Abstract
Lateral transfer of mobile DNA is a hallmark of bacteria with a free-living replicative stage; however, its significance in obligate intracellular bacteria and other heritable endosymbionts remains controversial. Comparative sequence analyses from laboratory stocks infected with Wolbachia pipientis provide some of the most compelling evidence that bacteriophage WO-B transfers laterally between infections of the same insect host. Lateral transfer between coinfections, however, has been evaluated neither in natural populations nor between closely related Wolbachia strains. Here, we analyze bacterial and phage genes from two pairs of natural sympatric field isolates, of Gryllus pennsylvanicus field crickets and of Neochlamisus bebbianae leaf beetles, to demonstrate WO-B transfers between supergroup B Wolbachia. N. bebbianae revealed the highest number of phage haplotypes yet recorded, hinting that lab lines could underestimate phage haplotype variation and lateral transfer. Finally, using the approximate age of insect host species as the maximum available time for phage transfer between host-associated bacteria, we very conservatively estimate phage WO-B transfer to occur at least once every 0–5.4 My within a host species. Increasing discoveries of mobile elements, intragenic recombination, and bacterial coinfections in host-switching obligate intracellular bacteria specify that mobile element transfer is common in these species.
Keywords: lateral gene transfer, Wolbachia, bacteriophage, obligate intracellular bacteria, recombination, coinfection
The restrictive lifestyle of obligate intracellular bacteria and inherited endosymbionts limits exposure to novel gene pools and enhances reductive evolution, often resulting in a low mobile DNA element composition (Moran and Plague 2004; Bordenstein and Reznikoff 2005); however, genera prone to lateral transmission between hosts tend to establish coinfections and integrate foreign DNA into their genomes (Bordenstein and Reznikoff 2005). For instance, in the reproductive parasite Wolbachia, up to 21% of its genes represent mobile DNA (Wu et al. 2004; Klasson et al. 2009). Similarly, defensive bacterial symbionts of aphids that are transmitted both maternally and horizontally have dynamic genomes exhibiting phage transfer, recombination, and toxin-gene acquisitions (Oliver et al. 2010).
In Wolbachia, prophage regions can comprise more than 20% of mobile DNA genes and account for the largest fraction of absent/divergent genes between closely related strains (Ishmael et al. 2009). Among these regions, temperate phage WO-B may inhibit Wolbachia’s capacity to adaptively modify invertebrate reproduction (Bordenstein et al. 2006) or enhance it by encoding ankyrin repeat–containing genes or effectors (Iturbe-Ormaetxe et al. 2005; Sinkins et al. 2005; Sanogo and Dobson 2006; Tanaka et al. 2009). Phage WO-B occurs in the majority of supergroup A and B Wolbachia, can integrate as a prophage and produce active phage particles, and rampantly transfers between Wolbachia infections (Masui et al. 2000; Bordenstein and Wernegreen 2004; Gavotte et al. 2007). Further, multiple and divergent WO-B haplotypes commonly co-occur in single Wolbachia infections, potentially by recurrent phage transfers (Masui et al. 2000; Bordenstein and Wernegreen 2004; Gavotte et al. 2004; Gavotte et al. 2007). Likewise, active transposition and frequent lateral transmission of the insertion sequence element ISWpi1 is evident in Wolbachia (Cordaux 2008). The lateral transfer of mobile elements in Wolbachia and other heritable symbionts has major implications for genome evolution, gene acquisition, and the ecological interactions between selfish elements and their bacterial hosts. However, the ecological contributions to these exchanges in natural populations remain unexplored.
In laboratory lines of Drosophila flies, Nasonia wasps, and Ephestia moths, insect hosts serve as incubators for phage WO-B exchange between Wolbachia coinfections (Masui et al. 2000; Bordenstein and Wernegreen 2004). For example, the rapidly evolving phage ORF7 gene that encodes the minor capsid protein has nearly complete sequence identity between coinfections of the divergent A and B Wolbachia in these animals. However, outstanding issues remain. It is not known if WO-B movement between Wolbachia infections occurs in nature, how frequent it is, or if it is possible between closely related coinfections.
Here, we infer recent lateral transfer events between supergroup B Wolbachia infections in field isolates. Specifically, Gryllus pennsylvanicus field crickets were sampled from an Ithaca, New York, population known to include individuals with coinfections of different B Wolbachia (Mandel et al. 2001). Neochlamisus bebbianae leaf beetles were collected from Salix bebbiana (Bebb's willow) in Caledonia County, Vermont. Crickets and beetles were screened for single Wolbachia infections using PCR and sequences from four multilocus sequence typing (MLST) genes, ftsZ, gatB, hcpA, and coxA (Baldo et al. 2006).
