Abstract
Neurodegenerative diseases are often associated with dysfunction in protein quality control. The endoplasmic reticulum (ER), a key site for protein synthesis, senses stressful conditions by activating the unfolded protein response (UPR). Here we report the creation of a novel mouse model where GRP78/BiP, a major ER chaperone and master regulator of UPR, is specifically eliminated in the Purkinje cells (PCs). GRP78 depleted PCs activate UPR including induction of GRP94, PDI, CHOP and GADD34, feedback suppression of eIF2α phosphorylation and apoptotic cell death. In contrast to current models of protein misfolding where abnormal accumulation of ubiquitinated protein is prominent, cytosolic ubiquitin staining is dramatically reduced in GRP78 null PCs. Ultrastructural evaluation reveals that the ER shows prominent dilatation with focal accumulation of electron-dense material within the ER. The mice show retarded growth and severe motor coordination defect by week 5 and cerebellar atrophy by week 13. Our studies uncover a novel link between GRP78 depletion and reduction in cytosolic ubiquitination and establish a novel mouse model of accelerated cerebellar degeneration with basic and clinical applications.
Keywords: GRP78/BiP, conditional knockout mice, Purkinje cell survival, unfolded protein response, neurodegeneration
Introduction
Neurodegenerative diseases are often associated with the formation of toxic intracellular protein aggregates; therefore, it is crucial to understand factors that regulate their formation and degradation.1 The unfolded protein response (UPR) is an evolutionarily conserved mechanism to allow cells to adapt to stress targeted to the endoplasmic reticulum (ER).2 Thus, when the protein load exceeds the protein folding capacity of the ER, the UPR signaling pathways are induced. ER protein quality control is critical for cerebellar Purkinje cell survival3–5 and is linked to the Marinesco-Sjogren syndrome in humans.6,7 ER chaperone GRP78, also referred to as BiP/HSPA5, is a central regulator of ER function and the UPR.2,8,9 Tissue culture studies revealed that upon ER stress, GRP78/BiP is released from ER transmembrane sensors (PERK, ATF6 and IRE1) resulting in the triggering of the UPR.10 However, the direct role of GRP78 in regulating the UPR in vivo and in neurodegeneration is unknown, in part because of lethality of Grp78 −/− embryos.11 Purkinje cells (PCs) are the principal neurons and the sole output of the computational circuitry of the cerebellar cortex, providing signals required for balance, motor coordination and cognitive learning.12 The recent discovery that mutations in SIL1, which encodes a nucleotide exchange factor for GRP78, cause progressive multisystem disorder including cerebellar ataxia in humans6,7 and protein accumulation and Purkinje cell death in the SIL1 deficient woozy mutant mice3 provides the first hint that ER protein quality control in terminally differentiated neurons is critical for their survival. Interestingly, in the woozy mutant mice, GRP78 is upregulated in the PCs,3 and other co-chaperones such as GRP170 (also known as ORP150) and P58IPK (also known as DNAJc3) may serve as alternative nucleotide exchange factors for GRP78.13–16 To test directly the role of GRP78 in the UPR in vivo and in neurodegeneration and the mechanism for its putative protective function, here we created a Purkinje cell-specific knockout mouse model of GRP78. Our studies reveal that complete elimination of GRP78 in PCs leads to specific modulation of the UPR and reduction in cytosolic ubiquitination. The GRP78 conditional knockout mice developed Purkinje cell degeneration by 4.5 week, associating with severe motor coordination defect by week 5 and cerebellar atrophy by week 13, thereby establishing the requirement of GRP78 for Purkinje cell survival and normal cerebellar function.
Results
Accelerated Purkinje cell degeneration and cerebellar atrophy in the Grp78 conditional knockout mice
The Grp78 Purkinje cell specific knockout mice (abbreviated below as F/-; pc-Cre) were generated by crossing Grp78 F/- mice17 with L7Cre-2 transgenic mice.18 In the Grp78 (−) allele, exons 5, 6 and 7 encoding the ATPase domain and peptide-binding domain essential for the chaperone function of GRP78 are deleted (Figure 1a), and no truncated protein was detected.11 The genotypes of mice harboring the various Grp78 alleles and the pc-Cre allele were determined by PCR (Figure 1b) and cohorts of mice with the following genotypes: (1) Grp78 +/+, (2) Grp78 F/F, (3) Grp78 F/F; pc-Cre, (4) Grp78 F/-, and (5) Grp78 F/-; pc-Cre were monitored for growth and development.
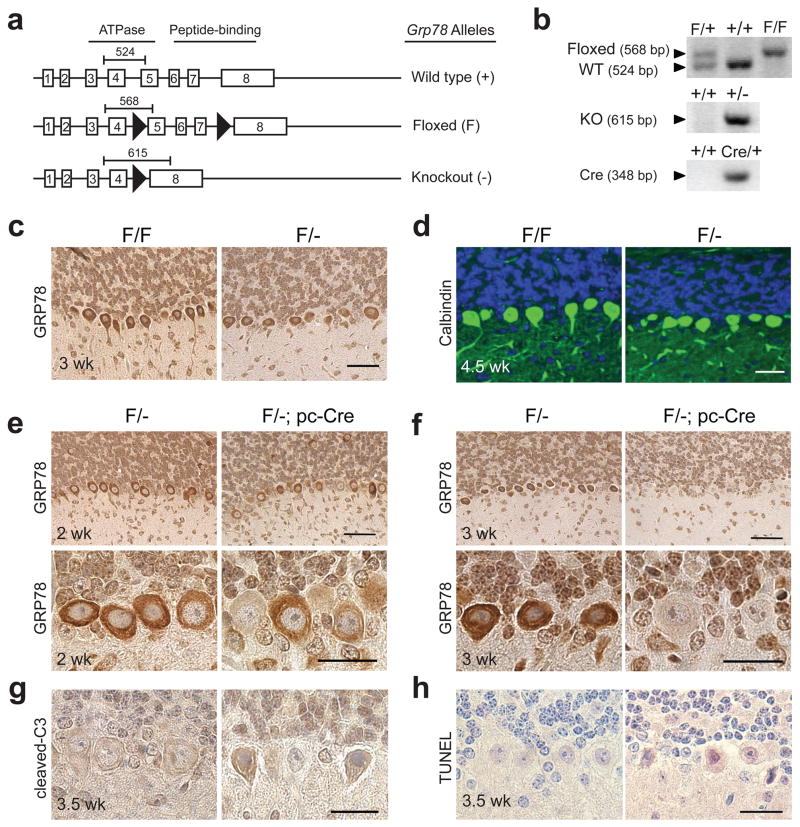
Figure 1.
