Abstract
Many fundamental questions remain regarding the cellular and molecular mechanisms of digestive lipid metabolism. One major impediment to answering important questions in the field has been the lack of a tractable and sufficiently complex model system. Until recently, most studies of lipid metabolism have been performed in vitro or in mice, yet each approach possesses certain limitations. The zebrafish (Danio rerio) offers an excellent model system in which to study lipid metabolism in vivo, owing to its small size, genetic tractability and optical clarity. Fluorescent lipid dyes and optical reporters of lipid-modifying enzymes are now being used in live zebrafish to generate visible readouts of digestive physiology. Here we review recent advances in visualizing intestinal lipid metabolism in live larval zebrafish.
Keywords: BODIPY, fatty acids, genetics, intestine, zebrafish
Given the numerous roles lipids play in cellular function, it is not surprising that defects in lipid metabolism underlie many human diseases [1–3]. More than a third of adults and 17% of children are currently classified as obese in the USA [4,5], with obesity and Type 2 diabetes on the rise in developing countries [6]. The prevalence of lipid disorders has generated a great need to advance our understanding of lipid metabolism as it pertains to the development of effective clinical treatments for these conditions. Only a handful of pharmacological drugs targeting intestinal lipid absorption have been approved by the US FDA in recent years, the most prominent of which are the pancreatic lipase inhibitor orlistat (Alli®, GlaxoSmithKline) and the cholesterol absorption inhibitor ezetimibe (Zetia®, Merck/Schering Plough). The development of effective therapeutics for lipid disorders is hindered by gaps in our understanding of the molecular mechanisms underlying lipid transport and processing. A detailed cell biological model of intestinal lipid absorption is therefore needed to identify key molecular targets for therapeutic development.
It has become increasingly apparent that studies of lipid metabolism are more powerful when performed in vivo. Although in vitro studies have laid much of the groundwork for our biochemical understanding of lipid metabolism, they cannot recreate the complex interplay of neural, chemical and hormonal cues known to regulate metabolic processes in vivo. The observation that ezetimibe’s potency as an inhibitor of cholesterol absorption greatly increases (400 times) after being metabolized by the liver, highlights the importance of utilizing whole animal models for initial drug discovery [7]. To this end, recent studies utilizing larval zebrafish employ various strategies for visualizing lipid metabolism and demonstrate the ability of in vivo based approaches to elucidate lipid metabolic pathways and screen for new therapeutics.
Lipophilic dyes and fluorescent lipid analogs are used in forward genetic screens to identify or characterize lipid-related genes. Optical reporters sensitive to cleavage by either lipases or proteases serve as readouts of enzymatic function in digestive organs and are used to identify mutants that would not have been detected by morphological screening criteria [8,9]. Fluorescent microspheres assess swallowing abilities, intestinal lumen integrity and peristalsis [10]. In total, these methods enable the visualization of lipid metabolism in vivo in a relatively easy, cost-effective way and are amenable to use in genetic and pharmacological screens to identity new lipid metabolism genes and pharmaceutical targets.
Importance of in vivo studies of lipid metabolism
Many studies of lipid metabolism were initially only tractable in vitro. Numerous laboratories utilized cultured cells to determine the basic steps of lipid metabolism, identifying and characterizing the proteins that activate, transport and β-oxidize fatty acids [11]. Although a fair number of experimental questions are best addressed initially in cultured cells (e.g., the enzymology of a lipase family), such an approach has limitations. Typically, cultured cell lines exhibit characteristics of less differentiated, more cancer-like cells. The Caco-2 cell line is a commonly used intestinal-like cultured cell; however, it is derived from colorectal adenocarcinoma cells and has characteristics of transformed cells. Villus structure is highly variable [12] and they possess altered levels of enzymes involved in glycerolipid synthesis when compared with intestinal enterocytes [13]. There is no currently available cell line that adequately captures all the cellular features of an intestinal enterocyte.
The most insurmountable limitation of many cultured cell models is that they are predominantly comprised of a single cell type and cannot replicate the complex environment of a multicellular organ. The intestine is composed of stem, enteroendocrine, immune and goblet cells. Furthermore, the intestine contains symbiotic organisms, bile and mucus that have been shown to influence lipid processing [14–19], yet these components are usually absent in a cultured cell environment. Owing to these shortcomings, cultured cell studies are unable to answer many long-standing questions in the lipid field, such as how a polarized intestinal epithelial cell absorbs lipid and transfers it to lipoproteins that are released at the basal cell surface. Whole-animal approaches using the mouse and zebrafish have been developed to address many of the limitations of cultured cells.
Mouse models have traditionally been the first choice of many researchers seeking a whole-animal system to study lipid metabolism. The genetic and anatomical similarities between mouse and human, combined with robust gene-targeting technology, have allowed human lipid disorders to be recreated and studied in the mouse model. Mouse models of lipid disorders include familial combined hyperlipidemia, a common inherited lipid disorder wherein individuals develop premature coronary heart disease (reviewed in [20]) [21], fatty liver disease (reviewed in [22]) and obesity (reviewed in [23]). Lipid studies undertaken in the mouse have also led to the verification of key components of lipid metabolic pathways.
As with any model organism, there are disadvantages to working with mice. To determine the lipid content of the digestive organs, animals must be sacrificed and internal organs surgically isolated. Additionally, their small litters, relatively large body size, and significant husbandry costs make mice more suited for reverse genetic studies wherein one knows what gene to disrupt. While forward genetic screens, where the entire genome is systematically mutated to assign gene functions, are possible in mice, they would cover only a limited portion of the genome and require exorbitant time and resources.
Advantages of the zebrafish model
Initially used for developmental and embryological research, the zebrafish has proven to be a powerful model organism for the study of vertebrate physiology and disease. A large body of evidence now demonstrates that zebrafish can serve as a ‘canonical vertebrate’ by using forward genetic approaches to assign functions to human genes [24]. Zebrafish are amenable to forward genetic studies because many genomes can economically fit into a relatively small space (~US $2 a fish/year at 30 fish/square foot vs $90 a mouse/year at 5 mice/square foot), enabling faster screening and mutant identification. Such screens have demonstrated that zebrafish carrying a mutation in a human disease gene often manifest a phenotype strikingly similar to those observed in humans (only a small sampling from the last few years is cited here [25–43]). For example, zebrafish homozygous for a mutation in the S-adenosylhomocysteine hydrolase (ahcy) gene develop hepatic steatosis and liver degeneration, similar to humans carrying heterozygous mutations in AHCY [44]. Other zebrafish models of human diseases include cardiovascular disease (reviewed in [45]), fatty liver disease [46], Duchenne muscular dystrophy [47], various cancers [48], blood disorders [49] and obesity [50]. The acceptance of zebrafish as disease models was further bolstered by the sequencing of the zebrafish genome, which revealed a high degree of genetic similarity to humans. Over 80% of the zebrafish genome is similar to those of mice and humans, with many of the biological systems and molecular pathways present in zebrafish closely resembling our own [51,52]. Additionally, most human genes have zebrafish orthologs organized into clusters that are syntenic with corresponding regions of the mammalian genome [53–55].
The reproductive and physical traits of zebrafish larvae are also favorable for phenotypic assessment of mutants and genetic screening. Many internal organs can be directly observed in the optically clear larvae without the need for surgery, and zebrafish fecundity (often more than 300 embryos/cross) guarantees an ample supply of material. In sum, their genetic similarity to humans, small size, rapid development and optical clarity make these vertebrates ideal for identifying genes involved in lipid metabolism and assaying for small molecules to treat lipid disorders [36].
Lipid metabolism is conserved among vertebrates
Zebrafish possess the same gastrointestinal organs as humans. The liver, intestine, exocrine and endocrine pancreas, and gallbladder are all present in the zebrafish [56]. The formation of these digestive organs and those of higher vertebrates is controlled by similar developmental programs to mammals [16,57–59]. The cellular composition of zebrafish digestive organs is also similar to those of mammals. The zebrafish intestinal epithelium is comprised of absorptive cells (enterocytes) (Figure 1), endocrine cells and goblet cells [16]. Zebrafish liver hepatocytes resemble what is observed in a mammalian liver, although there are some architectural differences [16]. Although the pancreas appears to be the least morphologically similar organ, it is made up of the same general array of pancreatic cell types found in humans: exocrine cells that secrete lipases and peptidases, the Islets of Langerhans and the acinar cells that secrete insulin and carboxypetidase A [16,61]. Zebrafish and other teleosts store fat in the liver as well as in adipocytes located primarily in intramuscular and subcutaneous tissues. Zebrafish develop adipocytes near the larval pancreas shortly after their yolk mass is depleted that ultimately distribute throughout the viscera as they mature [60]. The diverse cell types of the zebrafish digestive organs are largely visible in the optically clear larvae, facilitating their identification and phenotypic assessment. The conserved anatomical and cellular digestive structures, as well as the ability to easily observe them, make zebrafish particularly suitable for the study of human lipid metabolism.
Figure 1. Larval zebrafish digestive system anatomy.
