Abstract
Infection of domestic cats with virulent strains of the feline immunodeficiency virus (FIV) leads to an acquired immunodeficiency syndrome (AIDS), similar to the pathogenesis induced in humans by infection with human immunodeficiency virus type 1 (HIV-1). Thus, FIV is a highly relevant model for anti-HIV therapy and vaccine development. FIV is not infectious in humans, so it is also a potentially effective non-toxic gene therapy vector. To make better use of this model, it is important to define the cellular machinery utilized by each virus to produce virus particles so that relevant similarities can be identified. It is well understood that all replication-competent retroviruses encode gag, pol, and env genes, which provide core elements for virus replication. As a result, most antiretroviral therapy targets pol-derived enzymes (protease, reverse transcriptase, and integrase) or env-derived glycoproteins that mediate virus attachment and entry. However, resistance to drugs against these targets is a persistent problem, and novel targets must be identified to produce more effective drugs that can either substitute or be combined with current therapy. Elements of the gag gene (matrix, capsid, nucleocapsid, and “late” domains) have yet to be exploited as antiviral targets, even though the Gag precursor polyprotein is self-sufficient for the assembly and release of virus particles from cells. This process is far better understood in primate lentiviruses, especially HIV-1. However, there has been significant progress in recent years in defining how FIV Gag is targeted to the cellular plasma membrane, assembles into virions, incorporates FIV Env glycoproteins, and utilizes host cell machinery to complete virus release. Recent discoveries of intracellular restriction factors that target HIV-1 and FIV capsids after virus entry have also opened exciting new areas of research. This review summarizes currently known interactions involving HIV-1 and FIV Gag that affect virus release, infectivity, and replication.
Keywords: FIV, HIV-1, Gag, viral late domains, ESCRT, virus-cell interactions
1. Introduction: FIV is a relevant model for AIDS and lentiviral gene therapy
FIV is endemic in the wild and has evolved for millennia, along with its ancestral hosts (Pedersen et al., 1987; Troyer et al., 2005). FIV infection of its native host induces feline AIDS, characterized by a progressive decline in CD4+ T-cells that is clinically asymptomatic for years unless challenged with an opportunistic pathogen (Burkhard and Dean, 2003). Unlike SIV-induced simian AIDS, development of feline AIDS does not require a cross-species transmission, which provides some clear advantages for studying both horizontal and vertical transmission in relatively inexpensive cat colonies and feral cats (Coats, 2005; Willett et al., 1997). FIV pathology is similar to that of HIV-1 and is best described in domestic cats (Felis catus) infected with FIVfca, due to a high incidence of infection (~1 to 30%) with apparently no evolution of host resistance (Troyer et al., 2005; Winkler et al., 1999). Despite centuries of close contact, there is no evidence of FIV transmission to humans, possibly due to poor recognition of the FIV promoter (5'-LTR) in human cells (Mustafa et al., 2005). This block can be overcome for FIV-based gene therapy, applicable to both dividing and non-dividing cells of virtually any type, by substitution of the FIV 5'-LTR with a CMV promoter (Johnston et al., 1999). Thus, understanding FIV biology in human cells is also potentially relevant to clinical applications. FIV is the only lentivirus for which a vaccine is readily available (Hohdatsu et al., 1997), which is protective against a subset of known FIV subtypes. Mechanisms of protection appear to involve both humoral and cellular immunity (Pu et al., 1997). Thus, FIV is clearly a useful model for the development of AIDS vaccines, antiretroviral drugs, and non-pathogenic gene therapy vectors. Compared to primate lentiviruses, many fundamental aspects of FIV cellular biology are not well understood. However, significant progress has been made in recent years in identifying molecular mechanisms of infection, in part based on comparative studies with HIV-1 and other lentiviruses (Elder et al., 2008; Luttge et al., 2008).
2. FIV and HIV-1 genome homology
Both FIV and HIV-1 contain essential elements found in all retroviruses (gag, pol, env), possibly derived from a common ancestral lentivirus (Katzourakis et al., 2007), with the addition of accessory factors that enhance replication in vivo but are often dispensable in cell culture models [reviewed in (Elder et al., 2008)]. Since FIV has evolved independently in cats, it has virtually no sequence similarity to primate lentiviruses in homologous open reading frames and the collection of accessory factors is the least conserved (Olmsted et al., 1989; Pecon-Slattery et al., 2008). With the exception of Rev and Vif, most accessory factors encoded by HIV-1 (including Tat, Nef, Vpr, and Vpu) have not been clearly identified in FIV, although FIV Orf2/OrfA may have functions partially related to both Tat and Vpr (Sundstrom et al., 2008). Earlier studies with FIV Vif suggested a possible analogy with HIV-1 Vif, but their functional similarity has only recently been clearly demonstrated (Münk et al., 2008).
3. The Gag protein is a self-contained virus assembly “machine”
Expression of retroviral Gag polyprotein precursors alone, within a suitable host cell, is sufficient for the production of virus-like particles (VLPs) (Fig. 1A) [reviewed in (Adamson and Freed, 2007; Ganser-Pornillos et al., 2008)]. All retroviral Gag proteins contain domains required for membrane targeting, Gag-Gag interaction, and virus release (Fig. 1B). Through interactions and modifications of the membrane-targeting domain, lentiviral assembly typically occurs at the plasma membrane (PM) while budding away from the cytoplasm of the infected cell. Other steps in virus assembly and release are driven by interactions of Gag with itself and with host cell factors via “late” domains (Fig. 1C). After these domains have exerted their functions in assembly and release, the Gag precursor is cleaved by the virally encoded protease (PR), if present, into the final mature Gag proteins. For HIV-1 and FIV these include matrix (MA), capsid (CA), nucleocapsid (NC), spacer peptides (SP1, SP2) flanking NC, and a C-terminal peptide (p6 in HIV-1, p2 in FIV) (Elder et al., 1993; Ganser-Pornillos et al., 2008; Lin et al., 2006). Activation of PR, and the resulting cleavage of Gag, appears to coincide with final events in virus release. However, retrovirus release itself does not depend upon PR function; immature VLPs containing unprocessed Gag are released efficiently in the absence of PR (Calistri et al., 2009; Fu et al., 2006; Huang et al., 1995; Ono et al., 2004; Ono et al., 2000; Ono et al., 2005; Peng et al., 1989; Tomonaga et al., 1998).
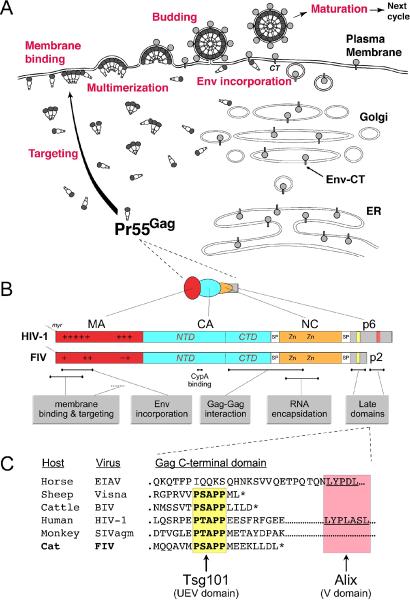
Figure 1. Overview of HIV-1 and FIV Gag domains and the virus assembly and release pathway.
A) Gag is synthesized as a precursor protein (HIV-1 Pr55Gag = FIV Pr50Gag) that exists in both monomeric and oligomeric forms in the cytoplasm. Interactions between Gag and host factors target Gag to the plasma membrane (PM) in most cell types. Membrane binding facilitates higher-order Gag multimerization that leads to virus assembly and budding. Release of virus particles is driven by interactions with host cell factors. Maturation of the virus occurs during or shortly after virus release and is required for virus infectivity. Env is a highly glycosylated protein, synthesized in the endoplasmic reticulum (ER), that follows the secretory pathway through the trans-Golgi network to the PM. Env is incorporated into budding virus particles via interactions between the matrix (MA) domain of Gag and the Env cytoplasmic tail (Env-CT). Adapted from (Joshi and Freed, 2007) with permission of Future Medicine Ltd. B) Domains of the Gag precursor protein from HIV-1 and FIV are shown, including known or predicted functions and interactions. The MA domain is covalently modified with myristate (myr) at the N-terminus. Two patches of basic residues (+) in MA are modestly conserved. Capsid (CA) folds into an N-terminal domain (NTD), which contains a conserved cyclophilin A (CypA) binding loop, and a C-terminal domain (CTD). Nucleocapsid (NC) contains two conserved zinc-coordinating motifs (Zn), which are important for interaction of Gag with viral RNA. NC is flanked by two spacer peptides (sp) in both HIV-1 and FIV, which must be cleaved from CA and NC during maturation for full viral infectivity. C-terminal domains of Gag in HIV-1 (p6) and FIV (p2) contain late domains important for virus release. C) An alignment of C-terminal domains of several lentiviral Gag proteins is shown, highlighting a highly conserved PSAPP motif that binds the ubiquitin E2-variant (UEV) domain of tumor-susceptibility gene 101 (Tsg101). Equine infectious anemia virus (EIAV) and HIV-1 share an LYPXnL motif that binds the V domain of apoptosis-linked-gene-2-interacting protein X (Alix). BIV, bovine immunodeficiency virus; SIVagm, simian immunodeficiency virus from African green monkey.