Generation of the Grp78 Purkinje cell-specific knockout mice. (a) Schematic drawing for the Grp78 wild-type (wt) allele (+), floxed allele (F) and knockout (KO) allele (−). The black arrowheads indicate loxP sites. The bars with numbers indicate the size and position of PCR products. (b) Representative genotyping results of Grp78 wt, floxed, and KO alleles, and Cre transgene with expected PCR product size. (c) Representative immunohistochemical (IHC) staining for GRP78 shows similar expression levels of GRP78 in the Purkinje cells (PCs) of Grp78 F/F mice and their F/-littermates at 3 wk of age. The staining was performed on the sagittal sections of the cerebellum of Grp78 F/F and F/- mice. Scale bar represents 50 μm. (d) Representative immunofluorescence (IF) staining for calbindin shows there is no Purkinje cell degeneration in Grp78 F/- mice compared with their F/F littermates at 4.5 wk of age. Green: Calbindin. Blue: DAPI. Scale bar represents 50 μm. (e–f) Representative IHC staining for GRP78 on the cerebellum of Grp78 F/- and their F/-; pc-Cre littermates at 2 wk (e) and 3 wk (f) of age. Scale bars represent 50 μm (upper panel) and 25 μm (lower panel). (g–h) Representative IHC staining for cleaved caspase-3 (g) and TUNEL (h) on PCs of Grp78 F/- and their F/-; pc-Cre littermates at 3.5 wk old. Scale bars represent 25μm. For panels c–h, n=3 mice per group were examined.
In agreement with previous studies that the Grp78 heterozygosity does not affect mouse development and normal organ function,11,19 the F/- mice are phenotypically normal (data not shown) and their PCs are intact and showed similar GRP78 staining as the F/F mice (Figure 1c and d). In the F/-; pc-Cre mice, Cre recombinase was expressed specifically in the PCs after postnatal day 6, causing deletion of exon 5 to exon 7 of Grp78. Immunohistochemical analysis revealed that at 2 week of age, GRP78 staining was partially reduced in about 10% of the PCs of the F/-; pc-Cre mice compared to the F/- control; and by week 3, GRP78 was undetectable in about 90% of the PCs (Figure 1e and f). Caspase-3 activation and positive TUNEL staining were detected in about 10% of the PCs at week 3.5 (Figure 1g and h), suggesting onset of Purkinje cell apoptosis.
Staining for the Purkinje cell specific marker calbindin confirmed partial loss of the PCs at week 4.5 (Figure 2a), and by week 13 all ten lobules of the cerebellum showed extensive Purkinje cell degeneration (Figure 2b). Correlating with Purkinje cell loss, cerebellar atrophy was evident in the F/-; pc-Cre mice by week 13 (Figure 2c). Quantification of the age-dependent Purkinje cell loss is summarized in Figure 2d. Collectively, these results establish that GRP78 is required for PC survival and cerebellum integrity.
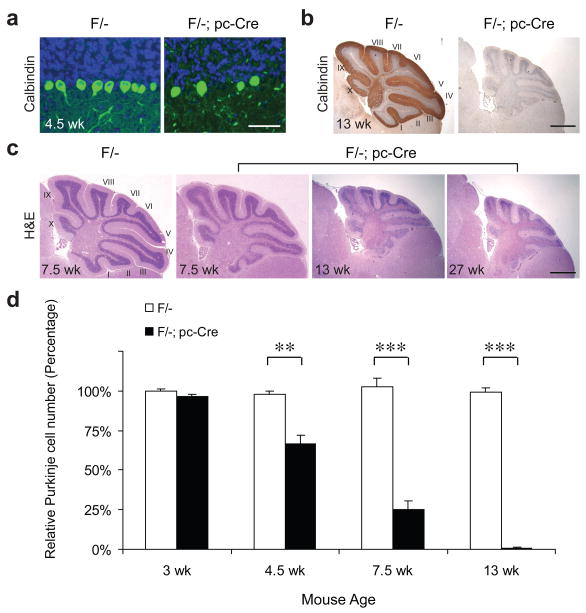
Figure 2.
The Grp78 Purkinje cell-specific knockout mice show accelerated Purkinje cell degeneration and cerebellar atrophy. (a) Representative immunofluorescence (IF) staining for calbindin shows Purkinje cell degeneration in the Grp78 F/-; pc-Cre mice compared with their F/- littermates at 4.5 wk of age. Green: Calbindin. Blue: DAPI. Scale bar represents 50 μm. (b) Representative immunohistochemical (IHC) staining for calbindin on the cerebellum sections of Grp78 F/- mice and their F/-; pc-Cre littermates at 13 wk old. Sections were counterstained with hematoxylin. Cerebellar lobules are indicated by Roman numerals. Scale bar represents 1.5 mm. (c) Hematoxylin and eosin (H&E) staining on the cerebellum sections of Grp78 F/- mice and their F/-; pc-Cre littermates at indicated ages, demonstrating the cerebellar atrophy. Scale bar represents 1.5 mm. (d) Age-dependent Purkinje cell degeneration in the Grp78 F/-; pc-Cre mice compared with their F/- littermates. The number of Purkinje cells determined by H&E staining was quantitated within the same areas of cerebellar sections and plotted against age of the mice. The number of PCs in 3 wk old F/- mouse cerebellum was set as 100%. Data are presented as mean±s.e.m. **p≤0.01, ***p≤0.001 (Student’s t-test). For all panels, n=3 mice per group were examined.
Severe motor coordination defects in the Grp78 Purkinje cell knockout mice
While the F/-; pc-Cre mice were born with the expected Mendelian ratio and survive as well as F/F or F/- siblings, both male and female mice developed postnatal growth retardation when compared to the F/- mice (Figure S1), such that there was significant decrease in overall body size (Figure 3a) and fasting body weight (Figure 3b). Food consumption of the F/-; pc-Cre mice was also less than the F/- mice, which might in part account for the lower body weight (Figure 3c). By week 5.5, both male and female F/-; pc-Cre mice exhibited motor defects indicative of cerebellar ataxia. They had difficulty walking in a straight line; even standing still the mice could not maintain balance and appeared “tipsy” (see Supplementary Information video). Rota rod test confirmed severely impaired motor coordination ability in both male and female cohorts of F/-; pc-Cre mice, consistently over multiple trials (Figure 3d). Footprint recording further revealed ataxic walking pattern (Figure 3e), with reduced stride/width ratio compared to the F/- mice (Figure 3f). Interestingly, none of these phenotypes were detected in mice with Purkinje cell specific knockout of GRP94, another major ER chaperone (Figure S2). Thus, not only that GRP78 function cannot be compensated by GRP94, GRP78 is uniquely required for Purkinje cell survival.
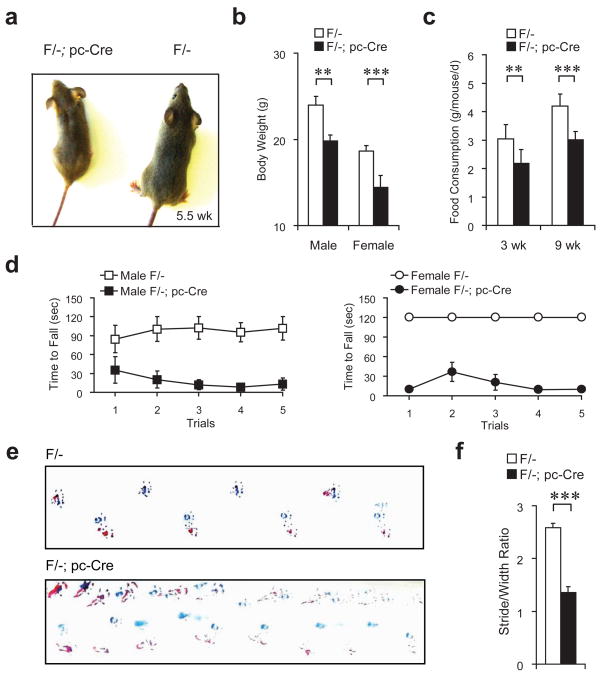
Figure 3.