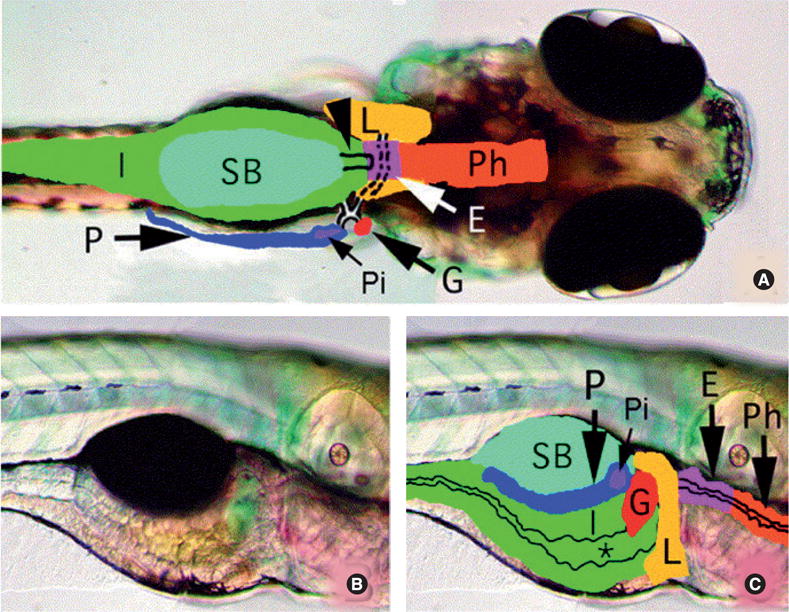
(A) Dorsal view of larval digestive organs (5 days postfertilization). Broken and solid lines outline the ducts of the liver, pancreas and gallbladder. Arrowhead indicates the pneumatic swimbladder duct. (B) and (C) Lateral views of the larval zebrafish (5 days postfertilization) digestive system. Intestinal lumen (*) is marked.
E: Esophagus; G: Gallbladder; I: Intestine; L: Liver; P: Pancreas; Ph: Pharynx; Pi: With solitary islet; SB: Swimbladder.
Reprinted from [55] with permission from Elsevier.
Lipid metabolism in zebrafish is similar to that of humans at the biochemical level as well. Zebrafish consume the same dietary lipids as most mammals and utilize analogous transport and lipolysis pathways, although there are some differences in both absorption and deposition [62]. Zebrafish and humans employ similar lipid signaling mechanisms, as evidenced by conserved prostanoid synthesis pathways, in which a high degree of homology is seen when comparing cyclooxygenases, which synthesize prostaglandins [63]. The high degree of genetic and functional homology between the zebrafish and mammalian metabolic pathways further validates zebrafish as an appropriate model system for studying lipid metabolism.
Lipid metabolism in developing zebrafish
During the first few days of development, a zebrafish embryo relies entirely on its finite yolk sac for the continuous supply of lipids needed to sustain its growth. Yolk lipids are the source of essential fat-soluble vitamins and cholesterol, a required component of cell membranes and a precursor for bile acids [64–66]. Lipids enter the developing embryo at the yolk–embryo interface, an area termed the yolk syncytial layer (YSL). In the YSL, lipoproteins (e.g., ApoE, ApoAI, ApoC-II and vitellogenin) and a host of lipid-modifying enzymes (e.g., microsomal trigylceride-transfer protein [mtp]) transport lipids from the yolk to the embryo [64,67]. Once the circulatory system forms, yolk, hepatic and intestinal lipids are transported by lipoproteins to specific target tissues throughout the organism via the bloodstream.
By 5–6 days postfertilization (dpf), the yolk has been depleted and larvae must now eat to acquire lipids. Both in the wild and the laboratory, zebrafish consume a lipid-rich diet (at least 10% by weight), high in triacylglycerol (TAG), phospholipids, fatty acids and sterols [68,69]. After entering the zebrafish intestine, TAG, cholesterol ester (CE) and phospholipids must be broken down by luminal lipases into free fatty acids and cholesterol before entering the specialized absorptive cells (enterocytes) that line the gut [70]. The main source of intestinal lipases in the zebrafish appears to be the exocrine pancreas [9], which is known in mammals to secrete lipase- and protease-rich pancreatic juice into the intestine to aid in digestion [71]. Zebrafish intestinal enterocytes are polarized epithelial cells that have an apical brush border membrane. The protruding microvilli of this membrane create a large absorptive surface and are highly similar to those of mammalian intestinal cells (Figure 2).
Figure 2. Zebrafish enterocytes exhibit similar cell morphology and organelle composition to mammalian enterocytes.
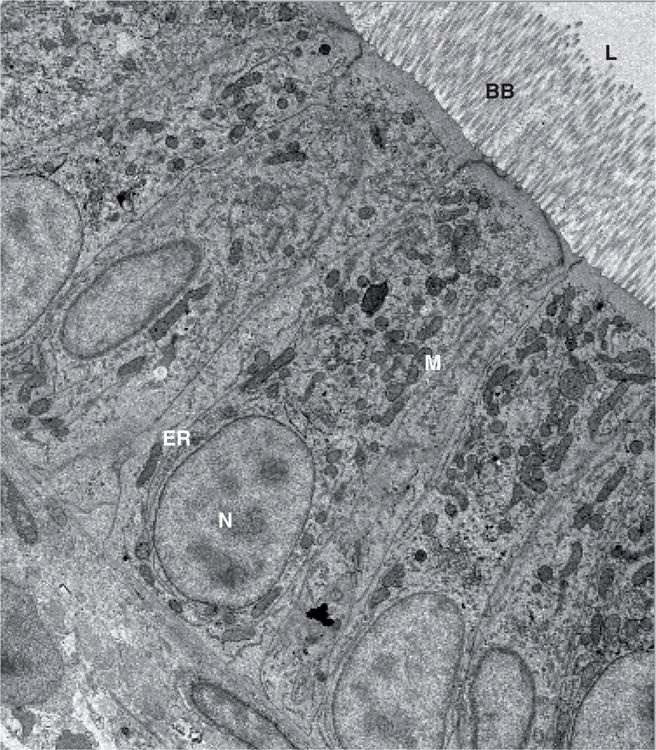
Subcellular structures of the enterocytes are labeled as endoplasmic reticulum (ER), brush border (BB), mitochondria (M) and nucleus (N). The intestinal lumen (L) is also shown.
Figure obtained from J Walters, Unpublished Data.
Following a meal, zebrafish accumulate lipid drops (LD) in the enterocytes of their proximal and medial intestine [67]. Owing to the appearance of LDs in many cell types and their relevance to human disease, techniques for visualizing these drops are important to develop for use in zebrafish. Previously, LDs were regarded solely as energy storage vesicles, a place where the cell stockpiled extra lipid it could not immediately transport. Recent studies now suggest that LDs are specialized organelles with multiple cellular functions and highly dynamic activities [72]. LDs are thought to form in the endoplasmic reticulum (ER) since they often appear closely associated with its cytoplasmic face [73–76]. The core of these drops is comprised predominantly of TAG and sterol esters, with diacylglycerol and fatty acid present in lesser amounts [77]. Surrounding this core assemblage of lipids is a phospholipid monolayer mediating interactions between the drop and droplet-binding proteins (perilipin, adipophilin and tail-interacting protein of 47kDa [TIP47]) [72]. LD-associated proteins are thought to help confer stability through structural reinforcement, serve as docking sites for trafficking proteins and regulate the size and cargo contents of the LDs [78]. It has been proposed that excessive LD accumulation is a result of the inability of lipoprotein-mediated secretion at the basal cell surface to keep up with the influx of neutral lipid at the brush border [67]. Furthermore, human patients with lipid-related malabsorption syndromes often exhibit excessive LD accumulation (steatosis) in their intestinal biopsies [79]. Thus, the appearance of LDs in intestinal enterocytes provides a basic, identifiable readout of lipid metabolism with relevance to human disease. For these reasons, various techniques for visualizing these drops in fixed tissue and live cells were developed and continue to be improved upon in the zebrafish for use in live animal studies of lipid metabolism.
Techniques for visualizing lipid metabolism: lipophilic dyes
In order to fully exploit the zebrafish model system for studies of lipid metabolism, a variety of lipid dyes, fluorescent lipid analogs and optical reporters have been developed. Lipophilic dyes, known as lysochromes, were one of the first methods used to visualize lipid. These dyes are capable of labeling a variety of lipids and lipid-containing structures including triglycerides, fatty acids and lipoproteins. Dyes, such as oil red O (ORO), Sudan black B and Nile red, were initially used to label LDs in tissue sections and cultured cells and continue to be used today. Marza and colleagues [67] recently utilized Sudan black B to identify LDs in histological sections of fed adult zebrafish (Figure 3). The authors found that the feeding of a high-fat meal increased expression of microsomal mtp in intestinal epithelial cells. This protein is required for proper assembly and secretion of liver and intestine ApoB-containing lipoproteins, chylomicrons and VLDLs [80]. Their observation that LDs are coincident with an upregulation of mtp expression is consistent with MTP’s known function in humans [67].
Figure 3. Histological semi-thin section of the anterior intestine of a fed adult zebrafish stained with Sudan black B.