3.1 Matrix (MA)
Lentiviral MA domains play important roles in the assembly of infectious particles by directing Gag to the PM, mediating the association between Gag and the inner leaflet of the PM lipid bilayer, and recruiting the viral envelope (Env) glycoproteins into virions (Freed, 1998; Freed and Martin, 1995, 1996) (Fig. 1B).
Membrane targeting
The ability of most retroviral MA domains to direct Gag-membrane binding requires the covalent modification of the MA N-terminus with myristic acid and is also promoted by a highly basic patch of amino acid residues (Dalton et al., 2007; Freed et al., 1994; Hill et al., 1996; Yuan et al., 1993; Zhou et al., 1994). For both HIV-1 and HIV-2, Gag targeting to the PM is facilitated by a direct interaction between some of these basic residues and the host factor phosphatidylinositol-4,5-bisphosphate [PI(4,5)P2], which is concentrated on the inner leaflet of the PM (Fig. 1B) (Chukkapalli et al., 2008; Ono et al., 2004; Ono et al., 2005; Saad et al., 2008; Saad et al., 2006). FIV MA appears to utilize a similar mechanism for targeting FIV assembly, since perturbation of PI(4,5)P2 levels inhibits FIV release (Luttge and Freed, unpublished results) and basic patches in FIV MA are required for particle production (Manrique et al., 2004a). SIV also relies on basic residues in MA for membrane-targeted assembly (González et al., 1993), and it has been shown that SIV MA can substitute for FIV MA to produce infectious virus (Manrique et al., 2004a). FIV MA, however, is generally less basic than SIV or HIV-1 MA (Fig. 1B), and cannot substitute for SIV MA in SIV to produce infectious particles. Interestingly, this block is readily overcome by site-directed mutagenesis of FIV MA to restore two critical lysine residues found in SIV and HIV-1 MA. Thus, as observed with many other domains of FIV Gag, these data suggest a high degree of conserved structure and function in FIV MA with MA from primate lentiviruses, despite the absence of significant amino acid sequence identity (Burkala and Poss, 2007; Elder et al., 1993).
Env incorporation
The N-terminal region of HIV-1 MA also functions to enhance incorporation of the Env glycoprotein into virions by a putative interaction with the cytoplasmic tail of HIV-1 Env (Env-CT, Fig. 1A) (Davis et al., 2006; Freed and Martin, 1996; Lambelé et al., 2007; Murakami and Freed, 2000). It has recently been proposed that the 47-kDa tail-interacting protein (TIP47; also known as mannose-6-phosphate receptor binding protein 1, M6PRBP1) may be an important adaptor protein that bridges this interaction by binding both the HIV-1 Env-CT and the N-terminal domain of HIV-1 MA (Lopez-Vergès et al., 2006). The requirement of the FIV Env-CT for virion incorporation of FIV Env is likely to be similar to that of HIV-1; however, a direct role for FIV MA has not yet been determined (Celma et al., 2007).
3.2 Capsid (CA)
Retroviral CA domains perform important functions both in promoting Gag-Gag interactions during assembly and in forming the outer shell of the viral core that houses the viral genome and pol-encoded enzymes in the mature virion. CA folds into two structural and functional domains, the N-terminal and C-terminal domains (NTD and CTD, respectively, Fig. 1B). The HIV-1 CTD contains a major interface for CA-CA dimerization that promotes Gag multimerization (Gamble et al., 1997). Both the NTD and the CTD bear interaction interfaces that are crucial for the assembly of the hexameric CA lattice that ultimately gives rise to the conical core characteristic of mature lentiviral particles (Ganser-Pornillos et al., 2007).
Structure
Compared to HIV-1 and several other retroviruses, very little is known about FIV CA (p24/p25) in terms of its structure, dimerization interface, and interactions with host cell factors. Several lines of evidence suggest that non-primate lentiviral CA proteins (from FIV and EIAV) retain a conserved fundamental structure that is similar to that of HIV-1 CA, despite sequence divergence. For example, vaccination of cats with HIV-1 CA provides protection against FIV infection, most likely through one or more cross-reactive T-cell epitopes (Coleman et al., 2005). Antibodies raised against CA from EIAV or HIV-1 are cross-reactive with FIV CA, and vice versa in the case of EIAV (Egberink et al., 1990; Egberink et al., 1991; Nath and Peterson, 2001). A three-dimensional structure for FIV CA is not yet available. However, secondary structural predictions of FIV CA NTD and CTD have been modeled against the crystal structures for EIAV and HIV-1 CA, and highlight many similarities (Burkala and Poss, 2007). Conserved structural features in each NTD include an N-terminal β-hairpin, tightly packed α-helices (seven in HIV-1 and five in both FIV and EIAV) arranged into an arrowhead shape, and an exposed flexible loop (Ganser-Pornillos et al., 2007; Jin et al., 1999). A highly conserved feature of this extended loop in each CA NTD is the exposure of proline residues, at least one of which binds peptidylprolyl isomerase A (cyclophilin A or CypA) (Gamble et al., 1996; Lin and Emerman, 2006). Structural highlights of the CTD include a strand-turn-helix motif known as the major homology region (MHR), which is found in many divergent retroviruses, and four highly conserved α-helices, one of which (helix 9 in HIV-1, helix 7 in FIV and EIAV) forms a critical dimerization interface between HIV-1 CA monomers in solution that is apparently retained in the mature capsid structure (Gamble et al., 1997; Ganser-Pornillos et al., 2007). Overall, these studies suggest that many elements of CA structure are highly conserved among lentiviruses.
Interactions with CypA and TRIM-family proteins
CA binding to CypA, described above, is a conserved feature of several retroviruses, including FIV and HIV-1 (Franke et al., 1994; Lin and Emerman, 2006). The precise role for this interaction in virus replication remains enigmatic. However, inhibition of CypA-CA binding by treatment with cyclosporin A inhibits the replication of both FIV and HIV-1 (Karpas et al., 1992; Mortola et al., 1998). Interestingly, factors expressed in monkey cells that naturally restrict HIV-1 infection have been shown to exploit the CypA-binding loop to target incoming viral capsids shortly after virus entry [reviewed in (Luban, 2007; Towers, 2007)]. Recent studies have shown that these restriction factors are based on derivatives of the tripartite motif 5 protein (TRIM5), which is part of a large family of TRIM proteins (~70 in humans) that possibly constitute a broad innate immune response (Ozato et al., 2008). The first example of a TRIM protein that restricts HIV-1, TRIM5αrh, was discovered in rhesus macaques (Stremlau et al., 2004), and is a natural inhibitor of HIV-1 infection that is stimulated by type 1 interferon. TRIM5α homologs, formerly referred to as Lv1 and Ref1 activities, have since been identified in African green monkey (AGM) and human (hu) cells, respectively (Keckesova et al., 2004). Perhaps not surprisingly, human TRIM5α has relatively weak anti-HIV-1 activity (Sokolskaja et al., 2006; Stremlau et al., 2004). The TRIM5α homologs described above (rh, hu, AGM) have also been shown to restrict FIV and EIAV, which suggests that this family of restriction factors may have evolved as broad-based lentiviral inhibitors (Hatziioannou et al., 2003; Hatziioannou et al., 2004; Saenz et al., 2005). Divergence of the TRIM-family proteins arises partly from alternative splicing of the RBCC tripartite motif (RING domain, B-Box domains, and coiled-coil motifs), and a variable C-terminal domain (CTD). Together these domains incorporate functions of E3 ubiquitin ligase activity (RING), TRIM protein multimerization (CC), and target-binding specificity (CTD). The antiretroviral activity of TRIM5αrh appears to require the RING and CC domains, resulting in a ubiquitin-mediated proteasomal degradation of the bound target, although TRIM multimerization may also be required at an earlier step to prematurely accelerate virion uncoating (Javanbakht et al., 2007; Rold and Aiken, 2008; Stremlau et al., 2006). Specificity for TRIM5α targeting to the CypA-binding loop of lentiviral capsids lies in the C-terminal B30.2/SPRY domain. In recent years, several laboratories have independently made similar discoveries of a TRIM5-related restriction factor in owl monkeys and Asian macaques (pig-tailed and rhesus), in which the B30.2/SPRY domain of TRIM5α is missing and contains a CypA domain instead, creating the TRIMCyp protein (Brennan et al., 2008; Newman et al., 2008; Nisole et al., 2004; Ribeiro et al., 2005; Sayah et al., 2004; Virgen et al., 2008; Wilson et al., 2008). Each of these TRIMCyp factors evolved entirely independently from each other and TRIM5α. Like HIV-1, FIV is sensitive to both owl monkey and macaque TRIMCyp proteins, which again argues that relatively broad-based restriction factors may have evolved through host resistance against ancient retroviral pathogens (Diaz-Griffero et al., 2007; Kratovac et al., 2008; Nath and Peterson, 2001; Virgen et al., 2008).
