSVP-TS) showed the weakest interaction with the UEV domain, suggesting that both ubiquitination and the PSVP late domain-like sequence of p33 contribute to interaction with UEV (Vps23p) (Fig. 3A). In contrast, all of the p33 mutants showed comparable levels of interaction with Ssa1p (Fig. 3B), indicating that the mutations did not affect the stability of p33 in yeast.
Accumulation of TBSV repRNA in yeast depends on the late domain-like sequence and ubiquitination region of p33
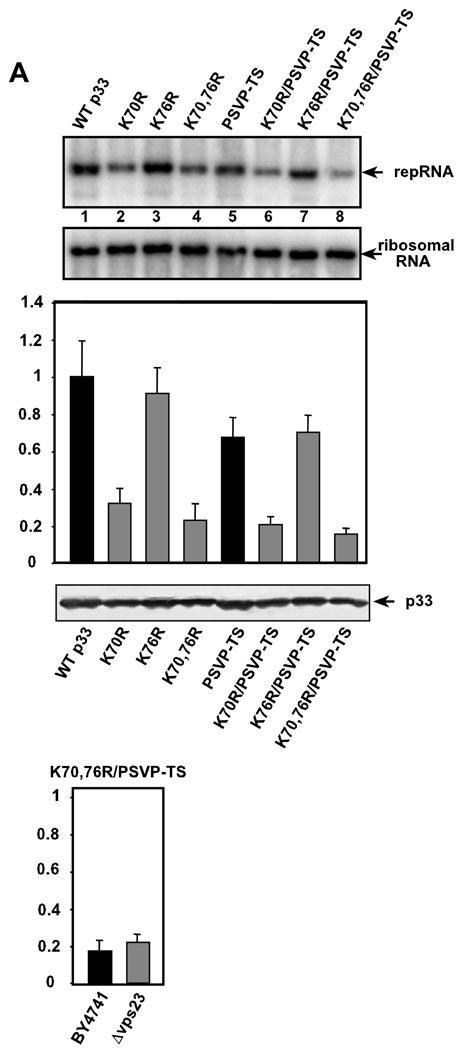
To test if ubiquitination of p33 and the interaction between p33 and Vps23p are important for the accumulation of TBSV repRNA, we expressed single and combined mutants of p33 carrying mutation(s) within the late domain and/or the ubiquitination region together with wt p92pol and repRNA in yeast. Measuring the level of TBSV replicon (rep)RNA accumulation in yeast 48 hours after the induction of repRNA replication revealed that mutating K70 had a moderate inhibitory effect, while changing K76 had no significant inhibitory effect (see K70R and K76R, Fig. 4A). Mutations in the late-domain-like PSVP motif had only 25% inhibitory effect on TBSV repRNA replication (mutant p33PSVP-TS, Fig. 4A, lane 5). Importantly, the combination of ubiquitination region and late-domain mutations had the most dramatic effect among the mutants tested by reducing repRNA accumulation by ~80% (mutant K70,76R/PSVP-TS Fig. 4A, lane 8). Overall, these data confirmed that the PSVP motif and the ubiquitination region in p33 affecting the interaction with Vps23p modulated TBSV repRNA accumulation in yeast, supporting the functional relevance of p33 - Vps23p interaction during viral repRNA replication.
Fig. 4.
Mutations of lysines 70 and 76 and the 'late domain'-like motif in p33 inhibit TBSV accumulation in yeast. (A) Top panel: Northern blot analysis of total RNA extracts obtained from yeast 48 hours after induction of repRNA replication. The given mutated p33 is shown on the top. Middle panel shows the ribosomal RNAs as loading controls for the Northern blot. The graph shows the level of TBSV repRNA accumulation in comparison with wt p33 (100%). Each mutant was tested in eight repeats. Bottom panel shows a Western blot analysis of total protein extracts for measuring p33HF levels in yeast using anti-6×His antibody. (B) The level of TBSV repRNA accumulation in the presence of K70, 76R/PSVP-TS p33 mutant and wt p92 in the parental and Δvps23 yeast. Note that the accumulation level of TBSV repRNA in the presence of wt p33/p92 in the parental yeast was chosen as 100%.
We performed additional tests if mutations in p33 within both ubiquitination region and late-domain are as detrimental to TBSV repRNA replication in vps23Δ yeast as in the parental yeast. The p33 mutant K70,76R/PSVP-TS supported as low level replication of TBSV repRNA in vps23Δ yeast as in the parental yeast (Fig. 4B). These data are consistent with the model that K70,76R/PSVP-TS mutation is mostly detrimental to p33 function by inhibiting p33 interaction with Vps23p, albeit pleiotropic effect of these mutations on p33 functions cannot be ruled out.
Reduced accumulation of tombusvirus RNA expressing p33 mutants deficient in binding to Vps23p in plant protoplasts and plant leaves
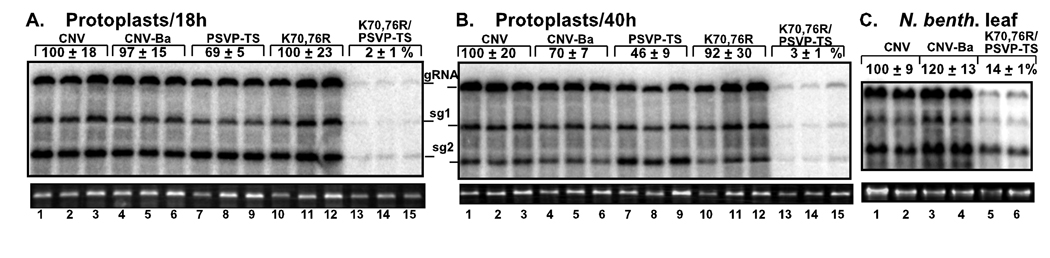
To validate our findings obtained in yeast on the role of p33 - Vps23p interaction in TBSV replication in a native host and with a fully infectious virus, we tested the effect of p33 mutations on the replication of the closely related Cucumber necrosis virus (CNV) genomic (g)RNA in Nicotiana benthamiana protoplasts. Similar to TBSV, CNV p33 also has the conserved K70, K76 and PSVP sequences. The three CNV mutants tested included (i) the PSVP late-domain-like mutant (p33PSVP-TS), (ii) the K K70, 76R ubiquitination-deficient mutant, and (iii) the combined K70, 76R/PSVP-TS mutant. Note that due to the overlapping expression strategy, p92 also carried the mutations present in p33. The accumulation of CNV gRNA in N. benthamiana protoplasts was measured by Northern blotting at 18 and 40 h time points. The largest inhibitory effect on CNV gRNA accumulation was observed with the combined K70, 76R/PSVP-TS mutant, which supported only 2–3% level of accumulation when compared to the control CNV (Fig. 5A–B, lanes 13–15). The K70, 76R or the PSVP-TS mutations had less detrimental effect on CNV accumulation under similar conditions (Fig. 5A–B, lanes 7–12).
Fig. 5.
