Abstract
Vaccines directed against the blood stages of Plasmodium falciparum malaria are intended to prevent the parasite from invading and replicating within host cells. No blood-stage malaria vaccine has shown clinical efficacy in humans. Most malaria vaccine antigens are parasite surface proteins that have evolved extensive genetic diversity, and this diversity could allow malaria parasites to escape vaccine-induced immunity. We examined the extent and within-host dynamics of genetic diversity in the blood-stage malaria vaccine antigen apical membrane antigen–1 in a longitudinal study in Mali. Two hundred and fourteen unique apical membrane antigen–1 haplotypes were identified among 506 human infections, and amino acid changes near a putative invasion machinery binding site were strongly associated with the development of clinical symptoms, suggesting that these residues may be important to consider in designing polyvalent apical membrane antigen–1 vaccines and in assessing vaccine efficacy in field trials. This extreme diversity may pose a serious obstacle to an effective polyvalent recombinant subunit apical membrane antigen–1 vaccine.
INTRODUCTION
The malaria parasite Plasmodium falciparum has a long evolutionary history with its human host and exhibits extensive genetic diversity, particularly in surface antigens that are subjected to selective pressure by the human immune system (1). Vaccines directed against these highly diverse antigens can only feasibly target a subset of the alleles circulating in a population, potentially compromising initial vaccine efficacy. Such allele-specific vaccines would be expected to exert directional selection that favors variants not targeted by the vaccine, leading to further waning of efficacy over time (2).
The P. falciparum apical membrane antigen–1 (AMA-1) is a polymorphic surface protein that is targeted by vaccines intended to prevent disease and death by inducing antibodies that block parasite invasion of host cells. At least three AMA-1–based vaccines are in clinical development, all based on AMA-1 sequences from one or both of two vaccine strains of P. falciparum: 3D7 and FVO (3-6). These strains were selected based on limited knowledge of natural genetic diversity in AMA-1.
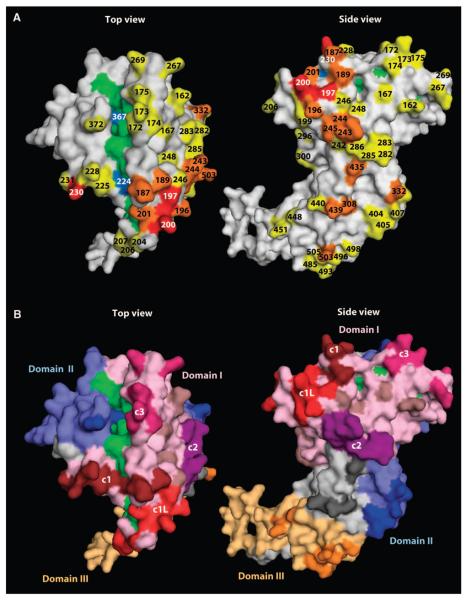
AMA-1 has three major domains stabilized by eight disulfide bonds (7). All of the sequence diversity in the gene encoding AMA-1 is in the form of single-nucleotide polymorphisms (SNPs), which are thought to be maintained by balancing selection (8-10).These polymorphisms are located predominantly on one face of the AMA-1 protein, with the most polymorphic residues concentrated near a hydrophobic trough in domain I that is hypothesized to be a binding site between AMA-1 and associated proteins that form the machinery for erythrocyte invasion (11, 12) (Fig. 1).
Fig. 1.
Polymorphic amino acids shown on the AMA-1 crystal structure. Polymorphisms are based on sequence data from P. falciparum infections acquired at a vaccine testing site in Mali, West Africa. (A) Polymorphic residues are numbered and highlighted. Yellow and blue residues are dimorphic, orange residues are trimorphic, and red residues have four to six possible amino acids. Residues highlighted in green and blue make up the hydrophobic pocket hypothesized to be a binding site between AMA-1 and the rest of the erythrocyte invasion machinery, with blue indicating polymorphic residues within the pocket (11, 12). (B) Conserved residues in AMA-1 domains I, II, and III are highlighted in light pink, light blue, and light orange, respectively. Polymorphic residues in domain I are highlighted in dark brown (c1), red (c1 and c1L), purple (c2), dark pink (c3), and light brown (not incorporated in a cluster). Polymorphic residues in domains II and III are highlighted in dark blue and dark orange, respectively. Light gray residues are not part of any of the three major domains, and dark gray residues are polymorphisms within the interdomain region.
Identification of the antigen polymorphisms that determine allele-specific immune protection, and use of this information to classify alleles into immunologically relevant groups (for example, serotypes), would inform the selection of parasite alleles for a broadly protective vaccine and facilitate assessment of allele-specific efficacy in clinical trials of malaria vaccines. In allelic exchange experiments using chimeric proteins with residues corresponding to either of the two leading P. falciparum vaccine strains, a cluster of residues surrounding the hydrophobic pocket in domain I had the greatest effect on antigenic escape (13). Others have used a clustering algorithm, commonly used for inferring population structure, to estimate the minimum number of AMA-1 alleles and reported that rabbit antibodies raised against recombinant AMA-1 differentially inhibited invasion by parasites from different haplotype groups (14). The relevance of these in vitro and animal experiments to natural and vaccine-induced immune protection against clinical malaria disease in humans is not known. We used a molecular epidemiological approach to assess how individual polymorphisms, clusters of polymorphisms, and haplotype groups in AMA-1 relate to the development of clinical symptoms in prospectively followed individuals who experienced repeated malaria infections at a vaccine testing site in Mali, West Africa.
RESULTS
AMA-1 diversity
We examined 748 AMA-1 sequences, corresponding to amino acid residues 149 to 552 of the AMA-1 ectodomain, generated from P. falciparum infections experienced by 100 individuals participating in a prospective longitudinal cohort study over 3 years at a vaccine testing site in Mali. On the basis of peak heights in the sequencing chromatograms, 506 sequences appeared to represent a single or predominant parasite clone, allowing resolution of AMA-1 haplotypes. Two hundred and forty-two sequences represented two or more parasite clones, precluding haplotype determination. Seventy-seven SNPs were detected among the 1212 amplified nucleotides, resulting in 62 polymorphic amino acid positions (fig. S1), including 46 dimorphic sites, 13 trimorphic sites, and 3 sites with four to six possible amino acids (Fig. 1A). The most highly polymorphic residues cluster around a hydrophobic pocket hypothesized to be a binding site between AMA-1 and other proteins that form the erythrocyte invasion complex (11, 12). Domain I was the most diverse of the three AMA-1 domains, with 36 of the 154 (23%) amino acid positions being polymorphic, followed by domain III [8 of 67 (12%) residues polymorphic] and domain II [11 of 99 (11%) residues polymorphic]. Average genetic diversity in each domain remained stable over the 3 years of the study (fig. S2).
On the basis of the 62 polymorphisms in the AMA-1 ectodomain, 214 unique haplotypes were observed among the 506 single- or predominant-clone infections. Individual haplotype prevalences were all low, ranging from 0.2 to 3.6%. Approximately half (50.9%) of the haplotypes were observed only once in the data set. The haplotype with amino acid sequence exactly matching that of the 3D7 vaccine strain of P. falciparum had a prevalence of 3.0%, whereas no haplotype had a sequence exactly matching that of the FVO vaccine strain (fig. S1). The implication of this extreme polymorphism is that, in the worst-case scenario that each haplotype is viewed by the immune system as immunologically distinct, a 214-valent vaccine might be required to protect against all of the variant parasites found in a single African town. We used molecular epidemiological approaches to try to reduce this large number of haplotypes to a more manageable number of groupings to be considered for inclusion in a broadly protective AMA-1 vaccine.
