Abstract
At least two human APOBEC3 proteins – APOBEC3F and APOBEC3G – are capable of inhibiting HIV-1 replication by mutation of the viral cDNA. HIV-1 averts lethal restriction through its accessory protein Vif, which targets these APOBEC3 proteins for proteasomal degradation. The life-or-death interaction between human APOBEC3 proteins and HIV-1 Vif has stimulated much interest in developing novel therapeutics aimed at altering the deaminase activity of the APOBEC3s, thus changing the virus’ mutation rate to either lethal or suboptimal levels. The current state of mechanistic information is reviewed and the possible risks and benefits of increasing (via hypermutation) or decreasing (via hypomutation) the HIV-1 mutation rate through APOBEC3 proteins are discussed.
Keywords: AIDS, APOBEC3F, APOBEC3G, Coffin’s razor, DNA cytidine deamination, HIV, host–pathogen interaction, hypermutation, hypomutation, retrovirus restriction, Vif
Basic concepts in antiviral therapy
The most straightforward and traditional approach to blocking viral replication is through the use of compounds that are engineered to be highly specific inhibitors of viral proteins. Such inhibitors are pervasive among therapeutics today, and are often used as standard treatments for chronic viral infections. The treatment of HIV, for example, generally includes one or more compounds that inhibit essential HIV-1 proteins, such as non-nucleoside reverse-transcriptase inhibitors, protease inhibitors and integrase inhibitors. In general, these compounds have few off-target effects and avoid most problems with cytotoxicity. Despite the numerous successes of this approach, a major problem with virus-specific compounds is the evolution of drug resistance. Rapidly evolving viruses, such as HIV-1, invariably mutate to alter their amino acid composition and resist the drug. The invariant correlate that all effective drugs eventually select drug-resistant viruses is known as Coffin’s razor.
A second general strategy takes advantage of the fact that viruses are dependent on host proteins for replication and pathogenesis. If inessential to the host, such proteins are good targets for antiviral compounds. Cellular proteins are many magnitudes more stable (less mutable) than viral proteins and, therefore, much less likely to contribute to the evolution of drug resistance. While this may increase the long-term efficacy of the drug, a potential drawback to this approach lies in the risk that disrupting a cellular process may have unintended consequences or off-target effects. Thus, a thorough knowledge of the human interactome within the realm of the targeted protein and extensive preclinical studies are required for any therapeutic compound in order to utilize this strategy. As with viral protein inhibitors, administration requires careful calculation and consideration of the cost–benefit ratio.
A third and relatively new antiviral strategy has been realized with the discovery of cellular proteins that function to inhibit viral replication. These ‘restriction factors’ include dominant-acting cellular proteins that block a specific stage of the retroviral life cycle, are under selective pressure and so more diverse than most other cellular proteins, are susceptible to neutralization (or evasion) by viral counter measures and often induced by interferon or by viral infection itself (i.e., as part of the innate immune system). Examples include APOBEC3G [1–5], TRIM5α [6–10] and BST2/TETHERIN [11,12]. The discovery of these antiviral factors has opened the door to novel therapeutic approaches intended to facilitate, supplement or improve the endogenous restriction strategies that are already in place. This article will focus on the APOBEC3 family, the impact of these proteins on HIV-1 biology and their potential influence on the future of HIV/AIDS treatments.
Attacking the viral genome: the APOBEC3 protein family
APOBEC3G is the archetype of the APOBEC3 subfamily of ssDNA cytidine deaminases. This seven-member group of DNA mutators – APOBEC3A, B, C, DE, F, G and H – plays a central role in innate immunity, defending the genome against mutation induced by the invasion of exogenous pathogens, such as retroviruses [4,13–24] and the movement of endogenous retroelements, such as human endogenous retrovirus, long interspersed repetitive element and Alu (Table 1) [25–34]. APOBEC3G was originally identified as a potent ssDNA mutator [1] with significant homology to the RNA0-editing enzyme and family namesake APOBEC1 [35]. Concurrently, APOBEC3G was identified as a dominant inhibitor of Vif-deficient HIV-1 replication, whose effects could be completely overcome by the neutralizing activity of the Vif accessory protein [2]. APOBEC3G’s potent DNA deaminase activity and its ability to restrict HIV-1 replication were quickly found to be linked [1,3–5]. The mutation and subsequent restriction of Vif-deficient HIV-1 by APOBEC3G has been extensively studied and has helped set the paradigm for continuing mechanistic studies on the role of DNA deamination in retroviral restriction and innate immunity.
Table 1.
The human APOBEC3 repertoire.
| A3A | A3B | A3C | A3DE | A3F | A3G | A3H | |
|---|---|---|---|---|---|---|---|
| Domains | Z1 | Z2Z1 | Z2 | Z2Z2 | Z2Z2 | Z2Z1 | Z3 |
| Subcellular localization |
Cell wide | Nuclear | Cell wide | Cytoplasmic | Cytoplasmic | Cytoplasmic | Cell wide |
| ΔVif HIV restriction |
No | Yes | Weak | Yes | Strong | Strong | Some variants |
| HIV Vif interaction |
No | No | Yes | Yes | Yes | Yes | Yes |
| Expression | PBLs, spleen, bone marrow and lung |
PBLs, bone marrow, lung and stem cells |
PBLs, thymus, spleen, lymph node, testis, ovary, small intestine, colon,liver, pancreas, heart, lung and adipose |
PBLs, thymus, spleen, lymph node, ovary, lung, liver and adipose |
PBLs, thymus, spleen, lymph node, testis, ovary, uterus, brain, lung, colon, liver, kidney and pancreas |
PBLs, thymus, spleen, lymph node, testis, ovary, uterus, brain, lung, small intestine, colon, liver, kidney and pancreas |
PBLs, thymus, testis, ovary, brain, small intestine, colon and skin |
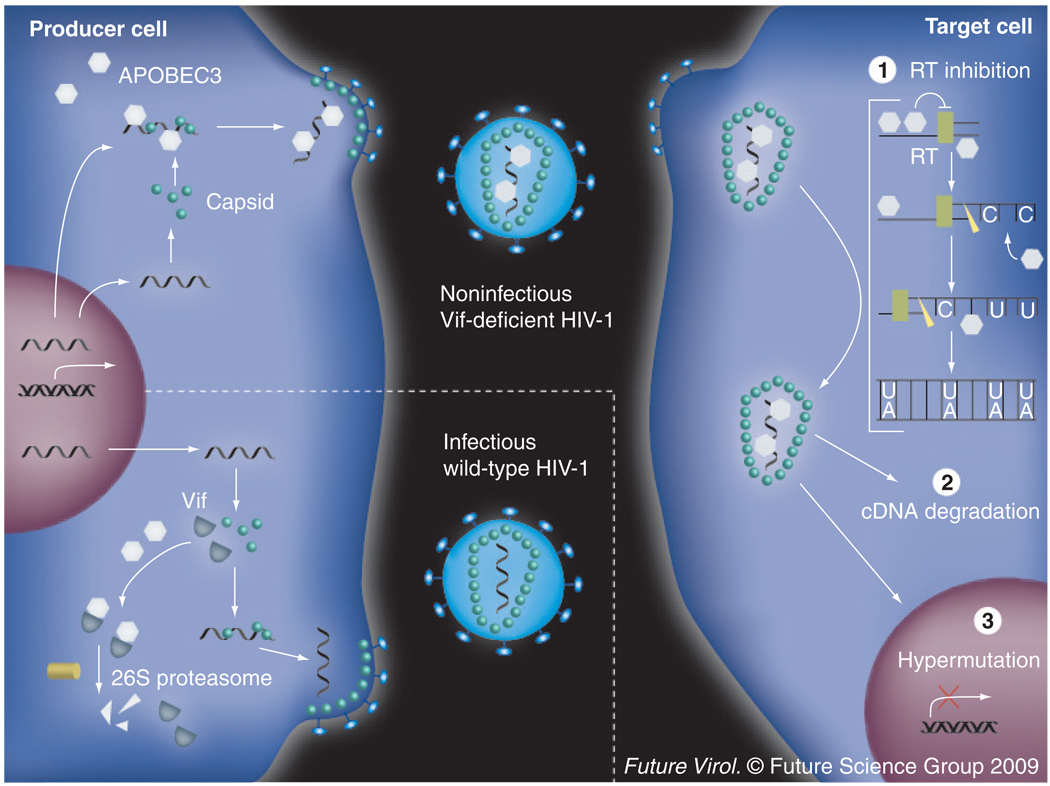
In the absence of Vif, APOBEC3G is encapsidated into the core of budding virions in a manner dependent on an interaction with both RNA and nucleocapsid protein (Figure 1) [36–42]. Once the virion fuses to a target cell and deposits its core, the availability of deoxyribonucleotide triphosphates enables reverse transcription. Within the deposited core, APOBEC3G is hypothesized to bind the viral gRNA and this action alone is thought to be sufficient to partially inhibit accumulation of reverse transcripts through a deaminase-independent mechanism(s) [43–50]. During first- (or minus-) strand synthesis, RNaseH degrades the gRNA to allow for second-strand synthesis, not only liberating APOBEC3G, but also exposing its ssDNA substrate [51]. APOBEC3G binds and deaminates cytidines to uridines on the exposed minus strand [3–5,13,15,17,52]. This heavily edited cDNA is highly susceptible to degradation, although the identity of the responsible factors remains unclear [45,53–56]. Edited viral cDNA copies that escape degradation template second-strand synthesis. The uridines code for plus-strand adenosines, thus, ultimately manifesting as G–A mutations [3–5,13]. The generation of frequent nonsense and missense mutations renders the resulting provirus incapable of further replication and infection.
Figure 1. Mechanism of APOBEC3G-mediated HIV-1 restriction.
The integrated provirus in the producer cell is transcribed and either packaged as gRNA or processed and translated to produce essential viral proteins, such as capsid, RT and integrase. In the absence of Vif (upper left), APOBEC3G can be encapsidated within the nucleic acid-containing core of the budding virus dependent on an interaction with both gRNA and nucleocapsid protein. This virion will be rendered replication deficient by three APOBEC3G-dependent mechanisms that occur after the viral core is deposited in the target cell. (1) APOBEC3G binds the gRNA-inhibiting RT in a deaminase-independent manner. (2) Closely following RT, the gRNA is degraded by RNaseH, both freeing APOBEC3G from its RNA-bound state and exposing the ssDNA of the minus strand. APOBEC3G deaminates available cytidines to uridines resulting in hypermutation. These mutations are fixed as G–A transitions upon second-strand synthesis. This heavily edited viral cDNA is highly susceptible to degradation by yet unidentified factors. (3) If the cDNA escapes degradation and is integrated, the frequent nonsense and missense mutations within the provirus prevent further replication. In the presence of Vif (lower left), APOBEC3G is polyubiquitinylated and degraded by the 26S proteasome. Thus, the budding virions are fully infectious.