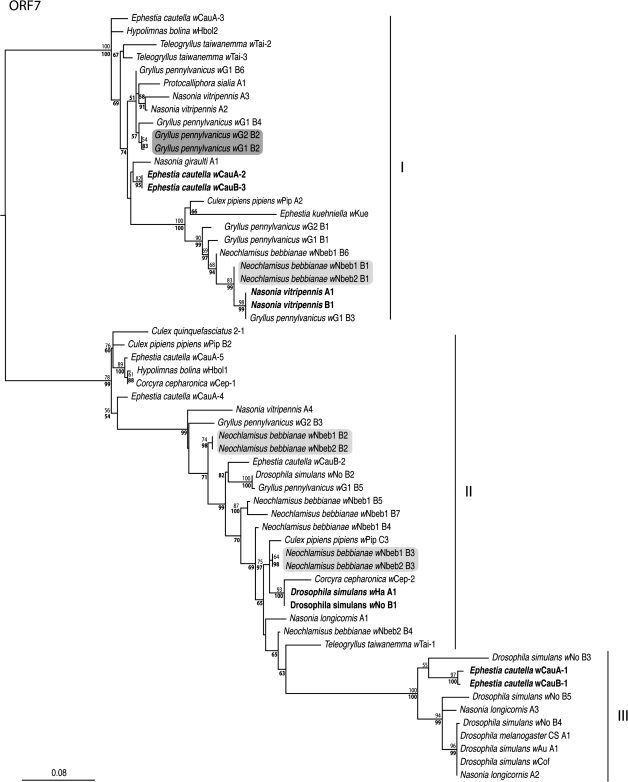
Comparative sequence analyses of MLST genes ftsZ, gatB, and hcpA from G. pennsylvanicus strains wG1 and wG2 showed 3.26% divergence (fig. 1). coxA was excluded from further consideration because we could not determine if its unusual sequence evolution reflected gene transfer to the host genome or recombination at the coxA locus. Cloned and sequenced PCR products of the ORF7 minor capsid gene from wG1 and wG2 revealed six and three distinct phage WO-B haplotypes, respectively, based on a cutoff criteria of 1.5% divergence (fig. 2). Variability in phage haplotype number and diversity is typical of Wolbachia (Bordenstein and Wernegreen 2004; Gavotte et al. 2007) and suggests that gene duplications and separate invasions of different phage haplotypes shape the evolution of phage WO-B in Wolbachia genomes. Notably, one of the cloned ORF7 haplotypes is shared between wG1 and wG2, differing by 1/358 bp (0.28%; fig. 2). The evolutionarily recent transfer of a given phage haplotype receives support if phage genetic divergence is lower than that of its two bacterial hosts despite the more rapid rate of phage nucleotide evolution (table 1). Indeed, a Fisher's exact test shows ORF7 divergence (1/358) to be significantly lower than that “expected” from the 3.26% divergence of its Wolbachia host genes (12/358; P = 0.001, one-tailed test).
FIG. 1.
ML phylogeny based on a concatenated nucleotide sequence alignment of ftsZ, gatB, and hcpA Wolbachia MLST loci. The tree is midpoint rooted, and numbers denote ML bootstrap values and Bayesian posterior probabilities (in bold). Taxa are denoted by host arthropod species and Wolbachia strain. Field specimens analyzed in this study are shown in shaded boxes. Wolbachia supergroup A and B clades are indicated to the right of each tree.
FIG. 2.
ML phylogeny of the bacteriophage ORF7 nucleotide sequences. The tree is midpoint rooted, and numbers denote ML bootstrap values and Bayesian posterior probabilities (in bold). Taxa names denote host arthropod species, Wolbachia strain, and ORF7 haplotype. Pairs of haplotypes in shaded boxes illustrate putative lateral transfer events in field specimens collected in this study. Bold taxa represent previously published examples of lateral transfer through laboratory segregation experiments. The three group labels (I–III) were assigned according to the nomenclature in Bordenstein and Wernegreen (2004).
Table 1.
Average Pairwise Nucleotide Diversity Per Site (π) in Five Genes.
| Taxa | Gene | Length of Alignment (bp)a | N | π |
| Wolbachia pipientis | ftsZ | 435 | 38 | 0.058 |
| gatB | 369 | 38 | 0.066 | |
| hcpA | 444 | 38 | 0.073 | |
| coxA | 402 | 38 | 0.080 | |
| Bacteriophage WO-B | ORF7 | 240 | 59 | 0.146 |
Length of the alignment is based on removal of indels.