Interactions with the cytoskeleton and associated cellular motors
Retroviral CA proteins have also been shown to interact with the host cell cytoskeleton, possibly to exploit retrograde cytoskeletal motors for directional transport of incoming intracellular viral cores, reverse transcription complexes, and pre-integration complexes toward the nucleus [reviewed in (Fackler and Krausslich, 2006; Naghavi and Goff, 2007)]. Similar interactions may also be involved with anterograde trafficking of virus assembly intermediates toward the plasma membrane. Early studies suggested direct interactions of HIV-1 Gag with filamentous actin (Bukrinskaya et al., 1998; Rey et al., 1996). Actin and actin-binding proteins (e.g., ezrin-radixin-moesin (ERM) family members, and cofilin) have also been shown to be incorporated into HIV-1 particles; however, attempts to define the role of actin in retrovirus release by treatment with actin-disrupting agents (latrunculin, cytochalasin D) have been complicated by the cytotoxic side effects of these drugs (Ott et al., 2000; Ott et al., 1996; Sasaki et al., 1995). The cortical actin layer just below the PM provides one of the first intracellular barriers to viral infection. Moesin, which is a cortical actin crosslinker, acts as a natural restriction factor against retroviral infection if highly expressed (Naghavi et al., 2007). The antiviral activity of moesin may be due to its ability to regulate the formation of stable microtubules (MT), which has also recently been shown for ezrin (Haedicke et al., 2008). Other reports suggest an indirect interaction of Gag with MT by binding MT-associated motors. For example, HIV-1 Gag binds KIF-4, an MT-associated anterograde cellular motor (Tang et al., 1999). Consistent with this finding, it has recently been shown that disruption of the function of endogenous KIF-4, either through siRNA-mediated knockdown of KIF-4 or expression of dominant-negative KIF-4, decreases HIV-1 virus-like particle production and Gag protein stability (Martinez et al., 2008). The relationship between Gag precursors, CA, cytoskeleton, and cellular motors still remains poorly understood for HIV-1, FIV, and most other retroviruses.
3.3 Nucleocapsid (NC)
NC domains serve several major roles during retroviral replication (Freed, 1998): 1) together with CA they mediate Gag-Gag interactions that lead to the assembly of the virus particle, 2) they bind the viral RNA genome and recruit it into the assembling virus particle, and 3) they serve as a nucleic acid chaperone that facilitates a variety of steps early in the virus replication cycle. FIV NC contains essentially all known structural components and functions of HIV-1 NC (Moscardini et al., 2002). Specifically, NC packages the FIV RNA genome through recognition of a specific packaging signal,Ψ, which partially overlaps the gag open reading frame (Kemler et al., 2002). Through this interaction, the cellular trafficking of viral RNA and Gag polyprotein precursors, as a ribonucleoprotein complex, are likely to be intimately linked; however, the precise cellular location at which Gag-RNA association begins is poorly understood and may differ between retroviruses. It has been well demonstrated that interactions between Gag and RNA, through the NC domain, are required for assembly of retrovirus particles (Campbell and Rein, 1999; Campbell and Vogt, 1995; Muriaux et al., 2001; Rulli et al., 2007). It is not yet clear why lentiviral NCs have two zinc (Zn) fingers, since most NC functions apparently rely on the proximal Zn finger. Nonetheless, the tandem arrangement of two CCHC-type Zn fingers in HIV-1 is conserved in FIV NC. The amino acid sequence of EIAV NC is more similar to that of FIV NC than that of HIV-1 NC; however, there is an unusually high degree of sequence similarity in the tandem Zn finger region among all lentiviruses. In particular, the spacing of Zn-coordinating Cys and His residues in each Zn finger is identical among NCs from FIV, HIV-1, and EIAV, with only a slightly longer basic linker between Zn fingers in HIV-1 NC. Three-dimensional structures for NC from both HIV-1 and EIAV are now available (Amodeo et al., 2006; De Guzman et al., 1998), and it remains to be seen what the structure of FIV NC will be.
3.4 Late (L) domains and FIV p2
Retroviral late domains
L domains are short peptide motifs found within the retroviral Gag protein that are required for efficient virus release. These motifs bind directly to components of the cellular endosomal sorting complexes required for transport (ESCRT-I, II, III), or bind cellular factors associated with this machinery. As the name implies, ESCRTs are required for the sorting and transport of ubiquitinated cargo proteins (associated with endosomal membranes) for delivery into late endosomes. The final action of ESCRT and associated proteins results in the budding of intraluminal vesicles (ILVs) away from the cytoplasm into late endosomes, hence these compartments are also known as multivesicular bodies (Fig. 2). The complete ESCRT and associated machinery is highly complex, and will not be described fully here, but has been detailed extensively in the literature, especially with regards to its role in virus release (Bieniasz, 2006; Demirov and Freed, 2004; Hurley and Emr, 2006; Morita and Sundquist, 2004; Williams and Urbe, 2007). Most if not all retroviruses, and many other enveloped viruses, utilize this machinery to drive the budding of virions from the PM, especially at the crucial “pinching off” step that completes virus release. There are currently three known retroviral L domain motifs (PT/SAP, YPXnL, and PPXY). The PT/SAP motif in HIV-1 and FIV Gag (Fig. 1C) binds Tsg101, one of four components of the human ESCRT-I (Garrus et al., 2001; Luttge et al., 2008; Martin-Serrano et al., 2001; VerPlank et al., 2001). The YPXnL motif in HIV-1 and EIAV Gag (Fig. 1C) binds Alix, which associates with ESCRT-I and III (Martin-Serrano et al., 2003; Strack et al., 2003). The PPXY motif (not found in FIV, HIV-1, or most other lentiviral Gag protein) binds Nedd4 or Nedd4-like ubiquitin E3 ligases (Kikonyogo et al., 2001). This motif is common in simple retroviruses (e.g., murine leukemia virus and Rous sarcoma virus), mouse mammary tumor virus, human T-cell lymphotropic virus, and some non-retroviruses like Ebola, often in combination with a PT/SAP motif. The mechanism by which viral proteins are able to divert ESCRT machinery for a budding process at the PM, rather than into late endosomes, is still unknown and somewhat controversial. Indeed, initial reports suggested that the initial release of HIV-1 may occur into an intracellular compartment in some cell types (Joshi and Freed, 2007). However, further studies now suggest that in fact these compartments, which were seemingly intracellular by two-dimensional electron microscopy, may actually be deeply invaginated extensions of the PM, from which virus is later released to the extracellular space formed between adjacent cells (Deneka et al., 2007; Groot et al., 2008). This form of release may be stimulated by cell-cell contact, resulting in a “virological synapse” with similarities to an immunological synapse between antigen-presenting cells and T-lymphocytes, to achieve a “cell-mediated” rather than “cell-free” infection (Sattentau, 2008).
Figure 2. Relationship between retrovirus release and ESCRT machinery.
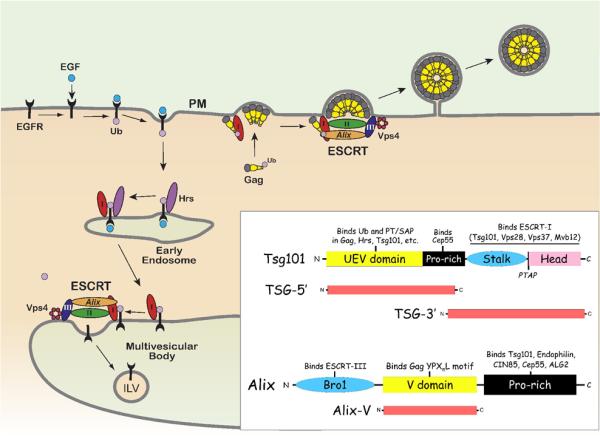
Retroviruses utilize late domains in Gag to recruit endosomal-sorting complexes required for transport (ESCRT-0, I, II, III), which facilitate virus release from the plasma membrane (PM). An example of the normal function of ESCRT is demonstrated by the downregulation of epidermal growth factor receptor (EGFR). Upon binding its ligand (EGF) at the PM, the EGFR is monoubiquitinated and internalized by endocytosis. Attachment of ubiquitin (Ub) to EGFR targets Ub-EGFR to the multivesicular body (MVB) through an endosomal sorting process, and results in the degradation of EGFR in lysosomes. First, Ub-EGFR bound to an endocytic vesicle is internalized into early endosomes by ESCRT-0 through a direct interaction with hepatocyte growth factor regulated tyrosine kinase substrate (Hrs). ESCRT-I (I) is recruited to Ub-EGFR through a PSAP motif in Hrs, which binds the UEV domain of Tsg101. The Tsg101 UEV domain binds Ub-EGFR to complete the transfer from Hrs. Tsg101 itself is bound to ESCRT-I through C-terminal Stalk and Head domains. ESCRT-I bound to Ub-EGFR then traffics from the early endosome to the MVB. Through a series of interactions with ESCRT-II, III, Alix, and Vps4, an intraluminal vesicle (ILV) containing EGFR is formed, which buds into the MVB. HIV-1 and FIV Gag are each ubiquitinated proteins with a PT/SAP motif, which mimic Hrs and recruit ESCRT machinery to stimulate virus release from the PM. TSG-5' and TSG-3' constructs are dominant-negative inhibitors of Tsg101 that inhibit HIV-1 and FIV release, because each lacks components found in the endogenous full-length protein. Alix is an ESCRT-associated protein that binds many cellular factors in the domains indicated, including Tsg101 and ESCRT-III. HIV-1 and EIAV Gag each contain a YPXnL motif that binds the Alix V domain. Association of Gag with Alix recruits ESCRT machinery, which enhances virus release. Expression of Alix-V alone, without ESCRT-(I,III)-binding domains, inhibits HIV-1 and EIAV Gag release but has no effect on FIV. Adapted from (Demirov and Freed, 2004) with permission from Elsevier.