RT Reverse transcriptase.
The APOBEC family is characterized by a conserved zinc(Z)-binding motif, H-x-E-x25–31-C-x2–4-C, required for deaminase activity [1,13,35,57–59]. These domains are organized in a modular fashion at the APOBEC3 locus to give rise to a series of single or double Z-domain proteins. The human APOBEC3 locus encodes four double Z-domain proteins (APOBEC3B, APOBEC3DE, APOBEC3F and APOBEC3G) and three single Z-domain proteins (APOBEC3A, APOBEC3C and APOBEC3H) from seven genes located in tandem on chromosome 22 (Table 1) [35,57]. These loci vary dramatically across species with for example, the sheep and cattle loci encoding three single Z-domain proteins and one double Z-domain protein from only three genes [59], or the cat locus encoding four single Z-domain proteins and one double Z-domain protein from four genes [60]. The Z-domains fall into three phylogenetic clusters based on conserved amino acid variations within the zinc-binding motif and are designated type Z1, Z2 or Z3 [58,59]. While no functional difference has yet been described to the different domain types, the Z-domain nomenclature has been useful for making cross-species comparisons and modeling the evolutionary history of the family [58,59].
The human, double-domain APOBEC3s have several properties that distinguish them from their single-domain family members, which may offer clues to their specific physiological functions. First, the double-domain APOBEC3s tend to display a separation of function between domains; one determines ssDNA cytidine deaminase activity and sequence specificity, while the other is responsible for subcellular localization, RNA-binding and encapsidation. For example, the N-terminal catalytic domain of APOBEC3G is responsible for viral RNA-binding and is required for encapsidation, but it lacks DNA deaminase activity [44,46,61,62]. By contrast, the C-terminal catalytic domain confers both DNA deaminase activity and sequence specificity (APOBEC3G prefers to deaminate cytidines in a 5′-CC context), but is unable to encapsidate on its own. APOBEC3F has a similar functional distribution, although it prefers cytidines in a 5′-TC context [15,57,61,63]. APOBEC3B mostly follows this rule, with the C-terminal domain being catalytically dominant and determining preference for cytidines in a 5′-TC context [64], but the N-terminal domain still elicits activity capable of mutating the HIV-1 genome [42]. The N-terminal domain of these three proteins also determines subcellular localization, nuclear for APOBEC3B and cytoplasmic for APOBEC3F and APOBEC3G [29,33,34,65–68]. It is worth noting that the active determinants of subcellular localization overlap with those that determine RNA-binding capacity, suggesting the two may go hand-in-hand, although protein–protein interactions have also been implicated in APOBEC3G trafficking and localization [67–69].
Second, the double-domain APOBEC3s are distinguishable from their single-domain family members in their capacity to homo and hetero-oligomerize to form higher order multimers in cells. APOBEC3G has long been known to be capable of homo-oligomerization, an interaction greatly facilitated by RNA [35,41,62,69,70]. It is also capable of assembling into much larger, heterogeneous high molecular mass (HMM) ribonucleoprotein complexes [71]. These HMM ribonucleoproteins lack definitive identification as either specific RNA granules, such as Staufen-containing RNA granules, or nonspecific stress granules and processing bodies [31,65,72,73]. Either way, APOBEC3G in HMM complexes has been shown to be catalytically inactive and incapable of HIV-1 restriction [71]. Similar to APOBEC3G, APOBEC3F is also known to self-oligomerize and assemble into HMM complexes [63,73,74]. The relevance of these higher order complexes is not yet known, although several reports provide clues that they may be functionally important to the restriction of retroelements, such as L1 and Alu [31,75]. Furthermore, APOBEC3G is not restricted to homo-oligomers, but has been shown to hetero-oligomerize with APOBEC3B, APOBEC3DE and APOBEC3F [23,63,76]. It is unknown whether or not these hetero-oligomers are incorporated into HMM complexes and what their physiological relevance may be.
Finally, and perhaps most notably, all of the double-domain APOBEC3s are capable of restricting Vif-deficient HIV-1 while the single-domain APOBEC3s appear to only do so weakly, if at all Table 1) [77,78]. Double-domain APOBEC3G and APOBEC3F have consistently demonstrated an impressive ability to restrict Vif-deficient HIV-1 in both single-cycle assays and spreading infections experiments performed by numerous laboratories [2–5,13,15,17,57,63,79]. Likewise, APOBEC3B and APOBEC3DE show strong inhibition by single-cycle assays, although there are a few conf licting reports [15,20,23,28,64,76]. By contrast, single-domain APOBEC3C, while highly efficient at restricting SIV, has only been reported to weakly inhibit HIV-1 [15,23,28,29,63,64,76,80]. Of the four APOBEC3H variants, only haplotype II appears capable of HIV-1 restriction, although to a lesser extent than either APOBEC3F or APOBEC3G [81–83]. APOBEC3A is also unable to restrict Vif-deficient HIV-1 [15,20,23,28–30,45,63,64]. While it is still unknown which APOBEC3s are relevant to HIV restriction in vivo, several APOBEC3s have been implicated. Analysis of proviral sequences obtained from clinical samples of HIV-1-infected patients demonstrates a mutational spectrum consisting of not only GG–AG mutations indicative of APOBEC3G, but also of GA–AA and GC–AC mutations indicative of APOBEC3F/APOBEC3B and APOBEC3DE, respectively [23,57,84,85]. While perhaps not an absolute correlation, the ability of the double-domain APOBEC3s to restrict HIV is striking and may be linked with one of the unique physical attributes delineated previously.
Besides HIV-1, the APOBEC3 subfamily has been shown to act on ssDNA-replication intermediates of other retroviruses, such as SIV, murine leukemia virus (MLV), human T-cell leukemia virus (HTLV), porcine endogenous retrovirus and foamy virus [4,13–16,18–24,64,86] and retroelements, such as human endogenous retro-virus, long interspersed repetitive element and Alu [25–34,56,87–89]. While working to promote the overall genomic integrity of the cell, it is unclear what specifically drove the maintenance and rapid expansion of the APOBEC3 locus from one to several unique genes. The single APOBEC3 homolog present in mice was found to be inessential, although the knockout mice were found to be more susceptible to mouse mammary tumor virus and MLV infection [90–92]. Did selective pressure from exogenous retrovirus transmission sculpt the locus and/or was the pressure derived from some endogenous retroelement replicating unrestrained in the genome? Regardless of factors responsible for shaping the APOBEC3 locus, the current APOBEC3 repertoire provides a potent, endogenous means by which to defend the host genome from both endogenous and exogenous parasitic elements.
Defending the viral genome: Vif & other inhibitors
Perhaps nothing speaks to the significance of the APOBEC3 family in viral restriction more clearly than the fact that nearly every relevant retroviral pathogen has evolved to be able to neutralize, bypass or otherwise overcome the APOBEC3 replication block. As any effective antiviral drug obeys Coffin’s razor and selects for resistance over time, so too do the APOBEC3s. The APOBEC3s have pressured viruses to evolve a variety of resistance mechanisms that include avoidance, sequestration and degradation (Figure 2). The emerging theme is that all of these mechanisms appear to work by preventing the encapsidation of APOBEC3s and, thereby, protecting the viral nucleic acid. The main paradigm for APOBEC3 antagonism, which has been set by the HIV-1 accessory protein Vif, is focused on here. All lentiviruses, except for equine infectious anemia virus, encode a Vif accessory protein that, while divergent in sequence, has a conserved function and a largely conserved mechanism of action.
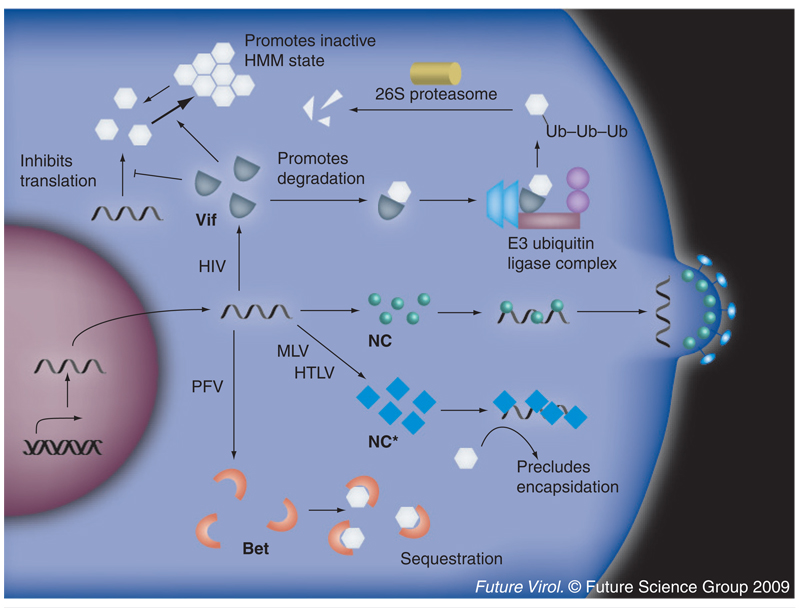
Figure 2. Mechanisms of natural APOBEC3 resistance.
Every relevant retroviral pathogen has evolved a mechanism to overcome APOBEC3-mediated restriction. For example, the HIV-1 accessory protein Vif acts as an adaptor between several human APOBEC3s and an ELOB/C–CUL5–RBX1 E3 ubiquitin ligase complex that polyubiquitinylates APOBEC3, targeting it for degradation by the 26S proteasome. This effectively decreases the steady-state levels of APOBEC3 in the cell, thus preventing sufficient encapsidation for viral inhibition. Vif is also hypothesized to have secondary, degradation-independent mechanisms to overcome the APOBEC3 replication block. These include translational inhibition and promotion of APOBEC3 accumulation in inactive HMM complexes. All of these mechanisms serve to inhibit the encapsidation of APOBEC3 and allow for the production of infectious viruses. The foamy viruses, such as PFV, similarly encode an accessory protein, Bet, that binds APOBEC3G and prevents it from encapsidating. However, Bet does not promote APOBEC3 degradation, but rather, is hypothesized to sequester APOBEC3 away from the sites of virion budding. MLV and HTLV do not require a specific accessory protein to overcome APOBEC3-mediated restriction. They both encode specialized nucleocapsid proteins that preclude APOBEC3 binding and encapsidation.
HMM: High molecular mass; HTLV: Human T-cell lymphotropic virus; MLV: Murine leukemia virus; NC: Nucleocapsid; NC*: Specialized nucleocapsid; PFV: Primate foamy virus.