Behavioral abnormalities of the Grp78 conditional knockout mice. (a) A representative male Grp78 F/-; pc-Cre mouse and its F/- littermate at 5.5 wk of age. (b) Fasting body weight of 10 wk old Grp78 F/- mice and their F/-; pc-Cre littermates, both male and female. n=10 mice per group. (c) Food consumption of male Grp78 F/- mice and their F/-; pc-Cre male littermates at 3 and 9 wk of age. The food intakes for each individual mouse were measured for seven consecutive days. n=4 mice per group. (d) Rota rod test of Grp78 F/- mice (open symbols) and their F/-; pc-Cre littermates (closed symbols) at 5.5 wk old. The time to fall off the rod (rotating at 30 rpm) at each trial is recorded. n=4 to 5 mice per group. p-value=0.016 (male mice) and p-value=0.008 (female mice) (randomization test). (e) Representative footprint records of Grp78 F/-male mice and their F/-; pc-Cre male littermates at 5.5 wk old. The forelimbs and hindlimbs were painted as blue and red respectively. (f) The ratios of stride lengths to paw-based widths from the footprint test. n=4 mice per group. Data in panel (b), (c), (d) and (f) are all presented as mean±s.e.m. **p≤0.01, ***p≤0.001 (Student’s t-test).
Modulation of UPR signaling pathways in GRP78 null Purkinje cells
The ability to genetically inactivate both Grp78 alleles in the PCs provides a unique experimental system to test directly how depletion of GRP78 affects the UPR in terminally differentiated neurons where ER protein quality control is central to their survival.20,21 One of the pro-survival branches of the UPR upregulates ER chaperones such as GRP78 and GRP94 and folding enzymes such as PDI to alleviate protein misfolding.22 We observed that, GRP94 and PDI were upregulated in almost all PCs of 3 week old F/-; pc-Cre mice (Figure 4a). Immunostaining revealed that CHOP, an ER stress-inducible transcription factor,23 was induced in about 5% of the PCs in 2 week old F/-; pc-Cre mice, however, by 3 week old, about 90% of the PCs were CHOP positive (Figure 4b). ER stress activates the PERK signaling pathway, which leads to transient global translational arrest through phosphorylation of the initiation factor eIF2α, however, this can be inhibited by a feedback mechanism mediated by GADD34, a downstream target of CHOP.24 Corresponding to CHOP induction and GADD34 upregulation in the PCs of the F/-; pc-Cre mice (Figure 4c), eIF2α phosphorylation, which was observed at 2 week, was dramatically reduced by 3 week with the overall level of eIF2α unaffected (Figure 4d and e). As summarized in Figure 4f, quantitation of GRP94, PDI, GADD34, phosphorylated eIF2α and eIF2α immunostaining indicates that GRP78 depletion significantly upregulates GRP94, PDI, and GADD34 and suppresses eIF2α phosphorylation without affecting eIF2α level in PCs of F/-; pc-Cre mice compared with their F/- littermates. In contrast, knockout of Grp94 did not induce UPR signaling pathways in mouse embryonic stem cells (mESCs) (Figure S3), correlating with no neurological phenotypes in Grp94 Purkinje cell specific knockout mice. Under thapsigargin-induced ER stress, Grp94−/− and +/+ mESCs exhibited no difference in phosphorylation of PERK and eIF2α, induction of GADD34 and splicing of XBP-1, further supporting that GRP94 did not affect UPR signaling (Figure S3).
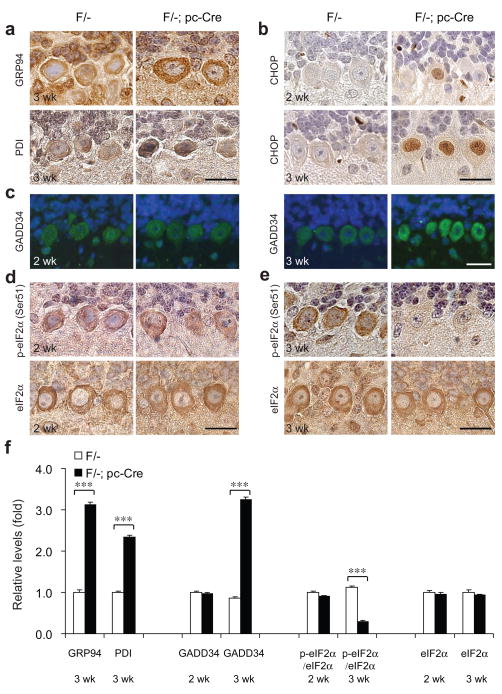
Figure 4.
Modulation of UPR pathways in the Purkinje cells of the Grp78 conditional knockout mice. (a–b) Representative immunohistochemical (IHC) staining for GRP94 and PDI (a) and CHOP (b) on the cerebellum sections of Grp78 F/- mice and their F/-; pc-Cre littermates at indicated ages. (c) Representative immunofluorescence (IF) staining for GADD34 on the cerebellum sections of Grp78 F/- mice and their F/-; pc-Cre littermates at 2 and 3 wk of age. Green: GADD34. Blue: DAPI. (d–e) Representative IHC staining for p-eIF2α (Ser51) and eIF2α on the cerebellum sections of Grp78 F/- mice and their F/-; pc-Cre littermates at 2 and 3 wk of age. IHC and IF staining were counterstained with hematoxylin and DAPI, respectively. All scale bars represent 25 μm. For panels (a) to (e), n=3 mice per group were examined. (f) Quantitation of the IHC and IF stainings shown in panel (a) to (e). The relative protein levels were quantitated by NIH software ImageJ. The mean level of staining in each group was determined by 40 randomly selected areas within Purkinje cells on the IHC or IF stained sections. The level of p-eIF2α was normalized to eIF2α. For GRP94 and PDI, the staining of PCs in F/- mice at 3 wk old was set as 1. For GADD34, p-eIF2α and eIF2α, the staining of PCs in F/- mice at 2 wk old was set as 1. Data are presented as mean±s.e.m. ***p≤0.001 (Student’s t-test).