Requirements for ESCRT in lentiviral release
The role of ESCRT and associated host factors in enhancing virus release can be shown by exogenous expression of either full-length or fragments of ESCRT-related proteins. These often have a dominant-negative effect on either ESCRT function in general or the association of Gag with ESCRT machinery. In either case, the ability of endogenous host factors to enhance virus release is abrogated. In some instances, inhibition is achieved simply by competing for a direct interaction with Gag. For example, overexpressing the N-terminal, ubiquitin E2-variant (UEV) domain of Tsg101 (TSG-5') has a dominant-negative effect on release of viruses that contain a PT/SAP motif (e.g. HIV-1, FIV, Fig. 1C), because the UEV domain binds to the PT/SAP motif and prevents it from interacting with endogenous Tsg101 (Fig. 2) (Demirov et al., 2002; Luttge et al., 2008). Similarly, overexpression of the central, Gag-binding V domain of Alix alone (Fig. 2) inhibits release of viruses that contain a YPXnL motif (HIV-1, EIAV) (Fig. 1C), because critical Bro1 and proline-rich domains of Alix are absent (Lee et al., 2007; Luttge et al., 2008; Munshi et al., 2007) [reviewed in (Fujii et al., 2007)]. Conversely, overexpression of ESCRT-associated factors that do not bind Gag late domains can also inhibit retroviral release by a variety of mechanisms. For example, overexpression of the C-terminal half of Tsg101 (TSG-3', Fig. 2) results in an aberrant aggresome-like accumulation of TSG-3' in the cytoplasm, within which other ESCRT components and possibly ubiquitin are sequestered (Goila-Gaur et al., 2003; Johnson et al., 2005; Luttge et al., 2008; Shehu-Xhilaga et al., 2004).
FIV p2
Most lentiviruses have a PT/SAP motif in the C-terminal domain of Gag (Fig. 1C). The PSAP motif in the p2 domain of FIV Gag is essential for FIV release in feline and human cells and for FIV replication in feline cell lines (Calistri et al., 2009; Luttge et al., 2008; Manrique et al., 2004b). Peptides derived from the sequence of FIV p2 bind directly to the UEV domain of human Tsg101 in vitro. Tsg101 is also required for efficient FIV release in human cells, as shown by siRNA-mediated knockdown of Tsg101 (Luttge et al., 2008). As is the case for HIV-1, FIV release is sensitive to ESCRT-associated dominant-negative inhibitors, and TSG-5' can be stably expressed in CrFK cells to constitutively inhibit FIV release and replication (Luttge et al., 2008). Unlike HIV-1 and EIAV, the ESCRT-associated protein Alix appears to have no role in facilitating FIV release in human cells. Specifically, the Alix V domain targets YPXnL late domains in HIV-1 and EIAV Gag (Fig. 1C) in vitro, but there is no apparent interaction with FIV Gag p2 (Luttge et al., 2008). Initial studies of site-directed mutants in FIV p2 suggested that a C-terminal LxxL motif was required for FIV release, using the FIV-14 molecular clone expressed in CrFK cells (Manrique et al., 2004b). However, this phenotype was not reproducible with the FIV-34TF10 clone in CrFK cells or with an FIV Gag-Pol expression vector in either feline or human (HeLa) cells (Luttge et al., 2008), even though Gag sequences used in each study were apparently identical.
4. Role of Gag ubiquitination in retroviral release
Covalent attachment of individual ubiquitin molecules (monoubiquitination or multiubiquitination) to lysine residues of intracellular proteins is a signal for recruitment of ESCRT and associated machinery, which results in the sorting of ubiquitinated cargo into MVBs (Fig. 2) [reviewed in (Hicke and Dunn, 2003; Stuffers et al., 2008)]. In contrast, attachment of a chain of ubiquitin molecules (polyubiquitination) to a single lysine residue is associated with targeting to the proteasome for protein degradation. Ubiquitination follows a pathway that involves a series of enzymes known as E1, E2, and E3. E1 activates individual ubiquitin molecules, which are then attached to an E2 ubiquitin-conjugating enzyme. E3 ligases transfer the activated ubiquitin from E2 to a bound substrate. Specificity for the E3 step is determined by adaptor proteins in a multimeric SCF complex (Skp1-Cullin-F box protein) [reviewed in (Ho et al., 2008)]. Some components of the ESCRT pathway, including Tsg101, utilize ubiquitin interacting motifs (UIMs) to facilitate interactions as ubiquitinated cargo is handed from one ESCRT complex to another (Hurley et al., 2006). The precise role of mono/multi-ubiquitination in retroviral release is not entirely clear [reviewed in (Martin-Serrano, 2007)]. However, most retroviral Gag proteins, including that of FIV, are ubiquitinated (Calistri et al., 2009). It has been speculated that ubiquitination of Gag enhances its interaction with ESCRT, through UIMs. For example, primary targets of the retroviral PPxY late domain motif (described above) are E3 ubiquitin ligases (Nedd4 and Nedd4-like proteins), yet viruses with this late domain still ultimately rely on ESCRT for release. Overexpression of Nedd4-like proteins has been shown to enhance ubiquitination of HIV-1 and FIV Gag, which appears to rescue virus release defects in PT/SAP-defective mutants (Calistri et al., 2009; Chung et al., 2008). One explanation for this phenomenon is that the ubiquitin moiety itself, attached to Gag, may be able to directly recruit ESCRT components that enhance virus release through UIMs, if the normal late domain is absent. Consistent with this hypothesis, release of EIAV Gag mutants that lack a late domain is rescued when Gag is artificially fused to ubiquitin (Joshi et al., 2008). Alternatively, the ubiquitination of unidentified host factors could enhance virus release, and ubiquitination of Gag may simply be a byproduct of its association with ESCRT machinery and proximal E3 Ub-ligases. Nedd4-like protein overexpression results in the monoubiquitination of many cellular proteins, in addition to Gag. This global increase in ubiquitination may enhance interactions between proteins with UIMs, such as the ESCRT machinery, which would likely favor virus release. In support of this hypothesis, foamy virus Gag naturally has very few lysine residues available for monoubiquitination, yet it can still be released efficiently in a ubiquitin-dependent manner when all sites for Gag ubiquitination are removed by site-directed mutagenesis (Zhadina et al., 2007).
5. Conclusions
Due to the fundamental importance of Gag and its many functions in the retroviral life cycle, the processes of virion assembly, release, and post-entry events remain attractive targets for antiviral intervention. Endogenous restriction factors appear to inhibit FIV or HIV-1 by targeting Gag or virions at each of these stages, some of which are counteracted by virally encoded accessory proteins. For example, APOBEC3G (a member of the APOBEC family of cytosine deaminases) is incorporated into virions during virus assembly, inhibits infectivity, and is counteracted by HIV-1 Vif (Goila-Gaur and Strebel, 2008; Münk et al., 2007). Recently it has been shown that feline APOBEC3CH inhibits the infectivity of HIV-1, SIV, and Vif-deficient FIV, which is counteracted by FIV Vif (Münk et al., 2008). Bst-2/tetherin, which inhibits fully infectious particles at the level of virus release, is counteracted by HIV-1 Vpu (Jouvenet et al., 2009; Neil et al., 2008; Van Damme et al., 2008) [reviewed in (Wolf and Goff, 2008)]. TRIM5-related proteins (TRIM-5α, TRIM-Cyp, described above) target the viral CA at a post-entry step (Ozato et al., 2008). Similarly, since Gag mediates many of its functions (membrane targeting, virus assembly, Env incorporation) while it is still relatively accessible prior to virus release and maturation, there may be an important window of opportunity for antiretroviral intervention during assembly. In this regard, CA assembly and virion maturation seem to be especially promising targets [reviewed in (Adamson and Freed, 2007; Ganser-Pornillos et al., 2008; Salzwedel et al., 2007) and (Adamson et al., 2009)]. FIV infectivity, like that of HIV-1, is highly sensitive to the efficiency and order of Gag processing during virion maturation, which encourages the continued use of FIV as a model for developing PR and maturation inhibitors (Lin et al., 2006).