Changes in clusters of polymorphic residues and risk of clinical malaria
Using longitudinal clinical and molecular data, we examined the within-host dynamics of AMA-1 polymorphisms in infected individuals over time. Our hypothesis was that, because of allele-specific immunity, individuals were more likely to get sick when infected with a parasite that was different from the parasite they were infected with previously in immunologically important regions of the protein. To test this hypothesis, we used logistic regression to estimate whether changes within specific clusters of AMA-1 polymorphisms were associated with the development of clinical symptoms in the second of individuals' paired consecutive infections. Infections with single- and multiple-clone infections were included in the analysis. The primary outcome in the regression model was whether the individual's next consecutive infection was a symptomatic episode (detected by passive surveillance when malaria was diagnosed at the time of presentation to the clinic with illness) or an asymptomatic infection (detected by active surveillance through collection of blood samples in monthly surveys). The primary predictor was the proportion of amino acids within a given cluster or domain that changed between the two consecutive infections, classified as an ordinal variable. The proportion of changes was examined separately for the whole AMA-1 ectodomain, for each of the three major domains, and for four subclusters within domain I, as defined by Dutta and colleagues (Fig. 1B) (13). We calculated adjusted odds ratios (ORs) and confidence intervals (CIs) comparing moderate and high proportions of change to the lowest proportion of change in intervals where individuals became ill versus remaining well (Table 1). Estimated effects were adjusted for age and for an interaction between the amount of genetic change and the time between consecutive infections. Any association between the amount of genetic change and the clinical outcome diminished in intervals longer than 4 to 6 weeks, possibly because of the short-lived nature of naturally acquired AMA-1 antibody responses, as documented in field immunological studies (15, 16). To account for this interaction, estimated effects in Table 1 are shown only for consecutive infections separated by 6 weeks or less, whereas estimated effects in both these and longer time intervals are shown in table S1. Because symptomatic infections were treated with a long-acting antimalarial drug (sulfadoxine-pyrimethamine) that prevents infection for about a month after treatment (17), the average time between consecutive infections was, as expected, longer when the first of the two episodes was symptomatic (52 days) than when the first of the two episodes was asymptomatic (34 days). After taking into account the interaction with time, the presence of symptoms in the first of two consecutive infections was not significantly associated with the outcome and this covariate was therefore left out of the final models. The presence of a mixed infection in the interval was also not significantly associated with the outcome and was removed from the final models. A dose-response relationship (the more changes in the AMA-1 cluster or domain, the stronger the association with symptoms) is consistent with the effect being driven by changes in AMA-1 rather than another locus.
Table 1.
Associations between changes in clusters of polymorphic AMA-1 amino acid residues and clinical malaria. OR, 95% CI, and P values comparing the odds of an individual's next consecutive infection being symptomatic to the odds of their next infection being asymptomatic during intervals when a medium or high proportion of amino acid changes at polymorphic sites occurred, with the lowest proportion of amino acid changes as the reference (medium versus low and high versus low, respectively). The same information is shown comparing intervals with a high proportion of amino acid changes to those with a medium proportion of changes (high versus medium). Categories of low, medium, and high proportions of change were based on cutoff points at the first and third quartiles. Estimated effects were adjusted for age and repeated measurements from the same individual and are shown for 133 consecutive infections separated by 6 weeks or less to account for a significant interaction between amount of genetic change and time between consecutive infections (table S1).
| Region of protein | Proportion of change in region |
Reference | OR | 95% CI | P value |
|---|---|---|---|---|---|
| Entire ectodomain | Medium | Low | 2.65 | 1.20–5.85 | 0.0158 |
| High | Low | 6.80 | 2.62–17.7 | <0.0001 | |
| High | Medium | 2.57 | 1.00–6.58 | 0.0501 | |
| Domain I | Medium | Low | 3.11 | 1.32–7.29 | 0.009 |
| High | Low | 3.32 | 1.22–9.03 | 0.019 | |
| High | Medium | 1.07 | 0.37–3.05 | 0.901 | |
| Domain I c1 | Medium | Low | 2.91 | 1.45–5.83 | 0.0027 |
| High | Low | 6.46 | 2.68–15.6 | <0.0001 | |
| High | Medium | 2.22 | 0.94–5.25 | 0.0685 | |
| Domain I c1L | Medium | Low | 2.48 | 1.15–5.33 | 0.0204 |
| High | Low | 5.98 | 2.58–13.9 | <0.0001 | |
| High | Medium | 2.41 | 1.01–5.79 | 0.0484 | |
| Domain I c2 | Medium | Low | 1.84 | 0.79–4.32 | 0.16 |
| High | Low | 1.24 | 0.53–2.87 | 0.62 | |
| High | Medium | 0.67 | 0.28–1.60 | 0.37 | |
| Domain I c3 | Medium | Low | 2.29 | 0.88–5.96 | 0.089 |
| High | Low | 2.64 | 1.02–6.80 | 0.045 | |
| High | Medium | 1.15 | 0.47–2.82 | 0.757 | |
| Domain II | Medium | Low | 3.15 | 1.21–8.16 | 0.019 |
| High | Low | 4.50 | 1.40–14.4 | 0.012 | |
| High | Medium | 1.43 | 0.58–3.53 | 0.438 | |
| Domain III | Medium | Low | 1.14 | 0.42–3.06 | 0.802 |
| High | Low | 3.06 | 1.32–7.13 | 0.009 | |
| High | Medium | 2.70 | 1.14–6.40 | 0.024 |
Overall, the proportion of amino acid changes within the entire ectodomain was associated with the development of symptoms in a strongly dose-response fashion (the strength of the effect increased significantly with each category of increased change) (Table 1). The proportion of changes in domain I cluster 1 (c1) and the c1 loop (c1L) were also significantly associated with the development of symptoms, with a strong dose response (Table 1). The effect of amino acid changes in c1 is likely to be driven primarily by the subset of residues included in c1L. These clusters contain the most polymorphic residues in the protein, which are located near the hydrophobic pocket [polymorphic positions 187 to 231 (c1) and 196 to 207 (c1L)] (Fig. 1). Changes in residues within domain I, domain II, and cluster 3 (c3) of domain I showed significant association with symptoms, but the strength of the association did not increase significantly in categories of increased change, suggesting that changes at these residues might have served as nonspecific markers for change occurring elsewhere in the protein. In domain III, only the highest proportion of changes showed a significant association with clinical symptoms.
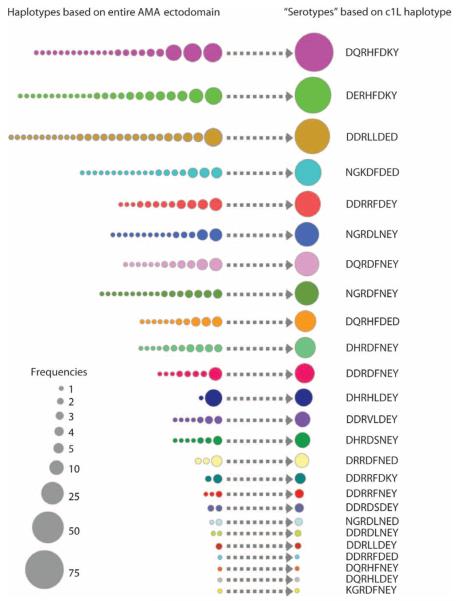
These results suggest that polymorphisms within domain I c1 or c1L could potentially be used to group parasites into immunologically relevant AMA-1 groups. The eight polymorphic residues in c1L defined 25 unique haplotypes among the 506 single- or predominant-clone infections from Mali. The c1L haplotypes ranged in prevalence from 0.2 to 15.0%, with just 16% of haplotypes being observed only once in the data set. The 10 most frequent c1L haplotypes, including those with sequence corresponding to the leading vaccine strains 3D7 (13.4%) and FVO (5.7%), accounted for 81% of the infections observed in the cohort (Fig. 2). Thus, this classification scheme could potentially reduce the number of AMA-1 variants needed for a broadly protective vaccine by a factor of ~20, from more than 200 to 10.
