Abstract
Cisplatin is the most prominent member of a series of platinum(II) antitumor drugs that demonstrate activity based on binding to adjacent purines on genomic DNA. The interactions between cisplatin and alternate biomolecules, including chemically similar RNA, are less understood than are those for DNA. In order to investigate potential implications of platinum(II) drug binding to a structurally complex RNA, we have characterized the reaction between cisplatin and the internal loop of a 41-nucleotide subdomain derived from the U2:U6 spliceosomal RNAs. This “BBD” RNA subdomain consists of a hairpin structure containing a purine-rich asymmetric internal loop. Aquated cisplatin is found to cross-link G nucleobases on opposing sides of the internal loop, forming an intramolecular internal loop cross-link in BBD and an analogous intermolecular cross-link in a two-piece construct containing the same internal loop sequence. The two opposing guanine residues involved in the cross-link were identified via limited alkaline hydrolysis. The kinetics of aquated cisplatin binding to the BBD RNA, a related RNA hairpin, and its DNA hairpin analogue were investigated in an ionic background of 0.1 M NaNO3 and 1 mM Mg(NO3)2. Both BBD and the RNA hairpin react 5–6-fold faster than the DNA hairpin, with calculated second-order rate constants of 2.0(2), 1.7(3), and 0.33(3) M−1 s−1, respectively, at 37 °C, pH 7.8. MALDI-MS data corroborate the biochemical studies and support a model in which kinetically preferred platinum binding sites compete with less reactive sites in these oligonucleotides. Taken together, these data indicate that cisplatin treatment has potential to create internal loop and other unusual cross-links in structurally complex RNAs, on a time scale that is relevant for RNA-dependent biological processes.
Introduction
Cisplatin (cis-diamminedichloroplatinum(II)) is the flagship compound for a series of platinum(II) antitumor agents employed in the treatment of a wide range of cancers.1–3 Cisplatin activity involves intracellular exchange of the labile chloride ligands and ultimate coordination to “soft” biomolecular donor sites. In vivo, cisplatin is known to bind to multiple targets including DNA, RNA, proteins, and small-molecule ligands. Drug binding to adjacent purines on genomic DNA has been linked to the induction of apoptosis, a foundation of antitumor activity. Despite their prevalent use, a comprehensive understanding of additional drug-related biological processes is still forming for the platinum antitumor compounds.
Early studies that are often cited in identifying DNA as a target for cisplatin reveal that, on a per nucleotide basis, drug binding to DNA and RNA is roughly equivalent.4,5 Additional studies have shown that platinum drug treatment is capable of interfering with transcription,6–8 and that critical RNA-dependent activities such as splicing9 and translation5,10,11 are inhibited when measured in cell extracts. Combined, these studies suggest that the binding of cisplatin-derived species to RNA may contribute to the drug’s in vivo effects. A limited number of studies have revealed further details concerning interactions of cisplatin with RNA. Elmroth, Chow, and co-workers have previously communicated enhanced reactivity and more pronounced dependence of reaction rate on ionic conditions for the reaction of a 13 nt RNA hairpin in comparison with a DNA analogue.12 Elmroth and co-workers have additionally suggested binding locations for cisplatin near the G•U wobble pair in a tRNAAla acceptor stem13,14 and have explored platinum–RNA adducts for directing RNA silencing.15 Cisplatin has been shown by Danenberg and co-workers to inhibit in vitro activity of a group I intron ribozyme.16 Very recently, Rijal and Chow have reported the use of cisplatin as a structural probe to identify accessible purine nucleobases in bacterial ribosomes.17 These studies suggest intriguingly selective cisplatin–RNA reactivity and call for more detailed kinetic analyses and comprehensive characterization of the nature of the platinated products in complex RNAs.