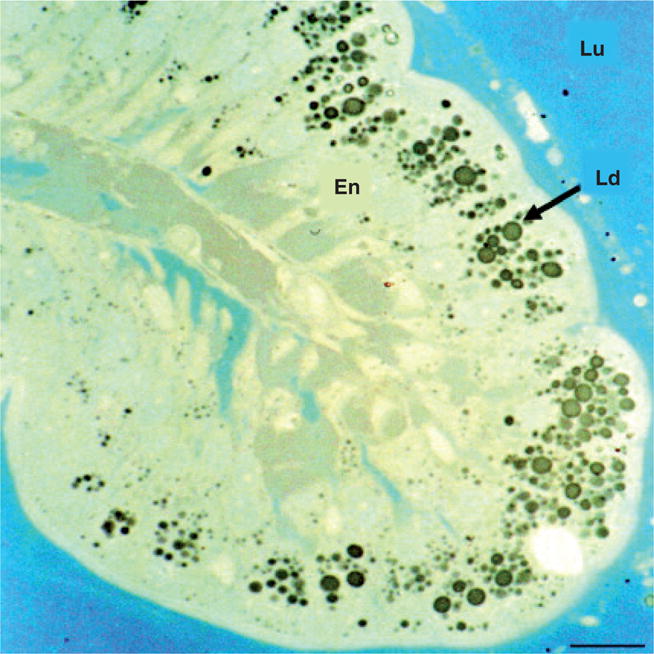
Large Ld are clearly observed in the En. The intestinal Lu is indicated. Scale bar is 50 μm.
En: Enterocytes; Ld: Lipid drops; Lu: Lumen.
Reprinted from [67] with permission of John Wiley & Sons Inc.
Lysochromes can also be used to visualize endogenous lipid stores in whole fixed zebrafish to generate an overall picture of neutral lipid dynamics during development. Schlegel et al. used ORO to assess the consequence of mtp knockdown, via targeted antisense morpholino (MO), on lipid absorption in whole zebrafish larvae [81]. mtp morphants exhibited decreased yolk consumption and an inability to absorb dietary neutral lipids, resulting in death by 6 dpf. Although lysochromes such as Sudan black B and ORO consistently label neutral lipids in tissue sections and fixed larvae, fixation techniques are laborious and staining procedures have been shown to cause artificial fusion of adjacent LDs and mislocalization of the LD marker, adipose differentiation-related protein (Adrp) [82]. More recent techniques to visualize LDs have focused on staining these drops in vivo.
Greenspan et al. first showed the utility of Nile red (9-diethylamino-5H-benzo[α] phenoxazine-5-one) to label intracellular lipid droplets in live cultured peritoneal macrophages and smooth muscle cells [83]. Nile red is an uncharged heterocyclic molecule that only fluoresces in a hydrophobic environment. Labeled neutral lipids fluoresce a yellow-gold to red color, depending on their relative hydrophobicity, with no detectable damage or deformation of dye-infused tissues [84]. More recently, Jones et al. used Nile red to visualize neutral lipid deposits in live zebrafish larvae. They initially demonstrated that daily exposure of larvae to Nile red-containing embryo media for 4 days (from 3 dpf to 7 dpf) consistently labeled lipid-rich tissues (Figure 4). The authors then sought to test the effects of known pharmacological inhibitors of triglyceride metabolism on total larval lipid content. Treatment with nicotinic acid, a potent pharmacological inhibitor of adipocyte lipolysis [85], resulted in an increase in total triglyceride content and decreased cholesterol levels. Treatment with resveratrol, a compound known to inhibit fatty acid synthase [18], resulted in a decrease in total triglyceride content as detected by Nile red staining. Total triglyceride content was further decreased when resveratrol was supplemented with norepinephrine [86]. The ability of the zebrafish to respond to small molecules in a similar fashion as humans validates its use for drug discovery.
Figure 4. Nile red staining visualizes deep tissue fat deposits in larval zebrafish.
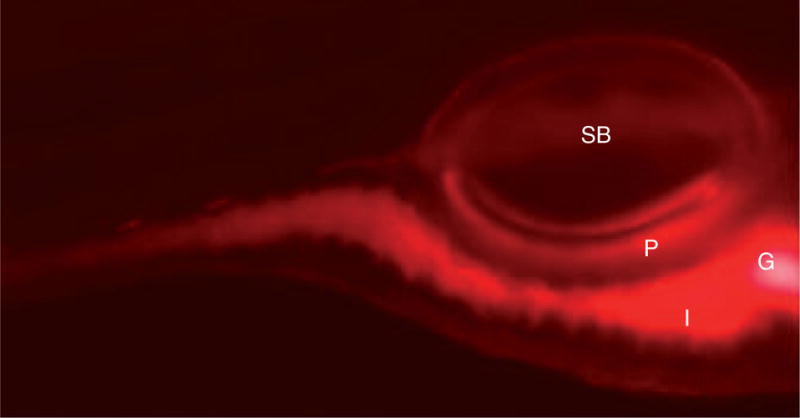
Extended incubation of larvae for at least 4 h in nile red (5 ng/ml) labels fat deposits. Nile red staining is present in the intestines (I), gall bladder (G) and pancreas (P). The swim bladder (SB) is also indicated.
Reprinted with permission from [87].
While fluorescent dyes and stains are useful for identifying lipid deposits in cells and tissues, issues arise regarding the distribution and affinity properties of these compounds. Nonspecific labeling of tissues devoid of lipid deposits may be observed and staining and destaining/washing procedures must then be carefully optimized to minimize this effect. Additionally, Nile red staining does not distinguish between fatty acids and cholesterol in vivo, although some discrimination based on staining intensity of tissue sections is possible [84]. Optical tools are now available that allow different lipids and lipid processing to be visualized in vivo.
Fluorescent lipids: BODIPY fluorophores
A common technique for visualizing lipid dynamics in living tissues is direct labeling of fatty acids or cholesterol with the borondipyrromethene (BODIPY) fluorescent moiety (4,4-difluoro-4-bora-3a, 4a-diaza-S-indacene). The labeling of fatty acids with BODIPY allows a number of lipids to be tracked, facilitating studies of intracellular lipid trafficking. BODIPY was initially used in cultured cell studies similar to the lysochrome dyes and is still used in current studies of fatty acid uptake [87]. First synthesized by Treibs and Kreuzer in 1968 [88], the BODIPY fluorophore possesses a number of advantageous qualities, including high photostability, strong and narrow wavelength emission in the visible spectrum and an overall uncharged state. Only recently has BODIPY gained recognition for its versatile applications in biological research [89].
Fluorescent phospholipase reporter (PED6) & NBD-cholesterol
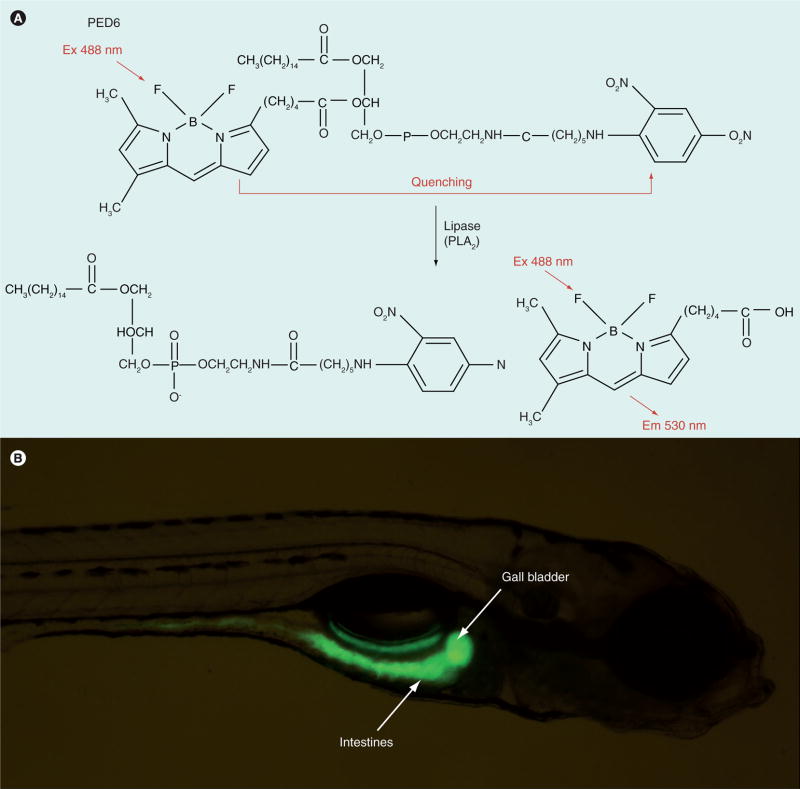
Fluorescent lipid reporters enable the direct observation of lipid metabolism in live zebrafish larvae by generating a visible readout of lipid processing. The reporters are designed such that cleavage by lipid-modifying enzymes results in altered spectral characteristics [90,91]. One reporter, N-([6-(2,4-dinitro-phenyl) amino]hexanoyl)-1-palmitoyl-2-BODIPY-FL-pentanoyl-sn-glycerol-3-phosphoethanolamine (PED6), is built from a lysophospholipid covalently linked to a BODIPY fatty acid at the sn2 position and a fluorescence quencher on the head group (Figure 5A). Cleavage of PED6 by phospholipase A2 (PLA2) releases the fluorescent BODIPY-labeled acyl-chain from the quencher, resulting in detectable green fluorescence at 488 nm [90]. PED6 therefore acts as a biosensor of lipase activity in live zebrafish.
Figure 5. PED6 is a biosensor of lipase activity in live larval zebrafish.
(A) PED6. Cleavage of PED6 by phospholipase A2 releases the quenched fluorescent BODIPY labeled acyl-chain, resulting in detectable green fluorescence at 488 nm. (B) Larvae (5 days postfertilization) soaked in PED6 (0.3 mg/ml) for 6 h exhibit fluorescent gall bladders and intestines.