Reduction of cytosolic ubiquitin staining and ER expansion in GRP78 null Purkinje cells
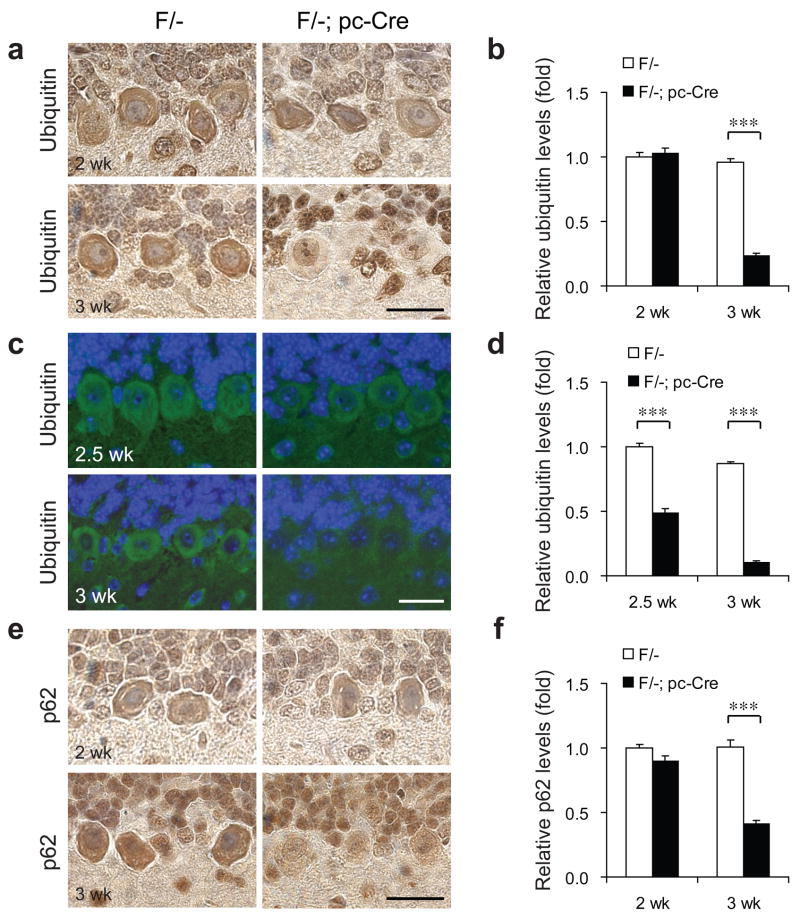
In the mammalian ER-associated protein degradation (ERAD) machinery, malfolded proteins released from calnexin are captured by a recognition complex containing EDEM, then passed on to GRP78 and PDI before unfolding and transport to the cytosol through the retrotranslocation machinery, polyubiquitinated by the E1-E2-E3 system, and degraded by the proteasome.25,26 For both the F/- and F/-; pc-Cre mice, ubiquitin staining was detectable in both the nucleus and cytoplasm in the 2 week old cerebellar PCs (Figure 5a). Strikingly, despite upregulation of GRP94, PDI, GADD34 and CHOP, and readily detectable level of other cellular proteins including calbindin, eIF2α, and β-actin (Figures 2a, 4 and data not shown), cystolic ubiquitin staining was nearly undetectable in the PCs of F/-; pc-Cre mice by 3 week (Figure 5a). Quantitation of the relative ubiquitin levels shows significantly reduced level of ubiquitin in F/-; pc-Cre mice at 3 week old compared with their F/- littermates (Figure 5b). The immunohistochemical staining results were confirmed by immunofluorescence analysis, which also showed that while ubiquitin staining was observed in the F/- PCs, its level was reduced in the PCs of F/-; pc-Cre mice by week 2.5 and barely detectable by week 3 (Figure 5c). The quantitation of ubiquitin immunofluorescence indicates the gradual loss of ubiquitin level in the PCs of F/-; pc-Cre mice compared with their F/- littermates (Figure 5d). Reduction of ubiquitin staining was further confirmed in GRP78 null prostate epithelial cells in another Grp78 conditional knockout mouse model (Figure S4).17 Since GRP78 knockout in these cells does not affect prostate function or gross morphology,17 reduction in ubiquitin staining as a result of GRP78 depletion is unlikely the consequence of cells undergoing the cell death process.
Figure 5.
Reduction of protein ubiquitination and p62 level in the Purkinje cells of the Grp78 conditional knockout mice. (a) Representative immunohistochemical (IHC) staining for ubiquitin on the cerebellum sections of Grp78 F/- mice and their F/-; pc-Cre littermates at 2 and 3 wk of age. (b) Quantitation of ubiquitin immunostaining, with the level of ubiquitin staining in the PCs of F/- mice at 2 wk old set as 1. (c) Representative immunofluorescence (IF) staining for ubiquitin on the cerebellum sections of Grp78 F/-mice and their F/-; pc-Cre littermates at 2.5 and 3 wk of age. (d) Quantitation of ubiquitin immunofluorescence with the level of ubiquitin staining in the PCs of F/- mice at 2.5 wk old set as 1. (e) Representative IHC staining for p62 on the cerebellum sections of Grp78 F/- mice and their F/-; pc-Cre littermates at 2 and 3 wk of age. (f) Quantitation of p62 immunostaining with level of p62 staining in the PCs of F/- mice at 2 wk old set as 1. All relative levels shown in panel (b), (d) and (f) were quantitated by NIH software ImageJ. The mean level of staining in each group was determined by 40 randomly selected areas within the Purkinje cells on the IHC or IF stained sections. For all panels, n=3 mice per group. Data in panel (b), (d) and (f) are all presented as mean±s.e.m. ***p≤0.001 (Student’s t-test). All scale bars represent 25 μm.
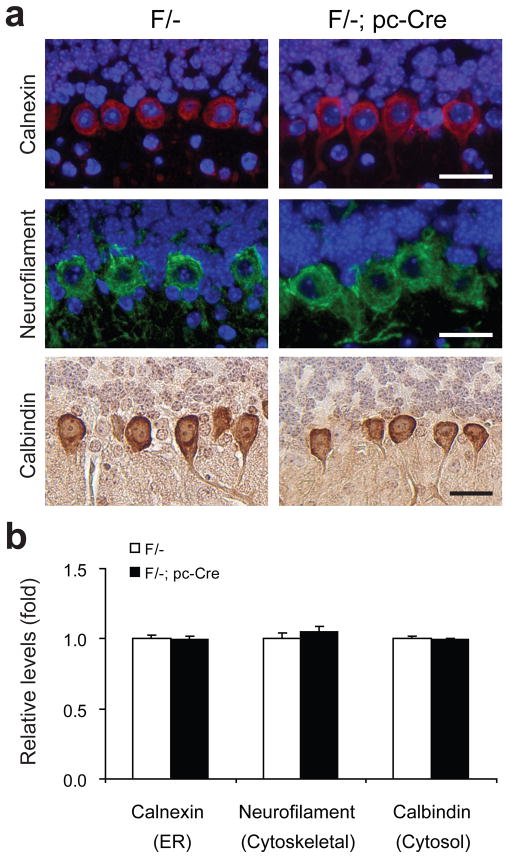
p62/SQSTM1 is a cytosolic protein which binds to polyubiquitinated proteins and targets them to the autophagy machinery for degradation.27 The polyubiquitinated protein aggregates stabilize p62.28 Consistent with the reduction in ubiquitinated protein level, p62 level was reduced in the 3 week old GRP78 null PCs, as compared to PCs from either 2 or 3 week old F/- mice, or 2 week old F/-; pc-Cre mice where GRP78 expression was still detected (Figure 5e). Quantitation of p62 immunostaining levels in F/- and F/-; pc-Cre mice showed significant reduction by week 3 (Figure 5f). In contrast, PCs from F/- and F/-; pc-Cre mice showed similar levels of calnexin, which is an ER protein, and neurofilament and calbindin, which are cytoskeletal and cytosolic proteins, respectively (Figure 6).
Figure 6.