Inhibition of tombusvirus RNA accumulation in plant protoplasts and plants by mutations in p33 known to affect its interaction with Vps23p. (A–B) The accumulation of CNV genomic and subgenomic RNAs is measured by Northern blotting in N. benthamiana protoplasts incubated for 18 (panel A) or 40 hours (panel B). The CNV genomic RNA had mutations in lysines 70 and 76 and/or within the late-domain PSVP region of p33 ORF as shown. CNV-Ba carries a BamHI site within the 5' noncoding region that was used to engineer the other mutations in p33. Ribosomal RNA (bottom panels) was used as a loading control. The experiments were repeated two times. (C) The accumulation of CNV RNAs is measured by Northern blotting in N. benthamiana leaves. CNV infection was launched from the 35S promoter in an Agrobacterium plasmid (introduced to the leaves via agroinfiltration). Samples were taken from the infiltrated leaves 3.5 days after infiltration.
To test the accumulation of mutated gCNV in N. benthamiana plants, we launched virus infection from an Agrobacterium plasmid carrying the full-length CNV cDNA, which was introduced to the leaves via agroinfiltration (Cheng et al., 2007). Measuring CNV RNA accumulation by Northern blotting in samples from the infiltrated leaves taken 3.5 days after infiltration demonstrated that the combined K70,76R/PSVP-TS mutant accumulated to ~15% level of the wt gRNA (Fig. 5C, lanes 5–6). This confirmed that the PSVP motif and the ubiquitination region of p33 involved in interaction with Vps23p are also important during tombusvirus infection of an experimental plant host. The higher level of RNA accumulation for K70,76R/PSVP-TS mutant in leaves than in protoplasts (Fig. 5A–C) is likely due to the continuous production of viral RNA transcripts from the 35S promoter in the Agrobacterium plasmid. This is not the case in protoplasts, in which the RNA transcripts are introduced only at the beginning of the infection.
Overall, the above data suggest that similar to TBSV repRNA replication in yeast, the ubiquitination region and late-domain-like PSVP motif together are important for p33 function and they are partly redundant. Moreover, these data reveal that tombusvirus accumulation depends on p33 sequences involved in binding to Vps23p, and this feature is similar in yeast and plant cells.
Ubiquitination of the p33 replication protein does not affect p33 degradation and stability in yeast
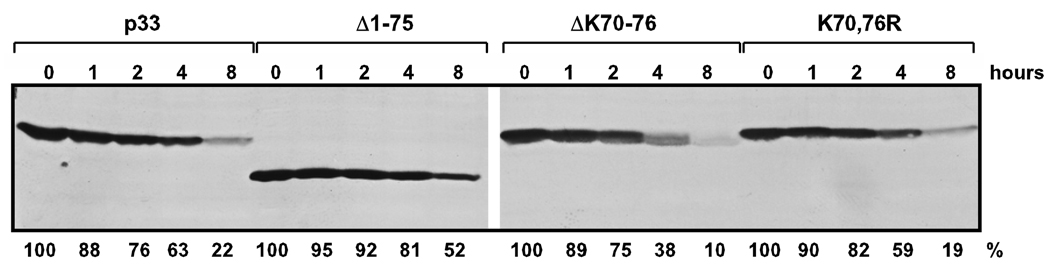
One of the major functions of ubiquitination, especially poly-ubiquitination, is to tag proteins for degradation by the 26S proteosome that is important for regulation in many cellular processes (Pickart, 2001; Pickart and Eddins, 2004). To test if ubiquitination of the N-terminal lysines (K70, 76) affects the stability of p33, we tested the half-life of p33 and its N-terminal mutants deficient in mono- and bi-ubiquitination (Fig. 6). After induction of expression of p33 and its derivatives from the GAL1 promoter, we turned off p33 mRNA synthesis by changing the media from galactose to glucose, followed by taking samples at given time points and Western blot analysis to measure the level of remaining p33 in yeast (Fig. 6). These experiments revealed that overall the half-life of p33 mutant deficient in mono- and bi-ubiquitination, such as K70,76R, was comparable to that of the wt p33 (Fig. 6), suggesting that the ability of wt p33 to be ubiquitinated within the N-terminal segment did not enhance its degradation.
Fig. 6.
Stability of p33 is not changed due to N-terminal deletions or mutations of lysines 70 and 76 involved in ubiquitination. Yeasts expressing wt p33HF or its mutants from plasmids with the inducible /repressible GAL1 promoter were grown in 2% galactose minimal media for 24 hours at 29°C to produce p33. Then, yeasts were subsequently transferred to 2% glucose minimal media and grown at 29°C to shut down p33 production. Total protein samples were taken at 0, 1, 2, 4 and 8 hours after the transfer to glucose and p33 accumulation was measured with Western blotting using anti-FLAG antibody. The data with K70,76R indicate that lysines 70 and 76 are unlikely to be involved in p33 stability/degradation.
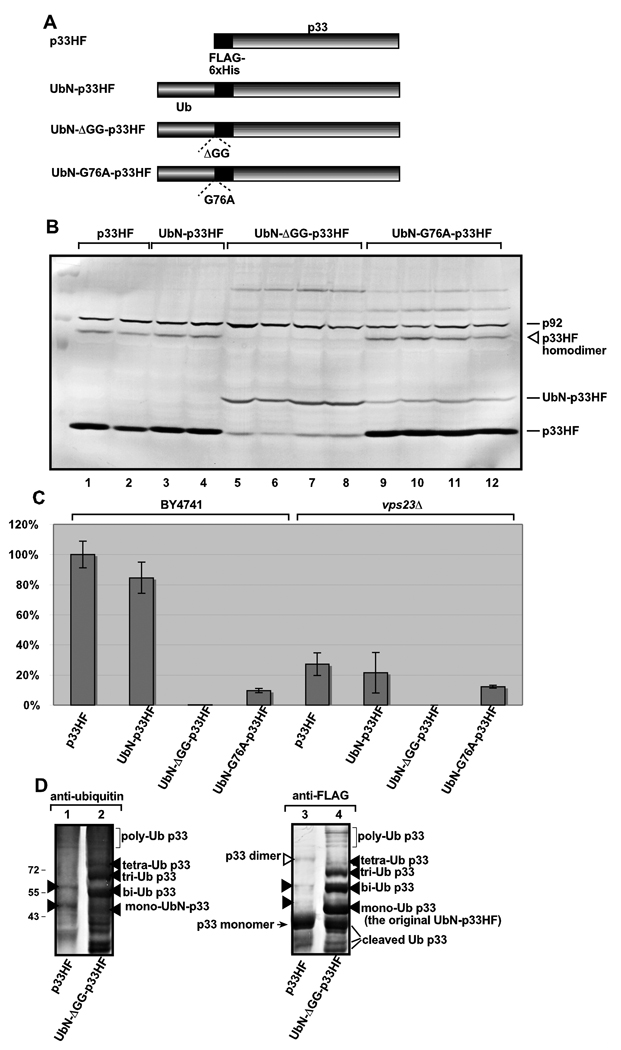
To further test the role and effect of p33 ubiquitination, we artificially introduced the 76 aa Ub to the N-terminus of p33. This approach mimics natural ubiquitination of lysines by covalently linking Ub to the target protein (Bossis et al., 2005; Morita and Sundquist, 2004). Expression of the resulting UbN-p33HF (carrying an N-terminal Ub plus 6×His and FLAG tags, Fig. 7A) resulted in wt-sized p33HF (Fig. 7B, compare lanes 3–4 with 1–2), suggesting that UbN-p33HF was processed rapidly and efficiently by a deubiquitinating protease (DUB). Replication of TBSV repRNA was only slightly decreased in wt yeast expressing UbN-p33HF (Fig. 7C) when compared with yeast expressing the wt p33HF. These data indicate that cleavage of UbN-p33HF by DUB is likely quick enough that the Ub tag did not affect the assembly of the viral replicase or did not result in increased degradation of p33. Overall, this result with the ubiquitination mimicking p33 mutant can be interpreted that deubiquitination of UbN-p33HF is rapid and efficient.