HIV-1 Vif is a highly basic, 23-kD accessory protein that, while not strictly required for viral replication, is essential for efficient pathogenesis on certain cell types, including primary CD4+ T cells and macrophages [93–95]. For over two decades, the field has known that a Vif-deficient virus could sustain a spreading infection on certain ‘permissive’ cell lines, but could not in other ‘nonpermissive’ cell lines [93– 95]. While Vif-deficient virions produced on permissive cells could infect nonpermissive cells, Vif-deficient virions produced on nonpermissive cells lacked the ability to efficiently infect any target cell [93,95]. This indicated that either Vif was required to overcome some viral restriction factor expressed by nonpermissive cells or that the absence of Vif required supplementation by a positive factor in permissive cells. Hybrids formed by the fusion of permissive and nonpermissive cells were unable to support Vif-deficient virion replication, arguing for the presence of a dominant negative factor in nonpermissive cells [96,97]. Subtractive hybridization between a parental nonpermissive line and a nearly isogenic, but permissive, daughter line identified APOBEC3G as this dominant restriction factor [2]. However, the ability of APOBEC3G to restrict required the absence of Vif. A Vif-proficient virus can overcome APOBEC3-mediated restriction and replicate almost equally well on either type of cell line.
Vif counters APOBEC3G-mediated restriction by primarily decreasing the steady-state level of APOBEC3G protein in an infected cell (Figure 2). Vif acts as an adaptor molecule, linking APOBEC3G to an ELONGINB/C–CULLIN5–RBX1 E3 ubiquitin ligase complex [98–100]. Bound APOBEC3G is then polyubiquitinated and degraded by the 26S proteasome [98,99,101–104]. In the presence of Vif, the half-life of APOBEC3G has been reported to fall from more than 8 h to anywhere between 5 min and 4 h [98,101,102,104]. While degradation is clearly a main contributor towards the exclusion of APOBEC3G from viral particles, several auxiliary mechanisms have been proposed based on reports demonstrating that the ability of Vif to degrade APOBEC3G does not necessarily correlate with its ability to restore infectivity or inhibit deaminase activity [105–107]. While the degradation-independent mechanism of APOBEC3G inhibition remains unclear, several hypotheses have been posited, including direct inhibition of enzymatic activity, indirect inhibition by promoting the incorporation of APOBEC3G into HMM complexes, steric hindrance of the interactions required for encapsidation and inhibition of mRNA translation [14,104,106–109].
Structural data on the HIV-1 Vif protein remain largely elusive owing to the difficulty of expressing high levels of the soluble, recombinant protein. Nevertheless, comparative studies between SIV, HIV-1 and -2 have identified several conserved interaction domains that subsequent mutagenesis studies proved essential for coordinating APOBEC3G degradation (reviewed in [110]). Two of the more conserved motifs serve to recruit the E3 ligase complex: a HCCH Z-coordinating motif and a SOCS-box motif. The HCCH motif consists of broadly conserved His/Cys pairs and several flanked hydrophobic residues of highly conserved spacing. Z-binding by the HCCH residues is thought to maintain a structural conformation that aligns the hydrophobic residues forming the CULLIN5 binding surface [111–114]. The SOCS-box (or BC-box) motif includes a highly conserved 144SLQ(Y/F)LA149 sequence responsible for the binding of ELONGINC [100,115–117]. Substitution of the SLQ residues with alanines results in a dramatic loss of HIV-1 infectivity on nonpermissive cell lines owing to the inability of this Vif variant to recruit the E3 ligase complex and degrade APOBEC3G [99,118]. In silico modeling of this conserved amino acid sequence, based on the structures of analogous BC-box motifs, predict an α-helical form with the conserved hydrophobic residues clustered, allowing for binding in the hydrophobic pocket of ELONGINC [100]. This prediction was confirmed by recent crystallographic studies showing the HIV-1 Vif BC-box peptide bound to CULLIN5 [119].
The N-terminal domain of HIV-1 Vif is largely responsible for binding APOBEC3s [102,120–125]. The Vif residues involved are arranged in a nonlinear fashion, indicating the involvement of multiple surfaces. For instance, APOBEC3G binding is dependent on a hydrophilic patch that includes the conserved 23SLVK26 and 40YRHHY44 motifs, as well as on a hydrophobic patch that includes four tryptophans at the very N-terminus and a 69YWxL72 cluster [120,121,123–125]. Thus, the binding of APOBEC3G by Vif requires the accurate arrangement of hydrophilic and hydrophobic residues on multiple surfaces. This complex binding scheme probably exists to ensure partial APOBEC3G binding and neutralization even if Vif has incurred one or more mutations in a binding motif. Vif also can bind and neutralize several other APOBEC3 family members, including APOBEC3C, APOBEC3DE and APOBEC3F (Table 1) [23,63,64,79,80,126]. The N-terminal domain of Vif is similarly responsible for the binding of these APOBEC3s, although through other residues. APOBEC3F binding, for example, is mediated by residues 14DRMR17 and an exclusive set of two tryptophans at the N-terminus [124,125,127].
Several other Vif domains are also crucial for HIV infectivity in nonpermissive cells. The central, hydrophilic 88EWRKKR93 motif is essential for protein stability, a mutation that causes a dramatic drop in Vif steady-state levels [128]. The 161PPLP164 proline-rich domain is required for Vif homomultimerization. Disruption of multimerization by either mutation of the domain or by expression of a peptide antagonist ablates the ability of Vif to prevent APOBEC3G encapsidation [129–131]. Finally, an RNA-binding domain exists at the N-terminus that mediates Vif interaction with viral gRNA [132–134]. Mutations of key residues within this domain also render HIV-1 incapable of replication on nonpermissive cells [134]. This RNA interaction is also required for efficient Vif incorporation into virions, although the role of Vif within virions is unclear [133,135–137]. It may be that Vif acts in the particle to inhibit APOBEC3G in a degradation-independent manner (as discussed earlier) or that Vif is required to fulfill some other role in the particle required for replication. For example, Vif is thought to play several roles that are similar to an RNA chaperone and it may function during genome folding and processing [138,139].
In contrast to APOBEC3G, which can effectively inhibit a broad array of retroviruses from other species, including SIV, equine infectious anemia virus, HTLV, porcine endogenous retro-virus and MLV [4,13–16,64,86], the potency of Vif and Vif-like molecules appears somewhat species specific. For example, HIV-1 Vif can neutralize human APOBEC3G, but not African green monkey APOBEC3. Similarly, SIVagm Vif can neutralize African green monkey APOBEC3G, but not human APOBEC3G [14,140]. This specificity has been traced to a single amino acid in human APOBEC3G at position 128, which as a D allows for the binding of HIV-1 Vif and as a K allows for the binding of SIVagm Vif [141–144]. This tendency towards species specificity probably reflects the strong positive selection that Vif is under to neutralize the particular APOBEC3 repertoire of its host species [14]. This is contrasted by the selective pressure exerted on the APOBEC3s to restrict a diverse set of targets, including not only human lentiviruses, but other viruses and retroelements that are both human and nonhuman. This mutual selection for broad antiviral activity by the APOBEC repertoire and for species specificity by the viral APOBEC antagonist may help explain why retroviral zoonotic transmissions are relatively rare [59,77,86].
While Vif-directed proteasomal degradation has provided a useful paradigm, several other mechanisms exist by which APOBEC3-mediated restriction is successfully avoided (Figure 2). For example, the foamy viruses are a family of complex viruses that infect a variety of mammals and whose replication can be inhibited by APOBEC3 proteins [19,21,24]. The primate foamy virus (PFV) accessory protein, Bet, functions similarly to Vif and can rescue infectivity of Vif-deficient HIV-1. Similarly, PFV Bet appears to function in a somewhat species-specific manner as it can bind to both human APOBEC3G and African green monkey APOBEC3G, but not to mouse APOBEC3 [21]. However, neither PFV Bet nor the related feline foamy virus Bet has been shown to decrease steady-state levels of their target APOBEC3s. Instead, it is hypothesized that Bet functions to sequester the APOBEC3s in the cell, thereby preventing encapsidation and restriction [19,21].
The ability of a virus to neutralize the APOBEC3s is not always dependent on the presence of a specialized accessory protein, such as Vif or Bet. For example, MLV is a simpler virus that lacks accessory genes. It has been found to be resistant to its host APOBEC3 (mouse APOBEC3), although not to human APOBEC3G [18]. This is owing to the failure of mouse APOBEC3 to encapsidate, probably due to its failure to efficiently bind MLV cap-sid protein (Figure 2). It has been hypothesized that MLV Gag has evolved to avoid mouse APOBEC3 binding, and that this may be aided by an inhibitory effect that the viral RNA has on this interaction [18,22]. This strategy appears similar to that used by HTLV-1 to evade human APOBEC3G. HTLV-1 similarly lacks a Vif- or Bet-like accessory protein, but rather, uses a novel motif at the C-terminus of the nucleocap-sid to prevent APOBEC3G encapsidation [145]. APOBEC3G packaging is dependent on RNA and on a direct or indirect association with viral nucleocapsid [36,39,41,69,146]. It is hypothesized that this C-terminal motif precludes or disrupts the crucial APOBEC3G/nucleocapsid association [145].
The prevailing trend is that every APOBEC3-susceptible retroelement has evolved a means to escape restriction. In addition to the mechanisms reviewed here – degradation, neutralization and avoidance – it is likely that viruses, as a whole, possess many other novel strategies for evading the APOBEC3 proteins. For example, recent data from our laboratory demonstrate that HIV-1 is capable of overcoming APOBEC3-mediated restriction by yet another means, tolerance [147]. Passaged continuously on CEM-SS cells stably expressing APOBEC3G, Vif-deficient virus can evolve APOBEC3G resistance by accumulating both a pyrimidine at position 200 and a null mutation in the accessory gene Vpr. While the role of the Vpr mutation is unknown, the A200C/T mutation was shown to dramatically increase viral titer by increasing translational efficiency [148]. This increase in viral titer serves to effectively titrate out available APOBEC3G, dropping mutational load back to tolerable levels [147]. As our knowledge of host–virus interactions continues to expand, other APOBEC3 evasion mechanisms are certain to emerge.