Similar expression levels of calnexin, non-phosphorylated neurofilament and calbindin in the Purkinje cells of wild type and mutant mice. (a) Representative immunofluorescence (IF) for calnexin and neurofilament and immunohistochemical (IHC) staining for calbindin on the cerebellum sections of Grp78 F/- mice and their F/-; pc-Cre littermates at 3 wk of age. All scale bars represent 25 μm. (b) Quantitation of the IF and IHC stainings shown in panel (a). The relative protein levels were quantitated by NIH software ImageJ. The mean level of staining in each group (n=3 mice per group) was determined by 40 randomly selected areas within the Purkinje cells on the IF or IHC stained sections. For all the quantitation, the staining of PCs in F/- mice at 3 wk old was set as 1. Data are presented as mean±s.e.m.
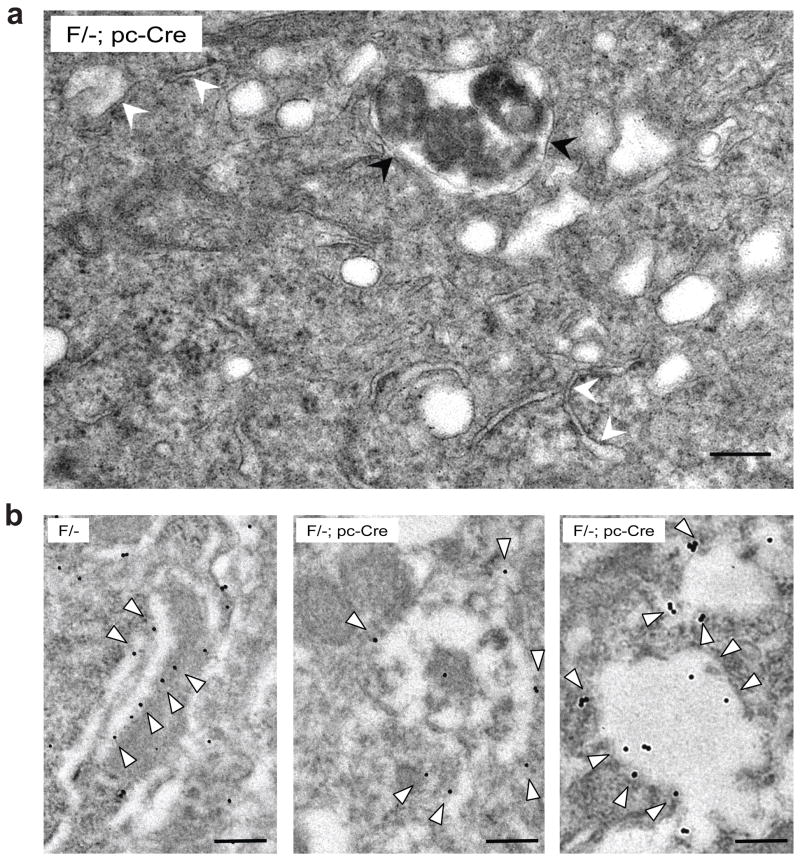
Transmission electron microscopy (EM) of PCs from 3.5 week old F/-; pc-Cre mice further revealed that the ER showed prominent dilatation with accumulation of flocculant material, and in some cases, focal accumulation of electron-dense material (Figure 7a and S5). No double membrane bound inclusions were seen that would be suggestive of autophagy. ImmunoEM analysis using antibody against calnexin, an ER transmembrane protein, showed that in the F/- mice, typical tubular ER structures were labeled; whereas in the F/-; pc-Cre mice, expanded ER structures enclosing electron dense or flocculent materials were labeled (Figure 7b). This confirms the ER identity of the expanded structures in Purkinje cells of the F/-; pc-Cre mice. The alterations in UPR pathways and pathology observed in the Grp78 F/-; pc-Cre mice are summarized in Figure 8.
Figure 7.
Altered ER structure in GRP78 null PCs. (a) Representative area of electron micrograph of a 3.5 wk old F/-; pc-Cre mouse cerebellum shows ER expansion and electron dense aggregates inside the ER in the PCs. White arrowheads indicate typical ER structure undergoing expansion. Grossly expanded ER contains diffuse accumulation of flocculent material, and in some cases, electron dense aggregates are prominent inside the ER (indicated by black arrowheads). Scale bar represents 0.5 μm. (b) Examples of electron micrographs showing immunogold labeling. Purkinje cells from 4.5 wk old F/- mice and their F/-; pc-Cre littermates were labeled for the ER marker calnexin. The 15 nm immunogold particles were indicated by open arrows. All scale bars represent 0.2 μm.
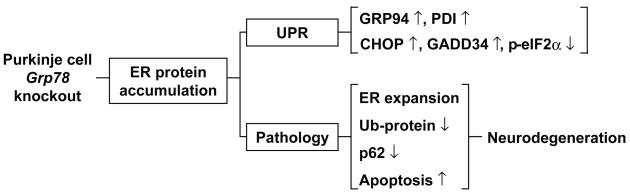
Figure 8.
Summary model of modulation of the UPR and pathology resulting from Grp78 conditional knockout in the PCs. GRP78 depletion in PCs leads to ER protein accumulation, abnormal ER expansion, block of protein ubiquitination and activation of the UPR and apoptosis, leading to neurodegeneration.
Discussion
The creation of the floxed Grp78 mouse model provides a valuable tool whereby the function of GRP78, a major molecular chaperone and key rheostat for ER homeostasis,29 can be examined in the context of mammalian development and in specific adult tissues and organs. Previous studies showed that homozygous knockout of Grp78 in mouse embryos results in proliferative defect and apoptosis of the inner cell mass, resulting in lethality at E3.5.11 Nonetheless, specific knockout of Grp78 in mouse prostate epithelial cells does not affect their viability17 and in tissue culture cell lines, knockdown of GRP78 by siRNA only induces apoptosis in 2 out of 7 human cell lines being tested.30 Thus, the requirement of GRP78 in adult organs and tissues remains to be determined. Here, through the creation of F/-; pc-Cre mouse where Grp78 is specifically knockout in the terminally differentiated, non-proliferating PCs and examination of the resulting mouse phenotypes, several novel observations are made. As evidenced in the phenotypically normal F/- mice, one functional allele of Grp78 is sufficient for PC survival and cerebellum integrity, as these mice maintain normal motor function up to the age 10 months. Interestingly, for mice over 10 months, we observed that while the female F/- mice maintain normal motor function, the male F/- mice exhibits mild signs of motor deficiency based on rotor rod tests (our unpublished results), suggesting an age-dependent requirement for GRP78 in maintaining cerebellum function in heterozygous males which warrants further confirmation and investigation.