Fig. 7.
The effect of UbN-p33 chimeras on p33 accumulation, polyubiquitination and TBSV repRNA accumulation. (A) Schematic representation of the UbN-p33 chimeras used in this study. We have expressed p33HF derivatives carrying (i) a single ubiquitin-tag at the N-terminus that had the cleavable wt Ub sequence, (ii) two glycine deletions to inhibit its cleavage, (iii) or a G76A mutation to partly inhibit cleavage by DUBs in yeast. (B) Western-blot analysis of p33HF and UbN-p33 chimeras expressed in yeast using anti-FLAG antibody and total protein extract. (C) Accumulation of TBSV repRNA in wt or vps23Δ yeast co-expressing the shown p33HF derivative, p92pol and repRNA based on Northern blot analysis. (D) Possible polyubiqitination of the cleavage-deficient derivative of UbN-p33 chimera in yeast cells. Western blotting of FLAG-affinity purified p33HF was done with either anti-ubiquitin (left panel) or anti-FLAG (right panel) antibodies. Unlike the original p33HF (lane 1), the cleavage-deficient UbN-HFp33 chimera showed many different ubiquitinated p33 forms (marked with arrowheads), including possibly polyubiquitinated forms (bracketed) as shown. The lack of these higher MW forms in the control p33HF, as seen with the anti-FLAG antibody on the right, suggests that p33 does not efficiently accumulate the higher MW forms, but mostly produces the mono- and bi-ubiquitinated forms at detectable levels. Note that these data indicate that the mono- and bi-ubiquitinated forms of p33HF ubiquitinated at K70 and K76 are unlikely to be intermediates for polyubiquitinated forms.
When we expressed a mutated UbN-p33HF (UbN-ΔGG-p33HF, Fig. 7A) deficient in cleavage by DUB due to deletion of the critical G75 and G76 within the Ub sequence (Bamezai, Tate, and Breslow, 1989), then we observed the accumulation of the full-length UbN-ΔGG-p33HF, but to a reduced level (Fig. 7B, lanes 5–8). This indicated that UbN-ΔGG-p33HF could be less stable than wt p33HF or UbN-p33HF, possibly due to poly-ubiquitination and degradation by the 26S proteosome. Therefore, we tested the FLAG-affinity purified UbN-ΔGG-p33HF by Western blotting (Fig. 7D). Indeed, we observed the higher MW ubiquitinated products of UbN-ΔGG-p33HF, including possible poly-ubiquitinated forms (Fig. 7D, lanes 2 and 4). Thus, it is likely that UbN-ΔGG-p33HF goes through the proteosome degradation pathway, resulting in reduced accumulation of UbN-ΔGG-p33HF when compared with wt p33HF. Even more importantly, UbN-ΔGG-p33HF did not support the accumulation of TBSV repRNA in wt yeast (Fig. 7C), suggesting that high level of ubiquitination of p33 interferes with TBSV replication.
To further test the effect of high-level p33 ubiquitination on TBSV replication using a ubiquitination mimicking p33 chimera, we expressed an additional UbN-p33 chimera that was partially deficient in cleavage by DUB due to mutation of G76A within the Ub sequence (UbN-G76A-p33HF, Fig. 7A) (Hodgins, Ellison, and Ellison, 1992). Interestingly, UbN-G76A-p33HF was efficiently cleaved by DUB in wt yeast and only about 20% remained uncleaved (Fig. 7B, lanes 9–12). Yet, in spite of the presence of almost wt level p33 in yeast, UbN-G76A-p33HF supported the accumulation of TBSV repRNA at ~10% level when compared with the wt p33HF (Fig. 7C). Based on the Ub mimicry data, we propose that the increased level of p33 ubiquitination is inhibitory for TBSV replication.
We also tested whether TBSV replication was similarly affected by the above UbN-p33 mutants in vps23Δ yeast. These experiments revealed reduced replication of TBSV repRNA in vps23Δ yeast (Fig. 7C). Thus, increased level of p33 ubiquitination is inhibitory for TBSV replication even in vps23Δ yeast. However, the difference in TBSV replication supported by wt p33HF versus UbN-G76A-p33HF was less in vps23Δ yeast (~50% decrease) than in wt yeast, in which UbN-G76A-p33HF supported only 10% level of the replication supported by wt p33HF (Fig. 7C). This could be partly due to the increased binding of UbN-p33HF to Vps23p, which might be inhibitory during replicase assembly, in wt yeast. Overall, the use of ubiquitination mimicking p33 mutants revealed the complex and dynamic nature of ubiquitination in p33 functions.
DISCUSSION
Post-translational modifications of tombusvirus replication proteins are emerging as important features to extend and/or regulate the multiple functions of these viral proteins during tombusvirus infections. For example, phosphorylation of threonine and serine in the proximity of the RNA-binding region of p33 interfered with the ability of p33 to bind to the viral RNA (Shapka, Stork, and Nagy, 2005; Stork, Panaviene, and Nagy, 2005). It was proposed that phosphorylation of p33 could facilitate the release of the viral RNA products during replication and/or prevent the assembly of new viral replicases at the late stage of infection (Shapka, Stork, and Nagy, 2005; Stork, Panaviene, and Nagy, 2005).
Similar to phosphorylation, protein ubiquitination is also a reversible, dynamic process. Yeast and other eukaryotes code for a legion of enzymes, including E1, E2 and E3 enzymes involved in ubiquitination as well as DUBs responsible for removing the covalently-linked Ub from the client proteins (Lindner, 2007; Pickart, 2001; Pickart and Eddins, 2004; Roos-Mattjus and Sistonen, 2004). Since a small fraction of p33 replication protein becomes ubiquitinated in yeast (Fig. 1B) (Li et al., 2008), we suggest that p33 ubiquitination could be involved in TBSV replication. Although in vitro data support the role of Cdc34p E2 Ub-conjugating enzyme in p33 ubiquitination (Li et al., 2008), genome-wide screens for host factors affecting TBSV replication (Jiang et al., 2006; Panavas et al., 2005b) and proteomics screen for yeast proteins interacting with p33 identified additional yeast E3 Ub-ligases, such as Rsp5p and Bre1p, or E2 Ub-conjugating enzymes, such as Rad6p, which could also be involved in p33 ubiquitination (Barajas, Li, and Nagy, 2009; Li et al., 2008). The same screens also identified DUBs, including Ubp3p, Ubp10p, Ubp15p, and Doa4p (Jiang et al., 2006; Li et al., 2008; Panavas et al., 2005b). Altogether, the identification of multiple ubiquitination enzymes and DUBs from these screens indicates the possible complexity of p33 ubiquitination in regulation of TBSV replication.
