Abstract
Evaluating the free energy landscape of proteins and the corresponding functional aspects presents a major challenge for computer simulation approaches. This challenge is due to the complexity of the landscape and the enormous computer time needed for converging simulations. The use of simplified coarse grained (CG) folding models offers an effective way of sampling the landscape but such a treatment, however, may not give the correct description of the effect of the actual protein residues. A general way around this problem that has been put forward in our early work (Fan et al, Theor Chem Acc (1999) 103:77-80) uses the CG model as a reference potential for free energy calculations of different properties of the explicit model. This method is refined and extended here, focusing on improving the electrostatic treatment and on demonstrating key applications. This application includes: evaluation of changes of folding energy upon mutations, calculations of transition states binding free energies (which are crucial for rational enzyme design), evaluation of catalytic landscape and simulation of the time dependent responses to pH changes. Furthermore, the general potential of our approach in overcoming major challenges in studies of structure function correlation in proteins is discussed.
Keywords: Coarse Grained model, free energy calculations, dielectric constants, proton transfer
1. Introduction
The elucidation of the landscape of proteins is an important element in attempts to explore the energetics and kinetics of the folding process. Furthermore, the ability to explore and sample large regions in the accessible conformational space can help in improving the description of functional properties and in exploring possible relationships between landscape and functions. Unfortunately, detailed sampling of protein landscapes requires enormous computational resources that are not generally available. Thus it is important to develop multiscale approaches that will allow one to effectively generate the free energy surface for folding and related processes. Simplified protein models have been in use since our early work in 19751 and are being used extensively in folding studies2-8. Furthermore, recent efforts have extended our early 1975 idea into membranes (see MARTINI force field ref9). However such coarse grained (CG) models are, by their nature, approximated and it is important to be able to move from the coarse-grained description to more realistic explicit models. A general way to resolve this problem has been introduced by us some time ago10, where we proposed the use of a simplified model as a reference potential for explicit calculations of folding free energies. This idea, which can be now classified under the umbrella name of multiscale modeling, became even more important in recent years in view of the interest in the relationship between landscape and function7,8,11-13.
The idea of using reference potential in multiscale modeling has been exploited by us in a wide range of problems including accelerating QM/MM calculations (ref14-16) and path integral calculations of nuclear quantum mechanical effects (ref17,18). The idea of using CG model as reference potential in folding studies10 and related processes has been also explored recently by other workers (e.g. refs8,19,20). Here we describe the details of the current version of our multiscale landscape modeling and provide instructive illustrations of its use in calculating the change in protein stability upon mutation. We also outline other promising applications of this powerful model.
2. Methods
2.1. The Simplified Protein Model
The simplified protein model used in this work is an extension of the model originally developed by Levitt and Warshel1 and modified in ref.10 In the current model we have made additional refinements of the original model, focusing on the introduction of the specialized solvation terms, designed to replicate the effects of the missing solvent molecules and on other factors that are not included explicitly.
Our simplified protein model, which is depicted schematically in Fig. 1, is created by replacing the side chain of each residue by an effective “atom” named (X) and an additional dummy atom named (D). The atom X is usually placed at the geometrical center of the heavy atoms of the corresponding side chains (with a residue dependent charge and van der Waals equilibrium radius), where, the distance between the newly placed X atom and the Cα atom, rCa−X, depends upon the type of the side chain of the residue it replaces. The dummy atoms are placed along the corresponding Cα - Cβ vectors and serve as tools for rotational transformations in the process of moving between the simplified and explicit models. That is, the Cα − D bond is used to define the Cα - Cβ bond of the explicit model. The actual rotational transformations are done by using simple rotation around the specific bond. The Dummy atoms do not have any charge or van der Waals interaction with the rest of the system. The backbone atoms of each residue are treated explicitly, and the interactions between main chain atoms are identical to those used in the explicit model but then modified to reflect the missing solvent terms. In the case of ionized residues, we shift the rCa−X of atom X to the charged center of the given residue.
Figure 1.
A schematic illustration of the conversion of an explicit side chain to its simplified equivalent. The position of the explicit Cβ is preserved, using a dummy atom, named D, and the geometrical center of the side chain heavy atoms is represented in the simplified model by the effective atom, named X.
The potential energy surface of the simplified (sp) model is written as
| (1) |
Where m and s designate, respectively, “main” and “side”, U0 is the gas phase potential of the indicated term, is the missing effect of the solvent and the protein. The more detailed expression on the right hand side involves the effective steric potential (Uef), the screened interaction between different charges (Q and q designate full charges of ionized groups and atomic residual charges, respectively), the ΔU terms represent the corrections to the indicated potential terms due to the solvent and is the self energy of the ionized groups. The nature of the different terms is described below.
2.1.1. The side-side interaction potential
The interaction between the simplified side chains is given by,
| (2) |
where is described by an “8-6” potential of the form,
| (3) |
where and . The parameters and define, respectively, the well depth and equilibrium distance. The unitless parameter Cscale has a value of 10 when is calculated for polar - non polar and ionized – non polar residues only. For side chain interactions between polar – polar and non polar – non polar residues, Cscale has a value of 1. These parameters were refined by minimizing the root-mean-square deviations between the calculated and observed values of both the atomic positions and the protein size (i.e., the radii of gyration) for a series of proteins. The corresponding refined parameters are given in Table I, together with the corresponding rCa−X parameters. The term represents the charge-charge interaction in the gas phase. This term is given by,
Table I.
Parameters for the Uef terma
| Residue Name | εi (kcal mol-1) | Polarity | ||||
|---|---|---|---|---|---|---|
| A | 4.26 | 0.05 | nonpolar | 1.52 | ||
| C | 4.79 | 1.17 | polar | 2.46 | ||
| bD | 4.45 | 1.17 | polar | 2.49 | ||
| bE | 4.68 | 1.17 | polar | 3.40 | ||
| F | 4.55 | 0.13 | nonpolar | 3.30 | ||
| H | 4.75 | 1.17 | polar | 3.20 | ||
| I | 4.68 | 0.18 | nonpolar | 2.60 | ||
| bK | 4.74 | 1.17 | polar | 3.92 | ||
| L | 4.72 | 0.18 | nonpolar | 2.56 | ||
| M | 4.76 | 0.17 | nonpolar | 3.24 | ||
| N | 4.59 | 1.17 | polar | 2.52 | ||
| P | 4.72 | 0.31 | nonpolar | 2.46 | ||
| Q | 4.79 | 1.17 | polar | 3.10 | ||
| bR | 4.47 | 1.17 | polar | 4.11 | ||
| S | 4.54 | 1.17 | polar | 2.46 | ||
| T | 4.62 | 1.17 | polar | 2.46 | ||
| V | 4.70 | 0.14 | nonpolar | 2.06 | ||
| W | 4.48 | 0.35 | nonpolar | 3.30 | ||
| Y | 4.51 | 1.17 | polar | 3.40 |
and εi are used for all interactions between simplified mode residues. is the equilibrium distance between the Cα and the X atom of the given simplified residue.