Upon knockout of both alleles of Grp78 in the PCs, our studies revealed that the mice show growth retardation, Purkinje cell apoptosis, ataxia and severe defect in motor coordination by week 5 and cerebral atrophy by week 13, directly demonstrating that GRP78 is required for survival of PCs and cerebellum integrity. Nonetheless, there are distinct differences between the Grp78 F/-; pc-Cre mice (referred below as the “tipsy” mice) and the SIL1 deficient “woozy” mice which also suffered from ER stress. A likely explanation is that SIL1 is a nucleotide exchange factor for GRP78 but is not the only one. In the GRP78 conditional knockout mouse model, Purkinje cell degeneration starts at week 4.5 and all ten lobules degenerate simultaneously and completely by 3 months, revealing that GRP78 function cannot be substituted by any other factor in all ten lobules. In contrast, onset of Purkinje cell degeneration in the woozy mice starts at 3 months and PCs in lobule X and caudal lobule IX are intact even at 1 year of age and older.3 The differences in Purkinje cell death kinetics and surviving lobule pattern suggest that there are other factors to maintain GRP78 function in the PCs of the woozy mice. Consistent with earlier reports that GRP170/ORP150 can serve as nucleotide exchange factor for GRP78,13,14 recently, it is discovered that brain-expression of the GRP170 transgene is able to fully suppress the Purkinje cell death phenotype caused by a SIL1 mutation (S. Ackerman, personal communication). Furthermore, the PCs of the woozy mice have highly elevated levels of GRP78 compared with wild-type control due to ER stress. Thus, despite SIL1 disruption, the residual GRP78 function in the PCs of the woozy mice could mitigate the pathology and rapid loss of cerebellum function due to complete lack of GRP78 in the tipsy mice.
Previous demonstration that GRP78 depletion results in ER stress signaling was derived from ER stress titration or siRNA studies in tissue culture cells.10,30,31 The ability to completely eliminate GRP78 in the PCs through genetic knockout in the tipsy mice offers a unique opportunity to interrogate how a non-proliferating, terminally differentiated neuron responds to the complete loss of this UPR regulator. While it is not possible to perform biochemical analysis since PCs constitute only a single cell layer in the cerebellum, the large size of the PC with distinct nuclear and cytosolic compartments enabled us to use immunostaining techniques to determine the status UPR kinetically corresponding to the gradual depletion of GRP78 from these cells following activation of the cre-recombinase about 1 week after birth. We noted that it took about 2 week for GRP78 to be completely depleted from the PCs. As compensatory measure to GRP78 deficiency, ER chaperone GRP94 and folding enzyme PDI are upregulated, consistent with previous reports in Grp78 heterozygous mouse embryo fibroblasts11 and in established cell lines where GRP78 was knockdown by siRNA.30,31 Also, as expected, CHOP is induced, confirming that GRP78 is a dominant suppressor of CHOP induction.32 While eIF2α level was not changed in the PCs of the tipsy mice, eIF2α phosphorylation was substantially reduced in the GRP78 null PCs. One contributing factor could be the upregulation of GADD34, which is a downstream target of CHOP and dominant inhibitor of eIF2α phosphorylation. Unfortunately, using the commercial available antibodies, we were unable to detect in tissue sections the phosphorylation level of PERK, which is an upstream regulator of eIF2α phosphorylation (data not shown). While further studies are required to determine whether alterations in the chaperone balance and the UPR markers in the GRP78 null PCs contribute to the pathology of the Grp78 F/-; pc-Cre mice, here we showed that knockout of Grp94 has no effect on ER stress signaling, correlating with no PC loss and no neurological phenotype.
An unusual observation in the PCs of the tipsy mice is that the level of cytosolic ubiquitin staining is dramatically reduced as GRP78 is depleted from the PCs. This is not a cell type specific or cell death associated observation as this was also observed in GRP78 depleted viable prostate epithelial cells in another Grp78 conditional knockout model.17 While we cannot exclude the possibility that PCs without GRP78 result in dramatic decrease in the cytosolic protein substrates for ubiquitination, the same cells show robust levels of GRP94, PDI, calnexin, GADD34, CHOP, eIF2α, calbindin, neurofilament and β-actin. A plausible explanation is that GRP78 is required for ER protein retrotranslocation to be ubiquitinated in the cytosol. In yeast, GRP78 facilitates the export of mutant proteins to the cytosol for degradation33; likewise, in mammalian cells, GRP78 stably binds to unfolded proteins and delivers them to the ER associated protein degradation system (ERAD).34 The molecular mechanism of GRP78 involvement is further clarified through the recent discovery that EDEM, ERdj5 and GRP78 form a functionally important ERAD complex.35 ERdj5 functions as a disulfide reductase which cleaves the disulfide bonds of misfolded proteins. ERdj5 binds to the ATP-bound form of GRP78 and activates ATP hydrolysis of GRP78, resulting in dissociation of GRP78 from ERdj5. In turn the ADP-bound form of GRP78 binds the protein substrates and holds them in a dislocation-competent state until they are transferred to the retrotranslocation channel in the ER membrane, for export to the cytosol and subsequent ubiquitination and degradation. Consistent with this model, ultrastructural images of the GRP78 null PCs show the accumulation of electron dense material inside the expanded ER, but not outside the ER. Nonetheless, the tipsy mouse model does not allow us to test this mechanism biochemically. Thus, the requirement of GRP78 in protein ubiquitination remains to be determined. Additionally, whether other functions of GRP78, such as binding of ER Ca2+, blocking the activation of ER associated pro-apoptotic factors such as BIK and caspase-722,36,37 and its requirement for stress-induced autophagy,31 contribute to Purkinje cell survival awaits further investigation. Besides the cerebellum, GRP78 is likely to be important for other critical neuronal functions as it is recently reported that knock-in mice expressing a mutant secreted form of GRP78 display disordered layer formation in the cerebral cortex and cerebellum, resulting in a neurological phenotype of reeler-mutant like malformation,38 which is linked to mental disorders.39 There is also accumulating evidence for age dependent decrease in GRP78 expression or activity in specific organs and tissues, including the brain, suggesting that aging defects may result from GRP78 depletion in senescent cells.40
In summary, our studies establish the Grp78 floxed mice as valuable tools for studying the physiological role of GRP78 and the UPR in specific organs, as exemplified for the creation of the tipsy mice. The latter differs from other models of Purkinje cell death since only the PCs, but not other neurons, are directly targeted. Considering that PCs are part of the core circuitry in the motor learning process, the tipsy mice provide a unique model for behavioral studies of Purkinje cell specific function. Further, the accelerated cerebellar degeneration in these mice will facilitate the development of therapeutic agents combating ER dysfunction in neurodegeneration and other diseases. Another exciting preclinical application is that this mouse model will provide an accelerated experimental system to access stem cell transplantation for rescue of the neurological phenotypes.
Materials and Methods
Generation of Purkinje cell (PC) specific and prostate specific Grp78 conditional knockout mice
The detailed information on generation of the Grp78 F/F, F/- mice and the prostate specific Grp78 conditional knockout mice was described previously.17 In brief, Grp78 T/+ mice11 carrying the targeted allele (T) were mated with the EIIA-Cre transgenic mice, and generated offspring carrying the floxed (F) or knockout (−) allele (Figure 1). The Purkinje cell specific Cre (pc-Cre) transgenic mice (line L7Cre-2) were obtained from the Jackson Laboratory.18 The Grp78 F/-; pc-Cre mice were generated through breeding between Grp78 F/- mice and the Purkinje cell specific Cre transgenic mice. All protocols for animal use and euthanasia were reviewed and approved by the University of Southern California Institutional Animal Care and Use Committee.