We have identified two lysines, K70 and K76, in p33, which are mono- and bi-ubiquitinated in yeast (Fig. 1–Fig 2). Mutations of these lysines, which are known to interfere with ubiquitination, reduced TBSV repRNA replication in yeast (Fig. 4) and affected its ability to interact with Vps23p (Fig. 3A), but not with Ssa1p HSP70 (Fig. 3B). Based on these data, we propose that one of the functions of p33 ubiquitination is to assist the recruitment of Vps23p ESCRT-I protein for TBSV replication. Since the interaction of p33 with Vps23p is also dependent on the late domain-like sequence in p33, it seems that p33 uses molecular mimicry by sharing features with Vps27p ESCRT-0 protein (i.e., late domain sequence-assisted binding to Vps23p) and the membrane-bound client proteins (mono-ubiquitination-assisted binding to Vps23p). Interestingly, the two regions, the N-terminal K70 and K76 and the 'late-domain-like' PSVP motif, are partly redundant in facilitating p33 binding to Vps23p and together they might optimize the binding of p33 to Vps23p. These features of p33 are important for tombusvirus replication in both yeast and plant cells, since combined mutations of the ubiquitination region and the late-domain-like sequence in p33 significantly inhibited tombusvirus accumulation (Fig. 5) without affecting p33 stability (Fig. 6).
Vps23p is a key ESCRT-I adaptor protein in the endosome pathway by binding to ubiquitinated cargo proteins and recruiting additional ESCRT-I/II/III proteins to the endosomes, which are involved in membrane deformation/invagination and vesicle formation (Bowers and Stevens, 2005; Hurley and Emr, 2006; Katzmann, Odorizzi, and Emr, 2002; Malerod and Stenmark, 2009; Saksena et al., 2009; Slagsvold et al., 2006). The endosome pathway is a major protein-sorting pathway in eukaryotic cells, which down regulates plasma membrane proteins via endocytosis; and sorts newly synthesized membrane proteins from trans-Golgi vesicles to the endosome, lysosome or the plasma membrane.
In addition, enveloped retroviruses, filo-, arena-, rhabdo-, and paramyxoviruses usurp the endosome pathway by redirecting ESCRT proteins to the plasma membrane, leading to budding and fission of the viral particles from infected cells (Morita and Sundquist, 2004; Perlman and Resh, 2006). Interestingly, HIV and other retroviruses co-opt the ESCRT pathway by interacting directly with Tsg101, the human homologue of Vps23p. This interaction requires the P(T/S)AP-type late domain sequence in the gag protein (Morita and Sundquist, 2004). Thus, different viruses seem to exploit the ESCRT proteins through co-opting Vps23p via direct protein-protein interaction based on ubiquitination and late-domain motifs.
Recruitment of Vps23p by p33 might also be important for tombusvirus replication in the plant host. This is based on the inhibitory effect of mutations within the PSVP motif and the lysines critical for ubiquitination in p33 on tombusvirus replication in plant protoplasts and leaves (Fig. 5). Since the separate mutagenesis of the PSVP motif and the critical lysines in p33 had only modest effect, whereas the combined mutations in both regions of p33 had more significant inhibitory effect on tombusvirus RNA accumulation, we propose that these regions play redundant roles, likely by recruiting Vps23p and other ESCRT factors. The observed differences in the extent of the inhibitory effect on tombusvirus replication caused by p33 mutations in yeast and in plant cells could be due to several factors, including (i) the presence of these mutations in both p33 and p92 proteins due to the overlapping expression strategy in plant cells, whereas only p33 carries the mutations in yeast cells since p33 and p92 are expressed separately; and (ii) the presence of two diverse VPS23 genes in plants (based on the sequenced Arabidopsis genome) (Winter and Hauser, 2006), which could make the interaction with p33 more complex than in yeast with a single VPS23 gene.
In addition to the proposed role of p33 ubiquitination in recruitment of Vps23p, ubiquitination could play a role in stability/degradation of p33. However, the data obtained during this work is not supportive of this model. First, mutations of K70 and K76 or their deletions in p33 did not significantly affected the half-life of p33 in yeast (Fig. 6), suggesting that these lysine residues or their ubiquitination are not critical for p33 degradation. Second, expression of a UbN-p33 chimera mimicking ubiquitination did not lead to reduced accumulation of p33 (Fig. 7B) and the UbN-p33 chimera was quickly processed to p33 by cleaving off Ub by DUBs. Indeed, TBSV repRNA replication was not affected by this chimera (Fig. 7C), suggesting that the cleavage of Ub from UbN-p33 is rapid and the generated p33 is active in supporting replication. In contrast, the partly cleavable or cleavage-deficient UbN-p33 chimeras inhibited TBSV repRNA replication to great extent (Fig. 7C). This suggests that high level ubiquitination of p33 is inhibitory, possibly by increased recruitment of Vps23p or other processes. Indeed, we likely observed polyubiquitination of the cleavage-deficient UbN-p33 chimera (Fig. 7D) and its amount was also greatly decreased in yeast (Fig. 7C), suggesting that the cleavage-deficient UbN-p33 chimera could be degraded by the host. Overall, these experiments with the UbN-p33 chimeras suggest that ubiquitination of p33 might play a complex role in p33 functions and during tombusvirus replication.
In summary, based on the data presented here, we propose that ubiquitination of a small percentage of p33 (Ub-p33) in combination with the late domain-like sequence of p33 facilitates the recruitment of the adaptor protein Vps23p. This in turn, might lead to the additional recruitment of other ESCRT proteins. The temporary recruitment of these factors might facilitate the optimal assembly of the replicase complex on membranous surfaces. Future experiments will address how ubiquitination of p33 and Vps23p-p33 interaction affect the structure/function of the TBSV replicase in infected cells.
Materials and Methods
Yeast strains and expression plasmids
The single-gene deletion yeast strains were purchased from Open Biosystems. pGBK-33HFH (Serva and Nagy, 2006) is a multicopy plasmid expressing a 6×His and FLAG-tagged CNV p33 under ADH1 promoter.
To make constructs expressing ubiquitin-tagged p33, ubiquitin was amplified by PCR from plasmid YEp105 (Ellison and Hochstrasser, 1991) with primers #2071 and #2072. To introduce mutations G76A and Δ75–76 (ΔGG), which interfere partially or largely with Ub cleavage by DUBs, PCRs were done with primers #2071/#2125 and #2071/#2126. The obtained PCR products were digested with NcoI and inserted into plasmid pGBK-33HFH in frame with the p33HF ORF.
To produce pGBK-33HFH-Δ1–75 and pGBK-33HFH-Δ1–150, PCRs were performed with primer pairs #1547/#992B and #633/992B, respectively, to amplify the truncated ORFs. The products were digested with BamHI and PstI and cloned into pGBK-33HFH. The lysine to alanine mutants of p33 were made using a two round PCR strategy. For the first round of PCR, portions of p33 ORF were amplified using the primers described in Table 1 (PCR1 and PCR2). PCR products were digested with NheI, ligated together and amplified with primers #424/#992B. These products were digested with BamHI and PstI and cloned into similarly digested pGBK-33HFH. To mutate lysine K5, we performed PCR using primers #2099 /#992B, followed by BamHI and PstI digestion and cloning into pGBK-33HFH.