When larvae (6 dpf) are bathed in PED6, bright green staining is observed in the intestine, gall bladder and liver (Figure 5B) [8]. This indicator of lipase activity enabled the first high-throughput forward genetic physiologic screen in zebrafish to be conducted, with the goal of identifying new genes that regulate lipid metabolism. Fat-free (ffr) was the first gene identified using the PED6 screening assay [8] and has been found to regulate a number of cellular processes, including golgi structure/maintenance, protein sorting, vesicle trafficking and intestinal lipid absorption and processing [92]. It is important to note that immediately after identifying the molecular nature of the zebrafish ffr mutation, the human ortholog was located by BLAST searches of public databases. While the human gene was present in these databases, there was no data on its possible function. This example demonstrates the power of forward genetic methods in zebrafish to assign functions to mammalian genes. Other reports of forward genetic screens using PED6 demonstrate the utility of this and similar synthetic analogs to identify genes involved in digestive processes. Wantanabe et al. used PED6 to identify genes that regulate digestive organ morphology and bile synthesis and secretion in Medaka fish [93].
ffr larvae were further characterized using an additional synthetic analog, 22-(N-[7-nitrobenz-2-oxa-1,3-dia-zol-4-yl] amino)-23,24-bisnor-5-cholen-3-ol (NBD)-cholesterol, to study the trafficking of sterol-like molecules in vivo. This reagent is different from PED6 in that it continuously fluoresces and, owing to its hydrophobicity, it is slightly more difficult to administer via feeding. ffr mutants were unable to concentrate NBD-cholesterol in their gall bladders, in contrast to wild-type larvae, which exhibited rapid accumulation of this lipid in their gall bladders within a few hours of feeding. This observation suggests ffr larvae have a serious defect in chylomicron formation and/or transport.
Fluorescent microspheres
Microspheres were first utilized to test swallowing ability in zebrafish by Farber et al. (Figure 6) [8], and have since been used in a number of developmental and toxicity studies. In one such investigation, Bolcome et al. were able to assess the structural integrity and permeability of zebrafish blood vessels following exposure to anthrax toxin, known to cause vascular dysfunction in human cases of anthrax exposure [94]. More recently, Field et al. utilized microspheres to visualize intestinal transit during peristaltic contractions in larval zebrafish [10]. The authors tested whether intestinal transit was defective in larvae with lower levels of or completely lacking the Ret protein. Ret is a receptor tyrosine kinase known to localize migrating neural crest cells to the gut where they eventually differentiate into enteric neurons. Using MO knockdown of ret transcripts, the authors showed that defective intestinal transit significantly correlated with the degree of enteric innervation (Figure 7).
Figure 6. Microspheres reveal normal swallowing activity in the fat-free mutants.
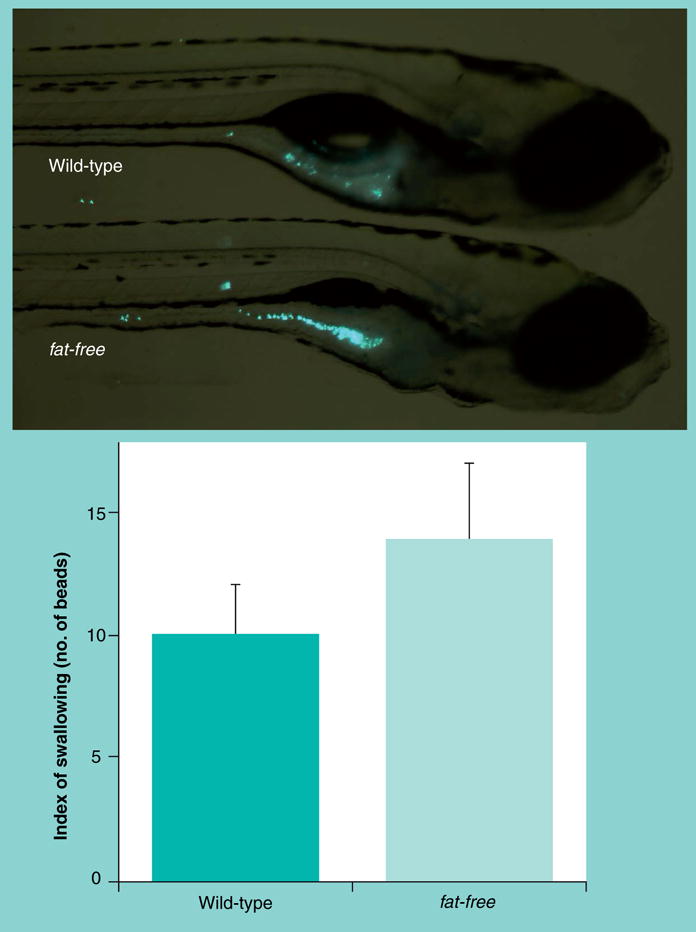
Larvae (6 dpf) were placed in embryo media containing fluorescent latex microspheres (0.0025% Fluoresbrite plain YG 2.0um, Polysciences Inc.) for 1 h, washed, and imaged. Numbers of beads were 10 ± 2 in the wild-type versus 14 ± 3 beads in fat-free mutant larvae (mean ± SEM, n = 9; p > 0.3).
Figure 7. The degree of enteric innervation correlates with intestinal transit.
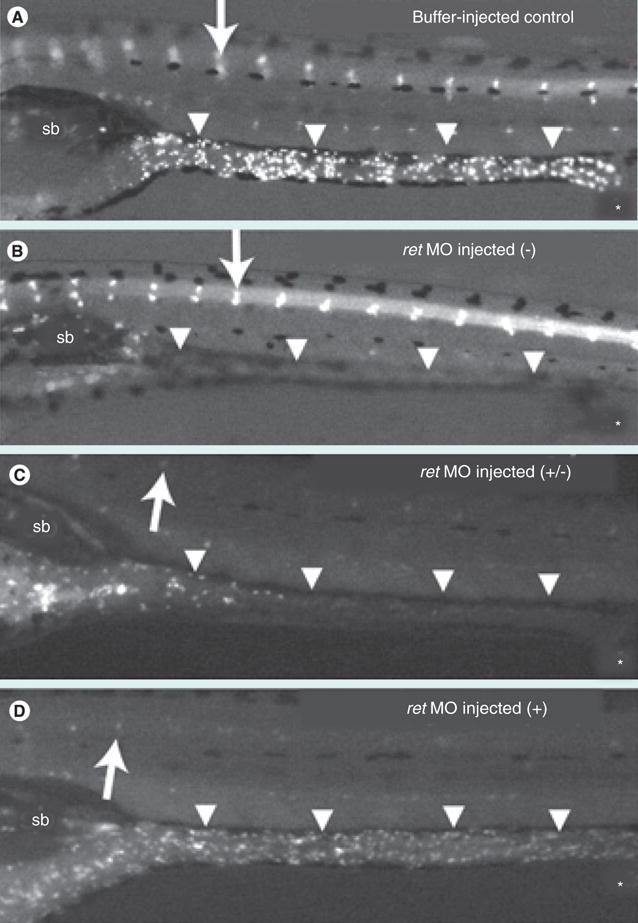
(A–D) Enteric neurons labeled with anti-HuC/D antibody are present in the cranial ganglia (arrow) in both buffer-injected control (A) and ret MO-injected (B–D) larvae. (A) In the buffer-injected control larvae, enteric neurons are present along the length of the GI tract (arrowheads). The majority of these larvae have completed or nearly completed transit after 24 h. (B–D) ret MO-injected larvae display variable degrees of enteric neuron loss along the length of the GI tract (arrowheads): no enteric neurons (−), reduced numbers of enteric neurons (+/−), wild-type enteric neurons (+). (C) Intestinal transit is delayed in ret MO-injected larvae with reduced enteric neurons. (D) ret MO-injected larvae with a normal complement of enteric innervation have a similar transit profile to the control larvae. The sb and anal opening (*) are indicated.
Figure obtained from Field et al. (2009) [100]. Reprinted from [100] with permission of John Wiley & Sons Inc.
MO: Morpholino; Sb: Swim bladder.
Triple screening: PED6, EnzChek & microspheres
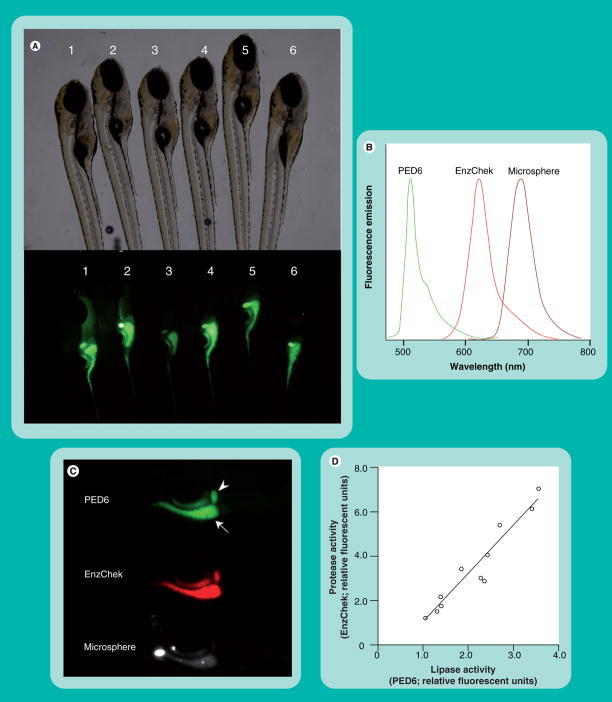
Although PED6 has proven successful in identifying abnormal lipase activity in zebrafish mutants, there can be significant variability in the digestive tract fluorescence observed between wild-type siblings (Figure 8A). Such inconsistent labeling makes the use of PED6 in genetic screens difficult, as an increase or decrease in fluorescence cannot be attributed solely to mutations. Variations in PED6 intensity may reflect interindividual differences in reporter ingestion. To address this issue, it was reasoned that PED6 could be used in conjunction with other reporters of digestive function to create a physiologically relevant readout of in vivo digestive processes.