Although this review is focused primarily on HIV-1 and FIV Gag, many other retrovirus-encoded components required for virus replication are still active targets for antiviral drug development. For example, enzymes encoded by the pol gene must be present for infectivity to be achieved. These enzymes include PR, reverse transcriptase (RT), integrase (IN), and an additional enzyme, dUTPase (DU), found only in non-primate lentiviruses. HIV-1 pol-encoded enzymes have been successfully targeted by PR, RT, and IN inhibitors, and many of these have recently been modeled against homologous FIV pol-encoded enzymes: PR (Heaslet et al., 2007; Lin et al., 2006; Norelli et al., 2008), RT (Martins et al., 2008), and IN (Savarino et al., 2007). Thus, FIV continues to be a useful model for understanding the cellular and molecular biology of HIV-1 replication and in developing more effective antiretroviral therapeutics.
Acknowledgements
We thank members of the Freed lab for critical review of the manuscript. Research in our lab is supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, and by the Intramural AIDS targeted Antiviral Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement We have no conflicts of interest with any persons or organizations that could inappropriately influence our work.
References
- Adamson CS, Freed EO. Human immunodeficiency virus type 1 assembly, release, and maturation. Adv Pharmacol. 2007;55:347–387. doi: 10.1016/S1054-3589(07)55010-6. [DOI] [PubMed] [Google Scholar]
- Adamson CS, Salzwedel K, Freed EO. Inhibiting viral maturation as a novel HIV-1 therapeutic target. Expert Opinion in Therapeutic Targets. 2009 doi: 10.1517/14728220903039714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amodeo P, Castiglione Morelli MA, Ostuni A, Battistuzzi G, Bavoso A. Structural features in EIAV NCp11: a lentivirus nucleocapsid protein with a short linker. Biochemistry. 2006;45:5517–5526. doi: 10.1021/bi0524924. [DOI] [PubMed] [Google Scholar]
- Bieniasz PD. Late budding domains and host proteins in enveloped virus release. Virology. 2006;344:55–63. doi: 10.1016/j.virol.2005.09.044. [DOI] [PubMed] [Google Scholar]
- Brennan G, Kozyrev Y, Hu SL. TRIMCyp expression in Old World primates Macaca nemestrina and Macaca fascicularis. Proc Natl Acad Sci USA. 2008 doi: 10.1073/pnas.0709511105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukrinskaya AG, Brichacek B, Mann A, Stevenson M. Establishment of a functional human immunodeficiency virus type 1 (HIV-1) reverse transcription complex involves the cytoskeleton. J Exp Med. 1998;188:2113–2125. doi: 10.1084/jem.188.11.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkala E, Poss M. Evolution of feline immunodeficiency virus Gag proteins. Virus Genes. 2007;35:251–264. doi: 10.1007/s11262-006-0058-8. [DOI] [PubMed] [Google Scholar]
- Burkhard MJ, Dean GA. Transmission and immunopathogenesis of FIV in cats as a model for HIV. Curr HIV Res. 2003;1:15–29. doi: 10.2174/1570162033352101. [DOI] [PubMed] [Google Scholar]
- Calistri A, Del Vecchio C, Salata C, Celestino M, Celegato M, Gottlinger H, Palu G, Parolin C. Role of the feline immunodeficiency virus L-domain in the presence or absence of Gag processing: involvement of ubiquitin and Nedd4-2s ligase in viral egress. J Cell Physiol. 2009;218:175–182. doi: 10.1002/jcp.21587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S, Rein A. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J Virol. 1999;73:2270–2279. doi: 10.1128/jvi.73.3.2270-2279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S, Vogt VM. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J Virol. 1995;69:6487–6497. doi: 10.1128/jvi.69.10.6487-6497.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celma CC, Paladino MG, González SA, Affranchino JL. Importance of the short cytoplasmic domain of the feline immunodeficiency virus transmembrane glycoprotein for fusion activity and envelope glycoprotein incorporation into virions. Virology. 2007;366:405–414. doi: 10.1016/j.virol.2007.05.019. [DOI] [PubMed] [Google Scholar]
- Chukkapalli V, Hogue IB, Boyko V, Hu WS, Ono A. Interaction between the human immunodeficiency virus type 1 Gag matrix domain and phosphatidylinositol-(4,5)-bisphosphate is essential for efficient gag membrane binding. J Virol. 2008;82:2405–2417. doi: 10.1128/JVI.01614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HY, Morita E, von Schwedler U, Müller B, Krausslich HG, Sundquist WI. NEDD4L overexpression rescues the release and infectivity of human immunodeficiency virus type 1 constructs lacking PTAP and YPXL late domains. Journal of Virology. 2008;82:4884–4897. doi: 10.1128/JVI.02667-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coats KS. The feline immunodeficiency virus-infected cat: a model for lentivirus-induced placental immunopathology and reproductive failure (mini-review) Am J Reprod Immunol. 2005;54:169–185. doi: 10.1111/j.1600-0897.2005.00296.x. [DOI] [PubMed] [Google Scholar]
- Coleman JK, Pu R, Martin M, Sato E, Yamamoto JK. HIV-1 p24 vaccine protects cats against feline immunodeficiency virus infection. AIDS. 2005;19:1457–1466. doi: 10.1097/01.aids.0000183627.81922.be. [DOI] [PubMed] [Google Scholar]
- Dalton AK, Ako-Adjei D, Murray PS, Murray D, Vogt VM. Electrostatic interactions drive membrane association of the human immunodeficiency virus type 1 Gag MA domain. J Virol. 2007;81:6434–6445. doi: 10.1128/JVI.02757-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MR, Jiang J, Zhou J, Freed EO, Aiken C. A mutation in the human immunodeficiency virus type 1 Gag protein destabilizes the interaction of the envelope protein subunits gp120 and gp41. Journal of Virology. 2006;80:2405–2417. doi: 10.1128/JVI.80.5.2405-2417.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- Demirov DG, Freed EO. Retrovirus budding. Virus Res. 2004;106:87–102. doi: 10.1016/j.virusres.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Demirov DG, Ono A, Orenstein JM, Freed EO. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc Natl Acad Sci USA. 2002;99:955–960. doi: 10.1073/pnas.032511899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deneka M, Pelchen-Matthews A, Byland R, Ruiz-Mateos E, Marsh M. In macrophages, HIV-1 assembles into an intracellular plasma membrane domain containing the tetraspanins CD81, CD9, and CD53. J Cell Biol. 2007;177:329–341. doi: 10.1083/jcb.200609050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Griffero F, Kar A, Lee M, Stremlau M, Poeschla EM, Sodroski J. Comparative requirements for the restriction of retrovirus infection by TRIM5alpha and TRIMCyp. Virology. 2007;369:400–410. doi: 10.1016/j.virol.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egberink HF, Ederveen J, Montelaro RC, Pedersen NC, Horzinek MC, Koolen MJ. Intracellular proteins of feline immunodeficiency virus and their antigenic relationship with equine infectious anaemia virus proteins. J Gen Virol. 1990;71(Pt 3):739–743. doi: 10.1099/0022-1317-71-3-739. [DOI] [PubMed] [Google Scholar]
- Egberink HF, Lutz H, Horzinek MC. Use of western blot and radioimmunoprecipitation for diagnosis of feline leukemia and feline immunodeficiency virus infections. J Am Vet Med Assoc. 1991;199:1339–1342. [PubMed] [Google Scholar]
- Elder JH, Schnolzer M, Hasselkus-Light CS, Henson M, Lerner DA, Phillips TR, Wagaman PC, Kent SB. Identification of proteolytic processing sites within the Gag and Pol polyproteins of feline immunodeficiency virus. J Virol. 1993;67:1869–1876. doi: 10.1128/jvi.67.4.1869-1876.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elder JH, Sundstrom M, de Rozieres S, de Parseval A, Grant CK, Lin YC. Molecular mechanisms of FIV infection. Vet Immunol Immunopathol. 2008;123:3–13. doi: 10.1016/j.vetimm.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fackler OT, Krausslich HG. Interactions of human retroviruses with the host cell cytoskeleton. Curr Opin Microbiol. 2006;9:409–415. doi: 10.1016/j.mib.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Franke EK, Yuan HE, Luban J. Specific incorporation of cyclophilin A into HIV-1 virions. Nature. 1994;372:359–362. doi: 10.1038/372359a0. [DOI] [PubMed] [Google Scholar]
- Freed EO. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology. 1998;251:1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- Freed EO, Martin MA. The role of human immunodeficiency virus type 1 envelope glycoproteins in virus infection. J Biol Chem. 1995;270:23883–23886. doi: 10.1074/jbc.270.41.23883. [DOI] [PubMed] [Google Scholar]