A common characteristic of naturally occurring metal sites in RNA is the involvement of ligands that are distant in primary sequence but brought into proximity in the folded RNA structure.18–20 By cross-linking two such ligands, cisplatin-induced chelation, whether in naturally occurring metal sites or novel target sites, has the potential to inhibit activities that depend on the dynamic nature of RNA. The spliceosome is an example of an RNA machine that is dependent on dynamic rearrangements for function.21,22 One key step in spliceosomal function is the formation of a complex between the U2 and U6 snRNAs that is implicated in the first step of pre-mRNA splicing.23 Here we provide an in vitro analysis of the reaction between aquated cisplatin and a 41 nt RNA construct termed BBD (branch-bulge domain) that contains the purine-rich internal loop formed from U2 and U6 RNA strands (Figure 1).24 We show that platinum forms a novel intrastrand cross-link across the internal loop of BBD and an interstrand cross-link in a two-piece construct. Additionally, we report that, under biologically pertinent ionic conditions, platination of both BBD and a related 40 nt RNA hairpin is ~5-fold faster than for a DNA hairpin analogue. MALDI-MS data are presented that complement the conclusions from our biochemical studies. Taken together, these results indicate facile cisplatin-induced adduct formation across an RNA internal loop and fast platination kinetics of RNA oligonucleotides.
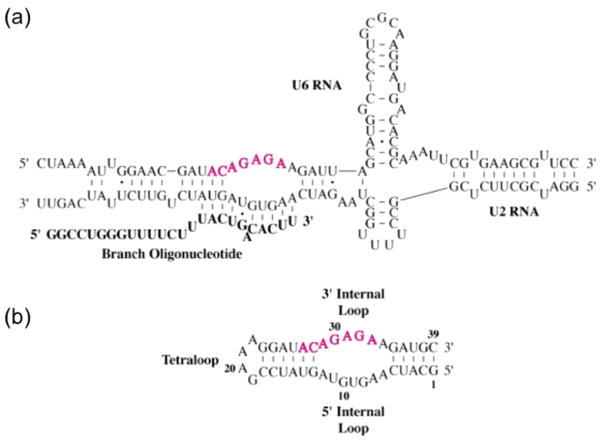
Figure 1.
(a) Proposed secondary structure of a human U2:U6 snRNA core complex including branch oligonucleotide.18 Conserved nucleotides are in pink. (b) Predicted secondary structure of the BBD RNA subdomain used in this study, with invariant nucleotides again highlighted in pink.
Results
Evidence of Cisplatin–RNA Cross-Linking
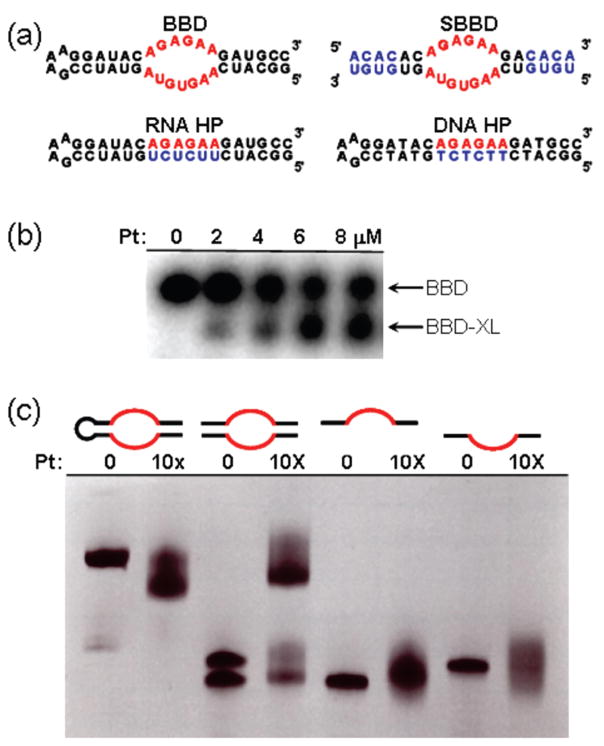
RNA and DNA oligonucleotides used in this study are shown in Figure 2a. The BBD RNA subdomain contains a purine-rich internal loop flanked by helical regions, whereas the RNA and DNA hairpin sequences (RNA HP and DNA HP) are fully base-paired. In previous studies, reaction of RNA or DNA with cisplatin has resulted in product species that are typically observed to migrate more slowly when analyzed by denaturing polyacrylamide gel electrophoresis (dPAGE).25,26 Slower migration of platinated oligonucleotides is likely due to added molecular weight and a decrease in the overall charge of the nucleic acid through binding of a [Pt(NH3)2]2+ fragment. By contrast, the reaction of cisplatin with BBD results in a product species with higher mobility (Figure 2b). Faster mobility under denaturing conditions may be caused by intrastrand cross-linking,27,28 which was hypothesized to occur across the purine-rich internal loop region of the BBD RNA. In order to test this hypothesis, the BBD internal loop sequence was embedded between two new duplex sequences, creating a two-piece “split” BBD construct (SBBD, Figure 2a). Platination of SBBD results in cross-linking of the two strands, as observed unambiguously by dPAGE (Figure 2c). Platination of the individual upper or lower SBBD strands does not result in a cross-linked species, although the presence of secondary platination sites is indicated by the dispersion of the dPAGE product bands (Figure 2c). These data indicate that cisplatin creates a cross-link across the internal loop of BBD RNA.