Modulating the APOBEC3s to facilitate virus eradication
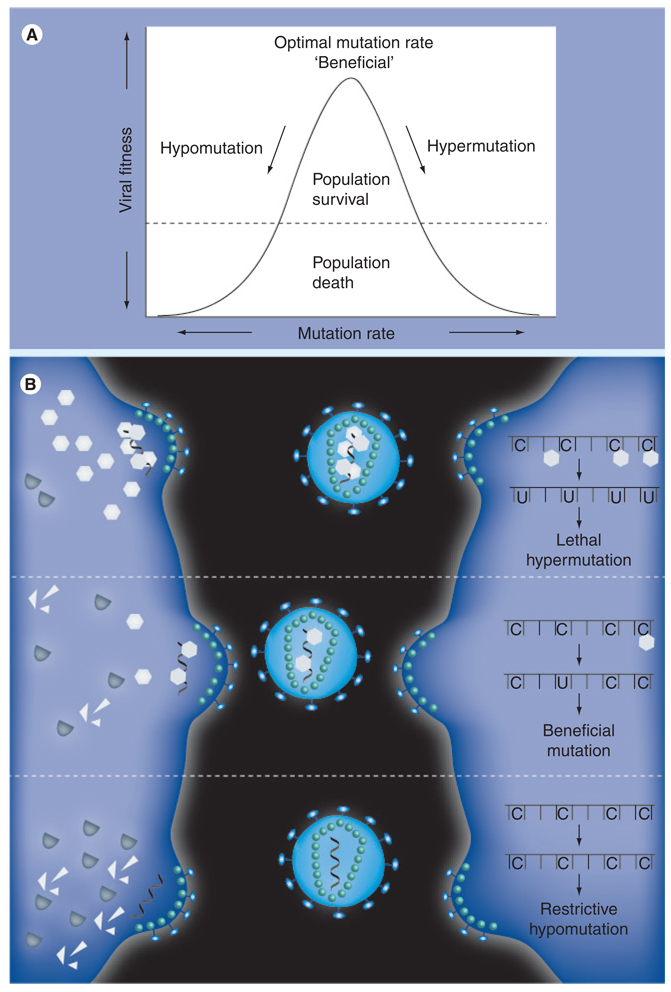
The remarkable success of HIV as a pathogen can largely be attributed to an optimal mutation rate that allows for stable transmission to successive generations while seeding sufficient diversity to allow for quick evolution in response to selective pressure (Figure 3A). Therapeutically, alteration of the viral mutation rate provides two conceivable means by which to restrict the virus in vivo. Either the mutation rate must be increased to a level that prohibits stable transmission (hypermutation), or the mutation rate must be decreased to inhibit the evolution of resistance and, thus, foster susceptibility to existent selective pressure (hypomutation). The APOBEC3G–Vif interaction provides us with a putative means by which to control the mutation rate and investigate both of these novel drug-design strategies.
Figure 3. Leveraging the activity of the APOBEC3s to alter the HIV-1 mutation rate.
(A) HIV-1 depends on an optimal mutation rate that allows for stable transmission to successive generations while seeding sufficient diversity to allow for quick evolution in response to selective pressure. Large increases in the mutation rate (hypermutagenesis) will prevent stable transmission of the genetic material to successive generations, decreasing viral fitness to lethal levels. By contrast, decreases in the mutation rate (hypomutagenesis) will impede the virus’ ability to evolve resistance to other sources of selective pressure and, hence, can be equally detrimental to viral fitness. (B) The optimal mutation rate of HIV-1 is modulated, at least partially, by the APOBEC3 mutators and their viral inhibitor, Vif, making them attractive targets for therapeutic compounds (middle panel). If Vif function could be inhibited or APOBEC3 activity augmented, high levels of APOBEC3 would be packaged into budding virions (top panel). This would increase the viral mutation rate and the resulting hypermutated viral cDNAs would either be degraded or code nonfunctional proteins. By contrast, if APOBEC3 activity could be abolished or Vif function enhanced, virtually no APOBEC3 would be encapsidated (bottom panel). This would decrease the viral mutation rate and inhibit accumulation of genetic diversity, impairing the virus’ ability to evolve resistance to the adaptive immune system and other antiviral compounds. Adapted from [151].
Hypermutation
APOBEC3G functions to increase the viral mutation rate and preclude stable transmission, while Vif functions to rein in APOBEC3G and restore the mutation rate to sublethal levels (middle panel, Figure 3B). Thus, if the APOBEC3G–Vif interaction could be disrupted or if Vif function could be disabled, the viral mutation rate would be predicted to soar to lethal levels (top panel, Figure 3B). Hypermutated viral cDNA would be subject to degradation or would encode frequent nonsense and missense mutations rendering the virus incapable of replication as seen ex vivo and detailed earlier. In other words, APOBEC3G-mediated hypermutagenesis would push the genome beyond the threshold for genomic stability, causing catastrophic error and replication failure. Recently, a screen to identify small-molecule inhibitors of the APOBEC3G–Vif interaction discovered a number of promising compounds [149]. One inhibitor, RN-18, functions to alter targeting of the APOBEC3G–Vif–E3 ligase complex to polyubiquitinylate Vif instead of APOBEC3G. This results in Vif degradation, increased encapsidation of APOBEC3G and inhibition of viral replication.
While an attractive strategy, the use of hyper-mutagenesis to destroy the virus is inherently risky. First, very little is known about the mechanisms controlling APOBEC3G’s discrimination of self from non-self DNA. APOBEC3G can act as a genomic DNA mutator in heterologous systems and poses an intrinsic hazard if not properly regulated [1,27]. While APOBEC3G is cyto-plasmic in human cells and sequestered from the genome, several redundant mechanisms most likely exist to ensure proper discrimination and must exist for those APOBEC3s that reside in the nucleus. Despite these systems, there is evidence that the nuclear APOBEC3s can pose problems for the host with maintenance dependent upon the cost–benefit ratio. For example, some human populations lack one or both copies of the APOBEC3B gene [150]. While there are no clear clinical manifestations of this event, it is possible that maintenance of the deletion is being driven by unintended, off-target deamination events whose costs outweigh the benefits of normal APOBEC3B function. Were any therapeutic to disrupt the enigmatic processes regulating APOBEC3 discrimination of self from non-self, the results could be catastrophic with the resultant genomic G-to-A hypermutation leading to massive cell death and/or to cancer.
Second, hypermutagenesis is also a risky strategy because it depends on reaching a lethal level of mutation in the viral genome sufficient to induce collapse of the population (Figure 3A). Failure to reach that threshold would promote further genetic diversification and could enhance the virus’ ability to become resistant to immune responses and/or therapeutics. If APOBEC3G was not completely neutralized by the virus, for example, it could be used to alter the mutation rate in the face of selective pressure and could contribute to the creation of a new resistant population. In this respect, Vif may act as a regulator of viral mutation rate and serve to modulate APOBEC3 levels rather than eliminate them (middle panel, Figure 3B) [151]. In other words, the optimal mutation rate of the virus may be dependent on the exploitation of the host’s APOBEC3 proteins to seed genetic diversity and, therefore, the APOBEC3 proteins may be beneficial to the virus in instances of sublethal mutation. Thus, any therapeutic designed to enhance or restore APOBEC3 function would have to do so fully and without altering its normal regulatory mechanisms.
Finally, the APOBEC3G–Vif interaction may not be an ideal target for disruption based on the genetic flexibility of the Vif gene itself. While other therapeutics inhibit the action of essential viral proteins with limited mutagenic potential, such as integrase, protease or reverse transcriptase, Vif is an accessory protein whose primary function is to overcome APOBEC3-mediated restriction. Thus, it is unclear how readily HIV-1 Vif will mutate to overcome small-molecule inhibition of the interaction. Moreover, as previously discussed, Vif-deficient HIV-1 has been shown to evolve in order to resist APOBEC3G in a Vif-independent manner [147]. Based on the wide range of mechanisms employed by various viruses to overcome the APOBEC3s (Figure 2), much more research is required on the ability of HIV-1 to evolve resistance before therapeutics can be developed and are clinically tested.
Hypomutation
As opposed to hypermutagenesis, which comes with a fair number of caveats, it is tempting to consider what would happen if one could render HIV-1 less mutable (Figure 3A). HIV-1 depends on an optimal mutation rate to ensure genetic variability in the face of an unending series of selective pressures. When selective pressure is exerted on the population in the form of an adaptive immune response or drug, there typically exist a number of resistant clones that survive and serve to repopulate the host. Hence, Coffin’s razor is dependent on a basal level of genetic variability in the population. By dropping the mutation rate of the virus, it has been hypothesized that that the genetic variation of the virus could be sufficiently diminished such that it is no longer able to evade the host immune system and antiviral compounds (bottom panel, Figure 3B) [151].
One contributing factor to the genetic variation of an HIV-1 population is likely to be the APOBEC3 family itself [152–155]. The main function of Vif is to rein in APOBEC3-mediated hypermutation and restore the mutation rate to sublethal levels. However, it is known that Vif does not always neutralize APOBEC3G completely. For example, early in infection, APOBEC3G levels have not yet been depleted and Vif levels are still on the increase resulting in a window where neutralization is incomplete [147]. In addition, defective Vif alleles, which would lead to incomplete neutralization of APOBEC3G, often arise in vivo and are detected in HIV-1 isolates from infected patients [156]. Furthermore, in clinical samples from patients infected with HIV-1, some proviral sequences show evidence of G-to-A hypermutation, demonstrating a failure by Vif to completely neutralize the APOBEC3 proteins [84]. While it is unclear to what extent the viruses depend on the APOBEC3s as sources of ‘beneficial’ mutations, there is some evidence that these mutations can lead to drug resistance. In a tabulation of all mutations associated with HIV-1 drug resistance, Berkhout and coworkers found G-to-A mutations are the most frequent [152]. While reverse transcriptase itself is responsible for a number of G-to-A mutations, even a minor dependency on the APOBEC3s for variation can be exploited by designing specific APOBEC3 inhibitors as therapeutics [151]. Even if APOBEC3 inhibitors do not considerably diminish the genetic variability of the virus to allow the adaptive immune system to clear infection by itself, they may still be useful as adjuvants that can enhance the long-term efficacy of other currently available therapies.
This approach is also not without its risks. Again, the normal physiological function of the APOBEC3s in the absence of viral infection is unknown and it is unclear what effects complete inhibition may have on the cell. Current data are consistent with an exclusive role in retroelement restriction, but more studies are needed to bolster this important point. For instance, while mouse APOBEC3 is inessential [90–92] and human APOBEC3B-null populations lack a definitive clinical symptom [150], unintended effects cannot be ruled out at this time. Furthermore, secondary viral infections in the absence of APOBEC3-mediated restriction may be significantly exacerbated. Such a therapy must also be careful to preserve the function of activation induced deaminase, an APOBEC3-related deaminase responsible for antibody gene diversification (reviewed in [157]). All therapeutics and antiviral drugs come with their own benefits and risks and so all require extensive preclinical research and rigorous clinical testing. However, the use of small compounds to alter the mutation rate of HIV-1 by leveraging APOBEC3 function is likely to provide, either alone or in conjunction with other antiviral drugs, a viable therapeutic option in the future.
