Abstract
In the title compound, C24H44N6 6+·6C7H7O3S−·2H2O, the macrocycle crystallizes in its hexaprotonated form, accompanied by six p-toluenesulfonate ions and two water molecules, and lies on an inversion center. The three independent p-toluenesulfonate anions and their inversion equivalents at (1 − x, 1 − y, 1 − z) are linked to the macrocyclic cation through N—H⋯O hydrogen bonds. Of these, two p-toluenesulfonate ions are located on opposite sides of the macrocyclic plane and are linked to bridgehead N atoms via N—H⋯O hydrogen bonds. The remaining four p-toluenesulfonate ions bridge two adjacent macrocyclic cationic units through N—H⋯O hydrogen bonding involving other N atoms, forming a chain along the a axis. The water molecules, which could not be located and may be disordered, do not interact with the macrocycle; however, they form hydrogen bonds with anions.
Related literature
For general background, see: Bianchi et al. (1997 ▶); Chen & Martell (1991 ▶); Hossain (2008 ▶); Llobet et al. (1994 ▶); Nagarajan & Ganem (1987 ▶); Ragunathan & Schneider (1996 ▶). For related structures, see: Bazzicalupi et al. (1995 ▶); Clifford et al. (2001 ▶); He et al. (2000 ▶); Li et al. (2009 ▶); Liu et al. (2008 ▶); Lu et al. (1995 ▶, 1998 ▶); Zhu et al. (2002 ▶).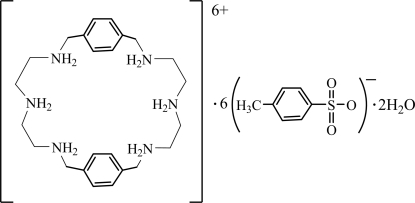
Experimental
Crystal data
C24H44N6 6+·6C7H7O3S−·2H2O
M r = 1479.80
Triclinic,
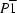
a = 11.513 (3) Å
b = 12.639 (5) Å
c = 13.556 (6) Å
α = 78.578 (16)°
β = 71.88 (2)°
γ = 89.30 (2)°
V = 1835.0 (12) Å3
Z = 1
Mo Kα radiation
μ = 0.26 mm−1
T = 90 K
0.17 × 0.10 × 0.03 mm
Data collection
Nonius KappaCCD diffractometer with an Oxford Cryosystems Cryostream cooler
Absorption correction: multi-scan (SCALEPACK; Otwinowski & Minor, 1997 ▶) T min = 0.947, T max = 0.992
27691 measured reflections
7213 independent reflections
2920 reflections with I > 2σ(I)
R int = 0.145
Refinement
R[F 2 > 2σ(F 2)] = 0.072
wR(F 2) = 0.156
S = 0.97
7213 reflections
446 parameters
H-atom parameters constrained
Δρmax = 0.39 e Å−3
Δρmin = −0.33 e Å−3
Data collection: COLLECT (Nonius, 1999 ▶); cell refinement: DENZO and SCALEPACK (Otwinowski & Minor, 1997 ▶); data reduction: DENZO and SCALEPACK; program(s) used to solve structure: SIR92 (Altomare et al., 1994 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 1997 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809035648/ci2898sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809035648/ci2898Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H11N⋯O7i | 0.92 | 1.86 | 2.738 (5) | 159 |
| N1—H12N⋯O8 | 0.92 | 2.10 | 2.923 (6) | 148 |
| N1—H12N⋯O9 | 0.92 | 2.33 | 3.130 (5) | 145 |
| N2—H21N⋯O5 | 0.92 | 1.85 | 2.745 (5) | 164 |
| N2—H22N⋯O1ii | 0.92 | 1.83 | 2.706 (4) | 159 |
| N3—H31N⋯O4ii | 0.92 | 1.84 | 2.748 (4) | 168 |
| N3—H32N⋯O2 | 0.92 | 1.92 | 2.842 (5) | 177 |
Symmetry codes: (i) 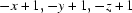 ; (ii)
; (ii) 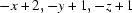 .
.
Acknowledgments
This work was supported by the National Institutes of Health, Division of National Center for Research Resources, under grant No. G12RR013459. The purchase of the diffractometer was made possible by grant No. LEQSF (1999–2000)-ENH-TR-13, administered by the Louisiana Board of Regents.
supplementary crystallographic information
Comment
Azamacrocyles mimic many natural polyamines including putrescine, spermidine and spermine, that are known to complex negatively charged nucleotides through hydrogen bonding interactions with phosphate groups (Nagarajan & Ganem, 1987; Bianchi et al., 1997)). Simple hexaazamacrocycles which are conveniently synthesized from Schiff base derived reaction (Chen & Martell, 1991) are known to complex both anions and cations (Llobet et al., 1994; Hossain, 2008). For example, m-xylyl macrocycle with six N atoms and rigid spacers were shown to form complexes with sulfate (Clifford et al., 2001), pyrophosphate (Lu et al., 1995), and triphosphate (Lu et al., 1998). A m-xylyl analogue with propylene chains, was recently reported to complex with chloride showing a chair-like structure, where chloride anions are bonded to protonated amines outside the cavity (Liu et al., 2008). The macrocycle in the title compound containing slightly larger cavity with p-xylyl groups, was reported to bind perchlorate (Bazzicalupi, et al., 1995) and benzene-1,2,4,5-tetracarboxylate ((Zhu et al., 2002) in the tetraprotonated form. Incorporating metal ions, this compound was found to catalyze the hydrolysis of bis(p-nitrophenyl)-phosphate (Ragunathan & Schneider, 1996), and was also shown to bind calf thymus DNA (Li et al., 2009). As a part of our work in the field of anion recognition, we synthesized the p-xylyl based macrocycle and isolated crystals of the p-toluenesulfonate salt. This report describes the synthesis and structural aspect of a ditopic anion complex of p-xylyl macrocycle.