Figure 2.
(a) Oligonucleotide sequences and predicted secondary structures. The BBD internal loop is highlighted in red. Differences in sequence relative to BBD are shown in blue. (b) Formation of a higher-mobility BBD product upon platination of BBD. (c) Confirmation of cisplatin-induced cross-linking in BBD internal loop sequence, showing products of platinum treatment with (i) BBD, (ii) SBBD hybrid, and (iii) individual strands of SBBD. Conditions: (b) 0.2 μM 5′ 32P-labeled BBD treated with indicated concentrations of cisplatin for 1.5 h in deionized water, analyzed by 18% dPAGE, and visualized by autoradiograph; (c) 20 μM (0.2 nmol) RNA reacted with 10× cisplatin (200 μM) in 5 mM TEA (pH 7.8), 12–15 h, 37 °C, analyzed by 20% dPAGE, and visualized by staining with methylene blue.
Identification of Cross-Linked Nucleobases
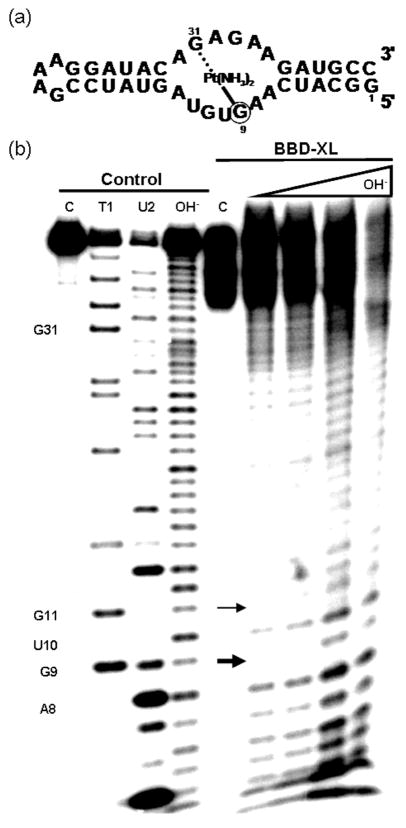
In order to identify the specific bases involved in formation of an intrastrand cross-link, the proposed cross-linked product (BBD-XL) was isolated following dPAGE and mapped by partial alkali hydrolysis. Using 5′ end-labeled RNA, normal hydrolysis products are expected to be observable up to the 5′ side of the cross-linked site.28–30 Hydrolysis products that contain the cross-linked site will result in significantly higher molecular weight species with unusual gel mobilities, leaving a gap in the hydrolysis ladder following the 5′ cross-linked site.28–30 As displayed in Figure 3, clear BBD-XL hydrolysis products are observed for nucleotides 3′ to A8 but not at G9, identifying G9 as the major 5′ site involved in the internal cross-link. An additional faint band at U10 suggests G11 as a minor secondary site for platinum-induced cross-linking. In the cross-linked species generated using the two-piece SBBD complex, the site equivalent to G9 was again identified as one major adduct site (Figure S1 in Supporting Information). Using this SBBD construct, hydrolysis mapping also identified the site corresponding to G31 as the cross-linking partner on the other side of the internal loop (Figure S2 in Supporting Information).
Figure 3.