Conclusion
At least two human APOBEC3 proteins, APOBEC3F and APOBEC3G, have the capacity to inhibit the infection of a broad number of retroelements, including the AIDS virus HIV-1. DNA cytidine deamination is a key part of most mechanisms of retrovirus restriction. Viruses circumvent this powerful innate defense in many different ways. HIV-1 encodes an accessory protein, Vif, which binds APOBEC3F and APOBEC3G, triggers their degradation and prevents their encapsidation. Numerous strategies are being explored currently to harness the therapeutic potential of these amazing proteins.
Future perspective
The discovery of APOBEC3G and the realization that retrovirus restriction factors can block the infectivity of many retroviruses, including HIV-1, has paved the way for new therapeutic ideas. We anticipate that one or more of these ideas will reach fruition within the next decade, most likely first for HIV/AIDS. Primarily, this will include the development of molecules, such as RN-18, which render viruses nonfunctional by hypermutation. Next, and perhaps even more innovative and paradigm shifting, will be therapies that work by diminishing viral mutation rates and rendering viruses susceptible to normal immune responses. This approach can be summarized as therapy by hypomutation. Also within the next decade, many other viruses will be proven susceptible to APOBEC3-dependent restriction. Probable examples already include HBV and human papillomavirus. Mechanistic information derived from studying the conflict between the human APOBEC3s and HIV-1 will undoubtedly facilitate efforts to combat these and other viruses as well. Overall, we expect antiviral therapies that work through the modulation of host-restriction factors will move from theory to reality and prove successful in the near future.
Executive summary
Basic concepts in antiviral therapy
Antiviral therapies typically target viral proteins.
Host proteins are also viable targets for antiviral therapy.
Attacking the viral genome: the APOBEC3 protein family
APOBEC3 proteins are DNA cytidine deaminases.
APOBEC3 proteins block the replication of a broad number of retroviruses and retrotransposons.
HIV is a prominent pathogen that is susceptible to APOBEC3-mediated restriction.
Defending the viral genome: Vif & other inhibitors
Viruses have evolved numerous mechanisms to prevent restriction.
HIV-1 prevents APOBEC3F and APOBEC3G encapsidation by triggering their proteosomal degradation.
Modulating the APOBEC3s to facilitate virus eradication
Strategies to enable the APOBEC3s to hypermutate retroviral substrates are therapeutically feasible, but may inadvertently facilitate viral evolution.
Strategies to block APOBEC3 function may increase viral genetic stability and, thereby, render the virus more susceptible to adaptive immune clearance and less likely to become drug resistant.
More research is required in order to discover and evaluate therapies that work through hypermutation or hypomutation
Acknowledgements
The authors thank J Albin, M Burns, L Lackey and M Li for thoughtful feedback on the manuscript and G Haché for helping inspire the phrase ‘Coffin’s razor’.
Footnotes
For reprint orders, please contact: reprints@futuremedicine.com
Financial & competing interests disclosure
JF Hultquist is supported in part by a National Science Foundation predoctoral fellowship. Work in the Harris laboratory is supported by grants from the NIH, GM090437, AI064046 and a GCE award from the Bill and Melinda Gates Foundation. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed
No writing assistance was utilized in the production of this manuscript
Contributor Information
Judd F Hultquist, Department of Genetics, Cell Biology & Development, University of Minnesota, Minneapolis, MN 55455, USA Tel.: +1 414 702 7232, Fax: +1 612 625 2163 hultq014@umn.edu.
Reuben S Harris, Department of Biochemistry, Molecular Biology & Biophysics, Department of Genetics, Cell Biology & Development, Institute for Molecular Virology and Center for Genome Engineering, University of Minnesota, Minneapolis, MN 55455, USA Tel.: +1 612 624 0457, Fax: +1 612 625 2163 rsh@umn.edu.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪of considerable interest
- 1. Harris RS, Petersen-Mahrt SK, Neuberger MS. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol. Cell. 2002;10(5):1247–1253. doi: 10.1016/s1097-2765(02)00742-6. ▪▪ Discovery of APOBEC3G as a DNA deaminase.
- 2. Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418(6898):646–650. doi: 10.1038/nature00939. ▪▪ Discovery of APOBEC3G as an HIV-1 restricition factor.
- 3.Lecossier D, Bouchonnet F, Clavel F, Hance AJ. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science. 2003;300(5622):1112. doi: 10.1126/science.1083338. [DOI] [PubMed] [Google Scholar]
- 4. Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424(6944):99–103. doi: 10.1038/nature01709. ▪▪ DNA deamination mechanism for retroviral restriction.
- 5. Zhang H, Yang B, Pomerantz RJ, Zhang C, Arunachalam SC, Gao L. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003;424(6944):94–98. doi: 10.1038/nature01707. ▪▪ DNA deamination mechanism for retroviral restriction.
- 6.Hatziioannou T, Perez-Caballero D, Yang A, Cowan S, Bieniasz PD. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5α. Proc. Natl Acad. Sci. USA. 2004;101(29):10774–10779. doi: 10.1073/pnas.0402361101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keckesova Z, Ylinen LM, Towers GJ. The human and African green monkey TRIM5α genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc. Natl Acad. Sci.USA. 2004;101(29):10780–10785. doi: 10.1073/pnas.0402474101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perron MJ, Stremlau M, Song B, Ulm W, Mulligan RC, Sodroski J. TRIM5α mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc. Natl Acad. Sci. USA. 2004;101(32):11827–11832. doi: 10.1073/pnas.0403364101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature. 2004;427(6977):848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 10.Yap MW, Nisole S, Lynch C, Stoye JP. Trim5α protein restricts both HIV-1 and murine leukemia virus. Proc. Natl Acad. Sci. USA. 2004;101(29):10786–10791. doi: 10.1073/pnas.0402876101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451(7177):425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 12.Van Damme N, Goff D, Katsura C, et al. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe. 2008;3(4):245–252. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Harris RS, Bishop KN, Sheehy AM, et al. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113(6):803–809. doi: 10.1016/s0092-8674(03)00423-9. ▪▪ DNA deamination mechanism for retroviral restriction.
- 14.Mariani R, Chen D, Schröfelbauer B, et al. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell. 2003;114(1):21–31. doi: 10.1016/s0092-8674(03)00515-4. [DOI] [PubMed] [Google Scholar]
- 15. Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, Malim MH. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 2004;14(15):1392–1396. doi: 10.1016/j.cub.2004.06.057. ▪Discovery of APOBEC3F as an HIV-1 restriction factor.
- 16.Kobayashi M, Takaori-Kondo A, Shindo K, Abudu A, Fukunaga K, Uchiyama T. APOBEC3G targets specific virus species. J. Virol. 2004;78(15):8238–8244. doi: 10.1128/JVI.78.15.8238-8244.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu Q, Konig R, Pillai S, et al. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat. Struct. Mol. Biol. 2004;11(5):435–442. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- 18.Doehle BP, Schäfer A, Wiegand HL, Bogerd HP, Cullen BR. Differential sensitivity of murine leukemia virus to APOBEC3-mediated inhibition is governed by virion exclusion. J. Virol. 2005;79(13):8201–8207. doi: 10.1128/JVI.79.13.8201-8207.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lochelt M, Romen F, Bastone P, et al. The antiretroviral activity of APOBEC3 is inhibited by the foamy virus accessory Bet protein. Proc. Natl Acad. Sci. USA. 2005;102(22):7982–7987. doi: 10.1073/pnas.0501445102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rose KM, Marin M, Kozak SL, Kabat D. Regulated production and anti-HIV type 1 activities of cytidine deaminases APOBEC3B, 3F, and 3G. AIDS Res. Hum. Retroviruses. 2005;21(7):611–619. doi: 10.1089/aid.2005.21.611. [DOI] [PubMed] [Google Scholar]
- 21.Russell RA, Wiegand HL, Moore MD, Schäfer A, McClure MO, Cullen BR. Foamy virus Bet proteins function as novel inhibitors of the APOBEC3 family of innate antiretroviral defense factors. J. Virol. 2005;79(14):8724–8731. doi: 10.1128/JVI.79.14.8724-8731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abudu A, Takaori-Kondo A, Izumi T, et al. Murine retrovirus escapes from murine APOBEC3 via two distinct novel mechanisms. Curr. Biol. 2006;16(15):1565–1570. doi: 10.1016/j.cub.2006.06.055. [DOI] [PubMed] [Google Scholar]
- 23.Dang Y, Wang X, Esselman WJ, Zheng YH. Identification of APOBEC3DE as another antiretroviral factor from the human APOBEC family. J. Virol. 2006;80(21):10522–10533. doi: 10.1128/JVI.01123-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delebecque F, Suspené R, Calattini S, et al. Restriction of foamy viruses by APOBEC cytidine deaminases. J. Virol. 2006;80(2):605–614. doi: 10.1128/JVI.80.2.605-614.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dutko JA, Schäfer A, Kenny AE, Cullen BR, Curcio MJ. Inhibition of a yeast LTR retrotransposon by human APOBEC3 cytidine deaminases. Curr. Biol. 2005;15(7):661–666. doi: 10.1016/j.cub.2005.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Esnault C, Heidmann O, Delebecque F, et al. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature. 2005;433(7024):430–433. doi: 10.1038/nature03238. [DOI] [PubMed] [Google Scholar]
- 27.Schumacher AJ, Nissley DV, Harris RS. APOBEC3G hypermutates genomic DNA and inhibits Ty1 retrotransposition in yeast. Proc. Natl Acad. Sci. USA. 2005;102(28):9854–9859. doi: 10.1073/pnas.0501694102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bogerd HP, Wiegand HL, Doehle BP, Lueders KK, Cullen BR. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res. 2006;34(1):89–95. doi: 10.1093/nar/gkj416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bogerd HP, Wiegand HL, Hulme AE, et al. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc. Natl Acad. Sci. USA. 2006;103(23):8780–8785. doi: 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen H, Lilley CE, Yu Q, et al. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr. Biol. 2006;16(5):480–485. doi: 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 31.Chiu YL, Witkowska HE, Hall SC, et al. High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc. Natl Acad. Sci. USA. 2006;103(42):15588–15593. doi: 10.1073/pnas.0604524103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Esnault C, Millet J, Schwartz O, Heidmann T. Dual inhibitory effects of APOBEC family proteins on retrotransposition of mammalian endogenous retroviruses. Nucleic Acids Res. 2006;34(5):1522–1531. doi: 10.1093/nar/gkl054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muckenfuss H, Hamdorf M, Held U, et al. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J. Biol. Chem. 2006;281(31):22161–22172. doi: 10.1074/jbc.M601716200. [DOI] [PubMed] [Google Scholar]