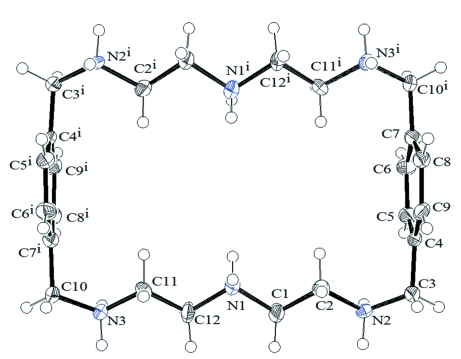
Single crystal analysis of the p-toluenesulfonate salt reveals that the ligand is hexaprotonated and crystallized with six p-toluenesulfonate anions and two water molecules. Each protonated amine of the macrocycle is involved in coordinating one p-toluenesulfonate with short (N···O) hydrogen bonds (Table 1). The cationic macrocycle lies on an inversion center. The macrocycle is found to adopt a rectangular shape in which two aromatic units are parallel to each other with a centroid-to-centroid distance of 9.990 Å (Fig. 1). The shape of the macrocycle is quite different from those observed in the neutral macrocycle (He et al., 2000) or its larger cationic analogue with m-xylyl spacers in the chloride complex (Liu et al., 2008), both of which adopt a chair conformation with two flip-flop aromatic moieties. In the solid state, the hydrogen atoms on the bridgehead amines direct toward the cavity, while those on the remaining nitrogen atoms point outwards from the cavity to minimize the electrostatic repulsion forces. The NCCN chains (N1—C1—C2—N2, N3—C11—C12—N1 and their symmetry related counterparts) linking the bridgehead N atoms, are essentially unstrained, with the torsion angles of -172.0 (4) and 170.9 (4)°, close to the trans conformation (ca 180°). On the other hand, gauche conformations (ca 60°) were found for the CNCC chains (C2—N2—C3—C4, C11—N3—C10—C7i and their counterparts) associated with the aromatic rings, with torsion angles of -51.8 (5) and 51.9 (5)°.
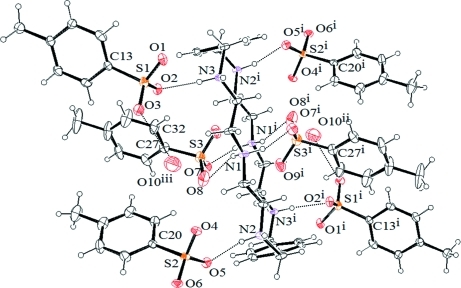
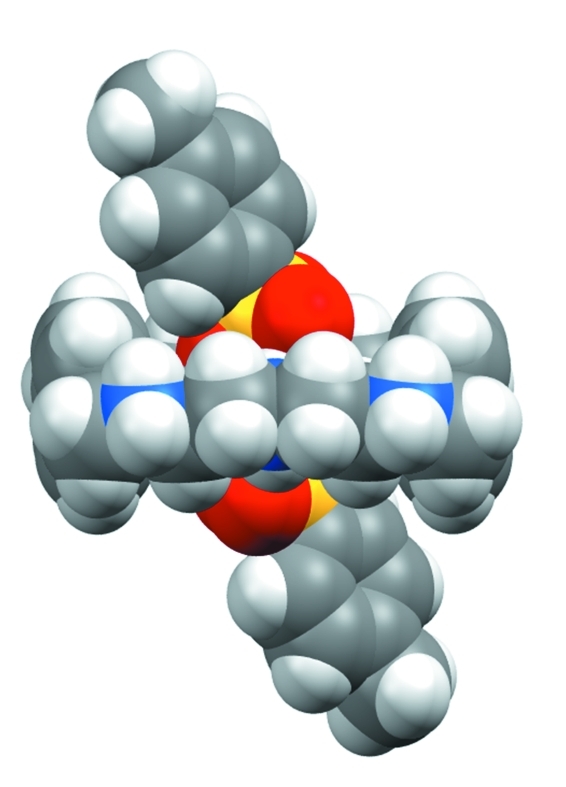
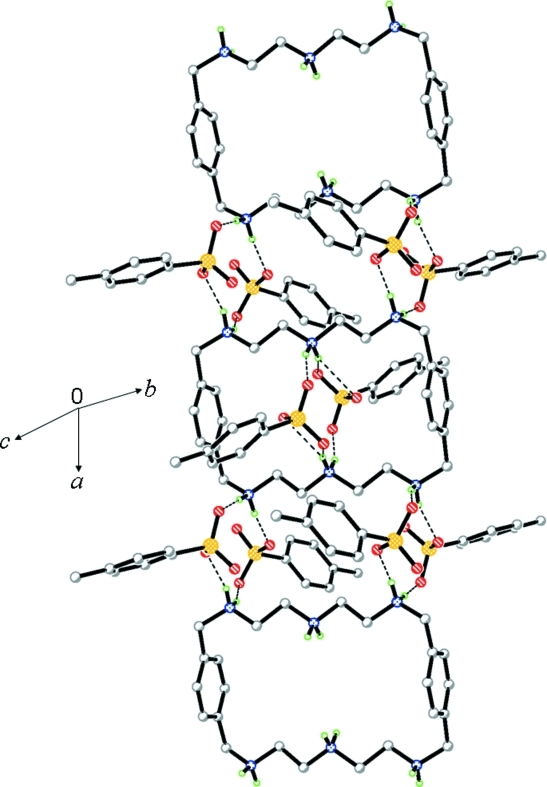
All six p-toluenesulfonate anions are strongly bonded to both faces of the macrocycle, with three groups at each face (Fig. 2). Four p-toluenesulfonates are singly bonded to the protonated amines (N2, N2i, N3 and N3i) linking the aromatic spacers, with almost linear N—H···O interactions (164 to 177 °). The distances in N···O bonds are in the range from 2.706 (4) to 2.842 (5) Å. Two other anionic groups are located above and below the macrocyclic plane, each with two N···O bonds with the central amines (N1 and N1i). In this case two oxygen atoms, O7 and O8 and their symmetry related O7ii and O8ii act as hydrogen bond acceptors and bridge the amines from the opposite sides. These two anions are partially included in the cavity with a ditopic binding mode (Fig. 3). In the crystal structure, the ionic units are linked by N—H..O hydrogen bonds forming a chain along the a axis. Along the c axis, the C4-C9 aromatic rings of the adjacent chains are stacked, with a centroid-to-centroid distance of 4.533 (3) Å (Fig. 4). The anions are found within the channels formed by the macrocycles, facing in opposite directions alternatively and are linked to the macrocycles with extensive hydrogen bonding frameworks.
In summary, a novel ditopic complex of p-toluenesulfonate anion has been synthesized, and its structure has been presented. The surrounding p-toluenesulfonates act as hydrogen bond acceptors for the nitrogen sites in the macrocycle, and play an important role in forming a hydrogen bonding network in the three dimensional structure. The water molecules do not interact with the macrocycle, however they form hydrogen bonds with anions.