For the ionizable residues, the equilibrium distance is shifted towards the geometrical center of the ionizable functional group.
| (4) |
where Qi designates the charge of the ith ionized residue, εeff is the effective dielectric for charge-charge interactions. Here we reflect the idea established in many of our works (e.g. refs21,22) and used a large effective dielectric, εeff (εeff = 40) even for protein interiors. This type of dielectric has been found recently to provide very powerful insight in studies of protein stability (see refs21,23) and thus expected to be very useful in modeling the electrostatic contribution to the stability of the simplified model.
2.1.2. Effective electrostatic terms that involve the main chain
The effective electrostatic interactions between the side chains and the main chain and between the main chain groups and themselves are given by
| (5) |
Here Qi and qk designate the charges of ionized residues and residual atomic charges on the main chain respectively (there are no residual charges on the simplified unionized side chains). The different dielectrics represent the compensation of the gas phase electrostatic interactions by the solvent and the protein. The dielectric for the Q and q terms is taken (following our previous studies22) as εeff = 10 for charge (Q)-residual (q) charge interaction and εeff = 4 for residual charge(q)-residual charge(q) interaction. The potential, U, is given in kcal/mole, where Q and q are given in atomic charges and r in Å. The factor 332 in Eq. (5) is used for these units (see Warshel and Russell early work24 for further details). Note that is the sum of the gas phase qq part of Umm plus the screening by the surrounding solvent and the protein. This treatment also includes the residual charge interactions that are involved in hydrogen bonding.
2.1.3 The self energy term
The self energy term represents the interaction of the side chain with its environment and accounts for the crucial free energy of transferring the side chain to the protein when all other groups are not ionized (note that the effect of interaction with other ionized groups is accounted separately by ). The overall term is given by,
| (6) |
where i runs over all ionized residues and and are the number of nonpolar and polar residues in the neighborhood of the ith residue. The functions Unp and Upolar are given by
| (7) |
and
| (8) |
The number of nonpolar residues neighboring the ith ionized residue is expressed by the analytical function
| (9) |
With
| (10) |
where rnp is the nonpolar radius defining the cutoff range of nonpolar neighboring residues (typically rnp = 7Å), and rij is the separation between the ionized residue i and nonpolar residue j. The same expression is used for neighboring polar residues (Npolar)., where rp = 7Å. This treatment is aimed at capturing the fact that an ionized group has to pay very large amount of energy for moving from water to a nonpolar environment24,25 and is usually surrounded by polar residues or water molecules22,24.
It should be noted that the calculations of Np, and Nnp are done while treating the ionizable residues as non polar residues. This is done since the effect of charging these groups is taken into account separately by the term.
The parameters in Eqs. (6-10) were determined (see section 3.1.3) by comparing the results obtained with these equations to the results of the actual solvation energy evaluated by the semimacroscopic version of the Protein Dipole Langevin Dipole (PDLD/S) in its linear response approximation (LRA) version (PDLD/S-LRA).
In principle it would be useful to include in the hydrophobic contribution of non polar residues and the effect of the solvent on the self energy of polar residues. However, these contributions are included implicitly in and thus we only consider explicitly the of ionized residues. This is done by using different in Eq. (3) for hydrophobic and polar residues.
2.1.4. The main chain torsional potential
The secondary structure of proteins depends strongly on the solvation of the main chains. Thus we added the correction potential that is used to modify the gas phase potential. This solvation potential is given by,
| (11) |
Where
| (12) |
The values of and are chosen to represent the minima of the α-helix and β-sheet regions of the Ramachandran plot, while Ai and have been selected to tune the simple model α-helix and β-sheet regions to match those of the explicit model (see section 3.1.1). The specific values of these parameters are listed in Table II.
Table II.
Parameters for the terma
| Region | Ai (kcal mol-1) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| α - helix | -1 | -95 | 8 | -5 | 16 | ||||
| β - sheet | -10 | -150 | 10 | 175 | 16 | ||||
| γ′-turn | -3 | -80 | 6 | 65 | 5 | ||||
| L-α - helix | -5 | 60 | 5 | 40 | 16 |
Angular values are given in degrees.
2.1.5. The effective hydrogen bonding potential
Solvation effects play a crucial role in determining hydrogen bonding. A part of this effect is taken into account by the of Eq. (5). However this uniform screening does not yield the correct physics of hydrogen bonding as established by our simulated studies where the RMS between the calculated and observed structure drift upward over time. Furthermore, even the introduction of have not resolved this problem.
In order to obtain better hydrogen bonding representation we introduced a potential , where a small Gaussian barrier was added to the potential well used to describe hydrogen bonds. At room temperature, this barrier is easily crossed during hydrogen bond formation while preventing the already formed bonds from breaking. This treatment gives the functional form
| (13) |
The inclusion of both and greatly increases the long term stability of the simple model, with a maximum RMS deviation of less than 3.0 Å over the course of a 100 ps trajectory.
2.1.6. The effective side chain-main chain potential
The effective non electrostatic interaction between the side chain and the main chain, , is represented by the same type of potential as the potential in Eq. (3) (see Table I).
2.2. Simple to Explicit Transformation
The greatest difficulty to multiscale simulations lies in accurately and efficiently recreating the original all atom system from the simplified model system. Our strategy is based on the approach introduced by Fan et. al.10 In this approach we considered the partition functions Qsp and Qep of the simplified and explicit models, respectively, as well as the ration between them, Qsp / Qep. This gives the free energy of transfer between the surfaces by
| (14) |
where β = 1/kBT, kB being the Boltzmann constant and T being the absolute Temperature. In order to evaluate Eq. (14), we start by expressing the simplified and explicit potentials as
| (15) |
and
| (16) |
where R̃ are the coordinates of the simplified model and r̃ are the coordinates of the explicit side chain atoms, relative to the centers of the corresponding side chains in the simplified model. Now our task is to evaluate the free energy of moving from the simplified to the explicit model, ΔGsp→ep, or −ΔGep→sp. This can be expressed as the ratio between the corresponding partition function
| (17) |
Thus we have
| (18) |
where the average is done formally over the combined R̃+r̃ coordinate set. Eq. (18) can be evaluated by using a free energy perturbation approach with a mapping potential:
| (19) |
so that we have,
| (20) |
with
| (21) |
In many applications we are not interested in the total partition function and the total free energy, but in the potential of mean force (PMF) of the simplified and explicit models. In such cases we define the PMF by considering a partial partition function
| (22) |
and
| (23) |
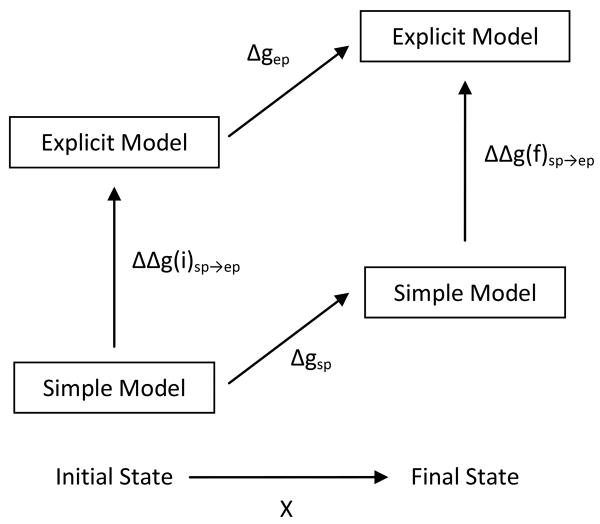
where X is the parameter that defines the PMF. With Δgsp(X) and Δgep(X) defined in the thermodynamic cycle of Fig. 2. The key for the transition between the simplified and explicit PMFs is the ratio
Figure 2.