Table 1.
The cloning strategy and the primers used for lysine-to-alanine mutagenesis in p33:
| First round PCR | PCR1 | PCR2 | Second round PCR |
|---|---|---|---|
| pGBK-33HFH-K5A | 2099 + 992B | - | - |
| pGBK-33HFH-K11A | 424 + 2101 | 2100 + 992B | 424 + 992B |
| pGBK-33HFH-K12A | 424 + 2103 | 2102 + 992B | 424 + 992B |
| pGBK-33HFH-K47A | 424 + 2105 | 2104 + 992B | 424 + 992B |
| pGBK-33HFH-K64A | 424 + 2107 | 2106 + 992B | 424 + 992B |
| pGBK-33HFH-K70A | 424 + 2109 | 2108 + 992B | 424 + 992B |
| pGBK-33HFH-K76A | 424 + 2111 | 2110 + 992B | 424 + 992B |
| pGBK-33HFH-Δ70–76 | 424 + 2109 | 2110 + 992B | 424 + 992B |
To produce pGBK-33HFH-ΔPSVP (shown as PSVP-TS in the text), the PSVP sequence in the p33 ORF was mutated to TS representing a SpeI site using a PCR-based strategy. Two portions of p33 ORF were amplified with primers #424/#2394 and #2393/#992B. The PCR products were digested with SpeI, ligated and amplified with primers #424 and #992B. The product was digested with BamHI and PstI and ligated into similarly digested pGBK-33HFH. To produce the lysine to arginine mutants of p33 N-proximal lysines (Fig. 2), a two-round mutagenic PCR using overlapping primers was employed. The primers and templates used for each mutant are summarized in Table 2. The final PCR products were digested with BamHI and PstI and cloned into pGBK-33HFH.
Table 2.
The cloning strategy and the primers used for lysine-to-arginine mutagenesis in p33:
| First round PCR | Second round PCR | ||||
|---|---|---|---|---|---|
| template | primers | template | primers | ||
| PCR1 | pGBK-33HFH | 424 + 2170 | K70R | PCR1 + PCR3 | 424 + 992B |
| PCR2 | pGBK-33HFH | 424 + 2172 | K76R | PCR2 + PCR4 | 424 + 992B |
| PCR3 | pGBK-33HFH | 2169 + 992B | K70,76R | PCR1 + PCR4 | 424 + 992B |
| PCR4 | pGBK-33HFH | 2171 + 992B | K70R/ΔPSVP | PCR1 + PCR5 | 424 + 992B |
| PCR5 | pGBK-33HFH- ΔPSVP |
2169 + 992B | K76R/ΔPSVP | PCR2 + PCR6 | 424 + 992B |
| PCR6 | pGBK-33HFH- ΔPSVP |
2171 + 992B | K70,76R/ΔPSVP | PCR1 + PCR6 | 424 + 992B |
Analysis of ubiquitination of p33 mutants
We used yeast strain InvSc1, which was transformed with pGBK-33HFH or derivatives, pYES2/NT/C (Invitrogen) and pYEp105 (Ellison and Hochstrasser, 1991). Analysis of CNV p33 ubiquitination was performed as described (Li et al., 2008). p33HF was detected using a monoclonal mouse anti-FLAG antibody (Sigma; 1/5,000 dilution) and an AP-conjugated anti-mouse antibody (1/5,000). As a negative control, we transformed yeast with pESC-His, pYEP105 and pYC-HF expressing a 6×His-FLAG peptide (Serva and Nagy, 2006).
Analysis of protein-protein interactions using the split-ubiquitin assay
This assay was based on the Dualmembrane kit 3 (Dualsystems biotech). The CNV p33 constructs used in this study are based on pGAD-BT2-N-His33, which has been described previously (Li et al., 2008). PCRs were performed with primers #2231 / #2262 using the following templates: plasmids pGBK-33HFH, pGBK-33HFH-ΔPSVP, pGBK-33HFH-K70,76R and pGBK-33HFH-K70,76R/ΔPSVP (Table 2). The PCR products were digested with XbaI and NcoI and ligated into pGAD-BT2-N-His33 digested with SpeI and NcoI, giving as a result plasmids pGAD-BT2-NX-His33, pGAD-BT2-NX-His33-ΔPSVP, pGAD-BT2-NX-His33-K70,76R and pGAD-BT2-NX-His33-K70,76R/ΔPSVP. To generate pPR-N-UEV, PCR was done using yeast genomic DNA and primers #2252 / #2292. The PCR product was digested with EcoRI and NheI and cloned into similarly digested pPR-N-RE (Li et al., 2008). NMY51 was transformed with pGAD-BT2-NX-His33, or derivatives and pPR-N-UEV or pPR-N-SSA1 (Li et al., 2008). Transformed colonies were selected in Trp-/Leu- plates. Yeast colonies were re-suspended in a small volume of water and streaked onto Trp-/Leu-/His-/Ade- plates to score interactions. Interactions were scored as described (Li et al., 2008).
Analysis of TBSV repRNA replication in yeast
Replication assay in yeast was performed as described (Panavas et al., 2008). For replication studies, S. cerevisiae strain BY4741 or vps24Δ yeast strains were co-transformed with plasmids pGBK-33HFH or derivatives, pGAD-His92 and pYC-DI72sat (Panavas and Nagy, 2003). Replication assay was performed as described (Panavas et al., 2005a). Accumulation of DI-72 repRNA was measured by Northern blot using RNA probes complementary to region III–IV of DI-72(+) and to 18S ribosomal RNA (Panavas et al., 2005a).
Analysis of tombusvirus replication in N. benthamiana protoplasts and leaves
The infectious RNA transcripts of CNV and its derivatives were obtained from plasmids linearized with SmaI and transcribed with T7 RNA polymerase (Nagy and Pogany, 2000). N. benthamiana protoplasts were produced from callus culture (Panaviene, Baker, and Nagy, 2003). The protoplasts were transfected by electroporation as described (Panaviene, Baker, and Nagy, 2003). Total RNA was extracted and analyzed by Northern blot using an RNA probe complementary to CNV RNA (Panaviene, Baker, and Nagy, 2003).
For these studies, plasmid pK2/M5-20Kstop (Rochon, 1991) was modified by PCR in order to remove the BamHI site from the polylinker and introduce it before the start codon of p33. PCRs were performed with primer pairs #2600/#2614 and #424/#2613, respectively. Both PCR products were digested with BamHI and PstI and ligated together, generating pK2/M5-20Kstop-(Ba). Fragments of p33 ORF consisting of sequences between the unique BamHI to EagI sites from plasmids pGBK-33HFH-ΔPSVP, pGBK-33HFH-K70,76R and pGBK-33HFH-K70,76R/ΔPSVP were removed by BamHI/EagI digestion and inserted into similarly digested pK2/M5-20Kstop-(Ba). The obtained plasmids were linearized with SmaI and transcribed with T7 RNA polymerase to generate infectious RNA transcripts.