Experimental
The synthesis was carried out following a modified literature procedure (Chen & Martell, 1991). An equimolar amount of diethylenetriamine (1.00 g, 9.70 x 10 -3 mol) in CH3OH (200 ml) and terephthalaldehyde (1.30 g, 9.70 x 10 -3 mol) in CH3OH (200 ml) were added to CH3OH (400 ml) over 4 h at 0°C. The mixture was stirred at room temperature for another 24 h. After the solvent was evaporated, the Schiff base was reduced to amine with NaBH4 (1.73 g, 45.7 x 10 -3 mol) in CH3OH (100 ml) at room temperature for 12 h. After evaporating the solvent under reduced pressure, the residue was dissolved in 1 M aq NaOH solution (100 ml), and the aqueous phase was extracted by CH2Cl2 (3x100 ml). The organic layers were combined and dried with MgSO4. Evaporation of the solvent gave a white powder of the neutral macrocycle. Yield: 60%. 1H NMR (300 MHz, CDCl3, TMS): δ 7.29 (s, 8H, ArH), 3.80 (s, 8H, ArCH2), 2.85 (t, 8H, CH2), 2.83 (t, 8H, CH2). The p-toluenesulfonate salt was obtained by titrating the macrocycle (50 mg) dissolved in CH3OH (2 ml) with TsOH. The addition of diethyl ether (2 ml) yielded a white precipitate of p-toluenesulfonate salt that was filtered and dried. Yield: 80%. 1H NMR (300 MHz, D2O, TSP): δ 7.70 (d, 12H, ArH), 7.42 (d, 12H, ArH), 7.26 (s, 8H, ArH), 4.21 (s, 8H, ArCH2), 3.27 (t, 8H, CH2), 3.01 (t, 8H, CH2), 2.43 (s, 12H, CH2). Single crystals suitable for X-ray analysis were obtained by slow evaporation of the solvent.
Refinement
H atoms on C were placed in idealized positions with C-H distances of 0.95-0.99 Å and thereafter treated as riding. Those on N were all visible in difference maps, but were placed in idealized positions with a N-H distance of 0.92 Å. Uiso(H) values were assigned as 1.2 times Ueq of the attached atom (1.5 for methyl). A torsional parameter was refined for each methyl group. H atoms on the water molecule could not be located.
Figures
Fig. 1.
The molecular structure of cationic unit in (1). Displacement ellipsoids are drawn at the 50% probability level. [Symmetry codes: (i) 1 - x, 1 - y, 1 - z].
Fig. 2.
The molecular structure of (1) showing the atom-numbering scheme and hydrogen bonding interactions. Displacement ellipsoids are drawn at the 50% probability level. [Symmetry codes: (i) 1 - x, 1 - y, 1 - z; (ii) 2 - x, 1 - y, 1 - z.]
Fig. 3.
Space-filling view of (1) showing two anions bonded with macrocycle at the both faces.
Fig. 4.
View of a hydrogen-bonded (dashed lines) chain in (1).
Crystal data
| C24H44N66+·6C7H7O3S−·2H2O | Z = 1 |
| Mr = 1479.80 | F(000) = 784 |
| Triclinic, P1 | Dx = 1.339 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 11.513 (3) Å | Cell parameters from 6760 reflections |
| b = 12.639 (5) Å | θ = 2.5–26.0° |
| c = 13.556 (6) Å | µ = 0.26 mm−1 |
| α = 78.578 (16)° | T = 90 K |
| β = 71.88 (2)° | Plate, colorless |
| γ = 89.30 (2)° | 0.17 × 0.10 × 0.03 mm |
| V = 1835.0 (12) Å3 |
Data collection
| Nonius KappaCCD diffractometer with an Oxford Cryosystems Cryostream cooler | 7213 independent reflections |
| Radiation source: fine-focus sealed tube | 2920 reflections with I > 2σ(I) |
| graphite | Rint = 0.145 |
| ω and φ scans | θmax = 26.3°, θmin = 2.7° |
| Absorption correction: multi-scan (SCALEPACK; Otwinowski & Minor, 1997) | h = −13→14 |
| Tmin = 0.947, Tmax = 0.992 | k = −15→15 |
| 27691 measured reflections | l = −16→16 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.072 | H-atom parameters constrained |
| wR(F2) = 0.156 | w = 1/[σ2(Fo2) + (0.0515P)2] where P = (Fo2 + 2Fc2)/3 |
| S = 0.97 | (Δ/σ)max = 0.001 |
| 7213 reflections | Δρmax = 0.39 e Å−3 |
| 446 parameters | Δρmin = −0.33 e Å−3 |
| 0 restraints | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.0019 (7) |
Special details
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| N1 | 0.7392 (3) | 0.5311 (3) | 0.4777 (3) | 0.0292 (10) | |
| H11N | 0.7039 | 0.5968 | 0.4705 | 0.035* | |