Figure 8. PED6, EnzChek and microspheres provide a readout of larval digestive function.
(A) Wild-type siblings exhibit variability in PED6 signal. Larvae (6 dpf) were placed in embryo media containing PED6 (0.3 mg/ml) for 3 h, washed and imaged. (B) PED6, EnzChek and microspheres each fluoresce at distinct wavelengths, allowing simultaneous screening for lipase, protease and swallowing activities, respectively. (C) PED6 and EnzChek signal in the gall bladder (arrowhead) and intestine (arrow) of wild-type zebrafish. Microspheres are present in the intestine following feeding and indicate normal swallowing activity. (D) Intestinal protease and phospholipase activities correlate, allowing the ratio of PED6 to EnzChek signal to serve as readout of digestive function.
Reprinted with permission from [9].
Hama et al. recently demonstrated that simultaneous feeding of PED6, the protease reporter EnzChek (Invitrogen Inc.) and nonhydrolyzable microspheres allows one to monitor lipase, protease and swallowing activities in larval zebrafish [9]. The protease reporter EnzChek was previously used to detect the activity of metallo-, serine, acid and thiol proteases in a number of biological systems [95–97, 201]. EnzChek consists of the phosphoprotein casein labeled with multiple red or green BODIPY fluorophores. Proteolytic cleavage of the quenched reporter generates highly fluorescent casein fragments, with total fluorescence proportional to enzyme activity [95]. Unlike PED6 and EnzChek, which report enzymatic function, nonhydrolyzable microspheres assess reporter swallowing and intestinal lumen integrity.
PED6, EnzChek and microspheres each fluoresce at distinct wavelengths, allowing simultaneous viewing of all three signals (Figures 8B & C). Use of the triple screening cocktail revealed a correlation between the intestinal protease and phospholipase activity, consistent with the hypothesis that the variance observed in PED6 fluorescence was partly due to differing amounts of PED6 consumed by each larva (Figure 8D). This work demonstrated that the ratio of PED6 to EnzChek fluorescence can serve as a readout of digestive function, since the variability in the PED6/EnzChek ratio observed in individual larvae is unaffected by differences in reporter ingestion (Figure 8D).
After validating the triple screening method, Hama et al. demonstrated the utility of this assay in evaluating the role of the exocrine pancreas in digestive function [9]. The exocrine pancreas secretes many of the gastric lipases and pro-teases needed for the breakdown and subsequent uptake of nutrients [71]. Previous work has demonstrated that MO knockdown of the pancreas transcription factor 1a (ptf1a) can selectively prevent exocrine pancreas development [61]. Analysis of ptf1a mutants (5 dpf) using the triple screen found that these larvae retain normal levels of lipase activity yet have reduced protease activity. Older larvae (6 dpf) exhibit decreased amounts of both protease and lipase activity, suggesting the exocrine pancreas begins providing gastric lipases at later stages of development.
The regulation of phospholipase and protease activity was also examined using the triple screening method. The peptide hormone cholecystokinin (CCK) facilitates digestion by causing the secretion of gastric enzymes into the small intestine after food consumption [98]. Release of CCK into the circulatory system activates the CCK receptor A (CCK-RA) in the exocrine pancreas. Larvae (5 dpf) treated with CCK-RA antagonist showed a reduction in protease activity but had unaffected lipase activity. Much like the ptf1a mutants, 6 dpf larvae had lower levels of both protease and lipase activity. Not surprisingly, the effect of CCK-RA antagonist was abolished in ptf1a morphants. This work suggests that CCK signaling regulates zebrafish secretion of exocrine pancreas-derived intestinal proteases earlier in development (5 dpf) and phospholipases later (6 dpf).
The triple screen is a versatile tool to visualize digestive function in live larval zebrafish. The reporters’ distinct emission wavelengths allow the simultaneous evaluation of multiple digestive processes. In addition, the intestinal phospholipase and protease activity correlate intraindividually, enabling the correction of significant inter-individual variation in digestive enzyme activity between wild-type larvae. As with any in vivo system, there is inherent variability (developmental timing, intestinal microenvironment), which will result in signal variation. Therefore, caution should be taken to minimize this variability by carefully scoring digestive organ morphology of larvae prior to and after reporter ingestion, discarding sickly larvae that lack swim bladders and using properly staged larvae.
A final cautionary note
It is important to recognize that when performing a forward genetic or small molecule screen to identify proteins that influence lipid metabolism, it is crucial to distinguish between primary and secondary lipid-associated defects. For example, one general problem in using any reagent requiring ingestion is that any perturbation in normal larval development (e.g., lower jaw formation, enteric neuron differentiation, esophagus formation) can result in attenuated fluorescence. Similarly, while an enlarged yolk sac may be indicative of a mutation specifically affecting lipid metabolism, unconsumed yolk is one of several abnormalities (e.g., degeneration of the brain, necrosis, under-development of the jaw, liver and gut, small eyes, enlarged heart cavity) that arise when overall development has gone awry [99]. Furthermore, dead or sick larvae fail to label when swallowing is the delivery method, such as with PED6. Thus, a failure of the gall bladder to become fluorescent may be due to a mutation that specifically alters lipid absorption (e.g., enterocyte chylomicron formation) or could result from a dying embryo with a mutation affecting tRNA synthesis. Therefore, additional secondary assays are required to check for viability and/or swallowing function, if one is to identify genes or small molecules that specifically influence lipid metabolism.
Conclusion
We have reviewed several strategies currently available to visualize lipid metabolism in larval zebrafish, presenting the advantages and limitations particular to each approach. Lysochrome dyes, such as Nile red, have proven beneficial for visualizing general neutral lipid distribution in vitro and in vivo; however, caution must be taken when using fixation techniques that may alter the localization and morphology of lipid-containing structures. Tagging specific lipids with fluorophores allows the tracking of lipids as they are transported and packaged through various steps of lipid metabolism. Fluorescent reporters such as PED6 and EnzChek serve as readouts of phospholipase and protease activity, respectively; in combination, the reporters can help address interindividual variation observed with single fluorescent labeling approaches. Fluorescent microspheres can be used to assess luminal integrity, swallowing activity and even intestinal transit. As seen with the PED6 work, a combined strategy utilizing multiple readouts of physiological processes, rather than one, better reflects the complexity of digestive processes.
Future perspective
Despite the immense potential of zebrafish as a therapeutic screening tool, this model organism is currently underutilized by both the academic and pharmaceutical research communities. The zebrafish has been primarily utilized to study embryologic questions specifically focused on early patterning; however, more academic researchers are discovering the system for studies of physiology and disease. This trend is likely to continue, with increasing numbers of groups exploiting the tractable genetics of the larval zebrafish to discover new genes that regulate important physiological processes. These efforts are bolstered by the ease with which human genes can be identified from zebrafish orthologs from searching publicly available databases. From a drug development perspective, zebrafish larvae possess a number of advantages; most notably the larvae exhibit the molecular complexity found in higher vertebrates and, thus, are ideally suited for new drug target identification. Zebrafish larvae readily absorb compounds from the water directly into their circulatory system without permeabilization. Despite these and other advantages, little attention has been focused on the study of metabolic processes in this organism. While pharmaceutical companies are beginning to consider the zebrafish model system, most ongoing high-throughput small molecule screens do not utilize cultured cells and instead assay for inhibitors of single molecule interactions. We advocate whole animal-based screens for drug discovery and expect to see the zebrafish system utilized more frequently for these efforts. As mentioned previously, in many animals certain compounds are metabolized into new compounds that can be significantly more bioactive than the starting compound (i.e., the intestinal cholesterol absorption inhibitor ezetimibe). It is entirely possible that compounds used in conventional screens are found to be inactive because they are not modified by host enzymes. A whole animal screening approach in zebrafish would potentially identify new bioactive compounds that will be critical for addressing the growing list of pandemics associated with abnormal lipid metabolism.
Executive summary
Digestive lipid physiology can be assayed in live zebrafish larvae.
Fluorescent lipids that alter their spectral properties can report lipase activities in vivo.
Nonhydrolyzable microspheres can be used to assay enteric neuron function and gut motility.
Fluorescently quenched protein can report digestive protease activities.
Forward genetic screens in zebrafish can assign new functions to vertebrate genes and provide insight into the function of poorly understood human proteins.
Screens can be based on fluorescent lipid processing (PED6).
Acknowledgments
The authors wish to thank Jennifer Anderson and Rosa Miyares for editorial advice. We are also grateful for the EM image provided by James Walters and Mike Sepanski.