Fig. 2.
Prevalence of AMA-1 domain I c1L haplotypes in Bandiagara, Mali. Haplotypes based on all polymorphic sites in the AMA-1 ectodomain are shown on the left, and putative serotypes defined by the eight polymorphic sites in domain I c1L (196, 197, 199, 200, 201, 204, 206, and 207) are shown on the right. The c1L haplotypes are indicated by the one-letter amino acid abbreviation at each of the eight polymorphic sites, with haplotypes corresponding to specific strains of P. falciparum indicated in parentheses. The areas of the circles correspond to haplotype prevalence. Abbreviations for the amino acid residues are as follows: D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; and Y, Tyr.
Changes in individual amino acids and risk of clinical malaria
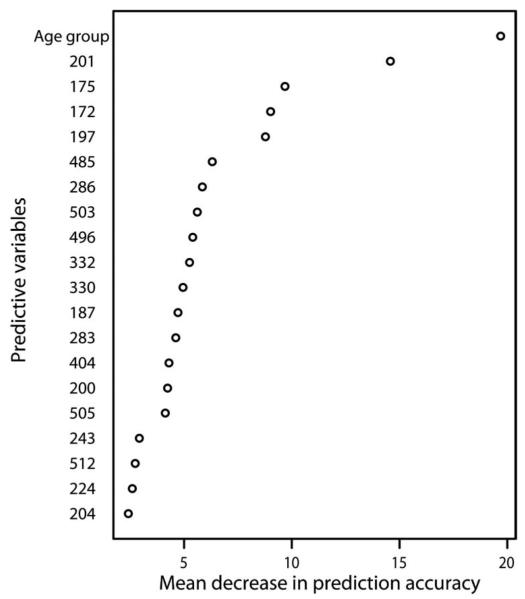
We used random forest (18), a statistical or machine-learning method, to determine whether changes in individual amino acids were associated with the development of clinical symptoms and to estimate the importance of variables in predicting clinical status. The outcome of interest was whether an individual's second of two consecutive infections was symptomatic, and the potential predictors were change at each polymorphic amino acid position in the AMA-1 ectodomain and age, which reflects naturally acquired immunity. We examined consecutive infections separated by 6 weeks or less and found that age was the best predictor of whether an individual's next infection was symptomatic. Of the polymorphic AMA-1 sites, changes at position 201 had the strongest predictive value, followed by sites 172, 175, and 197 (Fig. 3). There were 22 haplotypes based on these four amino acid positions, similar to the number of haplotypes based on the eight residues in domain I c1L, and the number of amino acid changes was associated with the development of symptoms (OR, 3.93; 95% CI, 1.87–8.27; P = 0.0003, comparing two changes to one or no changes; OR, 7.83; 95% CI, 2.83–21.7; P < 0.0001, comparing three or more changes to one or no changes; and OR, 1.99; 95% CI, 0.74–5.33; P = 0.17).
Fig. 3.
Prediction of clinical symptoms by amino acid changes at individual polymorphic sites in AMA-1. Plot of variable importance where the mean decrease in the accuracy of prediction of the outcome (a measure of variable importance) is plotted on the x axis and predictive variables are listed on the y axis. Change at each polymorphic site is represented by the amino acid position number. The variable age group represents the age group of the study participant. The random forest comprised 1 × 105 individual tree-based statistical models, with eight variables tried at each split. The overall error rate in predicting the outcome was 37.4%.
Global diversity and distribution of AMA-1 alleles
Our large data set of P. falciparum sequences from Mali shows extensive genetic diversity in AMA-1, even when focusing on subgroups based on putatively important amino acid residues. Is this diversity and distribution of alleles representative of the diversity observed in the rest of the world? To address this question, published AMA-1 sequences (8-10, 14, 19-26) were examined along with those reported in this study. Three hundred and eighteen AMA-1 sequences in GenBank included the complete AMA-1 ectodomain, and an additional 297 sequences included only domain I. The 615 GenBank sequences represented all major areas of the world with endemic malaria: 171 African sequences (152 West African), 380 Asian sequences (including 184 from Papua New Guinea, 71 from Thailand, and 99 from India), and 58 South American sequences.
Among the 824 AMA-1 sequences comprising positions 149 to 552, there were 62 polymorphic amino acid positions and 335 unique haplotypes. Haplotype prevalences ranged from 0.1 to 3.6%, with 58% of haplotypes being observed only once in the data set. Haplotypes with sequences matching the P. falciparum vaccine strains FVO and 3D7 were about equally frequent in the overall data set, with prevalences of 2.3 and 2.4%, respectively.
Among the 1121 AMA-1 sequences that included domain I of the protein, 48 unique haplotypes were observed based on the eight polymorphic residues within the c1L cluster of domain I. Haplotype prevalences based on c1L range from 0.09 to 13.5%, with 16 (33%) haplotypes observed only once, 7 of which came from India. As with haplotypes based on the entire ectodomain, c1L haplotypes with sequence matching the FVO and 3D7 vaccine strains were about equally frequent, with prevalences of 11.4 and 11.1%, respectively.
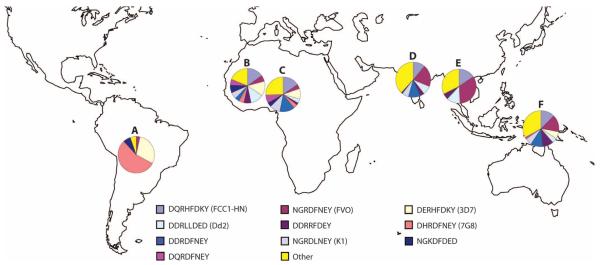
Distinct differences in AMA-1 allele prevalences were observed among parasites from different geographic regions. In Africa [based mostly on sequences from Mali (n = 570)], the c1L haplotype corresponding to the 3D7 strain (14%) was more than twice as prevalent as that corresponding to the FVO strain (6%) (Fig. 4). However, in Asia, the c1L haplotype corresponding to FVO (21%) was seven times more prevalent than that corresponding to 3D7 (3%) (Fig. 4). When Asian haplotype prevalences were examined by country, the haplotype corresponding to 3D7 was not observed at all among Thai sequences, whereas the FVO haplotype had a prevalence of 34%. An especially large number of haplotypes were observed among the Indian AMA-1 sequences, with 26 unique c1L haplotypes observed among only 99 sequences, none of which matched the sequence of 3D7. In South America, more than half of the c1L sequences matched that of the 7G8 strain of P. falciparum, 29% matched 3D7, and 3% matched FVO (Fig. 4).
Fig. 4.