| H12N | 0.6798 | 0.4779 | 0.4892 | 0.035* | |
| N2 | 0.8843 (3) | 0.5451 (3) | 0.1864 (3) | 0.0242 (10) | |
| H21N | 0.8920 | 0.4720 | 0.1907 | 0.029* | |
| H22N | 0.9589 | 0.5751 | 0.1815 | 0.029* | |
| N3 | 0.7276 (3) | 0.5161 (3) | 0.7591 (3) | 0.0222 (9) | |
| H31N | 0.8044 | 0.5469 | 0.7455 | 0.027* | |
| H32N | 0.7331 | 0.4423 | 0.7725 | 0.027* | |
| C1 | 0.8391 (4) | 0.5271 (4) | 0.3779 (3) | 0.0331 (13) | |
| H1A | 0.8667 | 0.4525 | 0.3792 | 0.040* | |
| H1B | 0.9097 | 0.5751 | 0.3710 | 0.040* | |
| C2 | 0.7921 (4) | 0.5632 (4) | 0.2853 (3) | 0.0263 (12) | |
| H2A | 0.7146 | 0.5220 | 0.2974 | 0.032* | |
| H2B | 0.7753 | 0.6409 | 0.2782 | 0.032* | |
| C3 | 0.8538 (4) | 0.5918 (4) | 0.0876 (4) | 0.0281 (12) | |
| H3A | 0.8608 | 0.6717 | 0.0750 | 0.034* | |
| H3B | 0.9132 | 0.5688 | 0.0264 | 0.034* | |
| C4 | 0.7271 (4) | 0.5565 (4) | 0.0956 (3) | 0.0219 (11) | |
| C5 | 0.6934 (4) | 0.4486 (4) | 0.1162 (3) | 0.0276 (12) | |
| H5 | 0.7526 | 0.3962 | 0.1200 | 0.033* | |
| C6 | 0.5747 (4) | 0.4155 (4) | 0.1315 (4) | 0.0286 (12) | |
| H6 | 0.5525 | 0.3404 | 0.1469 | 0.034* | |
| C7 | 0.4869 (4) | 0.4901 (4) | 0.1246 (3) | 0.0245 (12) | |
| C8 | 0.5205 (4) | 0.5989 (4) | 0.1009 (3) | 0.0253 (12) | |
| H8 | 0.4624 | 0.6513 | 0.0933 | 0.030* | |
| C9 | 0.6402 (4) | 0.6312 (4) | 0.0885 (3) | 0.0247 (12) | |
| H9 | 0.6625 | 0.7060 | 0.0748 | 0.030* | |
| C10 | 0.6439 (3) | 0.5474 (4) | 0.8560 (3) | 0.0239 (12) | |
| H10A | 0.6493 | 0.6270 | 0.8477 | 0.029* | |
| H10B | 0.6698 | 0.5149 | 0.9180 | 0.029* | |
| C11 | 0.6896 (4) | 0.5483 (4) | 0.6635 (3) | 0.0246 (12) | |
| H11A | 0.6815 | 0.6275 | 0.6486 | 0.030* | |
| H11B | 0.6092 | 0.5124 | 0.6751 | 0.030* | |
| C12 | 0.7840 (4) | 0.5161 (5) | 0.5708 (4) | 0.0389 (15) | |
| H12A | 0.8608 | 0.5607 | 0.5523 | 0.047* | |
| H12B | 0.8019 | 0.4394 | 0.5905 | 0.047* | |
| S1 | 0.86065 (10) | 0.27995 (10) | 0.83408 (10) | 0.0307 (4) | |
| O1 | 0.8820 (2) | 0.3732 (2) | 0.8763 (3) | 0.0301 (9) | |
| O2 | 0.7479 (2) | 0.2887 (2) | 0.8063 (2) | 0.0321 (9) | |
| O3 | 0.9661 (3) | 0.2628 (3) | 0.7468 (3) | 0.0405 (10) | |
| C13 | 0.8414 (4) | 0.1649 (4) | 0.9368 (4) | 0.0239 (12) | |
| C14 | 0.8237 (4) | 0.0639 (4) | 0.9169 (4) | 0.0368 (14) | |
| H14 | 0.8200 | 0.0575 | 0.8493 | 0.044* | |
| C15 | 0.8115 (4) | −0.0272 (4) | 0.9950 (4) | 0.0385 (14) | |
| H15 | 0.7995 | −0.0961 | 0.9805 | 0.046* | |
| C16 | 0.8164 (4) | −0.0201 (4) | 1.0952 (4) | 0.0298 (13) | |
| C17 | 0.8351 (4) | 0.0810 (4) | 1.1128 (4) | 0.0349 (13) | |
| H17 | 0.8388 | 0.0876 | 1.1803 | 0.042* | |
| C18 | 0.8485 (4) | 0.1730 (4) | 1.0349 (4) | 0.0370 (14) | |
| H18 | 0.8625 | 0.2417 | 1.0488 | 0.044* | |
| C19 | 0.8042 (4) | −0.1200 (4) | 1.1794 (4) | 0.0389 (14) | |
| H19A | 0.8052 | −0.0996 | 1.2452 | 0.058* | |
| H19B | 0.8725 | −0.1659 | 1.1561 | 0.058* | |
| H19C | 0.7267 | −0.1599 | 1.1919 | 0.058* | |
| S2 | 1.03881 (10) | 0.29487 (10) | 0.21857 (10) | 0.0261 (4) | |
| O4 | 1.0570 (2) | 0.3649 (2) | 0.2876 (2) | 0.0265 (8) | |
| O5 | 0.9472 (2) | 0.3379 (2) | 0.1689 (2) | 0.0310 (9) | |
| O6 | 1.1516 (2) | 0.2716 (2) | 0.1447 (2) | 0.0318 (9) | |
| C20 | 0.9732 (4) | 0.1722 (4) | 0.3034 (4) | 0.0243 (12) | |
| C21 | 1.0362 (4) | 0.0795 (4) | 0.3002 (4) | 0.0413 (14) | |
| H21 | 1.1164 | 0.0814 | 0.2517 | 0.050* | |
| C22 | 0.9831 (5) | −0.0177 (4) | 0.3677 (5) | 0.0515 (16) | |
| H22 | 1.0270 | −0.0817 | 0.3641 | 0.062* | |
| C23 | 0.8652 (5) | −0.0217 (5) | 0.4410 (4) | 0.0444 (15) | |
| C24 | 0.8042 (4) | 0.0714 (5) | 0.4437 (4) | 0.0429 (15) | |
| H24 | 0.7245 | 0.0699 | 0.4930 | 0.052* | |
| C25 | 0.8559 (4) | 0.1684 (4) | 0.3760 (4) | 0.0344 (13) | |
| H25 | 0.8113 | 0.2321 | 0.3791 | 0.041* | |
| C26 | 0.8091 (5) | −0.1286 (4) | 0.5137 (5) | 0.0640 (19) | |
| H26A | 0.7470 | −0.1148 | 0.5774 | 0.096* | |
| H26B | 0.8731 | −0.1706 | 0.5340 | 0.096* | |
| H26C | 0.7709 | −0.1694 | 0.4765 | 0.096* | |
| S3 | 0.51388 (11) | 0.31199 (11) | 0.53979 (12) | 0.0374 (4) | |