Financial & competing interests disclosure
The authors have no affiliations or financial arrangement with any organization that has a financial interest or stake in the material discussed in this manuscript. The Carnegie Institution does hold a patent together with the University of Pennslyvania on the invention of the author (Steven A Farber) that describes the use of fluorescent lipids in zebrafish for high-throughput screening Pub. No: US 2009/0136428 A1. The authors have no current consultancies, honoraria, stock ownership or options, expert testimony or royalties regarding the material described. However, the Carnegie Institution and/or University of Pennslyvania can license this technology in the future, potentially providing royalties to the author (Steven A Farber). Research performed by the authors and described in this manuscript was supported by the Carnegie Institution Endowment and with grants from the US NIH (RO1 GM63904 and RO1 DK060369). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Footnotes
For reprint orders, please contact: reprints@futuremedicine.com
Ethical conduct of research
All zebrafish care and experimental procedures were carried out as specified in our Institutional Animal Care and Use Committee Protocol (#647A). The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Joffe BI, Panz VR, Raal FJ. From lipodystrophy syndromes to diabetes mellitus. Lancet. 2001;357(9266):1379–1381. doi: 10.1016/S0140-6736(00)04616-X. [DOI] [PubMed] [Google Scholar]
- 2.McNeely MJ, Edwards KL, Marcovina SM, Brunzell JD, Motulsky AG, Austin MA. Lipoprotein and apolipoprotein abnormalities in familial combined hyperlipidemia: a 20-year prospective study. Atherosclerosis. 2001;159(2):471–481. doi: 10.1016/s0021-9150(01)00528-7. [DOI] [PubMed] [Google Scholar]
- 3.Watanabe S, Yaginuma R, Ikejima K, Miyazaki A. Liver diseases and metabolic syndrome. J Gastroenterol. 2008;43(7):509–518. doi: 10.1007/s00535-008-2193-6. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, Beydoun MA. The obesity epidemic in the United States – gender, age, socioeconomic, racial/ethnic, and geographic characteristics: a systematic review and meta-regression analysis. Epidemiologic Rev. 2007;29:6–28. doi: 10.1093/epirev/mxm007. [DOI] [PubMed] [Google Scholar]
- 5.Ford ES, Mokdad AH. Epidemiology of obesity in the Western Hemisphere. J Clin Endocrinol Metab. 2008;93(11 Suppl 1):S1–S8. doi: 10.1210/jc.2008-1356. [DOI] [PubMed] [Google Scholar]
- 6.Misra A, Khurana L. Obesity and the metabolic syndrome in developing countries. J Clin Endocrinol Metab. 2008;93(11 Suppl 1):S9–S30. doi: 10.1210/jc.2008-1595. [DOI] [PubMed] [Google Scholar]
- 7.Van Heek M, France CF, Compton DS, et al. In vivo metabolism-based discovery of a potent cholesterol absorption inhibitor, SCH58235, in the rat and rhesus monkey through the identification of the active metabolites of SCH48461. J Pharmacol Experimen Therap. 1997;283(1):157–163. [PubMed] [Google Scholar]
- 8▪.Farber SA, Pack M, Ho SY, et al. Genetic analysis of digestive physiology using fluorescent phospholipid reporters. Science. 2001;292(5520):1385–1388. doi: 10.1126/science.1060418. First forward genetic zebrafish screen to utilize physiological criteria. [DOI] [PubMed] [Google Scholar]
- 9.Hama K, Provost E, Baranowski TC, et al. In vivo imaging of zebrafish digestive organ function using multiple quenched fluorescent reporters. Am J Physiol Gastrointest Liver Physiol. 2009;296(2):G445–G453. doi: 10.1152/ajpgi.90513.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10▪.Field HA, Kelley KA, Martell L, Goldstein AM, Serluca FC. Analysis of gastrointestinal physiology using a novel intestinal transit assay in zebrafish. Neurogastroenterol Motil. 2009;21(3):304–312. doi: 10.1111/j.1365-2982.2008.01234.x. Nice utilization of fluorescent microspheres to assay intestinal transit. [DOI] [PubMed] [Google Scholar]
- 11.Coleman RA, Lee DP. Enzymes of triacylglycerol synthesis and their regulation. Prog Lipid Res. 2004;43(2):134–176. doi: 10.1016/s0163-7827(03)00051-1. [DOI] [PubMed] [Google Scholar]
- 12.Rousset M. The human colon carcinoma cell lines HT-29 and Caco-2: two in vitro models for the study of intestinal differentiation. Biochimie. 1986;68(9):1035–1040. doi: 10.1016/s0300-9084(86)80177-8. [DOI] [PubMed] [Google Scholar]
- 13.Trotter PJ, Storch J. Fatty acid esterification during differentiation of the human intestinal cell line Caco-2. J Biol Chem. 1993;268(14):10017–10023. [PubMed] [Google Scholar]
- 14.Field FJ. Regulation of intestinal cholesterol metabolism. In: Mansbach CM, Tso P, Kuksis A, editors. Intestinal Lipid Metabolism. Kluwer Academic; NY, USA: 2001. pp. 235–255. [Google Scholar]
- 15.Moschetta A, Xu F, Hagey LR, et al. A phylogenetic survey of biliary lipids in vertebrates. J Lipid Res. 2005;46(10):2221–2232. doi: 10.1194/jlr.M500178-JLR200. [DOI] [PubMed] [Google Scholar]
- 16.Pack M, Solnica-Krezel L, Malicki J, et al. Mutations affecting development of zebrafish digestive organs. Development. 1996;123:321–328. doi: 10.1242/dev.123.1.321. [DOI] [PubMed] [Google Scholar]
- 17.Kruit JK, Groen AK, van Berkel TJ, Kuipers F. Emerging roles of the intestine in control of cholesterol metabolism. World J Gastroenterol. 2006;12(40):6429–6439. doi: 10.3748/wjg.v12.i40.6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tian WX. Inhibition of fatty acid synthase by polyphenols. Curr Med Chem. 2006;13(8):967–977. doi: 10.2174/092986706776361012. [DOI] [PubMed] [Google Scholar]
- 19.Martin FP, Wang Y, Sprenger N, et al. Probiotic modulation of symbiotic gut microbial–host metabolic interactions in a humanized microbiome mouse model. Mol Syst Biol. 2008;4:157. doi: 10.1038/msb4100190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yutzey KE, Robbins J. Principles of genetic murine models for cardiac disease. Circulation. 2007;115(6):792–799. doi: 10.1161/CIRCULATIONAHA.106.682534. [DOI] [PubMed] [Google Scholar]
- 21.Masucci-Magoulas L, Plump A, Jiang XC, Walsh A, Breslow JL, Tall AR. Profound induction of hepatic cholesteryl ester transfer protein transgene expression in apolipoprotein E and low density lipoprotein receptor gene knockout mice. A novel mechanism signals changes in plasma cholesterol levels. J Clin Investig. 1996;97(1):154–161. doi: 10.1172/JCI118384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anstee QM, Goldin RD. Mouse models in non-alcoholic fatty liver disease and steatohepatitis research. Int J Exp Pathol. 2006;87(1):1–16. doi: 10.1111/j.0959-9673.2006.00465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carroll L, Voisey J, van Daal A. Mouse models of obesity. Clin Dermatol. 2004;22(4):345–349. doi: 10.1016/j.clindermatol.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 24.Fishman MC. Genomics. Zebrafish – the canonical vertebrate. Science. 2001;294(5545):1290–1291. doi: 10.1126/science.1066652. [DOI] [PubMed] [Google Scholar]
- 25.Amali AA, Rekha RD, Lin CJ, et al. Thioacetamide induced liver damage in zebrafish embryo as a disease model for steatohepatitis. J Biomed Sci. 2006;13(2):225–232. doi: 10.1007/s11373-005-9055-5. [DOI] [PubMed] [Google Scholar]
- 26.Bai Q, Mullett SJ, Garver JA, Hinkle DA, Burton EA. Zebrafish DJ-1 is evolutionarily conserved and expressed in dopaminergic neurons. Brain Res. 2006;1113(1):33–44. doi: 10.1016/j.brainres.2006.07.057. [DOI] [PubMed] [Google Scholar]
- 27.Berghmans S, Murphey RD, Wienholds E, et al. Tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc Natl Acad Sci USA. 2005;102(2):407–412. doi: 10.1073/pnas.0406252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fetcho JR. The utility of zebrafish for studies of the comparative biology of motor systems. J Exp Zoolog B Mol Dev Evol. 2006;308(5):550–562. doi: 10.1002/jez.b.21127. [DOI] [PubMed] [Google Scholar]
- 29.Fisher S, Jagadeeswaran P, Halpern ME. Radiographic analysis of zebrafish skeletal defects. Dev Biol. 2003;264(1):64–76. doi: 10.1016/s0012-1606(03)00399-3. [DOI] [PubMed] [Google Scholar]