- 34.Stenglein MD, Harris RS. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J. Biol. Chem. 2006;281(25):16837–16841. doi: 10.1074/jbc.M602367200. [DOI] [PubMed] [Google Scholar]
- 35.Jarmuz A, Chester A, Bayliss J, et al. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics. 2002;79(3):285–296. doi: 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- 36.Alce TM, Popik W. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J. Biol. Chem. 2004;279(33):34083–34086. doi: 10.1074/jbc.C400235200. [DOI] [PubMed] [Google Scholar]
- 37.Luo K, Liu B, Xiao Z, et al. Amino-terminal region of the human immunodeficiency virus type 1 nucleocapsid is required for human APOBEC3G packaging. J. Virol. 2004;78(21):11841–11852. doi: 10.1128/JVI.78.21.11841-11852.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schäfer A, Bogerd HP, Cullen BR. Specific packaging of APOBEC3G into HIV-1 virions is mediated by the nucleocapsid domain of the gag polyprotein precursor. Virology. 2004;328(2):163–168. doi: 10.1016/j.virol.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 39.Svarovskaia ES, Xu H, Mbisa JL, et al. Human apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs. J. Biol. Chem. 2004;279(34):35822–35828. doi: 10.1074/jbc.M405761200. [DOI] [PubMed] [Google Scholar]
- 40.Khan MA, Kao S, Miyagi E, et al. Viral RNA is required for the association of APOBEC3G with human immunodeficiency virus type 1 nucleoprotein complexes. J. Virol. 2005;79(9):5870–5874. doi: 10.1128/JVI.79.9.5870-5874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burnett A, Spearman P. APOBEC3G multimers are recruited to the plasma membrane for packaging into human immunodeficiency virus type 1 virus-like particles in an RNA-dependent process requiring the NC basic linker. J. Virol. 2007;81(10):5000–5013. doi: 10.1128/JVI.02237-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bogerd HP, Cullen BR. Single-stranded RNA facilitates nucleocapsid: APOBEC3G complex formation. RNA. 2008;14(6):1228–1236. doi: 10.1261/rna.964708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iwatani Y, Chan DS, Wang F, et al. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res. 2007;35(21):7096–7108. doi: 10.1093/nar/gkm750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iwatani Y, Takeuchi H, Strebel K, Levin JG. Biochemical activities of highly purified, catalytically active human APOBEC3G: correlation with antiviral effect. J. Virol. 2006;80(12):5992–6002. doi: 10.1128/JVI.02680-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mbisa JL, Barr R, Thomas JA, et al. Human immunodeficiency virus type 1 cDNAs produced in the presence of APOBEC3G exhibit defects in plus-strand DNA transfer and integration. J. Virol. 2007;81(13):7099–7110. doi: 10.1128/JVI.00272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Newman EN, Holmes RK, Craig HM, et al. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 2005;15(2):166–170. doi: 10.1016/j.cub.2004.12.068. [DOI] [PubMed] [Google Scholar]
- 47.Bishop KN, Holmes RK, Malim MH. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J. Virol. 2006;80(17):8450–8458. doi: 10.1128/JVI.00839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miyagi E, Opi S, Takeuchi H, et al. Enzymatically active APOBEC3G is required for efficient inhibition of human immunodeficiency virus type 1. J. Virol. 2007;81(24):13346–13353. doi: 10.1128/JVI.01361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bishop KN, Verma M, Kim EY, Wolinsky SM, Malim MH. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog. 2008;4(12):e1000231. doi: 10.1371/journal.ppat.1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holmes RK, Malim MH, Bishop KN. APOBEC-mediated viral restriction: not simply editing? Trends Biochem. Sci. 2007;32(3):118–128. doi: 10.1016/j.tibs.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 51.Soros VB, Yonemoto W, Greene WC. Newly synthesized APOBEC3G is incorporated into HIV virions, inhibited by HIV RNA, and subsequently activated by RNase H. PLoS Pathog. 2007;3(2):e15. doi: 10.1371/journal.ppat.0030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suspené R, Sommer P, Henry M, et al. APOBEC3G is a single-stranded DNA cytidine deaminase and functions independently of HIV reverse transcriptase. Nucleic Acids Res. 2004;32(8):2421–2429. doi: 10.1093/nar/gkh554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schröfelbauer B, Yu Q, Zeitlin SG, Landau NR. Human immunodeficiency virus type 1 Vpr induces the degradation of the UNG and SMUG uracil-DNA glycosylases. J. Virol. 2005;79(17):10978–10987. doi: 10.1128/JVI.79.17.10978-10987.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaiser SM, Emerman M. Uracil DNA glycosylase is dispensable for human immunodeficiency virus type 1 replication and does not contribute to the antiviral effects of the cytidine deaminase APOBEC3G. J. Virol. 2006;80(2):875–882. doi: 10.1128/JVI.80.2.875-882.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang B, Chen K, Zhang C, Huang S, Zhang H. Virion-associated uracil DNA glycosylase-2 and apurinic/apyrimidinic endonuclease are involved in the degradation of APOBEC3G-edited nascent HIV-1 DNA. J. Biol. Chem. 2007;282(16):11667–11675. doi: 10.1074/jbc.M606864200. [DOI] [PubMed] [Google Scholar]
- 56.Schumacher AJ, Haché G, Macduff DA, Brown WL, Harris RS. The DNA deaminase activity of human APOBEC3G is required for Ty1, MusD, and human immunodeficiency virus type 1 restriction. J. Virol. 2008;82(6):2652–2660. doi: 10.1128/JVI.02391-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Liddament MT, Brown WL, Schumacher AJ, Harris RS. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr. Biol. 2004;14(15):1385–1391. doi: 10.1016/j.cub.2004.06.050. ▪ Discovery of APOBEC3F as an HIV-1 restriction factor.
- 58.Conticello SG, Thomas CJ, Petersen-Mahrt SK, Neuberger MS. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol. Biol. Evol. 2005;22(2):367–377. doi: 10.1093/molbev/msi026. [DOI] [PubMed] [Google Scholar]
- 59.LaRue RS, Jónsson SR, Silverstein KA, et al. The artiodactyl APOBEC3 innate immune repertoire shows evidence for a multi-functional domain organization that existed in the ancestor of placental mammals. BMC Mol. Biol. 2008;9:104. doi: 10.1186/1471-2199-9-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Munk C, Beck T, Zielonka J, et al. Functions, structure, and read-through alternative splicing of feline APOBEC3 genes. Genome Biol. 2008;9(3):R48. doi: 10.1186/gb-2008-9-3-r48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haché G, Liddament MT, Harris RS. The retroviral hypermutation specificity of APOBEC3F and APOBEC3G is governed by the C-terminal DNA cytosine deaminase domain. J. Biol. Chem. 2005;280(12):10920–10924. doi: 10.1074/jbc.M500382200. [DOI] [PubMed] [Google Scholar]
- 62.Navarro F, Bollman B, Chen H, et al. Complementary function of the two catalytic domains of APOBEC3G. Virology. 2005;333(2):374–386. doi: 10.1016/j.virol.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 63. Wiegand HL, Doehle BP, Bogerd HP, Cullen BR. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 2004;23(12):2451–2458. doi: 10.1038/sj.emboj.7600246. ▪ Discovery of APOBEC3F as an HIV-1 restriction factor.
- 64.Yu Q, Chen D, Konig R, Mariani R, Unutmaz D, Landau NR. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. J. Biol. Chem. 2004;279(51):53379–53386. doi: 10.1074/jbc.M408802200. [DOI] [PubMed] [Google Scholar]
- 65.Wichroski MJ, Robb GB, Rana TM. Human retroviral host restriction factors APOBEC3G and APOBEC3F localize to mRNA processing bodies. PLoS Pathog. 2006;2(5):e41. doi: 10.1371/journal.ppat.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wichroski MJ, Ichiyama K, Rana TM. Analysis of HIV-1 viral infectivity factor-mediated proteasome-dependent depletion of APOBEC3G: correlating function and subcellular localization. J. Biol. Chem. 2005;280(9):8387–8396. doi: 10.1074/jbc.M408048200. [DOI] [PubMed] [Google Scholar]
- 67.Bennett RP, Presnyak V, Wedekind JE, Smith HC. Nuclear exclusion of the HIV-1 host defense factor APOBEC3G requires a novel cytoplasmic retention signal and is not dependent on RNA binding. J. Biol. Chem. 2008;283(12):7320–7327. doi: 10.1074/jbc.M708567200. [DOI] [PubMed] [Google Scholar]
- 68.Stenglein MD, Matsuo H, Harris RS. Two regions within the amino-terminal half of APOBEC3G cooperate to determine cytoplasmic localization. J. Virol. 2008;82(19):9591–9599. doi: 10.1128/JVI.02471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Friew YN, Boyko V, Hu WS, Pathak VK. Intracellular interactions between APOBEC3G, RNA, and HIV-1 Gag: APOBEC3G multimerization is dependent on its association with RNA. Retrovirology. 2009;6:56. doi: 10.1186/1742-4690-6-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bennett RP, Salter JD, Liu X, Wedekind JE, Smith HC. APOBEC3G subunits self-associate via the C-terminal deaminase domain. J. Biol. Chem. 2008;283(48):33329–33336. doi: 10.1074/jbc.M803726200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chiu YL, Soros VB, Kreisberg JF, Stopak K, Yonemoto W, Greene WC. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature. 2005;435(7038):108–114. doi: 10.1038/nature03493. [DOI] [PubMed] [Google Scholar]
- 72.Kozak SL, Marin M, Rose KM, Bystrom C, Kabat D. The anti-HIV-1 editing enzyme APOBEC3G binds HIV-1 RNA and messenger RNAs that shuttle between polysomes and stress granules. J. Biol. Chem. 2006;281(39):29105–29119. doi: 10.1074/jbc.M601901200. [DOI] [PubMed] [Google Scholar]
- 73.Gallois-Montbrun S, Kramer B, Swanson CM, et al. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J. Virol. 2007;81(5):2165–2178. doi: 10.1128/JVI.02287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang X, Dolan PT, Dang Y, Zheng YH. Biochemical differentiation of APOBEC3F and APOBEC3G proteins associated with HIV-1 life cycle. J. Biol. Chem. 2007;282(3):1585–1594. doi: 10.1074/jbc.M610150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huthoff H, Autore F, Gallois-Montbrun S, Fraternali F. Malim MH: RNA-dependent oligomerization of APOBEC3G is required for restriction of HIV-1. PLoS Pathog. 2009;5(3):e1000330. doi: 10.1371/journal.ppat.1000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Doehle BP, Schäfer A, Cullen BR. Human APOBEC3B is a potent inhibitor of HIV1 infectivity and is resistant to HIV-1 Vif. Virology. 2005;339(2):281–288. doi: 10.1016/j.virol.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 77.Chiu YL, Greene WC. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu. Rev. Immunol. 2008;26:317–353. doi: 10.1146/annurev.immunol.26.021607.090350. [DOI] [PubMed] [Google Scholar]
- 78.Goila-Gaur R, Strebel K. HIV-1 Vif, APOBEC, and intrinsic immunity. Retrovirology. 2008;5:51. doi: 10.1186/1742-4690-5-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zheng YH, Irwin D, Kurosu T, Tokunaga K, Sata T, Peterlin BM. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J. Virol. 2004;78(11):6073–6076. doi: 10.1128/JVI.78.11.6073-6076.2004. ▪ Discovery of APOBEC3F as an HIV-1 restriction factor.