Global distribution of AMA-1 domain I c1L haplotypes. Prevalence of the 10 most prevalent domain I c1L haplotypes in a data set of 1121 AMA-1 sequences, including AMA-1 sequences available in GenBank. Haplotypes are indicated by the one-letter amino acid abbreviation at each of the eight polymorphic sites in c1L: 196, 197, 199, 200, 201, 204, 206, and 207. Haplotypes corresponding to specific strains of P. falciparum are indicated in parentheses. Pie charts are shown only for regions with 50 or more available sequences. (A) South America, n = 58,7 haplotypes. (B) Mali, n = 570, 27 haplotypes. (C) Nigeria, n = 51, 16 haplotypes. (D) India, n = 99, 26 haplotypes. (E) Thailand, n = 71, 9 haplotypes. (F) Papua New Guinea, n = 184, 16 haplotypes.
Application of population structure algorithms to infer AMA-1 haplotype groups
Proteins that have similar amino acid sequences are expected to have similar tertiary structures; this expectation provides a rationale for using population structure clustering algorithms to categorize AMA-1 sequences that are genetically similar into haplotype groupings that may be associated with cross-reactive immune responses (14). These algorithms were designed for application to data from multiple, unlinked, neutral loci, however, rather than to potentially linked polymorphisms in a single gene under balancing selection. Violation of the model assumptions could affect the inference of haplotype groups, resulting in spurious results (27). To permit comparison with published results using this approach to analyze AMA-1 sequences from another site in Mali, we performed a population structure analysis of the AMA-1 sequences from this study (see figs.S3 and S4).
DISCUSSION
Our analysis of the diversity and dynamics of the P. falciparum AMA-1 protein within infected individuals in Mali provides direct evidence from human infections that agrees with the findings of a recent in vitro study (13) that showed that residues within the c1L cluster of domain I had the greatest effect on antigenic escape. Our results also corroborate the findings of an examination of the structure of AMA-1 in complex with an inhibitory monoclonal antibody that demonstrated that mutations at positions 197, 200, 201, 204, and 225 (all of which are located in c1L or c1) abrogated binding of this antibody (28). These combined findings suggest that residues within domain I c1L may be useful for categorizing parasites into groups for inclusion in a polyvalent AMA-1 vaccine and for monitoring vaccine-induced selection in clinical trials of AMA-1–based vaccines. Measures of vaccine-induced selection and allele-specific antibody responses in efficacy trials of AMA-1 vaccines will be required to confirm the importance of these residues because other factors, including immune responses to other antigens, could influence the dynamics of AMA-1 diversity. We plan to do such analyses on samples from phase 2 trials of both monovalent (6) and bivalent (5) AMA-1 vaccines in Mali. Further immunological investigations are also warranted to more comprehensively map the putative epitopes responsible for the associations observed in this study.
The inability to detect effects of AMA-1 type on risk of clinical illness after 6 weeks is consistent with the short-lived antibody responses reported in immunological studies (15, 16) and might be interpreted as evidence that protective AMA-1 antibody responses are too transient for AMA-1 vaccines to offer clinically meaningful protection. B cell memory may persist after antibody concentrations wane, however, and vaccine-induced immunity to AMA-1 may persist longer than naturally acquired humoral immunity: In a recent phase 1 study of an AMA-1–based vaccine in children, anti-AMA-1 antibody titers increased by at least 100-fold after immunization and remained high throughout the 1-year follow-up period (29). Indeed, a malaria vaccine must perform better than naturally acquired immunity, which confers only temporary protection against disease, but in doing so, vaccine-induced immunity may impose even greater selection pressure on the protein than does natural immunity. The strength, duration, and allele specificity of vaccine-induced immune responses, as well as their effect on the distribution of AMA-1 alleles in vaccinated individuals, are being evaluated in a phase 2 trial of the same vaccine that showed prolonged, high-antibody responses in the phase 1 trial (29).
The clusters of polymorphisms were defined on the basis of their proximity to one another in the crystal structure (13); however, it is possible that there were immunologically important polymorphisms outside of clusters c1 and c1L whose effects were masked because of their being grouped with less important residues. For example, based on the random forest analysis, amino acid changes at positions 172 and 175 were good predictors of the development of symptoms, and these findings are supported by results from growth-inhibitory assays that indicate that residue 175 (along with other residues) has a significant effect on erythrocyte invasion (14).
The prevalence of AMA-1 alleles differed among parasites from Africa, Asia, and South America. These results suggest that vaccine efficacy might also vary by geographic location, depending on the strain(s) targeted by the vaccine. For example, a vaccine based on the FVO strain might have higher efficacy in Asia than in West Africa, and a vaccine based on the 7G8 strain might have the highest efficacy in South America but low efficacy in Asia or Africa. Some malaria vaccines have shown varying efficacy by geography (30, 31); however, parasite genotyping was not done in these trials to determine whether geographic differences in allele prevalences contributed to the variable efficacy. The geographical differences in AMA-1 haplotype prevalences highlight the need to measure vaccine antigen allele prevalences in malaria endemic areas before testing and distribution of vaccines.
Examination of additional published AMA-1 sequences did not show evidence of a maximum amount of existing AMA-1 diversity. Analysis of 63% more AMA-1 ectodomain sequences resulted in a 57% increase in the number of observed AMA-1 ectodomain haplotypes. Likewise, a 122% increase in the number of domain I c1L sequences resulted in a 92% increase in the number of observed domain I c1L haplotypes. This seemingly limitless amount of diversity may pose a major obstacle for development of an effective polyvalent AMA-1–based vaccine.
A bivalent AMA-1 vaccine recently failed to demonstrate efficacy in Mali (5), although it is not yet known whether this resulted from a relatively poorly immunogenic formulation and/or inadequate cross-protection between the two vaccine strains and the diverse parasites in nature. An ongoing efficacy trial of a more highly immunogenic formulation of a monovalent AMA-1 vaccine will provide evidence of whether and to what extent vaccine-induced immunity is cross-protective or allele-restricted. Even a polyvalent recombinant subunit vaccine may not be feasible if 10 to 20 haplotypes are required to cover most of the diversity in a single geographic location. Nevertheless, it may be possible to select or engineer vaccine antigens that are more cross-protective. A recent study aligned published AMA-1 sequences to determine the location and extent of polymorphism in the protein and to identify linkage between specific residues, and this information was used to design three artificial AMA-1 constructs that share conserved amino acids while representing the greatest number of polymorphisms (32). Data from molecular epidemiological studies and/or analyses of protein structure can be used to identify subsets of residues (13, 33) that may be particularly important to take into consideration in engineering such artificial antigens, ensuring that the most important residues for protection are represented.
It may be desirable to avoid polymorphism altogether by engineering vaccine constructs that boost the immune response to protective epitopes in conserved regions of AMA-1 and other proteins. At least one epitope that inhibits parasite invasion has been identified in the domain II loop on the nonpolymorphic face of AMA-1 (34). If subunit vaccines based on conserved epitopes can divert the immune response away from highly polymorphic regions, they may be able to induce strain-transcending immunity. Alternatively, genomic and proteomic approaches are being used to identify new vaccine targets that are not immunodominant and likely to be more conserved than the current highly immunodominant and polymorphic candidates (35-37). Live attenuated whole-organism vaccines may also hold promise for circumventing the extreme polymorphism in individual antigens exemplified by this study (38).