- 30.Guyon JR, Steffen LS, Howell MH, Pusack TJ, Lawrence C, Kunkel LM. Modeling human muscle disease in zebrafish. Biochim Biophys Acta. 2007;1772(2):205–215. doi: 10.1016/j.bbadis.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 31.Khuchua Z, Yue Z, Batts L, Strauss AW. A zebrafish model of human Barth syndrome reveals the essential role of tafazzin in cardiac development and function. Circ Res. 2006;99(2):201–208. doi: 10.1161/01.RES.0000233378.95325.ce. [DOI] [PubMed] [Google Scholar]
- 32.Kunkel LM, Bachrach E, Bennett RR, Guyon J, Steffen L. Diagnosis and cell-based therapy for Duchenne muscular dystrophy in humans, mice, and zebrafish. J Hum Gen. 2006;51(5):397–406. doi: 10.1007/s10038-006-0374-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lam SH, Gong Z. Modeling liver cancer using zebrafish: a comparative oncogenomics approach. Cell Cycle. 2006;5(6):573–577. doi: 10.4161/cc.5.6.2550. [DOI] [PubMed] [Google Scholar]
- 34.Lam SH, Wu YL, Vega VB, et al. Conservation of gene expression signatures between zebrafish and human liver tumors and tumor progression. Nat Biotechnol. 2006;24(1):73–75. doi: 10.1038/nbt1169. [DOI] [PubMed] [Google Scholar]
- 35.Langenau DM, Zon LI. The zebrafish: a new model of T-cell and thymic development. Nat Rev Immunol. 2005;5(4):307–317. doi: 10.1038/nri1590. [DOI] [PubMed] [Google Scholar]
- 36.Langheinrich U, Vacun G, Wagner T. Zebrafish embryos express an orthologue of HERG and are sensitive toward a range of QT-prolonging drugs inducing severe arrhythmia. Toxicol Appl Pharmacol. 2003;193(3):370–382. doi: 10.1016/j.taap.2003.07.012. [DOI] [PubMed] [Google Scholar]
- 37.McKinley ET, Baranowski TC, Blavo DO, Cato C, Doan TN, Rubinstein AL. Neuroprotection of MPTP-induced toxicity in zebrafish dopaminergic neurons. Brain Res Mol Brain Res. 2005;141(2):128–137. doi: 10.1016/j.molbrainres.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 38.Norrby K. In vivo models of angiogenesis. J Cell Mol Med. 2006;10(3):588–612. doi: 10.1111/j.1582-4934.2006.tb00423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Obara T, Mangos S, Liu Y, et al. Polycystin-2 immunolocalization and function in zebrafish. J Am Soc Nephrol. 2006;17(10):2706–2718. doi: 10.1681/ASN.2006040412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sabaawy HE, Azuma M, Embree LJ, Tsai HJ, Starost MF, Hickstein DD. TEL-AML1 transgenic zebrafish model of precursor B cell acute lymphoblastic leukemia. Proc Natl Acad Sci USA. 2006;103(41):15166–15171. doi: 10.1073/pnas.0603349103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sayer JA, Otto EA, O’Toole JF, et al. The centrosomal protein nephrocystin-6 is mutated in Joubert syndrome and activates transcription factor ATF4. Nat Genet. 2006;38(6):674–681. doi: 10.1038/ng1786. [DOI] [PubMed] [Google Scholar]
- 42.Titus TA, Selvig DR, Qin B, et al. The Fanconi anemia gene network is conserved from zebrafish to human. Gene. 2006;371(2):211–223. doi: 10.1016/j.gene.2005.11.038. [DOI] [PubMed] [Google Scholar]
- 43.Williamson KA, Hever AM, Rainger J, et al. Mutations in SOX2 cause anophthalmia-esophageal-genital (AEG) syndrome. Hum Mol Gen. 2006;15(9):1413–1422. doi: 10.1093/hmg/ddl064. [DOI] [PubMed] [Google Scholar]
- 44.Matthews RP, Lorent K, Manoral-Mobias R, et al. TNFα-dependent hepatic steatosis and liver degeneration caused by mutation of zebrafish S-adenosylhomocysteine hydrolase. Development. 2009;136(5):865–875. doi: 10.1242/dev.027565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chico TJ, Ingham PW, Crossman DC. Modeling cardiovascular disease in the zebrafish. Trends Cardiovasc Med. 2008;18(4):150–155. doi: 10.1016/j.tcm.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 46▪.Sadler KC, Amsterdam A, Soroka C, Boyer J, Hopkins N. A genetic screen in zebrafish identifies the mutants vps18, nf2 and foie gras as models of liver disease. Development. 2005;132(15):3561–3572. doi: 10.1242/dev.01918. A forward genetic screen that identified novel genes required for liver development. [DOI] [PubMed] [Google Scholar]
- 47.Parsons MJ, Campos I, Hirst EM, Stemple DL. Removal of dystroglycan causes severe muscular dystrophy in zebrafish embryos. Development. 2002;129(14):3505–3512. doi: 10.1242/dev.129.14.3505. [DOI] [PubMed] [Google Scholar]
- 48.Feitsma H, Cuppen E. Zebrafish as a cancer model. Mol Cancer Res. 2008;6(5):685–694. doi: 10.1158/1541-7786.MCR-07-2167. [DOI] [PubMed] [Google Scholar]
- 49.Stainier DY, Weinstein BM, Detrich HW, 3rd, Zon LI, Fishman MC. Positional cloning of the zebrafish sauternes gene: a model for congenital sideroblastic anaemia. Nat Genet. 1998;20(3):244–250. doi: 10.1038/3049. [DOI] [PubMed] [Google Scholar]
- 50.Song Y, Cone RD. Creation of a genetic model of obesity in a teleost. FASEB J. 2007;21(9):2042–2049. doi: 10.1096/fj.06-7503com. [DOI] [PubMed] [Google Scholar]
- 51.Barbazuk WB, Korf I, Kadavi C, et al. The syntenic relationship of the zebrafish and human genomes. Genome Res. 2000;10(9):1351–1358. doi: 10.1101/gr.144700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 53.Postlethwait J, Amores A, Force A, Yan Y. The zebrafish genome. Methods Cell Biol. 1999;60:149–163. [PubMed] [Google Scholar]
- 54▪▪.Postlethwait JH, Yan YL, Gates MA, et al. Vertebrate genome evolution and the zebrafish gene map. Nat Genet. 1998;18(4):345–349. doi: 10.1038/ng0498-345. A comprehensive analysis of the zebrafish genome that revealed large conserved gene clusters between zebrafish and humans. [DOI] [PubMed] [Google Scholar]
- 55.Farber SA, De Rose RA, Olson ES, Halpern ME. The zebrafish annexin gene family. Genome Res. 2003;13(6A):1082–1096. doi: 10.1101/gr.479603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stainier DY, Weinstein BM, Detrich HW, 3rd, Zon LI, Fishman MC. Cloche, an early acting zebrafish gene, is required by both the endothelial and hematopoietic lineages. Development. 1995;121(10):3141–3150. doi: 10.1242/dev.121.10.3141. [DOI] [PubMed] [Google Scholar]
- 57▪▪.Wallace K, Pack M. Unique and conserved aspects of gut development in zebrafish. Dev Biol. 2003;255(1):12–29. doi: 10.1016/s0012-1606(02)00034-9. A thorough analysis of zebrafish gut development that revealed many similarities with mammals. [DOI] [PubMed] [Google Scholar]
- 58.Yee NS, Yusuff S, Pack M. Zebrafish pdx1 morphant displays defects in pancreas development and digestive organ chirality, and potentially identifies a multipotent pancreas progenitor cell. Genesis. 2001;30(3):137–140. doi: 10.1002/gene.1049. [DOI] [PubMed] [Google Scholar]
- 59.Wallace KN, Yusuff S, Sonntag JM, Chin AJ, Pack M. Zebrafish hhex regulates liver development and digestive organ chirality. Genesis. 2001;30(3):141–143. doi: 10.1002/gene.1050. [DOI] [PubMed] [Google Scholar]
- 60.Flynn EJ, 3rd, Trent CM, Rawls JF. Ontogeny and nutritional control of adipogenesis in zebrafish (Danio rerio) J Lipid Res. 2009 doi: 10.1194/jlr.M800590-JLR200. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin JW, Biankin AV, Horb ME, et al. Differential requirement for ptf1a in endocrine and exocrine lineages of developing zebrafish pancreas. Dev Biol. 2004;270(2):474–486. doi: 10.1016/j.ydbio.2004.02.023. [DOI] [PubMed] [Google Scholar]
- 62.Sheridan MA. Lipid dynamics in fish: aspects of absorption, transportation, deposition and mobilization. Comp Biochem Physio B. 1988;90(4):679–690. doi: 10.1016/0305-0491(88)90322-7. [DOI] [PubMed] [Google Scholar]
- 63.Grosser T, Yusuff S, Cheskis E, Pack MA, FitzGerald GA. Developmental expression of functional cyclooxygenases in zebrafish. Proc Natl Acad Sci USA. 2002;99(12):8418–8423. doi: 10.1073/pnas.112217799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Babin PJ, Thisse C, Durliat M, Andre M, Akimenko MA, Thisse B. Both apolipoprotein E and A-I genes are present in a nonmammalian vertebrate and are highly expressed during embryonic development. Proc Natl Acad Sci USA. 1997;94(16):8622–8627. doi: 10.1073/pnas.94.16.8622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bownes M. Why is there sequence similarity between insect yolk proteins and vertebrate lipases? J Lipid Res. 1992;33(6):777–790. [PubMed] [Google Scholar]
- 66.Munoz G, Donghi S, Cerisola H. Vitellogenesis in the crayfish Rhynchocinetes typus: role of hepatopancreas in lipid yolk biosynthesis. Cell Mol Biol. 1990;36(5):531–536. [PubMed] [Google Scholar]