To create plasmid pGD-35S-20Kstop, PCR was done using primers #532 / #720 and pK2/M5 20Kstop (Rochon, 1991) as template. The PCR product was digested with BamHI/XhoI and ligated into BamHI/XhoI digested pGD (Goodin et al., 2002). Plasmid pGD-35S-20Kstop-(Ba) was created by using PCR with primers #1803 and #952 and pK2/M5-20Kstop-(Ba) as a template. The obtained PCR product was digested with BglII and EcoRI and ligated into BamHI/EcoRI digested pGD-35S-20Kstop. To create pGD-35S-20Kstop-(Ba)K70,76R/ΔPSVP, the plasmid pK2/M5–20Kstop-(Ba)K70,76R/ΔPSVP was digested with BamHI/EcoRI and the portion comprising p33 and part of p92 was cloned into pGD-35S-20Kstop-(Ba) digested with BamHI/EcoRI. Plasmids were transformed into Agrobacterium tumefaciens strain C58C1. Agro-infiltrations of N. benthamiana and analysis of viral RNA accumulation in the infiltrated leaves 3.5 days after infiltration were done as described (Cheng et al., 2007; Wang and Nagy, 2008). Total RNA was extracted and analyzed by Northern blot using an RNA probe complementary to CNV (Panaviene, Baker, and Nagy, 2003).
Stability of p33 mutants deficient in ubiquitination
The CNV p33 mutants were cloned under the GAL1 promoter in plasmid pESC/DI72/His33 (Pathak, Sasvari, and Nagy, 2008). P33 mutants were amplified from plasmids pGBK-33HFH-Δ1–75, pGBK-33HFH-Δ1–150, pGBK-33HFH-Δ70–76 and pGBK-33HFH-K70,76R with primer pairs of #1547/#1403, #633/#1403, #424/#1403 and #424/#1403, respectively. The obtained PCR products were digested with BamHI and XhoI to clone into pESC/DI72/His33. The obtained plasmids pESC/DI72/His33-Δ1–75, pESC/DI72/His33-Δ1–150, pESC/DI72/His33-Δ70–76 and pESC/DI72/His33-K70,76R were transformed into S. cerevisiae InvSc1. Transformed yeasts were grown in minimal media supplemented with 2% galactose to induce p33 expression for 24h at 29°C. Cultures were centrifuged and resuspended in minimal media with 2% glucose to inhibit p33 expression. Samples were taken at different time points after switch to glucose media. p33 accumulation was analyzed by Western blotting using anti-6×His primary antibody and anti-mouse conjugated to alkaline phophatase secondary antibody.
Table 3.
List of primers used in this study
| 0633 | CGACGGATCCATGTACGCTACCCTACCCAGGGA |
| 0720 | CCCGCTCGAGGGGCTGCATTTCTGCAATGTTC |
| 0952 | CCCGCTCGAGTCATGCTACGGCGGAGTCAAGGA |
| 0992B | GAGCTGCAGCTATTTCACACCAAGGGA |
| 1402 | GCGGCAGATCTTACCATGGGGGGTTCTCA |
| 1403 | GCCGCTCGAGCTATTTCACACCAAGGGACTCA |
| 1547 | CGACGGATCCATGAAACGCAGGAGGGTTGGTGA |
| 1803 | GGCGAGATCTGGAAATTCTCCAGGATTTCTC |
| 2071 | GTCGGAATTCGCCATGGGGCAGATCTTCGTCAAGACG |
| 2072 | GAGCCCATGGATCCACCACCTCTTAGTCTTAAG |
| 2099 | GCTCGGATCCGATACCATCGCTAGCAGGATGCTGTGGCC |
| 2100 | GCTCGCTAGCAAAGAAATTTTTGTTGGCACGTTCG |
| 2101 | GCTCGCTAGCAGGCCACAGCATCCTC |
| 2102 | GCTCGCTAGCGAAATTTTTGTTGGCACGTTCGC |
| 2103 | GCTCGCTAGCCTTAGGCCACAGCATCCTC |
| 2104 | GCTCGCTAGCATCGAGAATAACACTGACAG |
| 2105 | GCTCGCTAGCCCCTGTCCTCATGTATCTC |
| 2106 | GCTCGCTAGCACAGATTGTGCAGCCAAATG |
| 2107 | GCTCGCTAGCGAGAAGTTCAACGATGAAATTTC |
| 2108 | GCTCGCTAGCTGGGAATGGTTCATGAAACG |
| 2109 | GCTCGCTAGCGGCTGCACAATCTGTCTTG |
| 2110 | GCTCGCTAGCCGCAGGAGGGTTGGTG |
| 2111 | GCTCGCTAGCCATGAACCATTCCCATTTGG |
| 2125 | GAGCGGATCCCATGGAAGCACCTCTTAGTCTTAAGAC |
| 2126 | GAGCGGATCCCATGGATCTTAGTCTTAAGACAAGATG |
| 2169 | ACAGATTGTGCAGCCAGATGGGAATGG |
| 2170 | CATGAACCATTCCCATCTGGCTGCAC |
| 2171 | GGGAATGGTTCATGAGACGCAGGAGG |
| 2172 | CCTCCTGCGTCTCATGAACCATTCC |
| 2231 | GCGTCTAGAATGGGAGGTTCTCATCATCATC |
| 2252 | GCCGAATTCATGTCTGCAAACGGCAAGATCTCTG |
| 2262 | GTCGCCATGGAGGCCTCTATTTCACACCAAGGGAC |
| 2292 | GCTCCTCGAGTTAGCTAGCCAATGGCGGTTTTAGGTGTGG |
| 2393 | GCCACTAGTAAGAAAGGCTTGCTACTGC |
| 2394 | CGGACTAGTCCTTGGAACCCTGATGTTG |
| 2600 | CGGAACATTGCAGAAATGCAG |
| 2613 | CGGCTGCAGCCCGGGCTGCATTTCTGC |
| 2614 | GCCGGATCCCATGTCGTCGTTTACTGGAAG |
ACKNOWLEDGEMENTS
We thank Drs. Judit Pogany and Zhenghe Li for critical reading of the manuscript and for very helpful suggestions. The authors thank Dr. Scott D. Emr, Cornell University, for yeast strains and plasmids. This work was supported by NIH-NIAID and by the Kentucky Tobacco Research and Development Center at the University of Kentucky, awarded to PDN. D. Barajas was supported in part by a postdoctoral fellowship from the Spanish Ministry of Education and Science.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bamezai S, Tate S, Breslow E. Inhibition of ubiquitin-dependent proteolysis by des-Gly-Gly-ubiquitin: implications for the mechanism of polyubiquitin synthesis. Biochem Biophys Res Commun. 1989;162(1):89–94. doi: 10.1016/0006-291x(89)91966-9. [DOI] [PubMed] [Google Scholar]
- Barajas D, Li Z, Nagy PD. The Nedd4-Type Rsp5p Ubiquitin Ligase Inhibits Tombusvirus Replication by Regulating Degradation of the p92 Replication Protein and Decreasing the Activity of the Tombusvirus Replicase. J Virol. 2009;83(22):11751–11764. doi: 10.1128/JVI.00789-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossis G, Malnou CE, Farras R, Andermarcher E, Hipskind R, Rodriguez M, Schmidt D, Muller S, Jariel-Encontre I, Piechaczyk M. Down-regulation of c-Fos/c-Jun AP-1 dimer activity by sumoylation. Mol Cell Biol. 2005;25(16):6964–6979. doi: 10.1128/MCB.25.16.6964-6979.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers K, Stevens TH. Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 2005;1744(3):438–454. doi: 10.1016/j.bbamcr.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Brigati C, Giacca M, Noonan DM, Albini A. HIV Tat, its TARgets and the control of viral gene expression. FEMS Microbiol Lett. 2003;220(1):57–65. doi: 10.1016/S0378-1097(03)00067-3. [DOI] [PubMed] [Google Scholar]
- Cereseto A, Manganaro L, Gutierrez MI, Terreni M, Fittipaldi A, Lusic M, Marcello A, Giacca M. Acetylation of HIV-1 integrase by p300 regulates viral integration. Embo J. 2005;24(17):3070–3081. doi: 10.1038/sj.emboj.7600770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CP, Jaag HM, Jonczyk M, Serviene E, Nagy PD. Expression of the Arabidopsis Xrn4p 5'-3' exoribonuclease facilitates degradation of tombusvirus RNA and promotes rapid emergence of viral variants in plants. Virology. 2007;368(2):238–248. doi: 10.1016/j.virol.2007.07.001. [DOI] [PubMed] [Google Scholar]