MATERIALS AND METHODS
Study site
Bandiagara is a rural town with 13,634 inhabitants in the Dogon Country of northeastern Mali. Mean annual rainfall is ~600 mm, and Anopheles gambiae s.l. is the primary vector for malaria. P. falciparum malaria has intense seasonal transmission corresponding to the July to October rainy season, with peak monthly entomological inoculation rates (EIRs) of up to 40 to 60 infected bites per person per month in September and total annual EIRs of 50 to 150. EIRs at the start and end of the transmission season (in June and December, respectively) are less than one infectious bite per person per month (39). P. falciparum accounts for 97% of malaria infections, with 3% due to Plasmodium malariae and rare infections with Plasmodium ovale. The disease burden is high, with children aged 10 years or less experiencing a mean of two clinical episodes of uncomplicated malaria a year (39) and a 2.3% annual incidence of severe malaria among children aged 6 years or less (40). Bandiagara has served as a vaccine testing site since 2003 (41) with four completed malaria vaccine trials. A phase 2 pediatric trial of an AMA-1–based vaccine derived from the 3D7 strain of P. falciparum has recently been completed at this site.
Study design
The samples used in this study were collected during a cohort study designed to measure the age-specific incidence of malaria infection and disease in children and young adults in Bandiagara and are the same samples included in a previous study of a different blood-stage malaria vaccine antigen (42). The study was conducted prospectively during the years 1999, 2000, and 2001 before clinical trials of malaria vaccines had been done at this site or elsewhere in Mali. Study subjects were aged ≥3 months to 20 years and were recruited from all eight sectors of Bandiagara town in proportion to the populations of those sectors. From July to January of each year, individuals were followed actively by weekly visits. Each visit included a brief clinical examination, and participants with symptoms consistent with malaria were given a full medical and laboratory assessment. Each participant contributed ~0.1 ml of blood, collected on 3MM Whatman filter paper, at least monthly and at the time of every episode of clinical malaria. Samples were collected under protocols reviewed and approved by institutional review boards of the University of Maryland School of Medicine and the University of Bamako Faculty of Medicine. Informed consent was obtained from all study participants or their guardians.
As described previously (42), 100 individuals with at least 2 years of follow-up during the malaria incidence study were randomly selected within three age strata. Thirty children aged ≤5 years, 32 aged 6 to 10 years, and 38 aged ≥11 years were selected. Blood samples (n = 1369) corresponding to monthly surveys and clinical episodes that were positive for parasites according to a previous polymerase chain reaction (PCR) (42) underwent PCR and sequencing of the ama-1 gene.
PCR
Nested PCR was used to amplify three overlapping fragments corresponding to the three domains of AMA-1. The domain I fragment was amplified with the following PCR primers (43): external forward, 5′-GAACCCGCACCACAAGAAC-3′; external reverse, 5′-TTGTTTAGGTTGATCCGAAGC-3′; internal forward, 5′-CCATGGACGGAATATATGGC-3′; and internal reverse, 5′-TTCCATCGACCCATAATCCG-3′. Primers for domains II and III were designed as follows: domain II (external forward, 5′-GAAACAGCATGTTTTGTTTTAG-3′; external reverse, 5′-GGGATGGGACAAAGCAGTAG-3′; internal forward, 5′-GGGAAAAAGTTTGCCCTAGAA-3′; and internal reverse, 5′-GGGATGGGACAAAGCAGTAG-3′) and domain III (external forward, 5′-TTCTTCCCACTGGTGCTTTT-3′; external reverse, 5′-TTTTTCAGCATTTCCTTTTCTTTT-3′; internal forward, 5′-AGCAGATAGATATAAAAGTC-3′; and internal reverse, 5′-ACCATTAAAATAGTTGCTAAT-3′). The external PCR consisted of 2.5 μl of 10× buffer, 0.75 μl of 50mM MgCl2, 0.2 μl of 25 mM deoxynucleoside triphosphate (dNTP), 0.125 μl of each 50-μm forward and reverse primer, 0.125 μl of Taq polymerase, 18.675 μl of water, and 2.5 μl of genomic DNA as template. All reagents were Invitrogen products (Life Technologies). Cycling conditions for the external PCR included 10 touchdown cycles beginning at a 60°C annealing temperature with −0.5°C per cycle, followed by 30 cycles with a 55°C annealing temperature. The internal PCR consisted of 5 μl of 10× buffer, 1.5 μl of 50mM MgCl2, 0.4 μl of 25 mM dNTP, 0.25 μl of each 50-μm forward and reverse primer, 0.25 μl of Taq polymerase, 37.35 μl of water, and 5 μl of external PCR product as template, run for 25 cycles at a 55°C annealing temperature. PCR products were visualized on a 2% agarose gel. In general, the domain II PCR had a higher efficiency (more PCR-positive samples) than the PCRs for the other two domains.
Sequencing
PCR products were purified with filter plates (Edge Biosystems) attached to a vacuum manifold and were eluted in water. Purified products (2 μl) were sequenced in a 10-μl reaction with BigDye v3.1 (Applied Biosystems), ethanol precipitated, and run in 7 μl of HiDi formamide on an ABI 3730XL 96-capillary sequencer. Sequences were edited and aligned with Sequencher 4.7 (Gene Codes) to form 1212–base pair contigs comprising codons 149 to 552 of the ama-1 gene. A total of 748 complete contigs were generated. Infections were classified as single or predominant allele infections or as multiple allele infections if the secondary peak height was >50% of the primary peak height at any polymorphic position. Sequences containing unique substitutions not observed in the rest of the data set were reamplified and sequenced from the original sample to rule out PCR error. Sequence alignment was performed with Bioedit version 7.0.1 (44).
GenBank sequences
All P. falciparum AMA-1 nucleotide sequences in GenBank as of 1 October 2008 were downloaded and aligned. Sequences containing mutations not observed in any other sequences were excluded from analysis, as were replicate sequences of laboratory strains (for example, 3D7). The geographic origin of each sequence was reported in GenBank for most sequences, and for those not documenting the sequence origin in GenBank, the origin was determined by consulting the associated publication.
Population structure analysis
Cluster analysis was performed with Structurama (45) and compared with results obtained from STRUCTURE (46). In Structurama, the number of populations can be fixed [as is assumed in STRUCTURE (46)] or can be treated as a random variable (47). Ten replicate runs were performed on sequences from the 506 single- or predominant-clone infections from the Malian cohort. For each run, the number of populations (haplotype groups) was treated as a random variable, with a Dirichlet process prior with α distributed according to a γ distribution with shape and scale both equal to 1. Both population assignment and the estimated number of populations were robust to the chosen values for the shape and scale parameters. For each replicate, the Markov chain was run for 500,000 generations, and the number of populations (haplotype groups) with the highest average posterior probability for the 10 replicates was reported in the Results section. The assignment of individual haplotypes to haplotype groups by Structurama was similar to the assignment by STRUCTURE when the model assumed the same number of groups, admixture, and correlated allele frequencies.