(a) Secondary structure of BBD representing the location of the cisplatin-induced cross-link found in BBD-XL. (b) Cleavage products produced by the alkali hydrolysis of isolated BBD-XL. Lanes from left to right. Control lanes (untreated BBD RNA) C: control 5′ end-labeled untreated BBD. T1: G-specific sequence ladder generated by partial nuclease digestion by RNase T1. U2: A-specific sequence ladder generated by partial nuclease digestion by RNase U2. OH−: Reference alkali hydrolysis ladder. BBD-XL lanes C: dPAGE-isolated BBD-XL. OH− lanes: dPAGE-isolated BBD-XL treated under alkali hydrolysis conditions for increasing amounts of time (see Methods). Arrows indicate major (thick arrow, G9) and minor (thin arrow, G11) sites of platinum coordination.
The identification from dPAGE-isolated products of the intrastrand cross-link in BBD and the analogous interstrand cross-link in the SBBD hybrid strongly suggests that internal loop cross-linking is a major structural determinant for altered gel mobility upon platinum coordination to BBD.
Cisplatin–RNA Reaction Rates
The in vivo relevance of cisplatin–RNA reactions, including the internal loop cross-linking reaction observed here, depends in part on their rates relative to adduct formation with DNA or other cellular targets. In evaluating the reaction rates of cisplatin with the oligonucleotides of Figure 2a, kinetic studies were performed at 37 °C and in a background of 0.1 M NaNO3/1 mM Mg(NO3)2 in order to approximate cation competition in vivo. Nitrate salts were used instead of chloride salts to prevent bias of the observed reaction rates due to an increase in the cisplatin anation rate.31 Because the aquation of cisplatin has been shown to be the rate-limiting step for reactions with oligonucleotides under similar conditions, cisplatin was aquated by reaction with AgNO3 immediately before use.32 RNA concentrations of 0.1 μM and platinum concentrations of at least 125-fold excess were used to ensure pseudo-first-order conditions for the reaction (Figure S4 in Supporting Information). Reaction products were analyzed following separation by dPAGE and autoradiography. Under these conditions, all data fit well to a single exponential function, indicating that a single rate-limiting step dominates the kinetics of product appearance in each case.
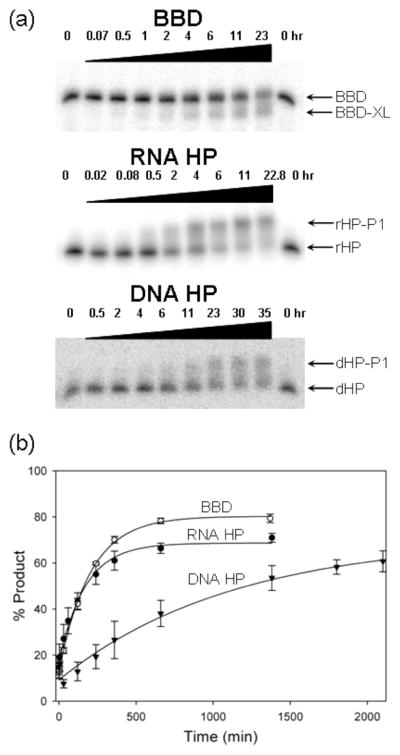
In addition to monitoring internal cross-link formation in BBD, reactions of cisplatin with two related hairpin structures were monitored. RNA HP and DNA HP (Figure 2a) retain the flanking helical and terminal loop sequences of BBD but replace the internal loop region with a fully base-paired sequence. These hairpin constructs provide control sequences having similar base composition, length, and terminal loops to those of BBD. Reaction of aquated cisplatin with both HP constructs results in products that migrate more slowly, in contrast to the faster-migrating cross-linked species produced with BBD (Figure 4a). As observed from Figure 4a,b, both RNA sequences (BBD and RNA HP) react at similar rates of kobs = 9.8(1.0) × 10−5 and 8.3(2) × 10−5 s−1, respectively, in 50 μM CP, pH 7.8 (Table S1 in Supporting Information). Product formation for the DNA construct is 5–6-fold slower, with an observed rate constant of 1.7(2) × 10−5 s−1 under identical conditions. The calculated second-order rate constants are krxn2 = 2.0(2), 1.7(3), and 0.33(3) M−1 s−1 for BBD, RNA HP, and DNA HP, respectively.