- 80.Langlois MA, Beale RC, Conticello SG, Neuberger MS. Mutational comparison of the single-domained APOBEC3C and double-domained APOBEC3F/G antiretroviral cytidine deaminases provides insight into their DNA target site specificities. Nucleic Acids Res. 2005;33(6):1913–1923. doi: 10.1093/nar/gki343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dang Y, Siew LM, Wang X, Han Y, Lampen R, Zheng YH. Human cytidine deaminase APOBEC3H restricts HIV-1 replication. J. Biol. Chem. 2008;283(17):11606–11614. doi: 10.1074/jbc.M707586200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Harari A, Ooms M, Mulder LC, Simon V. Polymorphisms and splice variants influence the antiretroviral activity of human APOBEC3H. J. Virol. 2009;83(1):295–303. doi: 10.1128/JVI.01665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.OhAinle M, Kerns JA, Malik HS, Emerman M. Adaptive evolution and antiviral activity of the conserved mammalian cytidine deaminase APOBEC3H. J. Virol. 2006;80(8):3853–3862. doi: 10.1128/JVI.80.8.3853-3862.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Janini M, Rogers M, Birx DR, McCutchan FE. Human immunodeficiency virus type 1 DNA sequences genetically damaged by hypermutation are often abundant in patient peripheral blood mononuclear cells and may be generated during near-simultaneous infection and activation of CD4+ T cells. J. Virol. 2001;75(17):7973–7986. doi: 10.1128/JVI.75.17.7973-7986.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Land AM, Ball TB, Luo M, et al. Human immunodeficiency virus (HIV) type 1 proviral hypermutation correlates with CD4 count in HIV-infected women from Kenya. J. Virol. 2008;82(16):8172–8182. doi: 10.1128/JVI.01115-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jónsson SR, LaRue RS, Stenglein MD, Fahrenkrug SC, Andresdottir V, Harris RS. The restriction of zoonotic PERV transmission by human APOBEC3G. PLoS One. 2007;2(9):e893. doi: 10.1371/journal.pone.0000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Esnault C, Priet S, Ribet D, Heidmann O, Heidmann T. Restriction by APOBEC3 proteins of endogenous retroviruses with an extracellular life cycle: ex vivo effects and in vivo “traces” on the murine IAPE and human HERV-K elements. Retrovirology. 2008;5:75. doi: 10.1186/1742-4690-5-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lee YN, Bieniasz PD. Reconstitution of an infectious human endogenous retrovirus. PLoS Pathog. 2007;3(1):e10. doi: 10.1371/journal.ppat.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee YN, Malim MH, Bieniasz PD. Hypermutation of an ancient human retrovirus by APOBEC3G. J. Virol. 2008;82(17):8762–8770. doi: 10.1128/JVI.00751-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Low A, Okeoma CM, Lovsin N, et al. Enhanced replication and pathogenesis of Moloney murine leukemia virus in mice defective in the murine APOBEC3 gene. Virology. 2009;385(2):455–463. doi: 10.1016/j.virol.2008.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mikl MC, Watt IN, Lu M, et al. Mice deficient in APOBEC2 and APOBEC3. Mol. Cell. Biol. 2005;25(16):7270–7277. doi: 10.1128/MCB.25.16.7270-7277.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Okeoma CM, Lovsin N, Peterlin BM, Ross SR. APOBEC3 inhibits mouse mammary tumour virus replication in vivo. Nature. 2007;445(7130):927–930. doi: 10.1038/nature05540. [DOI] [PubMed] [Google Scholar]
- 93.Gabuzda DH, Lawrence K, Langhoff E, et al. Role of vif in replication of HIV-1 in CD4+ T lymphocytes. J. Virol. 1992;66(11):6489–6495. doi: 10.1128/jvi.66.11.6489-6495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Strebel K, Daugherty D, Clouse K, Cohen D, Folks T, Martin MA. The HIV ‘A’ (sor) gene product is essential for virus infectivity. Nature. 1987;328(6132):728–730. doi: 10.1038/328728a0. [DOI] [PubMed] [Google Scholar]
- 95.von Schwedler U, Song J, Aiken C, Trono D. Vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J. Virol. 1993;67(8):4945–4955. doi: 10.1128/jvi.67.8.4945-4955.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Madani N, Kabat D. An endogenous inhibitor of human immunodeficiency virus in human lymphocytes is overcome by the viral Vif protein. J. Virol. 1998;72(12):10251–10255. doi: 10.1128/jvi.72.12.10251-10255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Simon JH, Gaddis NC, Fouchier RA, Malim MH. Evidence for a newly discovered cellular anti-HIV-1 phenotype. Nat. Med. 1998;4(12):1397–1400. doi: 10.1038/3987. [DOI] [PubMed] [Google Scholar]
- 98.Mehle A, Strack B, Ancuta P, Zhang C, McPike M, Gabuzda D. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J. Biol. Chem. 2004;279(9):7792–7798. doi: 10.1074/jbc.M313093200. [DOI] [PubMed] [Google Scholar]
- 99. Yu X, Yu Y, Liu B, et al. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif–Cul5–SCF complex. Science. 2003;302(5647):1056–1060. doi: 10.1126/science.1089591. ▪ Identification of the APOBEC3G degrading Vif–E3 ligation complex.
- 100.Yu Y, Xiao Z, Ehrlich ES, Yu X, Yu XF. Selective assembly of HIV-1 Vif–Cul5–elonginB–elonginC E3 ubiquitin ligase complex through a novel SOCS box and upstream cysteines. Genes Dev. 2004;18(23):2867–2872. doi: 10.1101/gad.1250204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Conticello SG, Harris RS, Neuberger MS. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol. 2003;13(22):2009–2013. doi: 10.1016/j.cub.2003.10.034. [DOI] [PubMed] [Google Scholar]
- 102.Marin M, Rose KM, Kozak SL, Kabat D. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 2003;9(11):1398–1403. doi: 10.1038/nm946. [DOI] [PubMed] [Google Scholar]
- 103.Sheehy AM, Gaddis NC, Malim MH. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 2003;9(11):1404–1407. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- 104.Stopak K, de Noronha C, Yonemoto W, Greene WC. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell. 2003;12(3):591–601. doi: 10.1016/s1097-2765(03)00353-8. [DOI] [PubMed] [Google Scholar]
- 105.Kao S, Goila-Gaur R, Miyagi E, et al. Production of infectious virus and degradation of APOBEC3G are separable functional properties of human immunodeficiency virus type 1 Vif. Virology. 2007;369(2):329–339. doi: 10.1016/j.virol.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Opi S, Kao S, Goila-Gaur R, et al. Human immunodeficiency virus type 1 Vif inhibits packaging and antiviral activity of a degradation-resistant APOBEC3G variant. J. Virol. 2007;81(15):8236–8246. doi: 10.1128/JVI.02694-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Santa-Marta M, da Silva FA, Fonseca AM, Goncalves J. HIV-1 Vif can directly inhibit apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3G-mediated cytidine deamination by using a single amino acid interaction and without protein degradation. J. Biol. Chem. 2005;280(10):8765–8775. doi: 10.1074/jbc.M409309200. [DOI] [PubMed] [Google Scholar]
- 108.Goila-Gaur R, Khan MA, Miyagi E, et al. HIV-1 Vif promotes the formation of high molecular mass APOBEC3G complexes. Virology. 2008;372(1):136–146. doi: 10.1016/j.virol.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kao S, Miyagi E, Khan MA, et al. Production of infectious human immunodeficiency virus type 1 does not require depletion of APOBEC3G from virus-producing cells. Retrovirology. 2004;1:27. doi: 10.1186/1742-4690-1-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Barraud P, Paillart JC, Marquet R, Tisne C. Advances in the structural understanding of Vif proteins. Curr. HIV Res. 2008;6(2):91–99. doi: 10.2174/157016208783885056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Luo K, Xiao Z, Ehrlich E, et al. Primate lentiviral virion infectivity factors are substrate receptors that assemble with cullin 5–E3 ligase through a HCCH motif to suppress APOBEC3G. Proc. Natl Acad. Sci. USA. 2005;102(32):11444–11449. doi: 10.1073/pnas.0502440102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mehle A, Thomas ER, Rajendran KS, Gabuzda D. A zinc-binding region in Vif binds Cul5 and determines cullin selection. J. Biol. Chem. 2006;281(25):17259–17265. doi: 10.1074/jbc.M602413200. [DOI] [PubMed] [Google Scholar]
- 113.Xiao Z, Ehrlich E, Yu Y, et al. Assembly of HIV-1 Vif–Cul5 E3 ubiquitin ligase through a novel zinc-binding domain-stabilized hydrophobic interface in Vif. Virology. 2006;349(2):290–299. doi: 10.1016/j.virol.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 114.Xiao Z, Xiong Y, Zhang W, et al. Characterization of a novel cullin5 binding domain in HIV-1 Vif. J. Mol. Biol. 2007;373(3):541–550. doi: 10.1016/j.jmb.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 115.Kamura T, Sato S, Haque D, et al. The Elongin BC complex interacts with the conserved SOCS-box motif present in members of the SOCS, Ras, WD-40 repeat, and ankyrin repeat families. Genes Dev. 1998;12(24):3872–3881. doi: 10.1101/gad.12.24.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mehle A, Goncalves J, Santa-Marta M, et al. Phosphorylation of a novel SOCS-box regulates assembly of the HIV-1 Vif–Cul5 complex that promotes APOBEC3G degradation. Genes Dev. 2004;18(23):2861–2866. doi: 10.1101/gad.1249904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhang JG, Farley A, Nicholson SE, et al. The conserved SOCS box motif in suppressors of cytokine signaling binds to elongins B and C and may couple bound proteins to proteasomal degradation. Proc. Natl Acad. Sci. USA. 1999;96(5):2071–2076. doi: 10.1073/pnas.96.5.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Simon JH, Sheehy AM, Carpenter EA, Fouchier RA, Malim MH. Mutational analysis of the human immunodeficiency virus type 1 Vif protein. J. Virol. 1999;3(4):2675–2681. doi: 10.1128/jvi.73.4.2675-2681.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Stanley BJ, Ehrlich ES, Short L, et al. Structural insight into the human immunodeficiency virus Vif SOCS box and its role in human E3 ubiquitin ligase assembly. J. Virol. 2008;82(17):8656–8663. doi: 10.1128/JVI.00767-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chen G, He Z, Wang T, Xu R, Yu XF. A patch of positively charged amino acids surrounding the human immunodeficiency virus type 1 Vif SLV×4Y×9Y motif influences its interaction with APOBEC3G. J. Virol. 2009;83(17):8674–8682. doi: 10.1128/JVI.00653-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dang Y, Wang X, Zhou T, York IA, Zheng YH. Identification of a novel W×SLVK motif in the N terminus of human immunodeficiency virus and simian immunodeficiency virus Vif that is critical for APOBEC3G and APOBEC3F neutralization. J. Virol. 2009;83(17):8544–8552. doi: 10.1128/JVI.00651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.He Z, Zhang W, Chen G, Xu R, Yu XF. Characterization of conserved motifs in HIV-1 Vif required for APOBEC3G and APOBEC3F interaction. J. Mol. Biol. 2008;381(4):1000–1011. doi: 10.1016/j.jmb.2008.06.061. [DOI] [PubMed] [Google Scholar]