The thermodynamic cycle used to calculate the change in free energy Δgep for a generic process. Having calculated the free energy change of the simple model, Δgsp, umbrella sampling can be used to calculate the free energy change ΔΔgsp→ep for the initial and final states to obtain Δgep.
| (24) |
Here we follow the same strategy as in Eq. (18) and obtain,
| (25) |
If we use the mapping potential of Eq. (19) we will have,
| (26) |
with,
| (27) |
In evaluating the free energy terms of Eqs. (18) or (27), we start typically from the simplified model in a given configuration and construct the explicit model by replacing the dummy atom with the explicit Cβ. Then, using a rotamer library26 (with random search followed by brief steepest decent torsional minimization) we select the side chain configurations that minimize the potential energy of the system, while also minimizing the displacement between the center of the explicit side chain and the position of the simplified atom. After this treatment that considers all the rotational degrees of freedom within the side chains, we apply a steepest descent minimization over the Cartesian degrees of freedom within each side chain to obtain the relaxed side chains structure. Once the explicit structure has been minimized we can use Molecular Dynamics (MD) or Monte Carlo (MC) for the evaluation of the free energy of moving from the simplified to the explicit model.
3. Results and Discussion
3.1. Validation and Parameterization
A crucial element of the introduction of any analytic potential surface is the parameterization and validation process. This section considers aspects of the refinements that were not considered in the previous sections.
3.1.1. Refining
The optimization of the parameters that determine the solvation contribution from and involved the use of the alanine dipeptide as a model system. The explicit free energy surface for this system was generated both for a gas phase model and for a solution model (using the surface constrained all atom solvent (SCAAS) model27,28). In order to recreate the hydrated landscape by the simplified model, it was necessary to add to Uphi−psi four potential wells at the minima of the α- helix (φ ∼ -90°, ψ ∼ -30°), β-sheet (φ ∼ -160°, ψ ∼ 160°), γ′-turn (φ ∼ -90°, ψ ∼ 60°), and L-α-helix (φ ∼ 60°, ψ ∼ 40°) regions of the Ramachandran (RC) diagram. The relevant parameters are given in Table II.
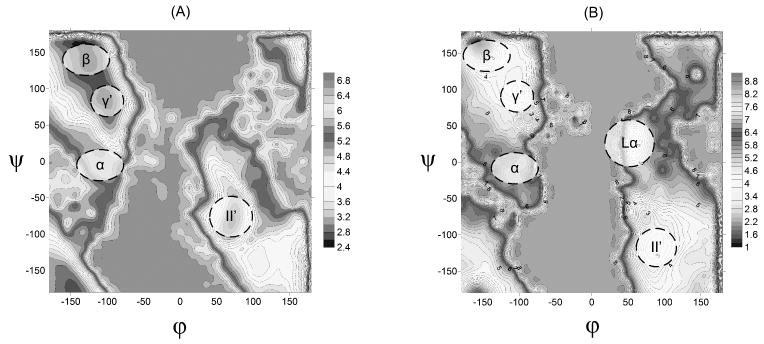
The addition of the term (and in some respects the ) to the simplified model created a surface with minima at the typical regions obtained in other works (e.g. Lovell et al.29). Our RC diagram is presented in Fig. 3 for both the explicit and simplified models. Apparently the simplified surface is not perfect, but allows us to capture the main features of the explicit model. The notations for the different regions are taken from the ref29.
Figure 3.
The Ramachandran diagram for alanine dipeptide, obtained by the explicit model (A) and the simplified model (B).
3.1.2. Examination of ΔUHB
The term is strongly coupled to and it is important to examine whether in addition to reproducing reasonable phi-psi map, we can retain a reasonable description of stable helices and other structures that are determined by hydrogen bonding. With the we find that a small barrier to hydrogen bond formation (∼1 kcal mol-1) is easily crossed at room temperature, but forms a significant barrier to the breaking of hydrogen bonds once they have been formed. By including ΔUHB in Eq. (1), we find that even full scale protein systems like ubiquitin or chorismate mutase21 are structurally stable (RMS displacement < 3 Å) for more than 100 ps of simulation.
3.1.3. Refining
The introduction of is perhaps the most important new element in our approach. This potential clearly improves the physical basis of our model but the reliability of the model still depends crucially on the parametization of the self energy potential. The refinement procedure was done by selecting different ionizable groups in several protein sites, evaluating their self energy using the PDLD/S-LRA approach28 and adjusting the parameters in to reproduce the best agreement between the PDLD/S results and those of the simplified model.
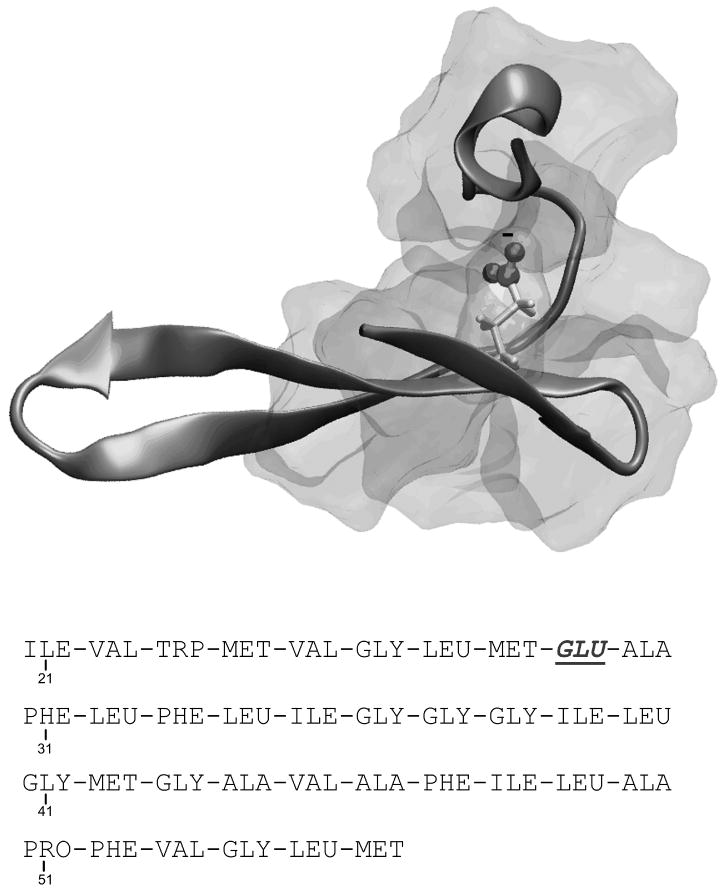
In the first step of this refinement procedure we started by considering in a fully hydrophobic, non polar, protein environment. For this purpose we created the relevant environment by using in-silico methods. That is, we mutated, in-silico, residues 21-56 (out of the total of 62 residues) of the protein SSO7d (PDB ID 1SSO) to all-hydrophobic residues, except Ile29 which we mutated into Glu. This created an all-hydrophobic environment surrounding the ionizable residue Glu29 (see Fig. 4), where Glu29 is buried, by residues 22 to 33 and 43 to 56. This model which is referred to as “the fully buried Glu 29” was then modified creating two additional environments for Glu29. In the second, which is referred to as a “semi-buried” environment, we deleted residues 49 to 56, making Glu29 more exposable to solvent. Finally for the third, which is refereed to here as “exposed” environment, we mutated Val22, Met24 and Phe47 into Glycine, making Glu29 fully exposed to solvent.