Statistical analysis
Generalized estimating equations were used to perform logistic regression to estimate whether changes in specific clusters of AMA-1 polymorphisms are associated with the development of clinical symptoms in individuals' consecutive infections. Both single- and multiple-clone infections were included in the analysis. The primary outcome was whether an individual's second of two paired consecutive infections was symptomatic or asymptomatic. The presence of infection was defined based on the results of a highly sensitive merozoite surface protein-1 PCR conducted as part of a previous study (42). For each polymorphic amino acid position, it was determined whether there was a change in amino acid in the next consecutive infection. The primary predictor for the analysis was the proportion of polymorphic residues within a cluster or domain that changed between the two consecutive infections. Because the proportion of amino acid changes was not normally distributed (that is, distributions tended to be skewed toward zero), this variable was classified as an ordinal variable with cut points at the first and third quartiles. The proportion of changes was examined in separate models for the whole AMA-1 ectodomain, each of the three major domains, and four subclusters within domain I. Domain I included polymorphic sites from positions 149 to 302, domain II positions 320 to 418, and domain III positions 443 to 509. The domain I subclusters, originally defined by Dutta and colleagues (48), were defined as follows (fig. S2): c1—187, 189, 190, 196, 197, 199, 200, 201, 204, 206, 207, 224, 225, 228, 230, and 231; c1L—196, 197, 199, 200, 201, 204, 206, and 207; cluster 2 (c2)—242, 243, 244, 245, 282, 283, 285, and 286; and c3—172, 173, 174, 175, 267, and 269. Estimated effects were adjusted for age and an interaction between the amount of genetic change and the time between consecutive infections. The time between consecutive infections was, on average, longer when the first of the two episodes was symptomatic, consistent with the fact that symptomatic individuals were treated with a long-acting drug (sulfadoxine-pyrimethamine) that prevents infection for about a month after treatment (39). After taking into account the interaction with time, the presence of symptoms in the first of two consecutive infections was not significantly associated with the outcome and was therefore left out of the final models. The presence of a mixed infection in the interval was also not significantly associated with the outcome and was removed from the final models. The interaction between the amount of genetic change and time between consecutive infections was explored by trying various cut points (for example, time strata of 1 to 4, 4 to 8, and >8 weeks, and strata of 1 to 6, 6 to 12, and >12 weeks) and indicated that any association with the outcome diminished in intervals longer than 4 to 6 weeks. As a result, time between consecutive infections was treated as a categorical variable with a cut point at 6 weeks. Consecutive infections spanning the dry season were not included in the analysis because no sampling was performed during the dry season when the rate of transmission of new infections approaches zero. Consecutive infections separated by ≤7 days were also excluded, as they likely represent the same infection, as were intervals preceding intervals ≤7 days. A compound symmetry covariance structure was used to model the correlation between measurements from the same individual, and estimates were robust to the choice of covariance structure. No corrections were made for multiple comparisons.
The same analysis was used to evaluate the association between change in AMA-1 haplotype group (determined by a population structure clustering algorithm) and risk of clinical malaria. Because the structure analysis can only be performed on single- or predominant-clone infections where a haplotype can be resolved, the within-host analyses can only evaluate consecutive infections that both contain a single clone, thus reducing the available sample size.
Random forests
The statistical or machine learning method known as random forest (18) was used with the randomForest package (49) for the R statistical programming system (50, 51) to classify whether an individual's second of two consecutive infections was symptomatic or asymptomatic for intervals <6 weeks (the response variable) based on changes at polymorphic amino acid positions in the AMA-1 ectodomain, age, and symptoms in the first infection (the predictor variables). Application of the random forest method in this study follows previous successful application of tree-based statistical models (52) and random forests to genetic data (53, 54). Here, the random forests comprised 1 × 105 individual tree-based statistical models. Variable importance was measured in terms of the increase in group purity when partitioning data on the basis of the permutation accuracy importance procedure (49). The procedure is broadly similar to other uses of permutation tests. A data set is analyzed and the prediction accuracy is quantified. A variable is permuted in the data set, the other variables remaining unpermuted, and the analysis is repeated. The difference in the prediction accuracy between the original (unpermuted) and the variable-permuted data sets is a measure of importance of the variable. If a predictor variable is strongly associated with response (that is, contributes substantially to the prediction), a marked decrease in prediction accuracy will result from the permutation. Among the several advantages of permutation accuracy importance is that it provides an assessment of the importance of the variable in interaction, positive or negative, with other variables.
Supplementary Material
55. Acknowledgments
We thank the population and the regional and district health authorities of Bandiagara, Mali for their continued support and the Bandiagara Malaria Project staff and managers for their dedication.
Funding: Supported by grant U19AI065683 and contract N01AI85346 from the National Institute of Allergy and Infectious Diseases of the U.S. NIH, the U.S. Agency for International Development Malaria Vaccine Program, the Doris Duke Charitable Foundation, and the Howard Hughes Medical Institute. S.L.T. is supported by the University of Maryland Multidisciplinary Clinical Research Career Development Program (NIH grant K12RR023250).
Footnotes
SUPPLEMENTARY MATERIAL
www.sciencetranslationalmedicine.org/cgi/content/full/1/2/2ra5/DC1
Competing interests: The authors have no competing interests to declare.
Accession numbers: GenBank FJ898536 to FJ899041.
REFERENCES AND NOTES
- 1.Escalante AA, Lal AA, Ayala FJ. Genetic polymorphism and natural selection in the malaria parasite Plasmodium falciparum. Genetics. 1998;149:189–202. doi: 10.1093/genetics/149.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takala SL, Plowe CV. Genetic diversity and malaria vaccine design, testing and efficacy: Preventing and overcoming “vaccine resistant malaria”. Parasite Immunol. 2009;31:560–573. doi: 10.1111/j.1365-3024.2009.01138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moran M, Guzman J, Ropars A, Jorgensen M, McDonald A, Potter S, Haile-Selassie H. The Malaria Product Pipeline: Planning for the Future. (The George Institute of International Health, London, 2007) www.thegeorgeinstitute.org/shadomx/apps/fms/fmsdownload.cfm?file_uuid=1ABDE79C-DA73-20B3-D6E1-F7AF25011EC9&siteName=iih [accessed July 2009]