- 123.Mehle A, Wilson H, Zhang C, et al. Identification of an APOBEC3G binding site in human immunodeficiency virus type 1 Vif and inhibitors of Vif–APOBEC3G binding. J. Virol. 2007;81(23):13235–13241. doi: 10.1128/JVI.00204-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Russell RA, Pathak VK. Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F. J. Virol. 2007;81(15):8201–8210. doi: 10.1128/JVI.00395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tian C, Yu X, Zhang W, Wang T, Xu R, Yu XF. Differential requirement for conserved tryptophans in human immunodeficiency virus type 1 Vif for the selective suppression of APOBEC3G and APOBEC3F. J. Virol. 2006;80(6):3112–3115. doi: 10.1128/JVI.80.6.3112-3115.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Marin M, Golem S, Rose KM, Kozak SL, Kabat D. Human immunodeficiency virus type 1 Vif functionally interacts with diverse APOBEC3 cytidine deaminases and moves with them between cytoplasmic sites of mRNA metabolism. J. Virol. 2008;82(2):987–998. doi: 10.1128/JVI.01078-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhang W, Chen G, Niewiadomska AM, Xu R, Yu XF. Distinct determinants in HIV-1 Vif and human APOBEC3 proteins are required for the suppression of diverse host antiviral proteins. PLoS One. 2008;3(12):e3963. doi: 10.1371/journal.pone.0003963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fujita M, Sakurai A, Yoshida A, et al. Amino acid residues 88 and 89 in the central hydrophilic region of human immunodeficiency virus type 1 Vif are critical for viral infectivity by enhancing the steady-state expression of Vif. J. Virol. 2003;77(2):1626–1632. doi: 10.1128/JVI.77.2.1626-1632.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Miller JH, Presnyak V, Smith HC. The dimerization domain of HIV-1 viral infectivity factor Vif is required to block virion incorporation of APOBEC3G. Retrovirology. 2007;4:81. doi: 10.1186/1742-4690-4-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yang B, Gao L, Li L, et al. Potent suppression of viral infectivity by the peptides that inhibit multimerization of human immunodeficiency virus type 1 (HIV-1) Vif proteins. J. Biol. Chem. 2003;278(8):6596–6602. doi: 10.1074/jbc.M210164200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yang S, Sun Y, Zhang H. The multimerization of human immunodeficiency virus type I Vif protein: a requirement for Vif function in the viral life cycle. J. Biol. Chem. 2001;276(7):4889–4893. doi: 10.1074/jbc.M004895200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Dettenhofer M, Cen S, Carlson BA, Kleiman L, Yu XF. Association of human immunodeficiency virus type 1 Vif with RNA and its role in reverse transcription. J. Virol. 2000;74(19):8938–8945. doi: 10.1128/jvi.74.19.8938-8945.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Khan MA, Aberham C, Kao S, et al. Human immunodeficiency virus type 1 Vif protein is packaged into the nucleoprotein complex through an interaction with viral genomic RNA. J. Virol. 2001;75(16):7252–7265. doi: 10.1128/JVI.75.16.7252-7265.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhang H, Pomerantz RJ, Dornadula G, Sun Y. Human immunodeficiency virus type 1 Vif protein is an integral component of an mRNP complex of viral RNA and could be involved in the viral RNA folding and packaging process. J. Virol. 2000;74(18):8252–8261. doi: 10.1128/jvi.74.18.8252-8261.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kao S, Khan MA, Miyagi E, Plishka R, Buckler-White A, Strebel K. The human immunodeficiency virus type 1 Vif protein reduces intracellular expression and inhibits packaging of APOBEC3G (CEM15), a cellular inhibitor of virus infectivity. J. Virol. 2003;77(21):11398–11407. doi: 10.1128/JVI.77.21.11398-11407.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Karczewski MK, Strebel K. Cytoskeleton association and virion incorporation of the human immunodeficiency virus type 1 Vif protein. J. Virol. 1996;70(1):494–507. doi: 10.1128/jvi.70.1.494-507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Liu H, Wu X, Newman M, Shaw GM, Hahn BH, Kappes JC. The Vif protein of human and simian immunodeficiency viruses is packaged into virions and associates with viral core structures. J. Virol. 1995;69(12):7630–7638. doi: 10.1128/jvi.69.12.7630-7638.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Henriet S, Mercenne G, Bernacchi S, Paillart JC, Marquet R. Tumultuous relationship between the human immunodeficiency virus type 1 viral infectivity factor (Vif) and the human APOBEC-3G and APOBEC-3F restriction factors. Microbiol. Mol. Biol. Rev. 2009;73(2):211–232. doi: 10.1128/MMBR.00040-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Henriet S, Sinck L, Bec G, Gorelick RJ, Marquet R, Paillart JC. Vif is a RNA chaperone that could temporally regulate RNA dimerization and the early steps of HIV-1 reverse transcription. Nucleic Acids Res. 2007;35(15):5141–5153. doi: 10.1093/nar/gkm542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Virgen CA, Hatziioannou T. Antiretroviral activity and Vif sensitivity of rhesus macaque APOBEC3 proteins. J. Virol. 2007;81(24):13932–13937. doi: 10.1128/JVI.01760-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bogerd HP, Doehle BP, Wiegand HL, Cullen BR. A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proc. Natl Acad. Sci. USA. 2004;101(11):3770–3774. doi: 10.1073/pnas.0307713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mangeat B, Turelli P, Liao S, Trono D. A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. J. Biol. Chem. 2004;279(15):14481–14483. doi: 10.1074/jbc.C400060200. [DOI] [PubMed] [Google Scholar]
- 143.Schröfelbauer B, Chen D, Landau NR. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif) Proc. Natl Acad. Sci. USA. 2004;101(11):3927–3932. doi: 10.1073/pnas.0307132101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Xu H, Svarovskaia ES, Barr R, et al. A single amino acid substitution in human APOBEC3G antiretroviral enzyme confers resistance to HIV-1 virion infectivity factor-induced depletion. Proc. Natl Acad. Sci. USA. 2004;101(15):5652–5657. doi: 10.1073/pnas.0400830101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Derse D, Hill SA, Princler G, Lloyd P, Heidecker G. Resistance of human T cell leukemia virus type 1 to APOBEC3G restriction is mediated by elements in nucleocapsid. Proc. Natl Acad. Sci. USA. 2007;104(8):2915–2920. doi: 10.1073/pnas.0609444104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Cen S, Guo F, Niu M, Saadatmand J, Deflassieux J, Kleiman L. The interaction between HIV-1 Gag and APOBEC3G. J. Biol. Chem. 2004;279(32):33177–33184. doi: 10.1074/jbc.M402062200. [DOI] [PubMed] [Google Scholar]
- 147.Haché G, Shindo K, Albin JS, Harris RS. Evolution of HIV-1 isolates that use a novel Vif-independent mechanism to resist restriction by human APOBEC3G. Curr. Biol. 2008;18(11):819–824. doi: 10.1016/j.cub.2008.04.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Haché G, Abbink TE, Berkhout B, Harris RS. Optimal translation initiation enables Vif-deficient human immunodeficiency virus type 1 to escape restriction by APOBEC3G. J. Virol. 2009;83(11):5956–5960. doi: 10.1128/JVI.00045-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Nathans R, Cao H, Sharova N, et al. Small-molecule inhibition of HIV-1 Vif. Nat. Biotechnol. 2008;26(10):1187–1192. doi: 10.1038/nbt.1496. ▪▪Discovery of the first small-molecule antagonists of Vif.
- 150.Kidd JM, Newman TL, Tuzun E, Kaul R, Eichler EE. Population stratification of a common APOBEC gene deletion polymorphism. PLoS Genet. 2007;3(4):e63. doi: 10.1371/journal.pgen.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Harris RS. Enhancing immunity to HIV through APOBEC. Nat. Biotechnol. 2008;26(10):1089–1090. doi: 10.1038/nbt1008-1089. [DOI] [PubMed] [Google Scholar]
- 152.Berkhout B, de Ronde A. APOBEC3G versus reverse transcriptase in the generation of HIV-1 drug-resistance mutations. AIDS. 2004;18(13):1861–1863. doi: 10.1097/00002030-200409030-00022. [DOI] [PubMed] [Google Scholar]
- 153.Haché G, Mansky LM, Harris RS. Human APOBEC3 proteins, retrovirus restriction, and HIV drug resistance. AIDS Rev. 2006;8(3):148–157. [PubMed] [Google Scholar]
- 154.Jern P, Russell RA, Pathak VK, Coffin JM. Likely role of APOBEC3G-mediated G-to-A mutations in HIV-1 evolution and drug resistance. PLoS Pathog. 2009;5(4):e1000367. doi: 10.1371/journal.ppat.1000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Smith RA, Loeb LA, Preston BD. Lethal mutagenesis of HIV. Virus Res. 2005;107(2):215–228. doi: 10.1016/j.virusres.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 156.Simon V, Zennou V, Murray D, Huang Y, Ho DD, Bieniasz PD. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog. 2005;1(1):e6. doi: 10.1371/journal.ppat.0010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 2007:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 158.Koning FA, Newman EN, Kim EY, Kunstman KJ, Wolinsky SM, Malim MH. Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. J. Virol. 2009;83(18):9474–9485. doi: 10.1128/JVI.01089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]