Figure 4.
The system used as a benchmark for the behavior of the self energy term in non polar environment. The backbone conformation of protein 1SSO residues 21-56 was taken, and the fully hydrophobic sequence after the mutations is reported in the Figure. The figure describes the hydrophobic environment generated around Glu29, buried by residues 22-33 and 43-56.
Next we calculated the solvation free energy of the ionized Glu29, in all 3 hydrophobic environments described above. We used the PDLD/S-LRA as well as simplified model's Eqs. (6 – 10), and adjusted the parameters in these equations to get the best agreement between the two values. The results for simplified and PDLD/S-LRA salvation energy for the artificial protein environments are given in the first three entries of Table III.
Table III.
Comparison of the self energy calculated by the CG and the PDLD/S-LRA models
| Test Systems | The CG model (kcal/mole) |
The PDLD/S-LRA (kcal/mole) |
|---|---|---|
| Glu29 burried | 3.9 | 5.2 |
| Glu29 semi buried | 3.2 | 3.8 |
| Glu29 exposed | -0.1 | 1.1 |
| Apoflavodoxin Glu 106 | 3.7 | 4.7 |
| Dimer DHFR ARG 28 | 2.9 | 2.3 |
| Dimer DHFR Asp 117 | 0.3 | -0.6 |
| Interleukin Glu 111 | 3.0 | 4.5 |
| Interleukin Glu 113 | 3.6 | 3.3 |
After refining the simplified model to obtain the best description of the effect of nonpolar residues on the self-energy, we turned to the refinement of the parameters that reflect the polar contributions. This was done by the same procedure described above but now for a number of real proteins sites (where we have significant number of polar residues). The protein sites were taken from three diverge proteins: Apoflavodoxin (PDB ID 1FTG) residue site Glu106, Tm DHFR (PDB ID 1CZ3) residue sites Arg28 and Asp117, and Interleukin (PDB ID 1IOB) residue sites Glu111 and Glu113. Following the same steps as with the artificial hydrophobic environments, we calculated by using both the PDLD/S-LRA and the simplified model's approach and then refined the parameters in Eqs. (6-10). The self energies obtained by both approaches are given in Table III. The final set of refined parameters for are given in Table IV. As seen from the tables, the agreement between simplified model , and PDLD/S-LRA is quite encouraging.
Table IV.
Refined distance rij parameters for CG model
| Distance parameters | Best fitted maximum valuesa |
|---|---|
| rij polar | 5 Å |
| rij non polar | 7 Å |
The best fitted values for the distances used in for the calculation of polar and non polar G(rij). The distances rij, polar and non polar, should not exceed the values shown on the right column
3.2. Evaluating the free energy of transition between the simplified and explicit surfaces
In considering the transition from simplified CG models to explicit models, we start by noting that Eq. (14) is not conceptually tied to a given type of reaction or process. Rather, it represents a general means of converting between two model representations of the same system. The thermodynamic cycle of this generic transformation is given in Fig. 2. Previously10, we have shown that this process can be exploited in calculating the PMF for unfolding processes, using the radius of gyration as a means of denaturing the system from the folded to the unfolded state. We have also shown that it is possible to use simplified model structures to obtain explicit structures of both folded and unfolded proteins12. In this work we explore other applications of our general strategy. This is done in the following subsection.
3.2.1 Evaluating changes in Folding Energy
One possible application of our model is the evaluation of the effect of mutation on protein stability. Although this can be done by evaluating the folding PMF for both systems, it is much simpler to do so by using the thermodynamic cycle of Fig. 5. More specifically, ultimately we are interested in evaluating:
Figure 5.
The thermodynamic cycles used to calculate the change in free energy of unfolding upon mutation (see text for details)
| (29) |
where and are the free energies of the mutant and native forms of the protein, respectively, evaluated by the explicit model. can be evaluated using the thermodynamic cycle of Fig. 5, yielding
| (30) |
where is the folding free energy of the mutant in the simplified model, while , are the changes in free energy of the transformation from the simplified model to the explicit model for the folded and unfolded mutant, respectively. The folding energy of the native system is produced through the same process as the mutant. However, rather than calculating the folding energy directly, we can use the thermodynamic cycle inside the grey box in Fig. 5 and obtain
| (31) |
It is therefore possible to calculate the change in folding free energy in the simple model by comparing the free energy of mutating the protein in the folded and unfolded systems. Ultimately, combining Eqs. (29 – 31), and the thermodynamic cycle of Fig. 5 gives,
| (32) |
While Eq. (32) appears more complicated than evaluating Eq. (29) directly, it has definite advantages. First, calculating ΔΔGuf−f via Eq. (32) requires only that we calculate the free energy change in the mutation process only in the simple model (this is done in the folded and unfolded states). With the exception of mutations to or from glycine, mutating residues within the simplified model is extremely straightforward. All that is required is the change of the parameters associated with the X atom for the residue being mutated. Furthermore, it is not necessary to force the folded system to unfold. Additionally, the unfolded state can be modeled using only the neighboring residues to the residue to be mutated, greatly simplifying the calculation of and reducing the computational cost involved. These computational savings can be reinvested by increasing the number and length of frames used to calculate and via the free energy perturbation method, which greatly improves the accuracy of the calculation.