- 4.Roestenberg M, Remarque E, de Jonge E, Hermsen R, Blythman H, Leroy O, Imoukhuede E, Jepsen S, Ofori-Anyinam O, Faber B, Kocken CH, Arnold M, Walraven V, Teelen K, Roeffen W, de Mast Q, Ballou WR, Cohen J, Dubois MC, Ascarateil S, van der Ven A, Thomas A, Sauerwein R. Safety and immunogenicity of a recombinant Plasmodium falciparum AMA1 malaria vaccine adjuvanted with Alhydrogel, Montanide ISA 720 or AS02. PLoS One. 2008;3:e3960. doi: 10.1371/journal.pone.0003960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sagara I, Dicko A, Ellis RD, Fay MP, Diawara SI, Assadou MH, Sissoko MS, Kone M, Diallo AI, Saye R, Guindo MA, Kante O, Niambele MB, Miura K, Mullen GE, Pierce M, Martin LB, Dolo A, Diallo DA, Doumbo OK, Miller LH, Saul A. A randomized controlled phase 2 trial of the blood stage AMA1-C1/Alhydrogel malaria vaccine in children in Mali. Vaccine. 2009;27:3090–3098. doi: 10.1016/j.vaccine.2009.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thera MA, Doumbo OK, Coulibaly D, Diallo DA, Kone AK, Guindo AB, Traore K, Dicko A, Sagara I, Sissoko MS, Baby M, Sissoko M, Diarra I, Niangaly A, Dolo A, Daou M, Heppner S. I. Diawara, D. G., Stewart VA, Angov E, Bergmann-Leitner ES, Lanar DE, Dutta S, Soisson L, Diggs CL, Leach A, Owusu A, Dubois MC, Cohen J, Nixon JN, Gregson A, Takala SL, Lyke KE, Plowe CV. Safety and immunogenicity of an AMA-1 malaria vaccine in Malian adults: Results of a phase 1 randomized controlled trial. PLoS One. 2008;3:e1465. doi: 10.1371/journal.pone.0001465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hodder AN, Crewther PE, Matthew ML, Reid GE, Moritz RL, Simpson RJ, Anders RF. The disulfide bond structure of Plasmodium apical membrane antigen-1. J. Biol. Chem. 1996;271:29446–29452. doi: 10.1074/jbc.271.46.29446. [DOI] [PubMed] [Google Scholar]
- 8.Cortés A, Mellombo M, Mueller I, Benet A, Reeder JC, Anders RF. Geographical structure of diversity and differences between symptomatic and asymptomatic infections for Plasmodium falciparum vaccine candidate AMA1. Infect. Immun. 2003;71:1416–1426. doi: 10.1128/IAI.71.3.1416-1426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Escalante AA, Grebert HM, Chaiyaroj SC, Magris M, Biswas S, Nahlen BL, Lal AA. Polymorphism in the gene encoding the apical membrane antigen-1 (AMA-1) of Plasmodium falciparum. X. Asembo Bay Cohort Project. Mol. Biochem. Parasitol. 2001;113:279–287. doi: 10.1016/s0166-6851(01)00229-8. [DOI] [PubMed] [Google Scholar]
- 10.Polley SD, Conway DJ. Strong diversifying selection on domains of the Plasmodium falciparum apical membrane antigen 1 gene. Genetics. 2001;158:1505–1512. doi: 10.1093/genetics/158.4.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bai T, Becker M, Gupta A, Strike P, Murphy VJ, Anders RF, Batchelor AH. Structure of AMA1 from Plasmodium falciparum reveals a clustering of polymorphisms that surround a conserved hydrophobic pocket. Proc. Natl. Acad. Sci. U.S.A. 2005;102:12736–12741. doi: 10.1073/pnas.0501808102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins CR, Withers-Martinez C, Hackett F, Blackman MJ. An inhibitory antibody blocks interactions between components of the malarial invasion machinery. PLoS Pathog. 2009;5:e1000273. doi: 10.1371/journal.ppat.1000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dutta S, Lee SY, Batchelor AH, Lanar DE. Structural basis of antigenic escape of a malaria vaccine candidate. Proc. Natl. Acad. Sci. U.S.A. 2007;104:12488–12493. doi: 10.1073/pnas.0701464104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duan J, Mu J, Thera MA, Joy D, Kosakovsky Pond SL, Diemert D, Long C, Zhou H, Miura K, Ouattara A, Dolo A, Doumbo O, Su XZ, Miller L. Population structure of the genes encoding the polymorphic Plasmodium falciparum apical membrane antigen 1: Implications for vaccine design. Proc. Natl. Acad. Sci. U.S.A. 2008;105:7857–7862. doi: 10.1073/pnas.0802328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akpogheneta OJ, Duah NO, Tetteh KK, Dunyo S, Lanar DE, Pinder M, Conway DJ. Duration of naturally acquired antibody responses to blood-stage Plasmodium falciparum is age dependent and antigen specific. Infect. Immun. 2008;76:1748–1755. doi: 10.1128/IAI.01333-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinyanjui SM, Conway DJ, Lanar DE, Marsh K. IgG antibody responses to Plasmodium falciparum merozoite antigens in Kenyan children have a short half-life. Malar. J. 2007;6:82. doi: 10.1186/1475-2875-6-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coulibaly D, Diallo DA, Thera MA, Dicko A, Guindo AB, Koné AK, Cissoko Y, Coulibaly S, Djimdé A, Lyke K, Doumbo OK, Plowe CV. Impact of preseason treatment on incidence of falciparum malaria and parasite density at a site for testing malaria vaccines in Bandiagara. Mali. Am. J. Trop. Med. Hyg. 2002;67:604–610. doi: 10.4269/ajtmh.2002.67.604. [DOI] [PubMed] [Google Scholar]
- 18.Breiman L. Random forests. Mach. Learn. 2001;45:5–32. [Google Scholar]
- 19.Eisen DP, Marshall VM, Billman-Jacobe H, Coppel RL. A Plasmodium falciparum apical membrane antigen-1 (AMA-1) gene apparently generated by intragenic recombination. Mol. Biochem. Parasitol. 1999;100:243–246. doi: 10.1016/s0166-6851(99)00054-7. [DOI] [PubMed] [Google Scholar]
- 20.Garg S, Alam MT, Das MK, Dev V, Kumar A, Dash AP, Sharma YD. Sequence diversity and natural selection at domain I of the apical membrane antigen 1 among Indian Plasmodium falciparum populations. Malar. J. 2007;6:154. doi: 10.1186/1475-2875-6-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kocken CH, Narum DL, 1, Massougbodji A, Ayivi B, Dubbeld MA, van der Wel A, Conway DJ, Sanni A, Thomas AW. Molecular characterisation of Plasmodium reichenowi apical membrane antigen-1 (AMA-1), comparison with P. falciparum AMA-1, and antibody-mediated inhibition of red cell invasion. Mol. Biochem. Parasitol. 2000;109:147–156. doi: 10.1016/s0166-6851(00)00250-4. [DOI] [PubMed] [Google Scholar]
- 22.Marshall VM, Zhang L, Anders RF, Coppel RL. Diversity of the vaccine candidate AMA-1 of Plasmodium falciparum. Mol. Biochem. Parasitol. 1996;77:109–113. doi: 10.1016/0166-6851(96)02583-2. [DOI] [PubMed] [Google Scholar]
- 23.Ord RL, Tami A, Sutherland CJ. ama 1 genes of sympatric Plasmodium vivax and P. falciparum from Venezuela differ significantly in genetic diversity and recombination frequency. PLoS One. 2008;3:e3366. doi: 10.1371/journal.pone.0003366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peterson MG, Marshall VM, Smythe JA, Crewther PE, Lew A, Silva A, Anders RF, Kemp DJ. Integral membrane protein located in the apical complex of Plasmodium falciparum. Mol. Cell. Biol. 1989;9:3151–3154. doi: 10.1128/mcb.9.7.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Polley SD, Chokejindachai W, Conway DJ. Allele frequency-based analyses robustly map sequence sites under balancing selection in a malaria vaccine candidate antigen. Genetics. 2003;165:555–561. doi: 10.1093/genetics/165.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rajesh V, Singamsetti VK, Vidya S, Gowrishankar M, Elamaran M, Tripathi J, Radhika NB, Kochar D, Ranjan A, Roy SK, Das A. Plasmodium falciparum: Genetic polymorphism in apical membrane antigen-1 gene from Indian isolates. Exp. Parasitol. 2008;119:144–151. doi: 10.1016/j.exppara.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 27.Pritchard JK, Wen X, Falush D. Documentation for Structure Software, Version 2.2. University of Chicago; Chicago: 2007. [Google Scholar]