Primary antibodies
Primary antibodies used include: rabbit anti-GRP78 (H129) (Santa Cruz Biotechnology), anti-cleaved caspase-3 (Asp175) and phospho-eIF2α (Ser51) (Cell Signaling), anti-GRP94 (SPA-851), PDI (SPA890) and calnexin (SPA-865) (Stressgen) and anti-p62 (SQSTM1) (BIOMOL), mouse anti-calbindin (Sigma-Aldrich), anti-CHOP (GADD153 B-3) and GADD34 (C-19) (Santa Cruz Biotechnology), anti-eIF2α (L57A5) and ubiquitin (P4D1) (Cell Signaling) and anti-neurofilament (non-phosphorylated) (SMI-32) (Sternberger Monoclonals).
Histological, immunohistochemical and immunofluorescence staining analyses
Mouse brains were exposed by removing the cranial bones and fixed in situ overnight in 10% buffered formalin. The entire brains were then removed from the skull and cut midsagittally before being embedded in paraffin using standard protocols. Immunohistochemical and immunofluorescence staining were carried out as described previously.19 In brief, Vectastain Elite avidin-biotin complex kit (Vector Laboratories) was used for immunohistochemistry. After antigen retrieval with retrivagen A (pH 6.0) (BD Pharmingen), paraffin sections were incubated with primary antibodies in blocking solution (1.5% serum in PBS) at 4°C overnight with antibodies against GRP78 (1:100), calbindin (1:100), cleaved caspase-3 (1:50), GRP94 (1:200), PDI (1:300), CHOP (1:50), eIF2α (1:50), phospho-eIF2α (Ser51) (1:50), ubiquitin (1:200) and p62 (1:1000). For immunofluorescence staining, paraffin sections were incubated with primary antibodies including calbindin (1:100), GADD34 (1:200), ubiquitin (1:100) and GRP78 (1:100), calnexin (1:200) and neurofilament (1:100) at 4°C overnight. For fluorescent detection of calbindin, GADD34, ubiquitin and neurofilament, Alexta Fluor 488-conjugated goat anti-mouse IgG (1:200) (Invitrogen) was used. For fluorescent detection of GRP78 and calnexin, Alexta Fluor 594-conjugated goat anti-rabbit IgG (1:200) (Invitrogen) were used. TUNEL assay was performed with Peroxidase In Situ Apoptosis Detection Kit (Chemicon International).
Motor coordination tests
Rota rod test and footprint test were performed as previously described.5 In brief, for the rota rod test, mice were placed on a rod rotating at 30 rpm, and the time taken for them to fall from the rod was measured. If a mouse stayed on the rod longer than the 2 min trial, a time of 120 sec was recorded. For the footprint test, the forelimbs and hindlimbs of mice were painted as blue and red with non-toxic ink respectively. Then their paw placement was recorded using a narrow tunnel (10 cm wide, 40 cm long and 10 cm high) with white paper on the bottom.
Electron microscopy (EM) and immunoEM analysis
For regular EM, mouse brain tissues were dissected from the skull and immersion fixed in half strength Karnovsky’s fixative (2% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.4) for 24 h at 4°C. Tissues were postfixed in 1% osmium tetroxide for 2 h on ice. Samples were dehydrated in graded alcohol and infiltrated with Eponate (Ted Pella Inc., Redding CA). Ultrathin sections were cut at 70 nm and stained with uranyl acetate and lead citrate. For immunoEM labeling, mouse brain tissues were dissected from the skull and immersion fixed in fixative (2% paraformaldehyde and 0.1% glutaraldehyde in 0.1M sodium cacodylate buffer) for 24 h at 4°C. Tissues were infiltrated with LR White acrylic resin (Ted Pella Inc.). After polymerization, ultrathin sections were cut at 70 nm. After incubation with blocking solution (1% BSA and 0.1% Triton-X100 in PBS) for 30 min, sections were stained with anti-calnexin antibody (1:40) at 4°C for overnight followed by 1 h incubation with 15 nm gold-conjugated goat anti-rabbit antibody (1:40) (Ted Pella Inc.). Sections were counter stained with uranyl acetate. All sections were examined on a JEOL JEM 2100 electron microscope (Peabody, MA) and photographed with the Orius SC1000B Gatan digital camera (Pleasanton, CA).
Body weight and food consumption measurements
Fasting body weights of the mice were measured weekly from 3- to 10-wk old. The food consumption was measured for seven successively days at 3- and 9-wk old.
Supplementary Material
Acknowledgments
We thank Dr. Robert Maxson and the Lee lab members for helpful discussion. We thank the University of Southern California/Norris Comprehensive Cancer Center Transgenic Core Facility, the Cell and Tissue Imaging Core Facility, the Translational Pathology Core Facility and the Biostatistics Core Facility for assistance and consultation. This work was supported by NIH grants R01-CA027607, R01-CA111700 and the Freeman Cosmetic Chair to ASL.
Abbreviations
- ER
endoplasmic reticulum
- ERAD
ER-associated protein degradation
- PCs
Purkinje cells
- UPR
unfolded protein response
References
- 1.Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 2.Malhotra JD, Kaufman RJ. The endoplasmic reticulum and the unfolded protein response. Semin Cell Dev Biol. 2007;18:716–731. doi: 10.1016/j.semcdb.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao L, Longo-Guess C, Harris BS, Lee JW, Ackerman SL. Protein accumulation and neurodegeneration in the woozy mutant mouse is caused by disruption of SIL1, a cochaperone of BiP. Nat Genet. 2005;37:974–979. doi: 10.1038/ng1620. [DOI] [PubMed] [Google Scholar]
- 4.Lee JW, Beebe K, Nangle LA, Jang J, Longo-Guess CM, Cook SA, et al. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443:50–55. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- 5.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 6.Anttonen AK, Mahjneh I, Hamalainen RH, Lagier-Tourenne C, Kopra O, Waris L, et al. The gene disrupted in Marinesco-Sjögren syndrome encodes SIL1, an HSPA5 cochaperone. Nat Genet. 2005;37:1309–1311. doi: 10.1038/ng1677. [DOI] [PubMed] [Google Scholar]
- 7.Senderek J, Krieger M, Stendel C, Bergmann C, Moser M, Breitbach-Faller N, et al. Mutations in SIL1 cause Marinesco-Sjögren syndrome, a cerebellar ataxia with cataract and myopathy. Nat Genet. 2005;37:1312–1314. doi: 10.1038/ng1678. [DOI] [PubMed] [Google Scholar]
- 8.Lee AS. The glucose-regulated proteins: stress induction and clinical applications. Trends Biochem Sci. 2001;26:504–510. doi: 10.1016/s0968-0004(01)01908-9. [DOI] [PubMed] [Google Scholar]
- 9.Hendershot LM. The ER function BiP is a master regulator of ER function. Mt Sinai J Med. 2004;71:289–297. [PubMed] [Google Scholar]
- 10.Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 11.Luo S, Mao C, Lee B, Lee AS. GRP78/BiP is required for cell proliferation and protecting the inner cell mass from apoptosis during early mouse embryonic development. Mol Cell Biol. 2006;26:5688–5697. doi: 10.1128/MCB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sillitoe RV, Joyner AL. Morphology, molecular codes, and circuitry produce the three-dimensional complexity of the cerebellum. Annu Rev Cell Dev Biol. 2007;23:549–577. doi: 10.1146/annurev.cellbio.23.090506.123237. [DOI] [PubMed] [Google Scholar]