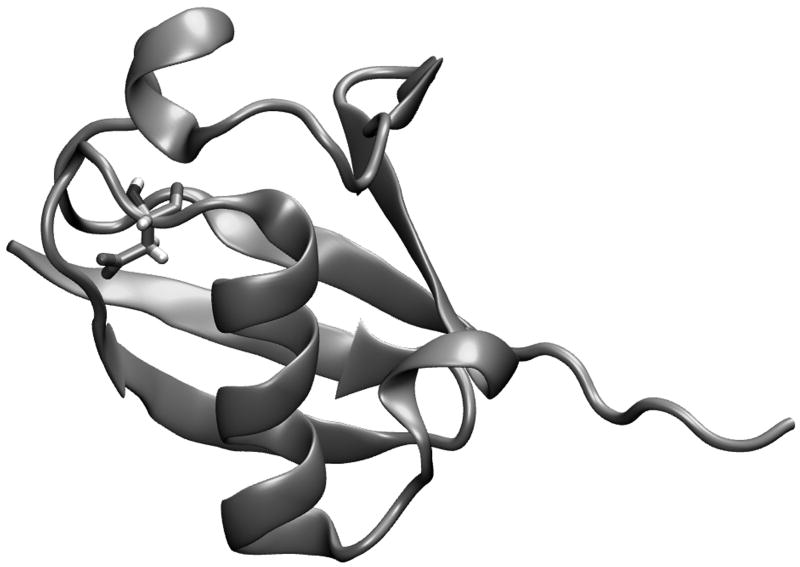
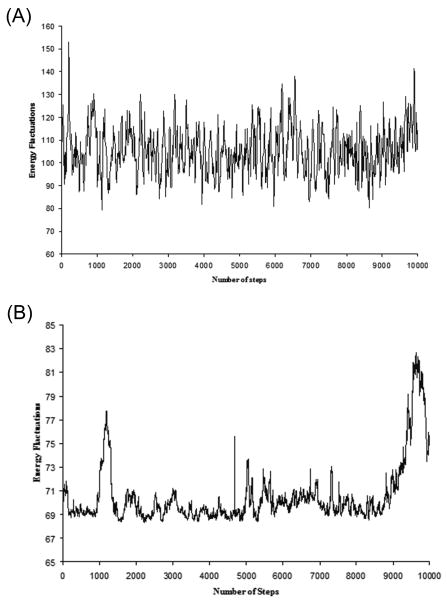
As a test case for the performance of Eq. (32), we have chosen to examine the pseudo wild-type ubiquitin and the Asp21Asn mutant discussed in our previous work21 (see also ref30). The three dimensional representation of this system is shown in Fig. 6. We have chosen this system because it has been well studied by other means21, and at 76 residues is large enough and has a compact enough secondary structure to represent an actual system of interest. Our FEP calculations were performed by both MD and Monte Carlo simulations. The calculations started with 10,000 MD relaxation steps at 300 K, using 1 fs for each step. The main chain and residues far from the mutated residues were fixed during this relaxation runs. The resulting explicit structure was then used to generate the simplified structure. Four such simplified structures were generated. Starting from the given simplified structure we simulated the motions of the explicit model by either MD or MC simulations at 300 K with the mapping potential of Eq. (19) and evaluated the free energy of moving from the simplified to the explicit model by a FEP approach. The key aspect of our procedure is demonstrated in Fig. 7, where we describe the fluctuations of the gap between the explicit and simplified potential for trajectories on the given mapping potential of the native protein (in this case the sixth window with λ = 0.5). The same procedure was repeated for 11 frames and the overall free energy of moving from the simplified to the explicit potential was calculated ( ). The same procedure was used to evaluate for the Asp 21Asn mutant. The calculations for the unfolded protein ( and ) are shown in Table V.
Figure 6.
A three dimensional representation of pseudo wild type ubiquitin. Asp21, which is involved in the mutational study, is represented explicitly.
Figure 7.
The fluctuations of the energy difference between the explicit and simplified potentials, obtained during (A) FEP/MD, (B) Monte Carlo, simulations
Table V.
The energetics of the Asp21Asn mutation in Ubiquatin.a
|
(calc) |
(obs) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (FEP/MD) | 67.40 | 64.36 | -179.89 | -172.94 | 13.77 | 8.57 | 0.73 | 0.85 | ||||||
| (MC) | 67.40 | 64.36 | -179.89 | -172.94 | 8.02 | 6.58 | 2.47 | 0.85 |
3.2.2 Evaluating transition state binding free energy
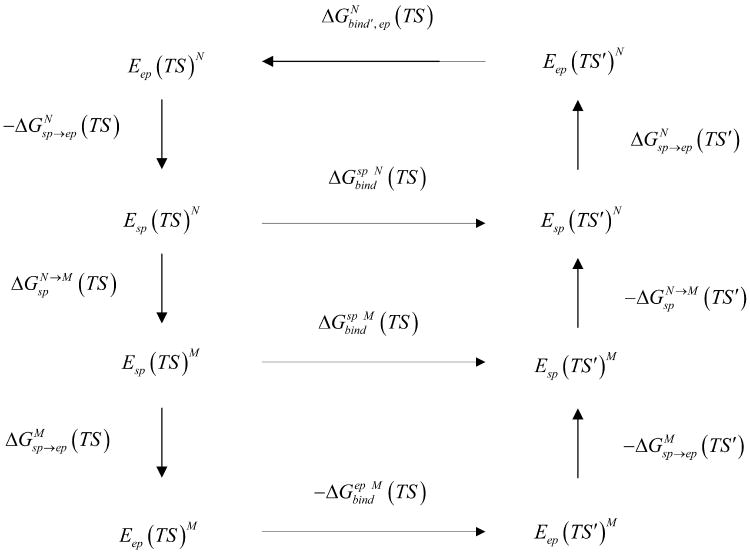
Another important use of our approach is in the field of enzyme design where it can be used to evaluate the binding free energy of rate determining transition states. This can be done by focusing on the electrostatic free energy contribution ΔGbind′, while using the cycle of Fig. 8, which leads to the following expression:
Figure 8.
The cycle used to evaluate mutational effects on transition states binding free energies.
| (33) |
Where and are the changes in the free energy of transforming the simplified model to the explicit model, for the fully charged TS in the native and mutant, respectively. is the difference in free energy between native and mutant simplified structures for the fully charged TS. and are the changes in the free energy of transforming the simplified model to the explicit model for the zero charged TS, in the native and mutant, respectively. is the difference in free energy between native and mutant simplified structures for the zero charged TS.
The calculations were performed on monomeric chorismate mutase (mMjCM) (whose active site is depicted in Fig. 9). The coordinates mMjCM (NMR) structure were taken from PDB access code 2GTV. The native protein preceded to 100ps MD simulations in order to obtain a relaxed structure. The resulting native explicit structure was then used to generate the simplified structure. Both the simplified and explicit models included an explicit empirical valence bond (EVB) description of the reacting system (see ref31 for details). Three such simplified structures were generated and used to simulate the fluctuations of the explicit model by MC moves of the torsional coordinates. The simulations were done at 300 K with the mapping potential of Eq. (19) and evaluated the free energy of moving from the simplified to the explicit model with full EVB transition state charges and zero EVB transition state charges. We have used a simulation length of 1 ps for each of the 11 mapping steps and the overall free energy of moving from the simplified to the explicit potential was calculated for the charged ( ) and zero charged transition state. We choose as test cases the mutations of Gln88 and Arg51 since both (particularly Gln88) were found to present a significant challenge in our recent exploration of the potential of the EVB in enzyme design31. Here we mutated Gln88 to Asn88 and repeated the calculation described above. We also mutated Arg51 to Gln51 and repeated the calculations. The results are shown in Table VI. Finally, we calculated by taking the corresponding change in average energy (see results in Table VI).
Figure 9.
A schematic description of the mMjCM active sites, depicting key residues that are involved in the binding of the transition-state analogue (bold).
Table VI.