| O7 | 0.4106 (3) | 0.2991 (3) | 0.5021 (3) | 0.0422 (10) | |
| O8 | 0.6303 (3) | 0.3321 (3) | 0.4573 (3) | 0.0696 (14) | |
| O9 | 0.4946 (3) | 0.3943 (3) | 0.6041 (3) | 0.0540 (11) | |
| C27 | 0.5184 (4) | 0.1876 (4) | 0.6248 (4) | 0.0338 (13) | |
| C28 | 0.4736 (4) | 0.0918 (4) | 0.6118 (5) | 0.0427 (15) | |
| H28 | 0.4402 | 0.0912 | 0.5560 | 0.051* | |
| C29 | 0.4784 (5) | −0.0029 (5) | 0.6816 (6) | 0.061 (2) | |
| H29 | 0.4467 | −0.0685 | 0.6733 | 0.073* | |
| C30 | 0.5274 (5) | −0.0056 (5) | 0.7628 (6) | 0.070 (2) | |
| C31 | 0.5708 (5) | 0.0922 (5) | 0.7738 (5) | 0.075 (2) | |
| H31 | 0.6030 | 0.0934 | 0.8302 | 0.089* | |
| C32 | 0.5681 (4) | 0.1871 (5) | 0.7050 (5) | 0.0520 (17) | |
| H32 | 0.6006 | 0.2526 | 0.7127 | 0.062* | |
| C33 | 0.5288 (6) | −0.1102 (5) | 0.8403 (7) | 0.118 (4) | |
| H33A | 0.5662 | −0.1657 | 0.8009 | 0.177* | |
| H33B | 0.4448 | −0.1346 | 0.8840 | 0.177* | |
| H33C | 0.5764 | −0.0978 | 0.8860 | 0.177* | |
| O10 | 1.0391 (4) | 0.6764 (4) | 0.4660 (4) | 0.0990 (16) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.024 (2) | 0.042 (3) | 0.019 (2) | 0.0028 (19) | −0.0035 (19) | −0.009 (2) |
| N2 | 0.0124 (19) | 0.037 (3) | 0.022 (2) | 0.0006 (17) | −0.0023 (17) | −0.007 (2) |
| N3 | 0.0195 (19) | 0.028 (2) | 0.018 (2) | −0.0020 (17) | −0.0027 (17) | −0.0059 (19) |
| C1 | 0.023 (3) | 0.056 (4) | 0.021 (3) | 0.007 (2) | −0.005 (2) | −0.013 (3) |
| C2 | 0.021 (2) | 0.033 (3) | 0.024 (3) | 0.000 (2) | −0.003 (2) | −0.011 (3) |
| C3 | 0.018 (2) | 0.048 (3) | 0.018 (3) | −0.001 (2) | −0.005 (2) | −0.006 (3) |
| C4 | 0.014 (2) | 0.036 (3) | 0.015 (3) | 0.001 (2) | −0.001 (2) | −0.011 (2) |
| C5 | 0.019 (3) | 0.039 (4) | 0.027 (3) | 0.006 (2) | −0.006 (2) | −0.013 (3) |
| C6 | 0.028 (3) | 0.027 (3) | 0.031 (3) | −0.007 (2) | −0.009 (2) | −0.005 (3) |
| C7 | 0.020 (3) | 0.035 (3) | 0.022 (3) | 0.004 (2) | −0.008 (2) | −0.010 (3) |
| C8 | 0.021 (3) | 0.034 (3) | 0.021 (3) | 0.005 (2) | −0.006 (2) | −0.007 (2) |
| C9 | 0.022 (3) | 0.030 (3) | 0.019 (3) | −0.007 (2) | 0.000 (2) | −0.006 (2) |
| C10 | 0.021 (3) | 0.034 (3) | 0.016 (3) | 0.000 (2) | −0.004 (2) | −0.006 (2) |
| C11 | 0.023 (2) | 0.033 (3) | 0.022 (3) | 0.002 (2) | −0.013 (2) | −0.005 (2) |
| C12 | 0.025 (3) | 0.075 (4) | 0.015 (3) | 0.009 (3) | −0.005 (2) | −0.010 (3) |
| S1 | 0.0238 (7) | 0.0303 (9) | 0.0379 (9) | 0.0036 (6) | −0.0102 (6) | −0.0060 (7) |
| O1 | 0.0245 (17) | 0.022 (2) | 0.047 (2) | −0.0024 (14) | −0.0159 (16) | −0.0081 (17) |
| O2 | 0.0287 (18) | 0.029 (2) | 0.042 (2) | 0.0011 (15) | −0.0177 (16) | −0.0050 (17) |
| O3 | 0.0298 (19) | 0.044 (2) | 0.038 (2) | 0.0123 (16) | −0.0013 (17) | −0.0027 (18) |
| C13 | 0.016 (2) | 0.024 (3) | 0.033 (3) | 0.004 (2) | −0.006 (2) | −0.009 (3) |
| C14 | 0.052 (3) | 0.033 (4) | 0.035 (4) | −0.003 (3) | −0.020 (3) | −0.017 (3) |
| C15 | 0.047 (3) | 0.026 (4) | 0.044 (4) | 0.000 (3) | −0.014 (3) | −0.013 (3) |
| C16 | 0.025 (3) | 0.028 (4) | 0.036 (4) | 0.003 (2) | −0.010 (2) | −0.007 (3) |
| C17 | 0.045 (3) | 0.029 (4) | 0.030 (3) | 0.002 (3) | −0.014 (3) | −0.003 (3) |
| C18 | 0.036 (3) | 0.032 (4) | 0.052 (4) | −0.003 (2) | −0.018 (3) | −0.022 (3) |
| C19 | 0.039 (3) | 0.039 (4) | 0.039 (4) | 0.002 (3) | −0.012 (3) | −0.006 (3) |
| S2 | 0.0209 (7) | 0.0306 (8) | 0.0250 (8) | −0.0015 (6) | −0.0034 (6) | −0.0079 (7) |
| O4 | 0.0207 (16) | 0.031 (2) | 0.030 (2) | −0.0027 (14) | −0.0065 (14) | −0.0141 (17) |
| O5 | 0.0295 (18) | 0.035 (2) | 0.034 (2) | 0.0029 (15) | −0.0168 (16) | −0.0073 (17) |
| O6 | 0.0240 (17) | 0.039 (2) | 0.024 (2) | 0.0006 (15) | 0.0047 (15) | −0.0082 (17) |
| C20 | 0.024 (3) | 0.027 (3) | 0.026 (3) | 0.001 (2) | −0.011 (2) | −0.010 (2) |
| C21 | 0.031 (3) | 0.035 (4) | 0.049 (4) | −0.001 (3) | 0.001 (3) | −0.011 (3) |
| C22 | 0.056 (4) | 0.025 (4) | 0.068 (5) | 0.004 (3) | −0.017 (3) | −0.001 (3) |
| C23 | 0.039 (3) | 0.041 (4) | 0.052 (4) | −0.016 (3) | −0.021 (3) | 0.006 (3) |