The two N. bebbianae leaf beetle infections wNbeb1 and wNbeb2 show 1.0% sequence divergence in combined MLST gene sequences. Figure 2 reveals seven and four ORF7 haplotypes within wNbeb1 and wNbeb2, respectively, with the former value representing the most haplotypes reported for any single Wolbachia infection (Gavotte et al. 2007). Three of the cloned ORF7 haplotypes are shared between wNbeb1 and wNbeb2, suggesting these sequences have experienced recent lateral transfer. The Fisher's exact test yields a marginally nonsignificant difference between the observed (0/358) and expected (4/358) divergence (P = 0.062), likely due to the lower 1% divergence in Wolbachia genes than in the crickets. Given the 2-fold higher sequence diversity in our phage gene compared with the Wolbachia ones (table 1) and the complete identity of the three shared haplotypes, this P value represents a conservative estimate of the likelihood that these three phage haplotypes transferred.
Support for lateral phage transfer between these closely related Wolbachia can also be bolstered if the two infections arose by independent acquisitions, rather than by descent and duplication from a common ancestor within N. bebbianae. Estimating absolute divergence times is problematic in the absence of fossil data, but molecular clocks can provide rough estimates. Synonymous divergence between wNbeb1 and wNbeb2 (3.56%) suggests their infections are 3.96 My old based on a rate derived for diverse bacteria (Ochman et al. 1999). Meanwhile, N. bebbianae is 0.48 My old based on mitochondrial DNA divergence (Brower 1994; Funk 1999). These estimates support independent acquisitions of these two Wolbachia strains and lateral transfer of three phage haplotypes within the last 480,000 years (table 2).
Table 2.
Summary of Phage WO-B Transfers in Five Host-Associated Cases.
| Insect Host | Wolbachia Strain | Wolbachia Supergroup | No. of Phage Haplotypesa | No. of Transferred Haplotypes between Host-Associated Wolbachia | Maximum Time Span of Phage Transfer (× 106 years) |
| Neochlamisus bebbianae | wNbeb1 | B | 7 | 3 | 0.48b |
| wNbeb2 | B | 4 | |||
| Gryllus pennsylvanicus | wG1 | B | 6 | 1 | – |
| wG2 | B | 3 | |||
| Nasonia vitripennis | wVitA | A | 4 | 1 | 1.0c |
| wVitB | B | 1 | |||
| Drosophila simulans | wHa | A | 5 | 1 | 5.4d |
| wNo | B | 1 | |||
| Ephestia cautella | wCauA | A | 5 | 2 | – |
| wCauB | B | 3 |
Haplotypes are defined as having greater than 1.5% nucleotide diversity in the ORF7 gene.
Estimated age of N. bebbianae is derived from Funk (1999) and Brower (1994).
Estimated age of N. vitripennis is based on Campbell et al. (1993).
Estimated age of D. simulans is based on Tamura et al. (2004).
Using insect isolates with different Wolbachia infections, these experiments support coinfection in the same insect host as a mechanism for lateral phage transfer in natural populations of obligate intracellular bacteria. Phage transfer has been documented in all five instances where it has been investigated, with three cases occurring between A and B Wolbachia coinfections in lab stocks of Hymenoptera, Diptera, and Lepidoptera and two cases (this study) occurring between different B Wolbachia lineages in natural populations of Orthoptera and Coleoptera (table 2). Using estimates of insect host age as the maximal age of phage transfer between independently acquired host-associated Wolbachia, all documented WO-B transfers have occurred within the last 5.4 My. In our study, the actual time since lateral phage transfer is expected to be considerably shorter than the maximum age estimate because the phage ORF7 genes exhibit complete or near complete nucleotide identity. N. bebbianae shows the most recent transfers, with each of the three haplotypes transferring within 480 Kyr (table 2). Our field samples identified more phage haplotypes for G. pennsylvanicus wG1 (six) and N. bebbianae wNbeb1 (seven) than any previously reported study. Perhaps such field samples provide improved snapshots of the extent of lateral transfer in heritable symbionts because laboratory lines may be subject to relaxed selection and phage loss over time (Oliver, et al. 2009). In this event, our data indicate that bacteriophage WO-B is more abundant and capable of lateral transfer than previously thought. Phages may thus play underappreciated roles as conduits for lateral transfer between coinfecting bacterial strains of arthropod hosts.