- Freed EO, Martin MA. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J Virol. 1996;70:341–351. doi: 10.1128/jvi.70.1.341-351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed EO, Orenstein JM, Buckler-White AJ, Martin MA. Single amino acid changes in the human immunodeficiency virus type 1 matrix protein block virus particle production. Journal of Virology. 1994;68:5311–5320. doi: 10.1128/jvi.68.8.5311-5320.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W, Dang Q, Nagashima K, Freed EO, Pathak VK, Hu WS. Effects of Gag mutation and processing on retroviral dimeric RNA maturation. J Virol. 2006;80:1242–1249. doi: 10.1128/JVI.80.3.1242-1249.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii K, Hurley J, Freed EO. Beyond Tsg101: the role of Alix in `ESCRTing' HIV-1. Nat Rev Microbiol. 2007;5:912–916. doi: 10.1038/nrmicro1790. [DOI] [PubMed] [Google Scholar]
- Gamble TR, Vajdos FF, Yoo S, Worthylake DK, Houseweart M, Sundquist WI, Hill CP. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell. 1996;87:1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- Gamble TR, Yoo S, Vajdos FF, von Schwedler UK, Worthylake DK, Wang H, McCutcheon JP, Sundquist WI, Hill CP. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;278:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- Ganser-Pornillos BK, Cheng A, Yeager M. Structure of full-length HIV-1 CA: a model for the mature capsid lattice. Cell. 2007;131:70–79. doi: 10.1016/j.cell.2007.08.018. [DOI] [PubMed] [Google Scholar]
- Ganser-Pornillos BK, Yeager M, Sundquist WI. The structural biology of HIV assembly. Current Opinion in Structural Biology. 2008;18:203–217. doi: 10.1016/j.sbi.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Côté M, Rich RL, Myszka DG, Sundquist WI. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- Goila-Gaur R, Demirov DG, Orenstein JM, Ono A, Freed EO. Defects in human immunodeficiency virus budding and endosomal sorting induced by TSG101 overexpression. J Virol. 2003;77:6507–6519. doi: 10.1128/JVI.77.11.6507-6519.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goila-Gaur R, Strebel K. HIV-1 Vif, APOBEC, and intrinsic immunity. Retrovirology. 2008;5:51. doi: 10.1186/1742-4690-5-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González SA, Affranchino JL, Gelderblom HR, Burny A. Assembly of the matrix protein of simian immunodeficiency virus into virus-like particles. Virology. 1993;194:548–556. doi: 10.1006/viro.1993.1293. [DOI] [PubMed] [Google Scholar]
- Groot F, Welsch S, Sattentau QJ. Efficient HIV-1 transmission from macrophages to T cells across transient virological synapses. Blood. 2008;111:4660–4663. doi: 10.1182/blood-2007-12-130070. [DOI] [PubMed] [Google Scholar]
- Haedicke J, de Los Santos K, Goff SP, Naghavi MH. The Ezrin-radixin-moesin family member ezrin regulates stable microtubule formation and retroviral infection. J Virol. 2008;82:4665–4670. doi: 10.1128/JVI.02403-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatziioannou T, Cowan S, Goff SP, Bieniasz PD, Towers GJ. Restriction of multiple divergent retroviruses by Lv1 and Ref1. EMBO J. 2003;22:385–394. doi: 10.1093/emboj/cdg042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatziioannou T, Perez-Caballero D, Yang A, Cowan S, Bieniasz PD. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5alpha. Proc Natl Acad Sci USA. 2004;101:10774–10779. doi: 10.1073/pnas.0402361101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaslet H, Lin YC, Tam K, Torbett BE, Elder JH, Stout CD. Crystal structure of an FIV/HIV chimeric protease complexed with the broad-based inhibitor, TL-3. Retrovirology. 2007;4:1. doi: 10.1186/1742-4690-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L, Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol. 2003;19:141–172. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- Hill CP, Worthylake D, Bancroft DP, Christensen AM, Sundquist WI. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc Natl Acad Sci USA. 1996;93:3099–3104. doi: 10.1073/pnas.93.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho MS, Ou C, Chan YR, Chien CT, Pi H. The utility F-box for protein destruction. Cell Mol Life Sci. 2008;65:1977–2000. doi: 10.1007/s00018-008-7592-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohdatsu T, Okada S, Motokawa K, Aizawa C, Yamamoto JK, Koyama H. Effect of dual-subtype vaccine against feline immunodeficiency virus infection. Vet Microbiol. 1997;58:155–165. doi: 10.1016/S0378-1135(97)00164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Orenstein JM, Martin MA, Freed EO. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH, Emr SD. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annual review of biophysics and biomolecular structure. 2006;35:277–298. doi: 10.1146/annurev.biophys.35.040405.102126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH, Lee S, Prag G. Ubiquitin-binding domains. Biochem J. 2006;399:361–372. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javanbakht H, Diaz-Griffero F, Yuan W, Yeung DF, Li X, Song B, Sodroski J. The ability of multimerized cyclophilin A to restrict retrovirus infection. Virology. 2007;367:19–29. doi: 10.1016/j.virol.2007.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Jin L, Peterson DL, Lawson CL. Model for lentivirus capsid core assembly based on crystal dimers of EIAV p26. J Mol Biol. 1999;286:83–93. doi: 10.1006/jmbi.1998.2443. [DOI] [PubMed] [Google Scholar]
- Johnson MC, Spidel JL, Ako-Adjei D, Wills JW, Vogt VM. The C-terminal half of TSG101 blocks Rous sarcoma virus budding and sequesters Gag into unique nonendosomal structures. J Virol. 2005;79:3775–3786. doi: 10.1128/JVI.79.6.3775-3786.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JC, Gasmi M, Lim LE, Elder JH, Yee JK, Jolly DJ, Campbell KP, Davidson BL, Sauter SL. Minimum requirements for efficient transduction of dividing and nondividing cells by feline immunodeficiency virus vectors. J Virol. 1999;73:4991–5000. doi: 10.1128/jvi.73.6.4991-5000.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A, Freed E. HIV-1 Gag trafficking. Future HIV Ther. 2007;1:427–438. [Google Scholar]
- Joshi A, Munshi U, Ablan SD, Nagashima K, Freed EO. Functional replacement of a retroviral late domain by ubiquitin fusion. Traffic. 2008;9:1972–1983. doi: 10.1111/j.1600-0854.2008.00817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N, Neil SJ, Zhadina M, Zang T, Kratovac Z, Lee Y, McNatt M, Hatziioannou T, Bieniasz PD. Broad-spectrum inhibition of retroviral and filoviral particle release by tetherin. J Virol. 2009;83:1837–1844. doi: 10.1128/JVI.02211-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpas A, Lowdell M, Jacobson SK, Hill F. Inhibition of human immunodeficiency virus and growth of infected T cells by the immunosuppressive drugs cyclosporin A and FK 506. Proc Natl Acad Sci USA. 1992;89:8351–8355. doi: 10.1073/pnas.89.17.8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzourakis A, Tristem M, Pybus OG, Gifford RJ. Discovery and analysis of the first endogenous lentivirus. Proc Natl Acad Sci USA. 2007;104:6261–6265. doi: 10.1073/pnas.0700471104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keckesova Z, Ylinen LM, Towers GJ. The human and African green monkey TRIM5alpha genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc Natl Acad Sci U S A. 2004;101:10780–10785. doi: 10.1073/pnas.0402474101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemler I, Barraza RA, Poeschla EM. Mapping the encapsidation determinants of feline immunodeficiency virus. J Virol. 2002;76:11889–11903. doi: 10.1128/JVI.76.23.11889-11903.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikonyogo A, Bouamr F, Vana ML, Xiang Y, Aiyar A, Carter C, Leis J. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc Natl Acad Sci USA. 2001;98:11199–11204. doi: 10.1073/pnas.201268998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratovac Z, Virgen CA, Bibollet-Ruche F, Hahn BH, Bieniasz PD, Hatziioannou T. Primate lentivirus capsid sensitivity to TRIM5 proteins. J Virol. 2008;82:6772–6777. doi: 10.1128/JVI.00410-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambelé M, Labrosse B, Roch E, Moreau A, Verrier B, Barin F, Roingeard P, Mammano F, Brand D. Impact of natural polymorphism within the gp41 cytoplasmic tail of human immunodeficiency virus type 1 on the intracellular distribution of envelope glycoproteins and viral assembly. J Virol. 2007;81:125–140. doi: 10.1128/JVI.01659-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Joshi A, Nagashima K, Freed EO, Hurley JH. Structural basis for viral late-domain binding to Alix. Nat Struct Mol Biol. 2007;14:194–199. doi: 10.1038/nsmb1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin TY, Emerman M. Cyclophilin A interacts with diverse lentiviral capsids. Retrovirology. 2006;3:70. doi: 10.1186/1742-4690-3-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Brik A, de Parseval A, Tam K, Torbett BE, Wong CH, Elder JH. Altered gag polyprotein cleavage specificity of feline immunodeficiency virus/human immunodeficiency virus mutant proteases as demonstrated in a cell-based expression system. J Virol. 2006;80:7832–7843. doi: 10.1128/JVI.00374-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Vergès S, Camus G, Blot G, Beauvoir R, Benarous R, Berlioz-Torrent C. Tail-interacting protein TIP47 is a connector between Gag and Env and is required for Env incorporation into HIV-1 virions. Proc Natl Acad Sci USA. 2006;103:14947–14952. doi: 10.1073/pnas.0602941103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luban J. Cyclophilin A, TRIM5, and resistance to human immunodeficiency virus type 1 infection. J Virol. 2007;81:1054–1061. doi: 10.1128/JVI.01519-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttge BG, Shehu-Xhilaga M, Demirov DG, Adamson CS, Soheilian F, Nagashima K, Stephen AG, Fisher RJ, Freed EO. Molecular characterization of feline immunodeficiency virus budding. J Virol. 2008;82:2106–2119. doi: 10.1128/JVI.02337-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manrique ML, González SA, Affranchino JL. Functional relationship between the matrix proteins of feline and simian immunodeficiency viruses. Virology. 2004a;329:157–167. doi: 10.1016/j.virol.2004.07.029. [DOI] [PubMed] [Google Scholar]