- 67▪.Marza E, Barthe C, Andre M, Villeneuve L, Helou C, Babin PJ. Developmental expression and nutritional regulation of a zebrafish gene homologous to mammalian microsomal triglyceride transfer protein large subunit. Dev Dyn. 2005;232(2):506–518. doi: 10.1002/dvdy.20251. A nice example of using the zebrafish model system to study the effect of dietary lipid on gene transcription. [DOI] [PubMed] [Google Scholar]
- 68.Spence R, Fatema MK, Ellis S, Ahmed ZF, Smith C. Diet, growth and recruitment of wild zebrafish in Bangladesh. J Fish Biol. 2007;71(1):304–309. [Google Scholar]
- 69.Enzler L, Smith V, Lin JS, Olcott HS. Lipids of Mono Lake, California, brine shrimp (Artemia salina) J Agricul Food Chem. 1974;22(2):330–331. doi: 10.1021/jf60192a017. [DOI] [PubMed] [Google Scholar]
- 70.Thomson AB, Schoeller C, Keelan M, Smith L, Clandinin MT. Lipid absorption: passing through the unstirred layers, brush-border membrane, and beyond. Can J Physiol Pharmacol. 1993;71(8):531–555. doi: 10.1139/y93-078. [DOI] [PubMed] [Google Scholar]
- 71.Layer P, Keller J. Pancreatic enzymes: secretion and luminal nutrient digestion in health and disease. J Clin Gastroenterol. 1999;28(1):3–10. doi: 10.1097/00004836-199901000-00002. [DOI] [PubMed] [Google Scholar]
- 72.Brasaemle DL. Thematic review series: adipocyte biology. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis. J Lipid Res. 2007;48(12):2547–2559. doi: 10.1194/jlr.R700014-JLR200. [DOI] [PubMed] [Google Scholar]
- 73.Cases S, Smith SJ, Zheng YW, et al. Identification of a gene encoding an acyl CoA:diacylglycerol acyltransferase, a key enzyme in triacylglycerol synthesis. Proc Natl Acad Sci USA. 1998;95(22):13018–13023. doi: 10.1073/pnas.95.22.13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cases S, Stone SJ, Zhou P, et al. Cloning of DGAT2, a second mammalian diacylglycerol acyltransferase, and related family members. J Biol Chem. 2001;276(42):38870–38876. doi: 10.1074/jbc.M106219200. [DOI] [PubMed] [Google Scholar]
- 75.Targett-Adams P, Chambers D, Gledhill S, et al. Live cell analysis and targeting of the lipid droplet-binding adipocyte differentiation-related protein. J Biol Chem. 2003;278(18):15998–16007. doi: 10.1074/jbc.M211289200. [DOI] [PubMed] [Google Scholar]
- 76.Novikoff AB, Novikoff PM, Rosen OM, Rubin CS. Organelle relationships in cultured 3T3-L1 preadipocytes. J Cell Biol. 1980;87(1):180–196. doi: 10.1083/jcb.87.1.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bartz R, Li WH, Venables B, et al. Lipidomics reveals that adiposomes store ether lipids and mediate phospholipid traffic. J Lipid Res. 2007;48(4):837–847. doi: 10.1194/jlr.M600413-JLR200. [DOI] [PubMed] [Google Scholar]
- 78.Goodman JM. The gregarious lipid droplet. J Biol Chem. 2008;283(42):28005–28009. doi: 10.1074/jbc.R800042200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Samson-Bouma ME, Verthier N, Ginsel LA, Feldmann G, Fransen JA, Aggerbeck LP. Ultrastructural immunogold labeling of lipid-laden enterocytes from patients with genetic malabsorption syndromes. Biol Cell. 1996;87(3):189–196. [PubMed] [Google Scholar]
- 80.Gordon DA, Wetterau JR, Gregg RE. Microsomal triglyceride transfer protein: a protein complex required for the assembly of lipoprotein particles. Trends Cell Biol. 1995;5(8):317–321. doi: 10.1016/s0962-8924(00)89054-6. [DOI] [PubMed] [Google Scholar]
- 81.Schlegel A, Stainier DY. Microsomal triglyceride transfer protein is required for yolk lipid utilization and absorption of dietary lipids in zebrafish larvae. Biochemistry. 2006;45(51):15179–15187. doi: 10.1021/bi0619268. [DOI] [PubMed] [Google Scholar]
- 82.Fukumoto S, Fujimoto T. Deformation of lipid droplets in fixed samples. Histochem Cell Biol. 2002;118(5):423–428. doi: 10.1007/s00418-002-0462-7. [DOI] [PubMed] [Google Scholar]
- 83.Greenspan P, Mayer EP, Fowler SD. Nile red: a selective fluorescent stain for intracellular lipid droplets. J Cell Biol. 1985;100(3):965–973. doi: 10.1083/jcb.100.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fowler SD, Greenspan P. Application of Nile red, a fluorescent hydrophobic probe, for the detection of neutral lipid deposits in tissue sections: comparison with oil red O. J Histochem Cytochem. 1985;33(8):833–836. doi: 10.1177/33.8.4020099. [DOI] [PubMed] [Google Scholar]
- 85.Carlson LA. Studies on the effect of nicotinic acid on catecholamine stimulated lipolysis in adipose tissue in vitro. Acta Medica Scandinavica. 1963;173:719–722. doi: 10.1111/j.0954-6820.1963.tb17457.x. [DOI] [PubMed] [Google Scholar]
- 86.Jones KS, Alimov AP, Rilo HL, Jandacek RJ, Woollett LA, Penberthy WT. A high throughput live transparent animal bioassay to identify non-toxic small molecules or genes that regulate vertebrate fat metabolism for obesity drug development. Nut Metab. 2008;5:23. doi: 10.1186/1743-7075-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sandoval A, Fraisl P, Arias-Barrau E, et al. Fatty acid transport and activation and the expression patterns of genes involved in fatty acid trafficking. Arch Biochem Biophysics. 2008;477(2):363–371. doi: 10.1016/j.abb.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 88.Treibs A, Kreuzer F-H. Difluorborylkomplexe von di- und tripyrrylmethenen. Liebigs Ann Chem. 1968;718:208–223. [Google Scholar]
- 89.Boldyrev IA, Zhai X, Momsen MM, Brockman HL, Brown RE, Molotkovsky JG. New BODIPY lipid probes for fluorescence studies of membranes. J Lipid Res. 2007;48(7):1518–1532. doi: 10.1194/jlr.M600459-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hendrickson HS, Hendrickson EK, Johnson ID, Farber SA. Intramolecularly quenched BODIPY-labeled phospholipid analogs in phospholipase A2 and platelet-activating factor acetylhydrolase assays and in vivo fluorescence imaging. Anal Biochem. 1999;276(1):27–35. doi: 10.1006/abio.1999.4280. [DOI] [PubMed] [Google Scholar]
- 91.Farber SA, Olson ES, Clark JD, Halpern ME. Characterization of Ca2+-dependent phospholipase A2 activity during zebrafish embryogenesis. J Biol Chem. 1999;274(27):19338–19346. doi: 10.1074/jbc.274.27.19338. [DOI] [PubMed] [Google Scholar]
- 92.Ho SY, Lorent K, Pack M, Farber SA. Zebrafish fat-free is required for intestinal lipid absorption and Golgi apparatus structure. Cell Metab. 2006;3(4):289–300. doi: 10.1016/j.cmet.2006.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Watanabe T, Asaka S, Kitagawa D, et al. Mutations affecting liver development and function in Medaka, Oryzias latipes, screened by multiple criteria. Mech Dev. 2004;121(7–8):791–802. doi: 10.1016/j.mod.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 94.Bolcome RE, 3rd, Sullivan SE, Zeller R, Barker AP, Collier RJ, Chan J. Anthrax lethal toxin induces cell death-independent permeability in zebrafish vasculature. Proc Natl Acad Sci USA. 2008;105(7):2439–2444. doi: 10.1073/pnas.0712195105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jones LJ, Upson RH, Haugland RP, Panchuk-Voloshina N, Zhou M, Haugland RP. Quenched BODIPY dye-labeled casein substrates for the assay of protease activity by direct fluorescence measurement. Anal Biochem. 1997;251(2):144–152. doi: 10.1006/abio.1997.2259. [DOI] [PubMed] [Google Scholar]
- 96.Menges DA, Ternullo DL, Tan-Wilson AL, Gal S. Continuous assay of proteases using a microtiter plate fluorescence reader. Anal Biochem. 1997;254(1):144–147. doi: 10.1006/abio.1997.2408. [DOI] [PubMed] [Google Scholar]
- 97.Sarment DP, Korostoff J, D’Angelo M, Polson AM, Feldman RS, Billings PC. In situ localization and characterization of active proteases in chronically inflamed and healthy human gingival tissues. J Periodontol. 1999;70(11):1303–1312. doi: 10.1902/jop.1999.70.11.1303. [DOI] [PubMed] [Google Scholar]
- 98.Raybould HE. Mechanisms of CCK signaling from gut to brain. Curr Opin Pharmacol. 2007;7(6):570–574. doi: 10.1016/j.coph.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Haffter P, Granato M, Brand M, et al. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- 100.Field HA, Kelley KA, Martell L, Goldstein AM, Serluca FC. Analysis of gastrointestinal physiology using a novel intestinal transit assay in zebrafish. Neurogastroenterol Motil. 2009;21(3):304–312. doi: 10.1111/j.1365-2982.2008.01234.x. [DOI] [PubMed] [Google Scholar]
Patents
- 201.Haugland RP, Kang HC. 1988 US Patent US4774339. [Google Scholar]