Figure 4.
(a) Time-dependent product band appearance following treatment of radiolabeled RNA and DNA substrates with aquated cisplatin and analysis by dPAGE. Product bands are labeled BBD-XL, rHP-P1, and dHP-P1 for BBD, RNA HP (rHP), and DNA HP (dHP), respectively. (b) Comparison of the reaction rates of aquated cisplatin with BBD (open circles), RNA HP (filled circles), and DNA HP (triangles). Data are fit to a pseudo-first-order rate expression as described in Materials and Methods. Conditions in (a): 0.1 μM oligonucleotide, 50 μM aquated cisplatin, 100 mM NaNO3, 1 mM Mg(NO3)2, and 5 mM TEA (pH 7.8) at 37 °C.
Reaction rates were also investigated, under identical buffer conditions, for the two-stranded SBBD hybrid substrate for which product formation results in a clearly separated cross-linked species (Figure 2c and Figure S3 in Supporting Information). For SBBD, a pseudo-first-order rate constant of kobs = 5.2(3) × 10−5 s−1 and a calculated second-order rate constant of krxn2 = 1.1(1) M−1 s−1 were obtained in 50 μM aquated cisplatin (Figure S3 and Table S1 in Supporting Information). These values are approximately 50% of those determined for BBD but still reflect faster reaction rates than observed for the DNA HP. A likely explanation for the slower reaction rate observed for intermolecular cross-linking of the two-piece RNA SBBD construct in comparison with intramolecular cross-linking in BBD is the incomplete hybridization of the SBBD strands under these reaction conditions (see Materials and Methods). Nonetheless, the fact that similar values are observed for cross-linking in both BBD and the two-piece SBBD suggests that similar rate-limiting steps guide the internal loop cross-linking reaction regardless of RNA construct.
The reactions of aquated cisplatin with BBD and SBBD are pH-dependent, increasing in rate as the pH is lowered. At pH 6.8, second-order rate constants of 8.5(7) and 6.8(2) M−1 s−1 are measured for BBD and SBBD, respectively (Table S1 in Supporting Information, data not shown). On the basis of known protonation equilibria for aquated cisplatin species, this rate enhancement likely reflects protonation of a cisplatin hydroxide ligand to aqua ligand on platinum(II) and is a closer approximation to rates that might be expected for in vivo conditions.
Analysis of Platinum–RNA Adducts by MALDI-MS
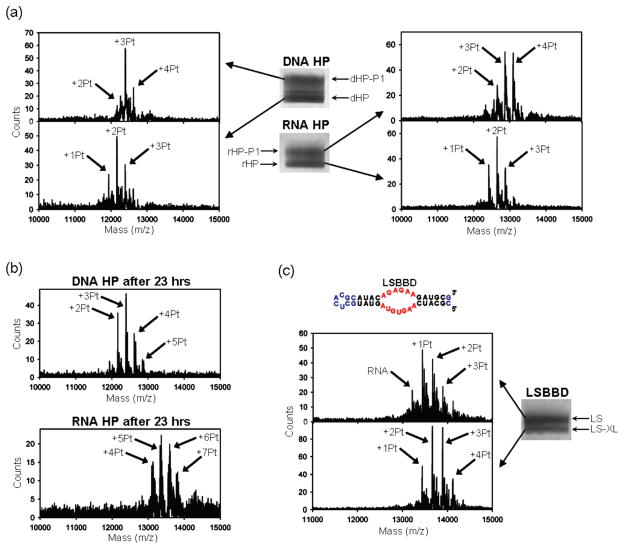
As described above, platination of the RNA and DNA domains used in this study results in products that have distinctly altered mobilities when analyzed by dPAGE. To further analyze these products, MALDI-MS was used to identify platinum–oligonucleotide species33 in samples isolated from the dPAGE gels. MALDI-MS data for the RNA and DNA HP sequences reacted for 5 h at a ratio of 5:1 platinum/oligonucleotide are shown in Figure 5a. Products containing [Pt(NH3)2]2+ adducts appear at the oligonucleotide mass plus increments of 229 amu. Additional lower-intensity peaks are often present at ~+17, +23, and +39 amu that are ascribed to residual H2O/OH/NH4+, Na+, and K+, respectively. These features appear in untreated RNA as well as platinated RNA, and their presence along with the breadth of the features precludes identification of [Pt(NH3)2X] (X = H2O, OH−, or Cl−) species. Although MALDI-MS is not precisely quantitative, comparison of relative intensities within identically treated samples provides qualitative information on relative populations of the major RNA–platinum adducts.