| C24 | 0.031 (3) | 0.047 (4) | 0.043 (4) | −0.011 (3) | −0.008 (3) | 0.002 (3) |
| C25 | 0.024 (3) | 0.035 (4) | 0.041 (4) | 0.002 (2) | −0.009 (3) | −0.004 (3) |
| C26 | 0.061 (4) | 0.046 (4) | 0.080 (5) | −0.022 (3) | −0.030 (3) | 0.014 (4) |
| S3 | 0.0244 (7) | 0.0398 (10) | 0.0454 (10) | 0.0028 (6) | −0.0109 (7) | −0.0034 (8) |
| O7 | 0.040 (2) | 0.046 (2) | 0.051 (3) | 0.0073 (17) | −0.0270 (18) | −0.0139 (19) |
| O8 | 0.032 (2) | 0.066 (3) | 0.075 (3) | 0.0132 (19) | 0.012 (2) | 0.021 (2) |
| O9 | 0.075 (3) | 0.033 (2) | 0.072 (3) | 0.0047 (19) | −0.045 (2) | −0.017 (2) |
| C27 | 0.022 (3) | 0.035 (4) | 0.046 (4) | −0.002 (2) | −0.014 (3) | −0.008 (3) |
| C28 | 0.031 (3) | 0.033 (4) | 0.063 (4) | −0.009 (3) | −0.010 (3) | −0.016 (3) |
| C29 | 0.039 (3) | 0.021 (4) | 0.114 (6) | −0.004 (3) | −0.014 (4) | −0.008 (4) |
| C30 | 0.051 (4) | 0.036 (4) | 0.120 (6) | −0.011 (3) | −0.047 (4) | 0.023 (4) |
| C31 | 0.071 (5) | 0.057 (5) | 0.102 (6) | −0.025 (4) | −0.059 (4) | 0.022 (4) |
| C32 | 0.052 (4) | 0.034 (4) | 0.070 (5) | −0.018 (3) | −0.032 (3) | 0.012 (3) |
| C33 | 0.106 (6) | 0.049 (5) | 0.193 (9) | −0.030 (4) | −0.087 (6) | 0.058 (6) |
| O10 | 0.116 (4) | 0.111 (4) | 0.087 (4) | 0.044 (3) | −0.047 (3) | −0.039 (3) |
Geometric parameters (Å, °)
| N1—C12 | 1.484 (5) | C15—C16 | 1.397 (7) |
| N1—C1 | 1.488 (5) | C15—H15 | 0.95 |
| N1—H11N | 0.92 | C16—C17 | 1.376 (6) |
| N1—H12N | 0.92 | C16—C19 | 1.501 (6) |
| N2—C2 | 1.486 (5) | C17—C18 | 1.381 (6) |
| N2—C3 | 1.496 (5) | C17—H17 | 0.95 |
| N2—H21N | 0.92 | C18—H18 | 0.95 |
| N2—H22N | 0.92 | C19—H19A | 0.98 |
| N3—C11 | 1.476 (5) | C19—H19B | 0.98 |
| N3—C10 | 1.490 (5) | C19—H19C | 0.98 |
| N3—H31N | 0.92 | S2—O6 | 1.440 (3) |
| N3—H32N | 0.92 | S2—O5 | 1.465 (3) |
| C1—C2 | 1.506 (6) | S2—O4 | 1.469 (3) |
| C1—H1A | 0.99 | S2—C20 | 1.754 (5) |
| C1—H1B | 0.99 | C20—C21 | 1.372 (6) |
| C2—H2A | 0.99 | C20—C25 | 1.397 (6) |
| C2—H2B | 0.99 | C21—C22 | 1.394 (7) |
| C3—C4 | 1.496 (5) | C21—H21 | 0.95 |
| C3—H3A | 0.99 | C22—C23 | 1.406 (7) |
| C3—H3B | 0.99 | C22—H22 | 0.95 |
| C4—C5 | 1.372 (6) | C23—C24 | 1.363 (7) |
| C4—C9 | 1.379 (6) | C23—C26 | 1.521 (7) |
| C5—C6 | 1.374 (5) | C24—C25 | 1.390 (6) |
| C5—H5 | 0.95 | C24—H24 | 0.95 |
| C6—C7 | 1.386 (6) | C25—H25 | 0.95 |
| C6—H6 | 0.95 | C26—H26A | 0.98 |
| C7—C8 | 1.380 (6) | C26—H26B | 0.98 |
| C7—C10i | 1.511 (5) | C26—H26C | 0.98 |
| C8—C9 | 1.391 (5) | S3—O8 | 1.440 (4) |
| C8—H8 | 0.95 | S3—O7 | 1.455 (3) |
| C9—H9 | 0.95 | S3—O9 | 1.457 (4) |
| C10—C7i | 1.511 (5) | S3—C27 | 1.766 (5) |
| C10—H10A | 0.99 | C27—C32 | 1.375 (7) |
| C10—H10B | 0.99 | C27—C28 | 1.386 (6) |
| C11—C12 | 1.506 (6) | C28—C29 | 1.383 (7) |
| C11—H11A | 0.99 | C28—H28 | 0.95 |
| C11—H11B | 0.99 | C29—C30 | 1.380 (8) |
| C12—H12A | 0.99 | C29—H29 | 0.95 |
| C12—H12B | 0.99 | C30—C31 | 1.391 (8) |
| S1—O2 | 1.457 (3) | C30—C33 | 1.521 (8) |
| S1—O3 | 1.457 (3) | C31—C32 | 1.373 (7) |
| S1—O1 | 1.463 (3) | C31—H31 | 0.95 |
| S1—C13 | 1.765 (5) | C32—H32 | 0.95 |
| C13—C18 | 1.381 (6) | C33—H33A | 0.98 |
| C13—C14 | 1.386 (6) | C33—H33B | 0.98 |
| C14—C15 | 1.376 (6) | C33—H33C | 0.98 |
| C14—H14 | 0.95 | ||
| C12—N1—C1 | 112.2 (3) | C15—C14—C13 | 120.1 (5) |
| C12—N1—H11N | 109.2 | C15—C14—H14 | 119.9 |
| C1—N1—H11N | 109.2 | C13—C14—H14 | 119.9 |
| C12—N1—H12N | 109.2 | C14—C15—C16 | 121.1 (5) |
| C1—N1—H12N | 109.2 | C14—C15—H15 | 119.4 |
| H11N—N1—H12N | 107.9 | C16—C15—H15 | 119.4 |
| C2—N2—C3 | 113.9 (3) | C17—C16—C15 | 117.7 (5) |
| C2—N2—H21N | 108.8 | C17—C16—C19 | 121.6 (5) |
| C3—N2—H21N | 108.8 | C15—C16—C19 | 120.7 (5) |
| C2—N2—H22N | 108.8 | C16—C17—C18 | 121.8 (5) |
| C3—N2—H22N | 108.8 | C16—C17—H17 | 119.1 |
| H21N—N2—H22N | 107.7 | C18—C17—H17 | 119.1 |
| C11—N3—C10 | 114.6 (3) | C17—C18—C13 | 119.9 (5) |
| C11—N3—H31N | 108.6 | C17—C18—H18 | 120.1 |
| C10—N3—H31N | 108.6 | C13—C18—H18 | 120.1 |
| C11—N3—H32N | 108.6 | C16—C19—H19A | 109.5 |