- Drugeon G, Jupin I. Stability in vitro of the 69K movement protein of Turnip yellow mosaic virus is regulated by the ubiquitin-mediated proteasome pathway. J Gen Virol. 2002;83(Pt 12):3187–3197. doi: 10.1099/0022-1317-83-12-3187. [DOI] [PubMed] [Google Scholar]
- Ellison MJ, Hochstrasser M. Epitope-tagged ubiquitin. A new probe for analyzing ubiquitin function. J Biol Chem. 1991;266(31):21150–21157. [PubMed] [Google Scholar]
- Evans MJ, Rice CM, Goff SP. Phosphorylation of hepatitis C virus nonstructural protein 5A modulates its protein interactions and viral RNA replication. Proc Natl Acad Sci U S A. 2004;101(35):13038–13043. doi: 10.1073/pnas.0405152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Aizaki H, He JW, Lai MM. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J Virol. 2004;78(7):3480–3488. doi: 10.1128/JVI.78.7.3480-3488.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Tu H, Shi ST, Lee KJ, Asanaka M, Hwang SB, Lai MM. Interaction with a ubiquitin-like protein enhances the ubiquitination and degradation of hepatitis C virus RNA-dependent RNA polymerase. J Virol. 2003;77(7):4149–4159. doi: 10.1128/JVI.77.7.4149-4159.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodin MM, Dietzgen RG, Schichnes D, Ruzin S, Jackson AO. pGD vectors: versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J. 2002;31(3):375–383. doi: 10.1046/j.1365-313x.2002.01360.x. [DOI] [PubMed] [Google Scholar]
- Hericourt F, Blanc S, Redeker V, Jupin I. Evidence for phosphorylation and ubiquitinylation of the turnip yellow mosaic virus RNA-dependent RNA polymerase domain expressed in a baculovirus-insect cell system. Biochem J. 2000;349(Pt 2):417–425. doi: 10.1042/0264-6021:3490417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgins RR, Ellison KS, Ellison MJ. Expression of a ubiquitin derivative that conjugates to protein irreversibly produces phenotypes consistent with a ubiquitin deficiency. J Biol Chem. 1992;267(13):8807–8812. [PubMed] [Google Scholar]
- Hurley JH, Emr SD. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu Rev Biophys Biomol Struct. 2006;35:277–298. doi: 10.1146/annurev.biophys.35.040405.102126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubiec A, Jupin I. Regulation of positive-strand RNA virus replication: the emerging role of phosphorylation. Virus Res. 2007;129(2):73–79. doi: 10.1016/j.virusres.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Serviene E, Gal J, Panavas T, Nagy PD. Identification of essential host factors affecting tombusvirus RNA replication based on the yeast Tet promoters Hughes Collection. J Virol. 2006;80(15):7394–7404. doi: 10.1128/JVI.02686-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonczyk M, Pathak KB, Sharma M, Nagy PD. Exploiting alternative subcellular location for replication: tombusvirus replication switches to the endoplasmic reticulum in the absence of peroxisomes. Virology. 2007;362(2):320–330. doi: 10.1016/j.virol.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Kapoor M, Zhang L, Ramachandra M, Kusukawa J, Ebner KE, Padmanabhan R. Association between NS3 and NS5 proteins of dengue virus type 2 in the putative RNA replicase is linked to differential phosphorylation of NS5. J Biol Chem. 1995;270(32):19100–19106. doi: 10.1074/jbc.270.32.19100. [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Odorizzi G, Emr SD. Receptor downregulation and multivesicular-body sorting. Nat Rev Mol Cell Biol. 2002;3(12):893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- Kim SH, Palukaitis P, Park YI. Phosphorylation of cucumber mosaic virus RNA polymerase 2a protein inhibits formation of replicase complex. Embo J. 2002;21(9):2292–2300. doi: 10.1093/emboj/21.9.2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Barajas D, Panavas T, Herbst DA, Nagy PD. Cdc34p Ubiquitin-Conjugating Enzyme Is a Component of the Tombusvirus Replicase Complex and Ubiquitinates p33 Replication Protein. J Virol. 2008;82(14):6911–6926. doi: 10.1128/JVI.00702-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Pogany J, Panavas T, Xu K, Esposito AM, Kinzy TG, Nagy PD. Translation elongation factor 1A is a component of the tombusvirus replicase complex and affects the stability of the p33 replication co-factor. Virology. 2009;385(1):245–260. doi: 10.1016/j.virol.2008.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner HA. Deubiquitination in virus infection. Virology. 2007;362(2):245–256. doi: 10.1016/j.virol.2006.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malerod L, Stenmark H. ESCRTing membrane deformation. Cell. 2009;136(1):15–17. doi: 10.1016/j.cell.2008.12.029. [DOI] [PubMed] [Google Scholar]
- Morita E, Sundquist WI. Retrovirus budding. Annu Rev Cell Dev Biol. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- Nagy PD, Pogany J. Partial purification and characterization of Cucumber necrosis virus and Tomato bushy stunt virus RNA-dependent RNA polymerases: similarities and differences in template usage between tombusvirus and carmovirus RNAdependent RNA polymerases. Virology. 2000;276(2):279–288. doi: 10.1006/viro.2000.0577. [DOI] [PubMed] [Google Scholar]
- Nagy PD, Pogany J. Yeast as a model host to dissect functions of viral and host factors in tombusvirus replication. Virology. 2006;344(1):211–220. doi: 10.1016/j.virol.2005.09.017. [DOI] [PubMed] [Google Scholar]
- Panavas T, Hawkins CM, Panaviene Z, Nagy PD. The role of the p33:p33/p92 interaction domain in RNA replication and intracellular localization of p33 and p92 proteins of Cucumber necrosis tombusvirus. Virology. 2005a;338(1):81–95. doi: 10.1016/j.virol.2005.04.025. [DOI] [PubMed] [Google Scholar]
- Panavas T, Nagy PD. Yeast as a model host to study replication and recombination of defective interfering RNA of Tomato bushy stunt virus. Virology. 2003;314(1):315–325. doi: 10.1016/s0042-6822(03)00436-7. [DOI] [PubMed] [Google Scholar]
- Panavas T, Serviene E, Brasher J, Nagy PD. Yeast genome-wide screen reveals dissimilar sets of host genes affecting replication of RNA viruses. Proc Natl Acad Sci U S A. 2005b;102(20):7326–7331. doi: 10.1073/pnas.0502604102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panavas T, Serviene E, Pogany J, Nagy PD. Genome-wide screens for identification of host factors in viral replication. Methods Mol Biol. 2008;451:615–624. doi: 10.1007/978-1-59745-102-4_41. [DOI] [PubMed] [Google Scholar]