Figure 5.
(a) Positive-ion mode MALDI mass spectra of products following aquated cisplatin treatment of the RNA HP and DNA HP and isolation via dPAGE. Product bands are labeled rHP-P1 and dHP-P1 for RNA HP (rHP) and DNA HP (dHP), respectively. (b) Positive-ion mode MALDI mass spectra of the products of 23 h reactions of RNA HP and DNA HP with cisplatin under the reaction conditions used for Figure 4. (c) Sequence and predicted secondary structure of LSBBD. The BBD internal loop is highlighted in red. Differences in sequence relative to BBD are shown in blue. Image and subsequent MALDI-MS of the two main electrophoretic bands resulting from cisplatin treatment of LSBBD (LS), with LS product band labeled as LS-XL. Conditions: (a) Reactions were performed with 30 μM oligonucleotide, 150 μM aquated cisplatin, 100 mM NaNO3, 1 mM Mg(NO3)2, and 5 mM MOPS (pH 6.8) at 37 °C for 5 h. The bands were separated by 20% dPAGE, stained with methylene blue, and then excised. The MALDI was performed in 3-hydroxypicolinic acid. (c) Same as in (a), except that the reaction contained 90 μM cisplatin.
The data in Figure 5 show that RNA and DNA HP samples, treated under the reaction conditions described above, separate into upper and lower dPAGE bands that both contain platinated oligomers. As expected from their slower electrophoretic mobility, samples isolated from the upper band of these gels show a higher extent of platination. At a ratio of 5:1 Pt/oligomer, species with bound 1–3 [Pt(NH3)2]2+ are present in the lowest-running dPAGE bands, whereas the higher bands contain oligomers with 2–4 bound [Pt(NH3)2]2+ ions (Figure 5a). Consistent with the observation that RNA exhibits a faster reaction time, a higher population of the +4 [Pt(NH3)2]2+ species is observed for the RNA HP sample than is found for the DNA analogue at the same time point. Faster reaction kinetics with RNA are also evident from bulk analysis of samples that are reacted for 23 h in the high platinum/oligonucleotide ratios used for kinetic studies. In this case, the bulk DNA HP sample shows an overall adduct distribution that has major populations of 2–3 bound [Pt(NH3)2]2+, distinctly fewer than the 5–6 [Pt(NH3)2]2+ in the highest amplitude MALDI-MS peaks for the RNA HP sample treated under identical conditions (Figure 5b).
From these data, it is apparent that the cisplatin-treated HP products separated by dPAGE all contain multiple platinum adducts. In the case of the mainly helical hairpin samples, the major products quantified for kinetics analysis are separated as lower-mobility species that contain on average at least one additional [Pt(NH3)2]2+ ion (Figure 5a). Although not a complete study, these data are broadly consistent with a model in which one site on the HP sequence both reacts more quickly with cisplatin and also causes a conformational change resulting in a distinctly lower-mobility species. Slower platination reactions occur at other sites, creating products that are not clearly separated by these dPAGE conditions.
The products of BBD platination were also analyzed by MALDI-MS. If a similar model holds in which one faster-reacting site results in altered gel mobility, then for the BBD reactions, it is predicted that the higher-mobility, cross-linked products will also contain on average one additional [Pt(NH3)2]2+ ion. An alternative model for reactions of platinum with the BBD sequence is that the cross-linking and non-cross-linking sites react at equal rates, which would result in an equal distribution of platinum in both dPAGE bands. To simplify the MALDI-MS data, a modified BBD sequence that maintains the internal loop but reduces the number of other purine sites was employed (Figure 5c). MALDI-MS spectra of platinated LSBBD shows that both dPAGE-isolated products contain multiple platinum adducts, but that peaks from the faster-migrating product band are indeed shifted by approximately +1 [Pt-(NH3)2]2+ fragment (Figure 5c). Additional evidence that the gel bands observed for platinated BBD differ by one [Pt-(NH3)2]2+ fragment is provided by the dependence of reaction rates on platinum concentration. With platinum in large excess, the observed reaction rates vary linearly with a slope of 1.1 (r2 = 1.0, Figure S4 in Supporting Information), consistent with a 1:1 ratio between platinum and product.