| C10—N3—H32N | 108.6 | C16—C19—H19B | 109.5 |
| H31N—N3—H32N | 107.6 | H19A—C19—H19B | 109.5 |
| N1—C1—C2 | 109.0 (3) | C16—C19—H19C | 109.5 |
| N1—C1—H1A | 109.9 | H19A—C19—H19C | 109.5 |
| C2—C1—H1A | 109.9 | H19B—C19—H19C | 109.5 |
| N1—C1—H1B | 109.9 | O6—S2—O5 | 113.90 (19) |
| C2—C1—H1B | 109.9 | O6—S2—O4 | 113.11 (17) |
| H1A—C1—H1B | 108.3 | O5—S2—O4 | 110.35 (19) |
| N2—C2—C1 | 109.8 (3) | O6—S2—C20 | 107.5 (2) |
| N2—C2—H2A | 109.7 | O5—S2—C20 | 105.69 (19) |
| C1—C2—H2A | 109.7 | O4—S2—C20 | 105.7 (2) |
| N2—C2—H2B | 109.7 | C21—C20—C25 | 119.2 (4) |
| C1—C2—H2B | 109.7 | C21—C20—S2 | 120.8 (4) |
| H2A—C2—H2B | 108.2 | C25—C20—S2 | 120.0 (4) |
| C4—C3—N2 | 111.6 (3) | C20—C21—C22 | 120.4 (5) |
| C4—C3—H3A | 109.3 | C20—C21—H21 | 119.8 |
| N2—C3—H3A | 109.3 | C22—C21—H21 | 119.8 |
| C4—C3—H3B | 109.3 | C21—C22—C23 | 120.5 (5) |
| N2—C3—H3B | 109.3 | C21—C22—H22 | 119.7 |
| H3A—C3—H3B | 108.0 | C23—C22—H22 | 119.7 |
| C5—C4—C9 | 118.8 (4) | C24—C23—C22 | 118.4 (5) |
| C5—C4—C3 | 120.4 (4) | C24—C23—C26 | 122.0 (5) |
| C9—C4—C3 | 120.7 (4) | C22—C23—C26 | 119.6 (5) |
| C4—C5—C6 | 120.6 (4) | C23—C24—C25 | 121.5 (5) |
| C4—C5—H5 | 119.7 | C23—C24—H24 | 119.2 |
| C6—C5—H5 | 119.7 | C25—C24—H24 | 119.2 |
| C5—C6—C7 | 120.9 (4) | C24—C25—C20 | 120.0 (5) |
| C5—C6—H6 | 119.6 | C24—C25—H25 | 120.0 |
| C7—C6—H6 | 119.6 | C20—C25—H25 | 120.0 |
| C8—C7—C6 | 119.0 (4) | C23—C26—H26A | 109.5 |
| C8—C7—C10i | 120.6 (4) | C23—C26—H26B | 109.5 |
| C6—C7—C10i | 120.3 (4) | H26A—C26—H26B | 109.5 |
| C7—C8—C9 | 119.4 (4) | C23—C26—H26C | 109.5 |
| C7—C8—H8 | 120.3 | H26A—C26—H26C | 109.5 |
| C9—C8—H8 | 120.3 | H26B—C26—H26C | 109.5 |
| C4—C9—C8 | 121.2 (4) | O8—S3—O7 | 114.2 (2) |
| C4—C9—H9 | 119.4 | O8—S3—O9 | 110.6 (2) |
| C8—C9—H9 | 119.4 | O7—S3—O9 | 111.7 (2) |
| N3—C10—C7i | 111.1 (4) | O8—S3—C27 | 107.7 (2) |
| N3—C10—H10A | 109.4 | O7—S3—C27 | 105.5 (2) |
| C7i—C10—H10A | 109.4 | O9—S3—C27 | 106.6 (2) |
| N3—C10—H10B | 109.4 | C32—C27—C28 | 120.0 (5) |
| C7i—C10—H10B | 109.4 | C32—C27—S3 | 118.6 (4) |
| H10A—C10—H10B | 108.0 | C28—C27—S3 | 121.3 (4) |
| N3—C11—C12 | 109.2 (3) | C29—C28—C27 | 118.7 (5) |
| N3—C11—H11A | 109.8 | C29—C28—H28 | 120.7 |
| C12—C11—H11A | 109.8 | C27—C28—H28 | 120.7 |
| N3—C11—H11B | 109.8 | C30—C29—C28 | 122.4 (5) |
| C12—C11—H11B | 109.8 | C30—C29—H29 | 118.8 |
| H11A—C11—H11B | 108.3 | C28—C29—H29 | 118.8 |
| N1—C12—C11 | 110.5 (4) | C29—C30—C31 | 117.3 (5) |
| N1—C12—H12A | 109.6 | C29—C30—C33 | 121.5 (6) |
| C11—C12—H12A | 109.6 | C31—C30—C33 | 121.2 (6) |
| N1—C12—H12B | 109.6 | C32—C31—C30 | 121.4 (6) |
| C11—C12—H12B | 109.6 | C32—C31—H31 | 119.3 |
| H12A—C12—H12B | 108.1 | C30—C31—H31 | 119.3 |
| O2—S1—O3 | 112.8 (2) | C31—C32—C27 | 120.1 (5) |
| O2—S1—O1 | 110.85 (18) | C31—C32—H32 | 119.9 |
| O3—S1—O1 | 111.88 (19) | C27—C32—H32 | 119.9 |
| O2—S1—C13 | 107.44 (19) | C30—C33—H33A | 109.5 |
| O3—S1—C13 | 106.6 (2) | C30—C33—H33B | 109.5 |
| O1—S1—C13 | 106.9 (2) | H33A—C33—H33B | 109.5 |
| C18—C13—C14 | 119.4 (5) | C30—C33—H33C | 109.5 |
| C18—C13—S1 | 121.5 (4) | H33A—C33—H33C | 109.5 |
| C14—C13—S1 | 119.1 (4) | H33B—C33—H33C | 109.5 |
| C12—N1—C1—C2 | −169.6 (4) | C14—C13—C18—C17 | −1.5 (7) |
| C3—N2—C2—C1 | −172.3 (4) | S1—C13—C18—C17 | −178.9 (3) |
| N1—C1—C2—N2 | −172.0 (4) | O6—S2—C20—C21 | 7.7 (4) |
| C2—N2—C3—C4 | −51.8 (5) | O5—S2—C20—C21 | 129.6 (4) |
| N2—C3—C4—C5 | −57.0 (6) | O4—S2—C20—C21 | −113.4 (4) |
| N2—C3—C4—C9 | 119.2 (4) | O6—S2—C20—C25 | −172.8 (4) |
| C9—C4—C5—C6 | −1.2 (7) | O5—S2—C20—C25 | −50.8 (4) |
| C3—C4—C5—C6 | 175.1 (4) | O4—S2—C20—C25 | 66.2 (4) |
| C4—C5—C6—C7 | 1.1 (7) | C25—C20—C21—C22 | 0.7 (8) |
| C5—C6—C7—C8 | 0.7 (7) | S2—C20—C21—C22 | −179.7 (4) |
| C5—C6—C7—C10i | −178.6 (4) | C20—C21—C22—C23 | −1.0 (8) |
| C6—C7—C8—C9 | −2.4 (7) | C21—C22—C23—C24 | 0.5 (8) |
| C10i—C7—C8—C9 | 176.9 (4) | C21—C22—C23—C26 | 179.9 (5) |
| C5—C4—C9—C8 | −0.5 (7) | C22—C23—C24—C25 | 0.2 (8) |