- 13.Weitzmann A, Volkmer J, Zimmermann R. The nucleotide exchange factor activity of Grp170 may explain the non-lethal phenotype of loss of Sil1 function in man and mouse. FEBS Lett. 2006;580:5237–5240. doi: 10.1016/j.febslet.2006.08.055. [DOI] [PubMed] [Google Scholar]
- 14.Weitzmann A, Baldes C, Dudek J, Zimmermann R. The heat shock protein 70 molecular chaperone network in the pancreatic endoplasmic reticulum - a quantitative approach. FEBS J. 2007;274:5175–5187. doi: 10.1111/j.1742-4658.2007.06039.x. [DOI] [PubMed] [Google Scholar]
- 15.Rutkowski DT, Kang SW, Goodman AG, Garrison JL, Taunton J, Katze MG, et al. The role of p58IPK in protecting the stressed endoplasmic reticulum. Mol Biol Cell. 2007;18:3681–3691. doi: 10.1091/mbc.E07-03-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petrova K, Oyadomari S, Hendershot LM, Ron D. Regulated association of misfolded endoplasmic reticulum lumenal proteins with P58/DNAJc3. EMBO J. 2008;27:2862–2872. doi: 10.1038/emboj.2008.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu Y, Wey S, Wang M, Ye R, Liao CP, Roy-Burman P, et al. Pten null prostate tumorigenesis and AKT activation are blocked by targeted knockout of the stress response chaperone GRP78/BiP in prostate epithelium. Proc Natl Acad Sci USA. 2008;105:19443–19448. doi: 10.1073/pnas.0807691105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barski JJ, Dethleffsen K, Meyer M. Cre recombinase expression in cerebellar Purkinje cells. Genesis. 2000;28:93–98. [PubMed] [Google Scholar]
- 19.Dong D, Ni M, Li J, Xiong S, Ye W, Virrey JJ, et al. Critical role of the stress chaperone GRP78/BiP in tumor proliferation, survival and tumor angiogenesis in transgene-induced mammary tumor development. Cancer Res. 2008;68:498–505. doi: 10.1158/0008-5472.CAN-07-2950. [DOI] [PubMed] [Google Scholar]
- 20.Rao RV, Bredesen DE. Misfolded proteins, endoplasmic reticulum stress and neurodegeneration. Curr Opin Cell Biol. 2004;16:653–662. doi: 10.1016/j.ceb.2004.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hetz CA, Soto C. Stressing out the ER: a role of the unfolded protein response in prion-related disorders. Curr Mol Med. 2006;6:37–43. doi: 10.2174/156652406775574578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ni M, Lee AS. ER chaperones in mammalian development and human diseases. FEBS Lett. 2007;581:3641–3651. doi: 10.1016/j.febslet.2007.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marciniak SJ, Yun CY, Oyadomari S, Novoa I, Zhang Y, Jungreis R, et al. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 2004;18:3066–3077. doi: 10.1101/gad.1250704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marciniak SJ, Ron D. Endoplasmic reticulum stress signaling in disease. Physiol Rev. 2006;86:1133–1149. doi: 10.1152/physrev.00015.2006. [DOI] [PubMed] [Google Scholar]
- 25.Molinari M, Galli C, Piccaluga V, Pieren M, Paganetti P. Sequential assistance of molecular chaperones and transient formation of covalent complexes during protein degradation from the ER. J Cell Biol. 2002;158:247–257. doi: 10.1083/jcb.200204122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshida H. ER stress and diseases. FEBS J. 2007;274:630–658. doi: 10.1111/j.1742-4658.2007.05639.x. [DOI] [PubMed] [Google Scholar]
- 27.Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seibenhener ML, Geetha T, Wooten MW. Sequestosome 1/p62--more than just a scaffold. FEBS Lett. 2007;581:175–179. doi: 10.1016/j.febslet.2006.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee AS. The ER chaperone and signaling regulator GRP78/BiP as a monitor of endoplasmic reticulum stress. Methods. 2005;35:373–381. doi: 10.1016/j.ymeth.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki T, Lu J, Zahed M, Kita K, Suzuki N. Reduction of GRP78 expression with siRNA activates unfolded protein response leading to apoptosis in HeLa cells. Arch Biochem Biophys. 2007;468:1–14. doi: 10.1016/j.abb.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 31.Li J, Ni M, Lee B, Barron E, Hinton DR, Lee AS. The unfolded protein response regulator GRP78/BiP is required for endoplasmic reticulum integrity and stress-induced autophagy in mammalian cells. Cell Death Differ. 2008;15:1460–1471. doi: 10.1038/cdd.2008.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pyrko P, Schonthal AH, Hofman FM, Chen TC, Lee AS. The unfolded protein response regulator GRP78/BiP as a novel target for increasing chemosensitivity in malignant gliomas. Cancer Res. 2007;67:9809–9816. doi: 10.1158/0008-5472.CAN-07-0625. [DOI] [PubMed] [Google Scholar]
- 33.Plemper RK, Bohmler S, Bordallo J, Sommer T, Wolf DH. Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature. 1997;388:891–895. doi: 10.1038/42276. [DOI] [PubMed] [Google Scholar]
- 34.Skowronek MH, Hendershot LM, Haas IG. The variable domain of nonassembled Ig light chains determines both their half-life and binding to the chaperone BiP. Proc Natl Acad Sci USA. 1998;95:1574–1578. doi: 10.1073/pnas.95.4.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ushioda R, Hoseki J, Araki K, Jansen G, Thomas DY, Nagata K. ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science. 2008;321:569–572. doi: 10.1126/science.1159293. [DOI] [PubMed] [Google Scholar]
- 36.Reddy RK, Mao C, Baumeister P, Austin RC, Kaufman RJ, Lee AS. Endoplasmic reticulum chaperone protein GRP78 protects cells from apoptosis induced by topoisomerase inhibitors: role of ATP binding site in suppression of caspase-7 activation. J Biol Chem. 2003;278:20915–20924. doi: 10.1074/jbc.M212328200. [DOI] [PubMed] [Google Scholar]
- 37.Fu Y, Li J, Lee AS. GRP78/BiP inhibits endoplasmic reticulum BIK and protects human breast cancer cells against estrogen-starvation induced apoptosis. Cancer Res. 2007;67:3734–3740. doi: 10.1158/0008-5472.CAN-06-4594. [DOI] [PubMed] [Google Scholar]
- 38.Mimura N, Yuasa S, Soma M, Jin H, Kimura K, Goto S, et al. Altered quality control in the endoplasmic reticulum causes cortical dysplasia in knock-in mice expressing a mutant BiP. Mol Cell Biol. 2008;28:293–301. doi: 10.1128/MCB.00473-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Impagnatiello F, Guidotti AR, Pesold C, Dwivedi Y, Caruncho H, Pisu MG, et al. A decrease of reelin expression as a putative vulnerability factor in schizophrenia. Proc Natl Acad Sci USA. 1998;95:15718–15723. doi: 10.1073/pnas.95.26.15718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Naidoo N. ER and aging-Protein folding and the ER stress response. Ageing Res Rev. 2009;8:150–159. doi: 10.1016/j.arr.2009.03.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.