Discussion
Understanding of the biological roles of RNA has vastly expanded over the last three decades.34 RNA-based regulation occurs through gene silencing and RNAi pathways,35,36 as well as through specific binding of small molecules in the structured regions of riboswitches.37–40 Intricate RNA structures support catalytic active sites,41 and dynamic RNA–protein rearrangements take place throughout regulatory pathways42–44 and in complex cellular machineries such as the spliceosome.22 These factors serve as the basis for the recent push in advancing RNA as a drug target and spur interest in understanding how existing nucleic-acid-targeted drugs might act in previously unidentified pathways.45,46 Cisplatin provides an example of a known DNA-binding compound that might have unique interactions with complex RNA structures. Factors that may be relevant to structurally diverse RNAs have been encountered in studies of platinated DNA hairpins, platinum cross-linked quadruplex structures, and platinated DNA–protein complexes.47–53 Additionally, preliminary investigations have already begun to address cisplatin’s use as a structural probe and as drug conjugate for targeting RNA.15,17,54,55
This report describes cisplatin cross-linking across the internal loop of a 41 nt RNA branch-bulge subdomain (BBD) that is derived from a U2–U6 snRNA complex proposed to form the active core of the spliceosome (Figure 1).23,24 Cisplatin-induced intramolecular cross-linking takes place between G bases located in opposing sides of the BBD internal loop. In a cellular context, cross-linking of this type could have the potential to disrupt binding of the branch oligonucleotide or dissociation of the U2: U6 complex. It is interesting to note that the 3′ side of the BBD internal loop corresponds to an invariant region of the U6 snRNA that is hypothesized to contain essential metal binding sites in the biological complex.56,57 The ability of cisplatin to compete for preorganized metal binding sites in RNA is unestablished and presents an interesting possibility for predicting in vivo drug binding locations. Further studies will also focus on the sequence requirements and generality of cisplatin-based drug interactions with structured RNAs. Initial experiments indicate that this cross-linking reaction is tolerant of single base substitutions in the BBD internal loop, but that substitution of G31, the 3′ partner in the major cross-link, with non metal-coordinating U results in a slightly different product as reflected by altered mapping data (data not shown). These limited studies indicate that platinum-induced cross-link formation is not strictly limited to internal loops with this exact BBD sequence and is likely to take place in other structured RNAs. In general, cisplatin cross-links in functional RNAs could disrupt a host of cellular processes that rely on RNA’s dynamic structure.
Drug-binding kinetics may be an important factor governing the significance of cisplatin–RNA adducts in vivo. In order to begin to address this topic, here we compared the in vitro reaction rates of similar RNA and DNA oligomers with cisplatin. Somewhat surprisingly, the RNA constructs exhibit reaction rates that are 5–6 times faster than those measured for the DNA construct. In a related study, Elmroth and co-workers observed faster binding by other platinum(II) compounds to a 13 nt RNA hairpin in comparison with its DNA analogue. Although that study used different reaction conditions than employed here, most notably lower ionic strength, the reported rate constants are within an order of magnitude of those in Table S1 in Supporting Information.12 Combined, these observations support RNA as a kinetically competitive target for cisplatin.
The application of polyelectrolyte theory to the platination of RNA or DNA suggests a model involving entry of a charged platinum species into the condensed cation atmosphere of a nucleic acid, followed by irreversible monoadduct and diadduct formation.12,58–60 The range of literature values for the rate of monoadduct formation on duplex DNA by [Pt(NH3)2Cl(OH2)]+ varies from ~0.1 to 1 M−1 s−1.61–63 From this study, the calculated second-order rate constant for the platination of the DNA HP is 0.33(3) M−1 s−1 and lies within this range, while the rates observed for the RNA constructs are somewhat faster at ~2 M−1 s−1 (Table S1 in Supporting Information). The observation that similar rates are obtained for monoadduct formation on two structurally distinct RNAs indicates that broader factors such as electrostatic potential and oligomer flexibility likely dictate enhanced reactivity when compared with DNA. Individual contributions of these factors are currently under investigation.