- Panaviene Z, Baker JM, Nagy PD. The overlapping RNA-binding domains of p33 and p92 replicase proteins are essential for tombusvirus replication. Virology. 2003;308(1):191–205. doi: 10.1016/s0042-6822(02)00132-0. [DOI] [PubMed] [Google Scholar]
- Panaviene Z, Panavas T, Nagy PD. Role of an internal and two 3'-terminal RNA elements in assembly of tombusvirus replicase. J Virol. 2005;79(16):10608–10618. doi: 10.1128/JVI.79.16.10608-10618.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaviene Z, Panavas T, Serva S, Nagy PD. Purification of the cucumber necrosis virus replicase from yeast cells: role of coexpressed viral RNA in stimulation of replicase activity. J Virol. 2004;78(15):8254–8263. doi: 10.1128/JVI.78.15.8254-8263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak KB, Sasvari Z, Nagy PD. The host Pex19p plays a role in peroxisomal localization of tombusvirus replication proteins. Virology. 2008;379(2):294–305. doi: 10.1016/j.virol.2008.06.044. [DOI] [PubMed] [Google Scholar]
- Perlman M, Resh MD. Identification of an intracellular trafficking and assembly pathway for HIV-1 gag. Traffic. 2006;7(6):731–745. doi: 10.1111/j.1398-9219.2006.00428.x. [DOI] [PubMed] [Google Scholar]
- Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695(1–3):55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- Pogany J, Nagy PD. Authentic replication and recombination of Tomato bushy stunt virus RNA in a cell-free extract from yeast. J Virol. 2008;82(12):5967–5980. doi: 10.1128/JVI.02737-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogany J, Stork J, Li Z, Nagy PD. In vitro assembly of the Tomato bushy stunt virus replicase requires the host Heat shock protein 70. Proc Natl Acad Sci U S A. 2008;105(50):19956–19961. doi: 10.1073/pnas.0810851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogany J, White KA, Nagy PD. Specific binding of tombusvirus replication protein p33 to an internal replication element in the viral RNA is essential for replication. J Virol. 2005;79(8):4859–4869. doi: 10.1128/JVI.79.8.4859-4869.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochon DM. Rapid de novo generation of defective interfering RNA by cucumber necrosis virus mutants that do not express the 20-kDa nonstructural protein. Proc Natl Acad Sci U S A. 1991;88(24):11153–11157. doi: 10.1073/pnas.88.24.11153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos-Mattjus P, Sistonen L. The ubiquitin-proteasome pathway. Ann Med. 2004;36(4):285–295. doi: 10.1080/07853890310016324. [DOI] [PubMed] [Google Scholar]
- Saksena S, Wahlman J, Teis D, Johnson AE, Emr SD. Functional reconstitution of ESCRT-III assembly and disassembly. Cell. 2009;136(1):97–109. doi: 10.1016/j.cell.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serva S, Nagy PD. Proteomics analysis of the tombusvirus replicase: Hsp70 molecular chaperone is associated with the replicase and enhances viral RNA replication. J Virol. 2006;80(5):2162–2169. doi: 10.1128/JVI.80.5.2162-2169.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serviene E, Jiang Y, Cheng CP, Baker J, Nagy PD. Screening of the yeast yTHC collection identifies essential host factors affecting tombusvirus RNA recombination. J Virol. 2006;80(3):1231–1241. doi: 10.1128/JVI.80.3.1231-1241.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serviene E, Shapka N, Cheng CP, Panavas T, Phuangrat B, Baker J, Nagy PD. Genome-wide screen identifies host genes affecting viral RNA recombination. Proc Natl Acad Sci U S A. 2005;102(30):10545–10550. doi: 10.1073/pnas.0504844102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapka N, Stork J, Nagy PD. Phosphorylation of the p33 replication protein of Cucumber necrosis tombusvirus adjacent to the RNA binding site affects viral RNA replication. Virology. 2005;343(1):65–78. doi: 10.1016/j.virol.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Si X, Gao G, Wong J, Wang Y, Zhang J, Luo H. Ubiquitination is required for effective replication of coxsackievirus B3. PLoS ONE. 2008;3(7):e2585. doi: 10.1371/journal.pone.0002585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slagsvold T, Pattni K, Malerod L, Stenmark H. Endosomal and non-endosomal functions of ESCRT proteins. Trends Cell Biol. 2006;16(6):317–326. doi: 10.1016/j.tcb.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Stork J, Panaviene Z, Nagy PD. Inhibition of in vitro RNA binding and replicase activity by phosphorylation of the p33 replication protein of Cucumber necrosis tombusvirus. Virology. 2005;343(1):79–92. doi: 10.1016/j.virol.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Thaminy S, Miller J, Stagljar I. The split-ubiquitin membrane-based yeast two-hybrid system. Methods Mol Biol. 2004;261:297–312. doi: 10.1385/1-59259-762-9:297. [DOI] [PubMed] [Google Scholar]
- Usami Y, Popov S, Popova E, Inoue M, Weissenhorn W, H GG. The ESCRT pathway and HIV-1 budding. Biochem Soc Trans. 2009;37(Pt 1):181–184. doi: 10.1042/BST0370181. [DOI] [PubMed] [Google Scholar]
- Vigerust DJ, Shepherd VL. Virus glycosylation: role in virulence and immune interactions. Trends Microbiol. 2007;15(5):211–218. doi: 10.1016/j.tim.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RY, Nagy PD. Tomato bushy stunt virus Co-Opts the RNA-Binding Function of a Host Metabolic Enzyme for Viral Genomic RNA Synthesis. Cell Host Microbe. 2008;3(3):178–187. doi: 10.1016/j.chom.2008.02.005. [DOI] [PubMed] [Google Scholar]
- Wang RY, Stork J, Nagy PD. A key role for heat shock protein 70 in localization and insertion of the tombusvirus replication proteins to intracellular membranes. J Virol. 2009 doi: 10.1128/JVI.02313-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RY, Stork J, Pogany J, Nagy PD. A temperature sensitive mutant of heat shock protein 70 reveals an essential role during the early steps of tombusvirus replication. Virology. 2009;394(1):28–38. doi: 10.1016/j.virol.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White KA, Nagy PD. Advances in the molecular biology of tombusviruses: gene expression, genome replication, and recombination. Prog Nucleic Acid Res Mol Biol. 2004;78:187–226. doi: 10.1016/S0079-6603(04)78005-8. [DOI] [PubMed] [Google Scholar]
- Winter V, Hauser MT. Exploring the ESCRTing machinery in eukaryotes. Trends Plant Sci. 2006;11(3):115–123. doi: 10.1016/j.tplants.2006.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]