- Manrique ML, Rauddi ML, González SA, Affranchino JL. Functional domains in the feline immunodeficiency virus nucleocapsid protein. Virology. 2004b;327:83–92. doi: 10.1016/j.virol.2004.06.019. [DOI] [PubMed] [Google Scholar]
- Martin-Serrano J. The role of ubiquitin in retroviral egress. Traffic. 2007;8:1297–1303. doi: 10.1111/j.1600-0854.2007.00609.x. [DOI] [PubMed] [Google Scholar]
- Martin-Serrano J, Yarovoy A, Perez-Caballero D, Bieniasz PD, Yaravoy A. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc Natl Acad Sci USA. 2003;100:12414–12419. doi: 10.1073/pnas.2133846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Serrano J, Zang T, Bieniasz PD. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat Med. 2001;7:1313–1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- Martinez NW, Xue X, Berro RG, Kreitzer G, Resh MD. Kinesin KIF4 regulates intracellular trafficking and stability of the human immunodeficiency virus type 1 Gag polyprotein. J Virol. 2008;82:9937–9950. doi: 10.1128/JVI.00819-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins AN, Medeiros SO, Simonetti JP, Schatzmayr HG, Tanuri A, Brindeiro RM. Phylogenetic and genetic analysis of feline immunodeficiency virus gag, pol, and env genes from domestic cats undergoing nucleoside reverse transcriptase inhibitor treatment or treatment-naïve cats in Rio de Janeiro, Brazil. J Virol. 2008;82:7863–7874. doi: 10.1128/JVI.00310-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E, Sundquist WI. Retrovirus budding. Annu Rev Cell Dev Biol. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- Mortola E, Endo Y, Ohno K, Watari T, Tsujimoto H, Hasegawa A. The use of two immunosuppressive drugs, cyclosporin A and tacrolimus, to inhibit virus replication and apoptosis in cells infected with feline immunodeficiency virus. Vet Res Commun. 1998;22:553–563. doi: 10.1023/a:1006197804888. [DOI] [PubMed] [Google Scholar]
- Moscardini M, Pistello M, Bendinelli M, Ficheux D, Miller JT, Gabus C, Le Grice SF, Surewicz WK, Darlix JL. Functional interactions of nucleocapsid protein of feline immunodeficiency virus and cellular prion protein with the viral RNA. J Mol Biol. 2002;318:149–159. doi: 10.1016/S0022-2836(02)00092-X. [DOI] [PubMed] [Google Scholar]
- Münk C, Beck T, Zielonka J, Hotz-Wagenblatt A, Chareza S, Battenberg M, Thielebein J, Cichutek K, Bravo I, O'Brien SJ, Löchelt M, Yuhki N. Functions, structure, and read-through alternative splicing of feline APOBEC3 genes. Genome Biol. 2008;9:R48. doi: 10.1186/gb-2008-9-3-r48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münk C, Zielonka J, Constabel H, Kloke BP, Rengstl B, Battenberg M, Bonci F, Pistello M, Löchelt M, Cichutek K. Multiple restrictions of human immunodeficiency virus type 1 in feline cells. J Virol. 2007;81:7048–7060. doi: 10.1128/JVI.02714-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munshi UM, Kim J, Nagashima K, Hurley JH, Freed EO. An Alix fragment potently inhibits HIV-1 budding: characterization of binding to retroviral YPXL late domains. J Biol Chem. 2007;282:3847–3855. doi: 10.1074/jbc.M607489200. [DOI] [PubMed] [Google Scholar]
- Murakami T, Freed EO. Genetic evidence for an interaction between human immunodeficiency virus type 1 matrix and alpha-helix 2 of the gp41 cytoplasmic tail. J Virol. 2000;74:3548–3554. doi: 10.1128/jvi.74.8.3548-3554.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muriaux D, Mirro J, Harvin D, Rein A. RNA is a structural element in retrovirus particles. Proc Natl Acad Sci USA. 2001;98:5246–5251. doi: 10.1073/pnas.091000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustafa F, Jayanth P, Phillip PS, Ghazawi A, Schmidt RD, Lew KA, Rizvi TA. Relative activity of the feline immunodeficiency virus promoter in feline and primate cell lines. Microbes Infect. 2005;7:233–239. doi: 10.1016/j.micinf.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Naghavi MH, Goff SP. Retroviral proteins that interact with the host cell cytoskeleton. Curr Opin Immunol. 2007;19:402–407. doi: 10.1016/j.coi.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naghavi MH, Valente S, Hatziioannou T, de Los Santos K, Wen Y, Mott C, Gundersen GG, Goff SP. Moesin regulates stable microtubule formation and limits retroviral infection in cultured cells. Embo J. 2007;26:41–52. doi: 10.1038/sj.emboj.7601475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath MD, Peterson DL. In vitro assembly of feline immunodeficiency virus capsid protein: biological role of conserved cysteines. Arch Biochem Biophys. 2001;392:287–294. doi: 10.1006/abbi.2001.2449. [DOI] [PubMed] [Google Scholar]
- Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- Newman RM, Hall L, Kirmaier A, Pozzi LA, Pery E, Farzan M, O'Neil SP, Johnson W. Evolution of a TRIM5-CypA splice isoform in old world monkeys. PLoS Pathog. 2008;4:e1000003. doi: 10.1371/journal.ppat.1000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisole S, Lynch C, Stoye JP, Yap MW. A Trim5-cyclophilin A fusion protein found in owl monkey kidney cells can restrict HIV-1. Proc Natl Acad Sci U S A. 2004;101:13324–13328. doi: 10.1073/pnas.0404640101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norelli S, El Daker S, D'Ostilio D, Mele F, Mancini F, Taglia F, Ruggieri A, Ciccozzi M, Cauda R, Ciervo A, Barreca ML, Pistello M, Bendinelli M, Savarino A. Response of feline immunodeficiency virus (FIV) to tipranavir may provide new clues for development of broad-based inhibitors of retroviral proteases acting on drug-resistant HIV-1. Curr HIV Res. 2008;6:306–317. doi: 10.2174/157016208785132527. [DOI] [PubMed] [Google Scholar]
- Olmsted RA, Hirsch VM, Purcell RH, Johnson PR. Nucleotide sequence analysis of feline immunodeficiency virus: genome organization and relationship to other lentiviruses. Proc Natl Acad Sci USA. 1989;86:8088–8092. doi: 10.1073/pnas.86.20.8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A, Ablan SD, Lockett SJ, Nagashima K, Freed EO. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc Natl Acad Sci USA. 2004;101:14889–14894. doi: 10.1073/pnas.0405596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A, Demirov DG, Freed EO. Relationship between human immunodeficiency virus type 1 Gag multimerization and membrane binding. J Virol. 2000;74:5142–5150. doi: 10.1128/jvi.74.11.5142-5150.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A, Waheed AA, Joshi A, Freed EO. Association of human immunodeficiency virus type 1 gag with membrane does not require highly basic sequences in the nucleocapsid: use of a novel Gag multimerization assay. J Virol. 2005;79:14131–14140. doi: 10.1128/JVI.79.22.14131-14140.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott DE, Coren LV, Johnson DG, Kane BP, Sowder RC, 2nd, Kim YD, Fisher RJ, Zhou XZ, Lu KP, Henderson LE. Actin-binding cellular proteins inside human immunodeficiency virus type 1. Virology. 2000;266:42–51. doi: 10.1006/viro.1999.0075. [DOI] [PubMed] [Google Scholar]
- Ott DE, Coren LV, Kane BP, Busch LK, Johnson DG, Sowder RC, 2nd, Chertova EN, Arthur LO, Henderson LE. Cytoskeletal proteins inside human immunodeficiency virus type 1 virions. J Virol. 1996;70:7734–7743. doi: 10.1128/jvi.70.11.7734-7743.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozato K, Shin DM, Chang TH, Morse HC. TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol. 2008;8:849–860. doi: 10.1038/nri2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecon-Slattery J, Troyer JL, Johnson WE, O'Brien SJ. Evolution of feline immunodeficiency virus in Felidae: implications for human health and wildlife ecology. Vet Immunol Immunopathol. 2008;123:32–44. doi: 10.1016/j.vetimm.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen NC, Ho EW, Brown ML, Yamamoto JK. Isolation of a T-lymphotropic virus from domestic cats with an immunodeficiency-like syndrome. Science. 1987;235:790–793. doi: 10.1126/science.3643650. [DOI] [PubMed] [Google Scholar]