- 28.Coley AM, Gupta A, Murphy VJ, Bai T, Kim H, Foley M, Anders RF, Batchelor AH. Structure of the malaria antigen AMA1 in complex with a growth-inhibitory antibody. PLoS Pathog. 2007;3:1308–1319. doi: 10.1371/journal.ppat.0030138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thera MA. Randomized, controlled, dose escalation phase 1 clinical trial to evaluate the safety and immunogenicity of Walter Reed Army Institute of Research's AMA-1 malaria vaccine (FMP 2.1) adjuvanted in GSKBio's AS02A vs. rabies vaccine in 1-6 year old children in Bandiagara. Mali. Am. J. Trop. Med. Hyg. 2007;77S:144. [Google Scholar]
- 30.D'Alessandro U, Leach A, Drakeley CJ, Bennett S, Olaleye BO, Fegan GW, Jawara M, Langerock P, George MO, Targett GAT, Greenwood BM. Efficacy trial of malaria vaccine SPf66 in Gambian infants. Lancet. 1995;346:462–467. doi: 10.1016/s0140-6736(95)91321-1. [DOI] [PubMed] [Google Scholar]
- 31.Valero MV, Amador LR, Galindo C, Figueroa J, Bello MS, Murillo LA, Mora AL, Patarroyo G, Rocha CL, Rojas M, Aponte JJ, Sarmiento LE, Lozada DM, Coronell CG, Ortega NM, Rosas JE, Patarroyo ME, Alonso PL. Vaccination with SPf66, a chemically synthesised vaccine, against Plasmodium falciparum malaria in Colombia. Lancet. 1993;341:705–710. doi: 10.1016/0140-6736(93)90483-w. [DOI] [PubMed] [Google Scholar]
- 32.Remarque EJ, Faber BW, Kocken CH, Thomas AW. A diversity-covering approach to immunization with Plasmodium falciparum apical membrane antigen 1 induces broader allelic recognition and growth inhibition responses in rabbits. Infect. Immun. 2008;76:2660–2670. doi: 10.1128/IAI.00170-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takala SL, Coulibaly D, Thera MA, Dicko A, Smith DL, Guindo AB, Kone AK, Traore K, Ouattara A, Djimde AA, Sehdev PS, Lyke KE, Diallo DA, Doumbo OK, Plowe CV. Dynamics of polymorphism in a malaria vaccine antigen at a vaccine-testing site in Mali. PLoS Med. 2007;4:e93. doi: 10.1371/journal.pmed.0040093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Collins CR, Withers-Martinez C, Bentley GA, Batchelor AH, Thomas AW, Blackman MJ. Fine mapping of an epitope recognized by an invasion-inhibitory monoclonal antibody on the malaria vaccine candidate apical membrane antigen 1. J. Biol. Chem. 2007;282:7431–7441. doi: 10.1074/jbc.M610562200. [DOI] [PubMed] [Google Scholar]
- 35.Chaudhuri R, Ahmed S, Ansari FA, Singh HV, Ramachandran S. MalVac: Database of malarial vaccine candidates. Malar. J. 2008;7:184. doi: 10.1186/1475-2875-7-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanoi BN, Egwang TG. New concepts in vaccine development in malaria. Curr. Opin. Infect. Dis. 2007;20:311–316. doi: 10.1097/QCO.0b013e32816b5cc2. [DOI] [PubMed] [Google Scholar]
- 37.Scarselli M, Giuliani MM, Adu-Bobie J, Pizza M, Rappuoli R. The impact of genomics on vaccine design. Trends Biotechnol. 2005;23:84–91. doi: 10.1016/j.tibtech.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 38.Luke TC, Hoffman SL. Rationale and plans for developing a non-replicating, metabolically active, radiation-attenuated Plasmodium falciparum sporozoite vaccine. J. Exp. Biol. 2003;206:3803–3808. doi: 10.1242/jeb.00644. [DOI] [PubMed] [Google Scholar]
- 39.Coulibaly D, Diallo DA, Thera MA, Dicko A, Guindo AB, Koné AK, Cissoko Y, Coulibaly S, Djimdé A, Lyke K, Doumbo OK, Plowe CV. Impact of preseason treatment on incidence of falciparum malaria and parasite density at a site for testing malaria vaccines in Bandiagara, Mali. Am. J. Trop. Med. Hyg. 2002;67:604–610. doi: 10.4269/ajtmh.2002.67.604. [DOI] [PubMed] [Google Scholar]
- 40.Lyke KE, Dicko A, Kone A, Coulibaly D, Guindo A, Cissoko Y, Traoré K, Plowe CV, Doumbo OK. Incidence of severe Plasmodium falciparum malaria as a primary endpoint for vaccine efficacy trials in Bandiagara, Mali. Vaccine. 2004;22:3169–3174. doi: 10.1016/j.vaccine.2004.01.054. [DOI] [PubMed] [Google Scholar]
- 41.Thera MA, Doumbo OK, Coulibaly D, Diallo DA, Sagara I, Dicko A, Diemert DJ, Heppner DG, Jr., Stewart VA, Angov E, Soisson L, Leach A, Tucker K, Lyke KE, Plowe; Mali FMP1 Working Group CV. Safety and allele-specific immunogenicity of a malaria vaccine in Malian adults: Results of a phase I randomized trial. PLoS Clin. Trials. 2006;1:e34. doi: 10.1371/journal.pctr.0010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takala SL, Coulibaly D, Thera MA, Dicko A, Smith DL, Guindo AB, Kone AK, Traore K, Ouattara A, Djimde AA, Sehdev PS, Lyke KE, Diallo DA, Doumbo OK, Plowe CV. Dynamics of polymorphism in a malaria vaccine antigen at a vaccine-testing site in Mali. PLoS Med. 2007;4:e93. doi: 10.1371/journal.pmed.0040093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cortés A, Mellombo M, Mueller I, Benet A, Reeder JC, Anders RF. Geographical structure of diversity and differences between symptomatic and asymptomatic infections for Plasmodium falciparum vaccine candidate AMA1. Infect. Immun. 2003;71:1416–1426. doi: 10.1128/IAI.71.3.1416-1426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 45.Huelsenbeck JP, Andolfatto P. Inference of population structure under a Dirichlet process model. Genetics. 2007;175:1787–1802. doi: 10.1534/genetics.106.061317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pella J, Masuda M. The Gibbs and split–merge sampler for population mixture analysis from genetic data with incomplete baselines. Can. J. Fish. Aquat. Sci. 2006;63:576–596. [Google Scholar]
- 48.Dutta S, Lee SY, Batchelor AH, Lanar DE. Structural basis of antigenic escape of a malaria vaccine candidate. Proc. Natl. Acad. Sci. U.S.A. 2007;104:12488–12493. doi: 10.1073/pnas.0701464104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liaw A, Wiener M. Classification and regression by randomForest. R News. 2002;2:18–22. [Google Scholar]
- 50.Ihaka R, Gentleman R. R: A language for data analysis and graphics. J. Comput. Graph. Stat. 1996;5:299–314. [Google Scholar]
- 51.R Development Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna: 2008. [Google Scholar]
- 52.Segal MR, Cummings MP, Hubbard AE. Relating amino acid sequence to phenotype: Analysis of peptide-binding data. Biometrics. 2001;57:632–642. doi: 10.1111/j.0006-341x.2001.00632.x. [DOI] [PubMed] [Google Scholar]
- 53.Cummings MP, Segal MR. Few amino acid positions in rpoB are associated with most of the rifampin resistance in Mycobacterium tuberculosis. BMC Bioinformatics. 2004;5:137. doi: 10.1186/1471-2105-5-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cummings MP, Myers DS. Simple statistical models predict C-to-U edited sites in plant mitochondrial RNA. BMC Bioinformatics. 2004;5:132. doi: 10.1186/1471-2105-5-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takala SL, Coulibaly D, Thera MA, Batchelor AH, Cummings MP, Escalante AA, Ouattara A, Traoré K, Niangaly A, Djimdé AA, Doumbo OK, Plowe CV. Extreme polymorphism in a vaccine antigen and risk of clinical malaria: Implications for vaccine development. Sci. Transl. Med. 2009;1:2ra5. doi: 10.1126/scitranslmed.3000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.