Materials and Methods
Bacteriophage and Wolbachia DNA were amplified using PCR. The phage ORF7 capsid locus (Masui et al. 2000) was amplified along with 16S ribosomal RNA; wsp; and the MLST genes, ftsZ, gatB, coxA, and hcpA (Baldo et al. 2006). Bidirectional sequencing was performed directly from PCR products or from products cloned into plasmid vectors using the GC Cloning Kit (Lucigen). Sequences were assembled, manually edited, and aligned using MUSCLE in Geneious v. 4.6.5 (Drummond et al. 2009) and MacClade version 4.08 (Maddison D and Maddison W 2005). Cloned sequences showing >1.5% nucleotide divergence from each other were selected for analysis. K2p (Kimura 1980) genetic distances were calculated using PHYLIP (Felsenstein 1993), and average nucleotide diversities and synonymous divergences were determined with DnaSP v.5. (Rozas et al. 2003).
Maximum likelihood (ML) and Bayesian methods were carried out using PAUP 4.0b10 and the Bayes plugin in Geneious (Huelsenbeck and Ronquist 2001). Model selection for the ML analysis was estimated using the Akaike information criterion in Modeltest v3.7. The DNA substitution model selected for the concatenated MLST loci was the general time reversible (GTR) model in which the gamma distribution and invariant sites were estimated from the data (GTR + I + G). ML bootstrap values were generated from 100 bootstrap replicates, each using 10 random taxon addition replications. Data sets were also analyzed with Bayesian phylogenetic methods using GTR + I + G. Four chains were run for 1.1 million generations. Trees were sampled every 200 generations and a consensus tree was built with a burn-in of 200,000. All new sequences were deposited in GenBank under the accession numbers GUZ06996--GUZ07027.
Acknowledgments
This work was supported by grants National Science Foundation (NSF) IOS-0852344 and National Institute of Health (NIH) R01 GM085163-01 to S.R.B. This paper's contents are solely the responsibility of the authors and do not necessarily represent the official view of the NSF or NIH.
References
- Baldo L, Dunning Hotopp JC, Jolley KA, Bordenstein SR, Biber SA, Choudhury RR, Hayashi C, Maiden MC, Tettelin H, Werren JH. Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl Environ Microbiol. 2006;72:7098–7110. doi: 10.1128/AEM.00731-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordenstein SR, Marshall ML, Fry AJ, Kim U, Wernegreen JJ. The tripartite associations between bacteriophage, Wolbachia, and arthropods. PLoS Pathog. 2006;2:e43. doi: 10.1371/journal.ppat.0020043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordenstein SR, Reznikoff WS. Mobile DNA in obligate intracellular bacteria. Nat Rev Microbiol. 2005;3:688–699. doi: 10.1038/nrmicro1233. [DOI] [PubMed] [Google Scholar]
- Bordenstein SR, Wernegreen JJ. Bacteriophage flux in endosymbionts (Wolbachia): infection frequency, lateral transfer, and recombination rates. Mol Biol Evol. 2004;21:1981–1991. doi: 10.1093/molbev/msh211. [DOI] [PubMed] [Google Scholar]
- Brower AV. Rapid morphological radiation and convergence among races of the butterfly Heliconius erato inferred from patterns of mitochondrial DNA evolution. Proc Natl Acad Sci USA. 1994;91:6491–6495. doi: 10.1073/pnas.91.14.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell BC, Steffen-Campbell JD, Werren JH. Phylogeny of the Nasonia species complex (Hymenoptera: pteromalidae) inferred from an internal transcribed spacer (ITS2) and 28S rDNA sequences. Insect Mol Biol. 1993;2:225–237. doi: 10.1111/j.1365-2583.1994.tb00142.x. [DOI] [PubMed] [Google Scholar]
- Cordaux R. ISWpi1 from Wolbachia pipientis defines a novel group of insertion sequences within the IS5 family. Gene. 2008;409:20–27. doi: 10.1016/j.gene.2007.10.035. [DOI] [PubMed] [Google Scholar]
- Drummond A, Ashton B, Cheung M, Heled J, Kearse M, Moir R, Stones-Havas S, Thierer T, Wilson A. Geneious v.4.7. 2009. Auckland, New zealand. Available from: http://www.geneious.com/ [Google Scholar]
- Felsenstein J. PHYLIP (phylogeny inference package) Version 3.567. Distributed by the author. 1993. Department of Genetics, Seattle (WA): University of Washington. [Google Scholar]