| C3—C4—C9—C8 | −176.8 (4) | C26—C23—C24—C25 | −179.2 (5) |
| C7—C8—C9—C4 | 2.3 (7) | C23—C24—C25—C20 | −0.4 (8) |
| C11—N3—C10—C7i | 51.9 (5) | C21—C20—C25—C24 | −0.1 (7) |
| C10—N3—C11—C12 | 177.8 (4) | S2—C20—C25—C24 | −179.6 (4) |
| C1—N1—C12—C11 | 165.6 (4) | O8—S3—C27—C32 | 84.8 (5) |
| N3—C11—C12—N1 | 170.9 (4) | O7—S3—C27—C32 | −152.9 (4) |
| O2—S1—C13—C18 | −119.5 (4) | O9—S3—C27—C32 | −33.9 (5) |
| O3—S1—C13—C18 | 119.3 (4) | O8—S3—C27—C28 | −95.0 (4) |
| O1—S1—C13—C18 | −0.5 (4) | O7—S3—C27—C28 | 27.4 (5) |
| O2—S1—C13—C14 | 63.1 (4) | O9—S3—C27—C28 | 146.3 (4) |
| O3—S1—C13—C14 | −58.1 (4) | C32—C27—C28—C29 | 1.0 (7) |
| O1—S1—C13—C14 | −177.8 (3) | S3—C27—C28—C29 | −179.3 (4) |
| C18—C13—C14—C15 | 1.0 (7) | C27—C28—C29—C30 | −0.8 (8) |
| S1—C13—C14—C15 | 178.4 (3) | C28—C29—C30—C31 | 1.2 (9) |
| C13—C14—C15—C16 | 0.1 (7) | C28—C29—C30—C33 | 178.5 (6) |
| C14—C15—C16—C17 | −0.6 (7) | C29—C30—C31—C32 | −1.7 (10) |
| C14—C15—C16—C19 | −179.4 (4) | C33—C30—C31—C32 | −179.1 (6) |
| C15—C16—C17—C18 | 0.1 (7) | C30—C31—C32—C27 | 2.0 (9) |
| C19—C16—C17—C18 | 178.9 (4) | C28—C27—C32—C31 | −1.5 (8) |
| C16—C17—C18—C13 | 1.0 (7) | S3—C27—C32—C31 | 178.7 (5) |
Symmetry codes: (i) −x+1, −y+1, −z+1.
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H11N···O7i | 0.92 | 1.86 | 2.738 (5) | 159 |
| N1—H12N···O8 | 0.92 | 2.10 | 2.923 (6) | 148 |
| N1—H12N···O9 | 0.92 | 2.33 | 3.130 (5) | 145 |
| N2—H21N···O5 | 0.92 | 1.85 | 2.745 (5) | 164 |
| N2—H22N···O1ii | 0.92 | 1.83 | 2.706 (4) | 159 |
| N3—H31N···O4ii | 0.92 | 1.84 | 2.748 (4) | 168 |
| N3—H32N···O2 | 0.92 | 1.92 | 2.842 (5) | 177 |
| N3—H32N···O1 | 0.92 | 2.58 | 3.081 (4) | 115 |
Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) −x+2, −y+1, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: CI2898).
References
- Altomare, A., Cascarano, G., Giacovazzo, C., Guagliardi, A., Burla, M. C., Polidori, G. & Camalli, M. (1994). J. Appl. Cryst.27, 435.
- Bazzicalupi, A., Bencini, A., Bianchi, A., Fusi, V., Giorgi, C., Paoletti, P., Stefani, A. & Valtancoli, B. (1995). J. Chem. Soc. Perkin Trans. 2, pp. 275–280.
- Bianchi, A., García-España, E. & Bowman-James, K. (1997). Supramolecular Chemistry of Anions New York: Wiley-VCH.
- Chen, D. & Martell, A. E. (1991). Tetrahedron, 47, 6895–6902.
- Clifford, T., Danby, A., Llinares, J. M., Mason, S., Alcock, N. W., Powell, D., Aguilar, J. A., García-España, E. & Bowman-James, K. (2001). Inorg. Chem.40, 4710–4720. [DOI] [PubMed]
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- He, W.-J., Ye, Z.-F., Xu, Y., Guo, Z.-J. & Zhu, L.-G. (2000). Acta Cryst. C56, 1019–1020. [DOI] [PubMed]
- Hossain, M. A. (2008). Curr. Org. Chem.12, 1231–1256.
- Li, T., Lin, H., Li, T., He, W., Li, Y., Yu Zhang, Y., Zhu, Y. & Zijian Guo, Z. (2009). Inorg. Chim. Acta, 362, 967–974.
- Liu, H.-Y., Wei, G.-H. & Ma, J.-F. (2008). Acta Cryst. E64, o126.
- Llobet, A., Reibenspies, J. & Martell, A. E. (1994). Inorg. Chem.33, 5946–5951.
- Lu, Q., Motekaitis, R. J., Reibenspies, J. & Martell, A. E. (1995). Inorg. Chem.34, 4958–4964.
- Lu, Q., Riebenspies, J. H., Caroll, R. I., Martell, A. E. & Clearfield, A. (1998). Inorg. Chim. Acta, 270, 207–215.
- Nagarajan, S. B. & Ganem, B. (1987). J. Org. Chem.52, 5044–5046.
- Nonius (1999). COLLECT. Nonius BV, Delft, The Netherlands.
- Otwinowski, Z. & Minor, W. (1997). Methods in Enzymology, Vol. 276, Macromolecular Crystallography, Part A, edited by C. W. Carter Jr & R. M. Sweet, pp. 307–326. New York: Academic Press.
- Ragunathan, K. G. & Schneider, H.-S. (1996). Angew. Chem. Int. Ed. Engl.35, 1219–1221.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Zhu, L.-G., Ellern, A. M. & Kostić, N. M. (2002). Acta Cryst. C58, o129–o130. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809035648/ci2898sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809035648/ci2898Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report