The energetics of the Gln88Asn and Arg51Gln mutations in monomeric chorismate mutase (mMjCM).a
| Gln88Asn | Arg51Gln | ||
|---|---|---|---|
| Free energies values | |||
|
|
64.50 | 57.46 | |
|
|
77.30 | 23.50 | |
|
|
16.69 | 3.57 | |
|
|
27.14 | 105.93 | |
|
|
37.89 | 71.21 | |
|
|
12.80 | 0.5 | |
|
|
5.94 | 3.83 | |
|
|
4.76 | 3.27 |
Energies in kcal/mol.
TS and TS′ designate, respectively, the transition state with its full charges and without any residual charges.
experimental values are taken from on Woycechowsky et al42. The observed value of is given by
3.3 Other applications
The use of the CG model as a reference potential can facilitate studies of other long-standing problems and here we list several new key applications.
3.3.1 Studies of the coupling between conformational and chemical motions
The proposal that slow conformational motions on the millisecond time scale play a major role in enzyme catalysis has been featured in many recent high profile works (see discussion in ref32). Although there is no direct experimental support, and there are very clear conceptual and logical reasons why this idea is invalid (see ref32 and reference in that work), it is crucial to explore the dynamical proposal by simulation studies that can explore the relevant milliseconds time range. Furthermore, there are other fundamental functional problems whose analysis can be greatly helped from the ability to explore slow protein motions.
Recently we introduced a renormalization method were we move from the full model to the simplified model and then to an even simpler 2-D model in representing the landscape and dynamics in the space defined by the conformational and chemical coordinate in enzymes. In this treatment we force the CG model to have the same dynamical properties as the full model by a strategy similar to the one used in our modeling of ion channels33. The main element of our approach is the refinement of the friction in the CG model so that the response of this model to the application of a strong constraint will be similar to that of the full model. The use of this model and in particular its transfer to the 2-D model allows us to explore the time dependence of processes that occur on a long time scale (e.g. ref34 and ref32). The study of ref provided the first direct proof that the dynamical proposal is invalid and further progress in the use of the renormalization model is clearly expected.
3.3.2 Studies of constant pH dynamics
There is a significant current interest in MD simulations that take into account changes in ionizations states during the simulated process (ref35 and ref36,37). However, the current model does not consider the time dependence of the proton transport (PTR) process and thus cannot reproduce the proper time dependence of the response of a protein to changes in pH. To advance on this challenging field we combined our approach of time dependent Monte Carlo (MC) of PTR processes (ref38) and the simplified protein model described here, in studies of pH dependent MD
Our model represents the energetics of the system by a simplified version of the EVB using the modified Marcus equation (see ref39) for the energetic of any possible PTR step, where the free energies for the different protonation states is obtained by the electrostatic energy of the CG model. The MC moves are based on the electrostatic energies of the CG model and then scaled by the characteristic PT time to correspond to the rate constant predicted by transition state theory. More specifically (ref40 and ref41), the free energy of each protonation state is expressed by
| (34) |
where m designates the vector of the charge states of the given configuration (i.e. m = q1(m), q2(m),… qn(m)). Here qk(m) is the actual charge of the kth group at the mth configuration. The Wkl qk(m) ql(m) term represents the charge-charge interactions.
The barrier for the PT moves is given by39
| (35) |
where is the free energy of the reaction, and Hij is the EVB off-diagonal term that mixes the two relevant states whose average values at the transition state x≠ and at the reactant state , are designated by the corresponding H̄. The first term is the expression of the regular Marcus equation (see ref39), which corresponds to the intersection of Δg1 and Δg2 at x≠. The second and third terms represent, respectively, the effect of H12 at x≠ and .
Using Eq. (35) with simplified modifications38, we consider random jumps of the proton to any possible site, but accept only jumps to sites i+1 and i-1 (or in the more general case to sites that are less than 4.0 from site i (usually we use 3.5 Å cutoff but here we use a simplified model for the protein side chains). Furthermore, these jumps are accepted only if they satisfy the standard Metropolis criterion with regard to the free energy change (ΔGn+1 − ΔGn) or the closely related activation barrier , which is defined by the individual PT steps in this transition. The MC procedure is converted to a time-dependent simulation by exploiting the isomorphism between the probability obtained from the MC procedure and the probability factor of the transition state theory (TST). Since the probability of the MC jump satisfies the Boltzmann probability we can write:
| (36) |
Where τi→j is the real time required for a move from site i to site j and Ni→j is the number of MC steps that were required for this move. The factor S for PT between internal groups is given by38:
| (37) |
Where 0.165 is the average time, in picoseconds, it takes for a productive trajectory to reach the TS at room temperature (see supplementary information of ref38). The factor δ represents the solute and solvent reorganization barrier for a transfer between states of similar energy, i.e. . The MC approach guarantees that the rate of the PT steps will follow the relevant Boltzmann probability in TST (see ref38).
The PT to and from the bulk is determined by the same considerations but now using the free energy of Eq. (34) for the given pH. The MC move for ΔGN→N+1 is scaled by the time of H3O+ diffusion by the same considerations as in ref41 but now for pH = -1 (which corresponds to the pKa of H3O+). This allows us to express the time of transferring proton from the bulk to a water molecule on the surface of the protein and then to a given ionizable group, as:
| (38) |
Since some ionized groups are not in direct contact with the bulk solvent, we scale the MC move for PT to and from the bulk by a factor, F(R), that reflects the distance, Ri,bulk, between the given residue and the closest bulk water molecule.
| (39) |
Where a is a parameter of positive value. In this preliminary work, a is set to a = 10 Å. The distances R, are determined by using a 4 Å grid and excluding grid points that are within 4 Å from the protein (this large exclusion radius is due to the fact that we use a simplified protein model). The function F(R) was chosen to represent the chance of a bulk molecule to approach the protein residues. The grid is then updated once in every 1000 MD steps. Since the exponential factor of F(R) is included in the criteria of the MC move that determines the acceptance of the PT moves we can write:
| (40) |
Thus we can write for the PT from and to the bulk
| (41) |
The scaling by 0.16 allows us to use the same scaling for the MC for the PT from bulk and the MC for the PT between the protein groups and between the protein groups and the bulk.
The next issue is the coupling between the PT steps and the protein fluctuations. One option is to run Langevin dynamics (LD) of the CG model. This option requires a significant computational time, since the typical LD steps are about 10fs, where the MC steps correspond to 200fs. Another option is to also simulate the protein motion by an MC treatment, and then estimate the characteristic time for the MC moves. Finally, considering the fact that protein's structural changes are of interest mainly when they appear after the changes in its ionization state, it is possible to simply turn-on the protein relaxation, only after reaching new stable ionization states, then allow the protein to relax (with fixed ionization state) and finally turn on again the MC for the PT processes. While such approach may not be fully consistent, it will converge to the same final state as the more consistent and much more expensive fully coupled LD - MC approach. All the above options will be considered in the future. Regardless of the method used for simulating the protein fluctuations, we expect the overall method to be very instrumental in modeling pH induced conformational changes.