- Funk DJ. Molecular systematics of cytochrome oxidase I and 16S from Neochlamisus leaf beetles and the importance of sampling. Mol Biol Evol. 1999;16:67–82. doi: 10.1093/oxfordjournals.molbev.a026039. [DOI] [PubMed] [Google Scholar]
- Gavotte L, Henri H, Stouthamer R, Charif D, Charlat S, Bouletreau M, Vavre F. A survey of the bacteriophage WO in the endosymbiotic bacteria Wolbachia. Mol Biol Evol. 2007;24:427–435. doi: 10.1093/molbev/msl171. [DOI] [PubMed] [Google Scholar]
- Gavotte L, Vavre F, Henri H, Ravallec M, Stouthamer R, Bouletreau M. Diversity, distribution and specificity of WO phage infection in Wolbachia of four insect species. Insect Mol Biol. 2004;13:147–153. doi: 10.1111/j.0962-1075.2004.00471.x. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Ishmael N, Hotopp JC, Ioannidis P, et al. (11 co-authors) Extensive genomic diversity of closely related Wolbachia strains. Microbiology. 2009;155:2211–2222. doi: 10.1099/mic.0.027581-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturbe-Ormaetxe I, Burke GR, Riegler M, O'Neill SL. Distribution, expression, and motif variability of ankyrin domain genes in Wolbachia pipientis. J Bacteriol. 2005;187:5136–5145. doi: 10.1128/JB.187.15.5136-5145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Klasson L, Westberg J, Sapountzis P, et al. (12 co-authors) The mosaic genome structure of the Wolbachia wRi strain infecting Drosophila simulans. Proc Natl Acad Sci USA. 2009;106:5725–5730. doi: 10.1073/pnas.0810753106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison D, Maddison W. MacClade 4.08: Analysis of Phylogeny and Character Evolution. Sunderland (MA): Sinauer Associates; 2005. [Google Scholar]
- Mandel MJ, Ross CL, Harrison RG. Do Wolbachia infections play a role in unidirectional incompatibilities in a field cricket hybrid zone? Mol Ecol. 2001;10:703–709. doi: 10.1046/j.1365-294x.2001.01213.x. [DOI] [PubMed] [Google Scholar]
- Masui S, Kamoda S, Sasaki T, Ishikawa H. Distribution and evolution of bacteriophage WO in Wolbachia, the endosymbiont causing sexual alterations in arthropods. J Mol Evol. 2000;51:491–497. doi: 10.1007/s002390010112. [DOI] [PubMed] [Google Scholar]
- Moran NA, Plague GR. Genomic changes following host restriction in bacteria. Curr Opin Genet Dev. 2004;14:627–633. doi: 10.1016/j.gde.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Ochman H, Elwyn S, Moran NA. Calibrating bacterial evolution. Proc Natl Acad Sci USA. 1999;96:12638–12643. doi: 10.1073/pnas.96.22.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver KM, Degnan PH, Burke GR, Moran NA. Facultative symbionts of aphids and the horizontal transfer of ecologically important traits. Annu Rev Entomol. 2010;55:247–266. doi: 10.1146/annurev-ento-112408-085305. [DOI] [PubMed] [Google Scholar]
- Oliver KM, Degnan PH, Hunter MS, Moran NA. Bacteriophages encode factors required for protection in a symbiotic mutualism. Science. 2009;325:992–994. doi: 10.1126/science.1174463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Sanchez-DelBarrio J, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Sanogo YO, Dobson SL. WO bacteriophage transcription in Wolbachia-infected Culex pipiens. Insect Biochem Mol Biol. 2006;36:80–85. doi: 10.1016/j.ibmb.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Sinkins SP, Walker T, Lynd AR, Steven AR, Makepeace BL, Godfray HC, Parkhill J. Wolbachia variability and host effects on crossing type in Culex mosquitoes. Nature. 2005;436:257–260. doi: 10.1038/nature03629. [DOI] [PubMed] [Google Scholar]
- Tamura K, Subramanian S, Kumar S. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol Biol Evol. 2004;21:36–44. doi: 10.1093/molbev/msg236. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Furukawa S, Nikoh N, Sasaki T, Fukatsu T. Complete WO phage sequences reveal their dynamic evolutionary trajectories and putative functional elements required for integration into the Wolbachia genome. Appl Environ Microbiol. 2009;75:5676–5686. doi: 10.1128/AEM.01172-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Sun LV, Vamathevan J, et al. (30 co-authors) Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2004;2:E69. doi: 10.1371/journal.pbio.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]