- Peng C, Ho BK, Chang TW, Chang NT. Role of human immunodeficiency virus type 1-specific protease in core protein maturation and viral infectivity. J Virol. 1989;63:2550–2556. doi: 10.1128/jvi.63.6.2550-2556.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu R, Tellier MC, Yamamoto JK. Mechanism(s) of FIV vaccine protection. Leukemia. 1997;11(Suppl 3):98–101. [PubMed] [Google Scholar]
- Rey O, Canon J, Krogstad P. HIV-1 Gag protein associates with F-actin present in microfilaments. Virology. 1996;220:530–534. doi: 10.1006/viro.1996.0343. [DOI] [PubMed] [Google Scholar]
- Ribeiro IP, Menezes AN, Moreira MA, Bonvicino CR, Seuanez HN, Soares MA. Evolution of cyclophilin A and TRIMCyp retrotransposition in New World primates. J Virol. 2005;79:14998–15003. doi: 10.1128/JVI.79.23.14998-15003.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rold CJ, Aiken C. Proteasomal degradation of TRIM5alpha during retrovirus restriction. PLoS Pathog. 2008;4:e1000074. doi: 10.1371/journal.ppat.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulli SJ, Hibbert CS, Mirro J, Pederson T, Biswal S, Rein A. Selective and nonselective packaging of cellular RNAs in retrovirus particles. J Virol. 2007;81:6623–6631. doi: 10.1128/JVI.02833-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad JS, Ablan SD, Ghanam RH, Kim A, Andrews K, Nagashima K, Soheilian F, Freed EO, Summers MF. Structure of the myristylated human immunodeficiency virus type 2 matrix protein and the role of phosphatidylinositol-(4,5)-bisphosphate in membrane targeting. J Mol Biol. 2008;382:434–447. doi: 10.1016/j.jmb.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad JS, Miller J, Tai J, Kim A, Ghanam RH, Summers MF. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc Natl Acad Sci USA. 2006;103:11364–11369. doi: 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenz DT, Teo W, Olsen JC, Poeschla EM. Restriction of feline immunodeficiency virus by Ref1, Lv1, and primate TRIM5alpha proteins. J Virol. 2005;79:15175–15188. doi: 10.1128/JVI.79.24.15175-15188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzwedel K, Martin DE, Sakalian M. Maturation inhibitors: a new therapeutic class targets the virus structure. AIDS reviews. 2007;9:162–172. [PubMed] [Google Scholar]
- Sasaki H, Nakamura M, Ohno T, Matsuda Y, Yuda Y, Nonomura Y. Myosin-actin interaction plays an important role in human immunodeficiency virus type 1 release from host cells. Proc Natl Acad Sci U S A. 1995;92:2026–2030. doi: 10.1073/pnas.92.6.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattentau Q. Avoiding the void: cell-to-cell spread of human viruses. Nat Rev Microbiol. 2008;6:815–826. doi: 10.1038/nrmicro1972. [DOI] [PubMed] [Google Scholar]
- Savarino A, Pistello M, D'Ostilio D, Zabogli E, Taglia F, Mancini F, Ferro S, Matteucci D, De Luca L, Barreca ML, Ciervo A, Chimirri A, Ciccozzi M, Bendinelli M. Human immunodeficiency virus integrase inhibitors efficiently suppress feline immunodeficiency virus replication in vitro and provide a rationale to redesign antiretroviral treatment for feline AIDS. Retrovirology. 2007;4:79. doi: 10.1186/1742-4690-4-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayah DM, Sokolskaja E, Berthoux L, Luban J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 2004;430:569–573. doi: 10.1038/nature02777. [DOI] [PubMed] [Google Scholar]
- Shehu-Xhilaga M, Ablan SD, Demirov DG, Chen C, Montelaro RC, Freed EO. Late domain-dependent inhibition of equine infectious anemia virus budding. J Virol. 2004;78:724–732. doi: 10.1128/JVI.78.2.724-732.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolskaja E, Berthoux L, Luban J. Cyclophilin A and TRIM5alpha independently regulate human immunodeficiency virus type 1 infectivity in human cells. J Virol. 2006;80:2855–2862. doi: 10.1128/JVI.80.6.2855-2862.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack B, Calistri A, Craig S, Popova E, Göttlinger HG. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003;114:689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H, Diaz-Griffero F, Anderson DJ, Sundquist WI, Sodroski J. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci U S A. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuffers S, Brech A, Stenmark H. ESCRT proteins in physiology and disease. Exp Cell Res. 2008 doi: 10.1016/j.yexcr.2008.10.013. [DOI] [PubMed] [Google Scholar]
- Sundstrom M, Chatterji U, Schaffer L, de Rozières S, Elder JH. Feline immunodeficiency virus OrfA alters gene expression of splicing factors and proteasome-ubiquitination proteins. Virology. 2008;371:394–404. doi: 10.1016/j.virol.2007.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Winkler U, Freed EO, Torrey TA, Kim W, Li H, Goff SP, Morse HC. Cellular motor protein KIF-4 associates with retroviral Gag. J Virol. 1999;73:10508–10513. doi: 10.1128/jvi.73.12.10508-10513.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomonaga K, Itagaki SI, Kashiwase H, Kawaguchi Y, Inoshima Y, Ikeda Y, Mikami T. Characterization of an integrase mutant of feline immunodeficiency virus. Arch Virol. 1998;143:1–14. doi: 10.1007/s007050050263. [DOI] [PubMed] [Google Scholar]
- Towers GJ. The control of viral infection by tripartite motif proteins and cyclophilin A. Retrovirology. 2007;4:40. doi: 10.1186/1742-4690-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyer JL, Pecon-Slattery J, Roelke ME, Johnson W, VandeWoude S, Vazquez-Salat N, Brown M, Frank L, Woodroffe R, Winterbach C, Winterbach H, Hemson G, Bush M, Alexander KA, Revilla E, O'Brien SJ. Seroprevalence and genomic divergence of circulating strains of feline immunodeficiency virus among Felidae and Hyaenidae species. J Virol. 2005;79:8282–8294. doi: 10.1128/JVI.79.13.8282-8294.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme N, Goff D, Katsura C, Jorgenson RL, Mitchell R, Johnson MC, Stephens EB, Guatelli JC. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host & Microbe. 2008;3:245–252. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VerPlank L, Bouamr F, LaGrassa TJ, Agresta B, Kikonyogo A, Leis J, Carter CA. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag) Proc Natl Acad Sci USA. 2001;98:7724–7729. doi: 10.1073/pnas.131059198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgen CA, Kratovac Z, Bieniasz PD, Hatziioannou T. Independent genesis of chimeric TRIM5-cyclophilin proteins in two primate species. Proc Natl Acad Sci USA. 2008;105:3563–3568. doi: 10.1073/pnas.0709258105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willett BJ, Flynn JN, Hosie MJ. FIV infection of the domestic cat: an animal model for AIDS. Immunol Today. 1997;18:182–189. doi: 10.1016/s0167-5699(97)84665-8. [DOI] [PubMed] [Google Scholar]
- Williams RL, Urbe S. The emerging shape of the ESCRT machinery. Nat Rev Mol Cell Biol. 2007;8:355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- Wilson SJ, Webb BL, Ylinen LM, Verschoor E, Heeney JL, Towers GJ. Independent evolution of an antiviral TRIMCyp in rhesus macaques. Proc Natl Acad Sci USA. 2008;105:3557–3562. doi: 10.1073/pnas.0709003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler IG, Löchelt M, Flower RL. Epidemiology of feline foamy virus and feline immunodeficiency virus infections in domestic and feral cats: a seroepidemiological study. J Clin Microbiol. 1999;37:2848–2851. doi: 10.1128/jcm.37.9.2848-2851.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D, Goff SP. Host restriction factors blocking retroviral replication. Annu Rev Genet. 2008;42:143–163. doi: 10.1146/annurev.genet.42.110807.091704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Yu X, Lee TH, Essex M. Mutations in the N-terminal region of human immunodeficiency virus type 1 matrix protein block intracellular transport of the Gag precursor. J Virol. 1993;67:6387–6394. doi: 10.1128/jvi.67.11.6387-6394.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhadina M, McClure MO, Johnson MC, Bieniasz PD. Ubiquitin-dependent virus particle budding without viral protein ubiquitination. Proc Natl Acad Sci U S A. 2007;104:20031–20036. doi: 10.1073/pnas.0708002104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Parent LJ, Wills JW, Resh MD. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J Virol. 1994;68:2556–2569. doi: 10.1128/jvi.68.4.2556-2569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]