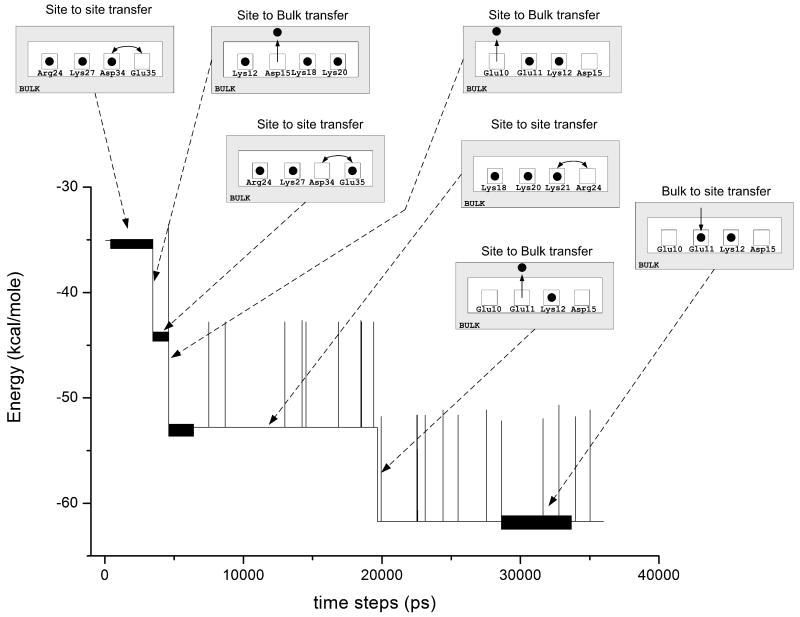
The preliminary implementation of the model demonstrated that calculated ionization state at the equilibrated system appeared to be quite similar to those obtained with the PDLD/S-LRA treatment. We also performed preliminary simulations of the time dependent PTR in protein SSO7d (PDB ID: 1SSO) following pH change from 0 to 7. The residue sites used for these simulations are: Lys4, Lys6, Lys8, Glu10, Glu11, Lys12, Asp15, Lys18, Lys20, Lys21, Lys24, Lys27, Asp34 and Glu35. The corresponding results are given in Fig. 10. The approach is promising but more validation is needed.
Figure 10.
Simulation of the proton transport process in protein 1SSO, upon change of the bulk pH from 0 to 7. The graph describes the relaxation of the overall free energy, while the insert figures describe the migration of the “active” protons. Residue sites are depicted in this figure as squares, which are sometimes occupied by a proton (black circle), and sometimes they are empty. Protons can move between residue sites, or between residue sites and the surrounding bulk.
4. Conclusions
This work presented the current version of our multiscale approach that uses a simplified folding model as a reference potential for all-atom simulations. The simplified model used here includes some innovations in terms of the electrostatic treatment and in particular in terms of the representation of the self-energy. However, the main point in our treatment is that the simplified potential does not have to be perfect since it is only used as a reference for the explicit potential. Of course, the closer are the simplified and the explicit potential, the faster will be the convergence of the calculations of the free energy for the transfer between these potentials.
The power of our treatment was illustrated in calculations of mutational effects, in the calculations of transition state binding energies and in calculations of time dependent response to pH changes. Another application that was demonstrated in our previous work involves the evaluation of the potential of mean force (PMF). We also outlined other applications of our approach, including its implementation in studies of long time scale dynamical effects and the proposal that dynamical effects contribute to catalysis32. More systematic options of using the reference potential for exploring the dynamics of the full model has not been fully formalized but developments in this crucial direction are clearly expected.
The exact time saving of the CG model is an issue that requires careful analysis, which is left to subsequent studies. That is, some of the key questions like obtaining a PMF for protein unfolding are hard to explore with explicit models since performing the corresponding calculations, where the proper sampling is extremely challenging. Similarly the effect of proper sampling calculations of binding energies is not yet fully clear. Thus we are not trying to give here time estimates, and hope to address the issue in subsequent works. However we can give here a rather trivial example, noting that a structural relaxation of the protein 1SSO took 100ps took 7 hours on a 64bit Dual Intel Xeon node, with 20Gb memory, while the equivalent relaxation of the simplified model on the same node took less than 2 minutes.
Overall it is clear that multiscale modeling of proteins has advanced significantly from its early days in 1975 and that a more rigorous treatments should focus on minimizing the difference between the average of the difference between the simplified and explicit potential <(Usp-Uexp)>. In fact, minimizing this functional with respect to the parameters of the simplified model is probably the most promising ways of refining the CG model.
Acknowledgments
This work was supported by NIH grants GM 24492 and GM40283. M.R. thanks the Generalitat Valenciana from Spain for the postdoctoral fellowship. We gratefully acknowledge the University of Southern California's High Performance Computing and Communications Center for computer time.
References
- 1.Levitt M, Warshel A. Computer-Simulation of Protein Folding. Nature. 1975;253(5494):694–698. doi: 10.1038/253694a0. [DOI] [PubMed] [Google Scholar]
- 2.Bryngelson JD, Wolynes PG. Spin glasses and the statistical mechanics of protein folding. Proc Natl Acad Sci U S A. 1987;84(21):7524–7528. doi: 10.1073/pnas.84.21.7524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dill KA. Dominant Forces in Protein Folding. Biochemistry. 1990;29(31):7133–7155. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- 4.Hinds DA, Levitt M. A lattice model for protein structure prediction at low resolution. Proc Natl Acad Sci U S A. 1992;89(7):2536–2540. doi: 10.1073/pnas.89.7.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olszewski KA, Kolinski A, Skolnick J. Folding simulations and computer redesign of protein A three-helix bundle motifs. Proteins. 1996;25(3):286–299. doi: 10.1002/(SICI)1097-0134(199607)25:3<286::AID-PROT2>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 6.Shakhnovich E, Abkevich V, Ptitsyn O. Conserved residues and the mechanism of protein folding. Nature. 1996;379(6560):96–98. doi: 10.1038/379096a0. [DOI] [PubMed] [Google Scholar]
- 7.Cheung MS, Chavez LL, Onuchic JN. The energy landscape for protein folding and possible connections to function. Polymer. 2004;45(2):547–555. [Google Scholar]
- 8.Heath AP, Kavraki LE, Clementi C. From coarse-grain to all-atom: toward multiscale analysis of protein landscapes. Proteins. 2007;68(3):646–661. doi: 10.1002/prot.21371. [DOI] [PubMed] [Google Scholar]
- 9.Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, de Vries AH. The MARTINI force field: coarse grained model for biomolecular simulations. J Phys Chem B. 2007;111(27):7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 10.Fan ZZ, Hwang JK, Warshel A. Using simplified protein representation as a reference potential for all-atom calculations of folding free energy. Theor Chem Acc. 1999;103(1):77–80. [Google Scholar]
- 11.Benkovic SJ, Hammes GG, Hammes-Schiffer S. Free-energy landscape of enzyme catalysis. Biochemistry. 2008;47(11):3317–3321. doi: 10.1021/bi800049z. [DOI] [PubMed] [Google Scholar]
- 12.Roca M, Messer BM, Hilvert D, Warshel A. On the relationship between folding and chemical landscapes in enzyme catalysis. Proc Natl Acad Sci U S A. 2008;105:13877–13882. doi: 10.1073/pnas.0803405105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiang Y, Goodman MF, Beard WA, Wilson SH, Warshel A. Exploring the role of large conformational changes in the fidelity of DNA polymerase beta. Proteins. 2008;70(1):231–247. doi: 10.1002/prot.21668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luzhkov V, Warshel A. Microscopic Models for Quantum Mechanical Calculations of Chemical Processes in Solutions: LD/AMPAC and SCAAS/AMPAC Calculations of Solvation Energies. J Comp Chem. 1992;13:199–213. [Google Scholar]
- 15.Rosta E, Klahn M, Warshel A. Towards Accurate Ab Initio QM/MM Calculations of Free-Energy Profiles of Enzymatic Reactions. J Phys Chem B. 2006;110(6):2934–2941. doi: 10.1021/jp057109j. [DOI] [PubMed] [Google Scholar]
- 16.Wesolowski TA, Warshel A. Frozen Density Functional Approach for Ab Initio Calculations of Solvated Molecules. J Phys Chem. 1993;97:8050–8053. [Google Scholar]
- 17.Hwang JK, Warshel A. A quantized classical path approach for calculations of quantum mechanical rate constants. J Phys Chem. 1993;97(39):10053–10058. [Google Scholar]
- 18.Hwang JK, Warshel A. How important are quantum mechanical nuclear motions in enzyme catalysis? J Am Chem Soc. 1996;118:11745–11751. [Google Scholar]
- 19.Brandt A. Principles of systematic upscaling. In: Fish J, editor. Bridging the scales in Science and Engineering. Oxford University Press; 2008. [Google Scholar]
- 20.Matysiak S, Clementi C. Minimalist protein model as a diagnostic tool for misfolding and aggregation. J Mol Biol. 2006;363(1):297–308. doi: 10.1016/j.jmb.2006.07.088. [DOI] [PubMed] [Google Scholar]
- 21.Roca M, Messer B, Warshel A. Electrostatic contributions to protein stability and folding energy. FEBS Lett. 2007;581(10):2065–2071. doi: 10.1016/j.febslet.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 22.Warshel A, Sharma PK, Kato M, Parson WW. Modeling electrostatic effects in proteins. Biochim Biophys Acta. 2006;1764(11):1647–1676. doi: 10.1016/j.bbapap.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 23.Vicatos S, Roca M, Warshel A. Effective Approach for Calculations of Absolute Stability of Proteins Using Focused dielectric Constants. Proteins. 2009 doi: 10.1002/prot.22481. In Print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warshel A, Russell ST. Calculations of electrostatic interactions in biological systems and in solutions. Q Rev Biophys. 1984;17(3):283–422. doi: 10.1017/s0033583500005333. [DOI] [PubMed] [Google Scholar]
- 25.Warshel A, Russell ST, Churg AK. Macroscopic models for studies of electrostatic interactions in proteins: limitations and applicability. Proc Natl Acad Sci U S A. 1984;81(15):4785–4789. doi: 10.1073/pnas.81.15.4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lovell SC, Word JM, Richardson JS, Richardson DC. The penultimate rotamer library. Proteins. 2000;40(3):389–408. [PubMed] [Google Scholar]
- 27.King G, Warshel A. A surface constrained all-atom solvent model for effective simulations of polar solutions. J Chem Phys. 1989;91(6):3647–3661. [Google Scholar]
- 28.Lee FS, Chu ZT, Warshel A. Microscopic and semimicroscopic calculations of electrostatic energies in proteins by the POLARIS and ENZYMIX programs. J Comp Chem. 1993;14:161–185. [Google Scholar]
- 29.Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins. 2003;50(3):437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 30.Went HM, Jackson SE. Ubiquitin folds through a highly polarized transition state. Protein Eng Des Sel. 2005;18(5):229–237. doi: 10.1093/protein/gzi025. [DOI] [PubMed] [Google Scholar]
- 31.Roca M, Vardi-Kilshtain A, Warshel A. Toward Accurate Screening in Computer-Aided Enzyme Design. Biochemistry. 2009;48(14):3046–3056. doi: 10.1021/bi802191b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pisliakov AV, Cao J, Kamerlin SCL, Warshel A. Enzyme millisecond conformational dynamics do not catalyze the chemical step. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0909150106. Early edition. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burykin A, Kato M, Warshel A. Exploring the origin of the ion selectivity of the KcsA potassium channel. Proteins. 2003;52(3):412–426. doi: 10.1002/prot.10455. [DOI] [PubMed] [Google Scholar]
- 34.Liu H, Shi Y, Chen XS, Warshel A. Simulating the electrostatic guidance of the vectorial translocations in hexameric helicases and translocases. Proc Natl Acad Sci U S A. 2009;106(18):7449–7454. doi: 10.1073/pnas.0900532106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khandogin J, Brooks CL., 3rd Constant pH molecular dynamics with proton tautomerism. Biophys J. 2005;89(1):141–157. doi: 10.1529/biophysj.105.061341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baptista AM, Martel PJ, Petersen SB. Simulation of protein conformational freedom as a function of pH: constant-pH molecular dynamics using implicit titration. Proteins. 1997;27(4):523–544. [PubMed] [Google Scholar]
- 37.Baptista AM, Martel PJ, Soares CM. Simulation of electron-proton coupling with a Monte Carlo method: application to cytochrome c3 using continuum electrostatics. Biophys J. 1999;76(6):2978–2998. doi: 10.1016/S0006-3495(99)77452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olsson MH, Warshel A. Monte Carlo simulations of proton pumps: on the working principles of the biological valve that controls proton pumping in cytochrome c oxidase. Proc Natl Acad Sci U S A. 2006;103(17):6500–6505. doi: 10.1073/pnas.0510860103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Braun-Sand S, Burykin A, Chu ZT, Warshel A. Realistic simulations of proton transport along the gramicidin channel: demonstrating the importance of solvation effects. J Phys Chem B. 2005;109(1):583–592. doi: 10.1021/jp0465783. [DOI] [PubMed] [Google Scholar]
- 40.Warshel A. Conversion of light energy to electrostatic energy in the proton pump of halobacterium halobium. Photochem Photobiol. 1979;30:285–290. doi: 10.1111/j.1751-1097.1979.tb07148.x. [DOI] [PubMed] [Google Scholar]
- 41.Sham YY, Muegge I, Warshel A. Simulating proton translocations in proteins: probing proton transfer pathways in the Rhodobacter sphaeroides reaction center. Proteins. 1999;36(4):484–500. [PubMed] [Google Scholar]
- 42.Woycechowsky KJ, Choutko A, Vamvaca K, Hilvert D. Relative Tolerance of an Enzymatic Molten Globule and Its Thermostable Counterpart to Point Mutation. Biochemistry. 2008;47(51):13489–13496. doi: 10.1021/bi801108a. [DOI] [PubMed] [Google Scholar]