MALDI-MS allows identification of nucleic acids with bound [Pt(NH3)2]2+ fragments33 and was used to analyze bulk reaction mixtures and dPAGE isolated oligomers in this study. MALDI-MS analysis of platinated oligomers isolated from dPAGE shows an average of one additional [Pt(NH3)2]2+ fragment in the product bands. This observation supports a model in which specific sites within each oligomer react quickly with cisplatin and are responsible for the structural distortions leading to altered gel mobility. Previous observations of kinetically preferred sites for platinum adduct formation on DNA have been reported. Enhanced reactivity can be based on electronegativity and target site geometry, as influenced by the nucleotide identity as well as oligonucleotide length, and secondary and tertiary structure.49,53,64–71 Particularly relevant to this study is the observation of cross-strand adduct formation by [Pt(NH3)2-(OH2)2]2+ in telomeric DNA model sequences.48,53 An even greater variety of platinum adducts might be expected for RNA based on its structural diversity and ability to specifically chelate divalent metals.18–20
The biological lifetime of each nucleic acid is an important factor in translating the faster in vitro rates observed for RNA in this study into a cellular context. In normal human cells, the most rapidly turned-over class of RNA is mRNA. Median mRNA lifetimes in human cell lines have been reported to be ~10 h, with a wide range of decay rates that vary by ~500-fold.72 Both tRNA and rRNAs have significantly slower turnover rates and lifetimes predicted to be on the order of days for average adults.73 The lifetimes and abundance of cellular RNAs suggest that platinum drug binding could occur on a time scale that may affect RNA processes within treated cells. The types of cellular RNAs that are sufficiently accessible for platinum adduct formation, and the range of cellular consequences that could result from cisplatin interactions with RNA including interstrand cross-linking such as is observed in this study, are topics that remain to be addressed.
Materials and Methods
Nucleic Acid Substrates
All RNAs, except BBD, were purchased from Dharmacon, Inc. DNA was purchased from Integrated DNA Technologies. BBD was transcribed in vitro from a plasmid template using T7 RNA polymerase. All nucleic acid substrates were purified by 20% dPAGE, eluted, then desalted and concentrated using Millipore YM-3 Centricon tubes. Subsequent buffer exchange and desalting was often accomplished using GE Healthcare G-25 microspin columns.
5′ End-Labeling
Prior to radiolabeling, the 5′ end of BBD was dephosphorylated using Antarctic phosphatase (New England Biolabs). 5′-OH Oligonucleotides were end-labeled with T4 poly-nucleotide kinase (USB) using γ32P-ATP (Perkin-Elmer). End-labeled oligonucleotides were purified by 20% dPAGE followed by overnight elution from excised gel bands. The resulting eluent was ethanol-precipitated and desalted or buffer-exchanged as described above.
Cisplatin Aquation
Cisplatin (Sigma-Aldrich) was stored as a 1 mM solution in 10 mM NaCl in the dark at 4 °C. Immediately before use, cisplatin was aquated with 0.95 equiv of 12 mM AgNO3 (stored in the dark). The aquation reaction was incubated at 50 °C for 1 h, at which time AgCl was precipitated by centrifugation. The supernatant solution was removed and diluted accordingly. On the basis of 195Pt NMR (data not shown), the main platinum species varied by pH: [Pt(NH3)2Cl(OH2)]+ for pH 6.8 and [Pt(NH3)2ClOH] for pH 7.8.
Platination of BBD (Figure 2b)
A trace amount of 5′ end-labeled BBD with 0.2 μM unlabeled BBD was annealed by heating to 90 °C for 90 s followed by cooling to room temperature, then reacted with 0–40 μM cisplatin in deionized water for 1.5 h at 37 °C. The bulk reaction mixtures were mixed with formamide loading buffer and immediately applied to 18% dPAGE. Results were imaged using a Molecular Dynamics phosphor screen and scanned on a Molecular Dynamics Storm 860.
Comparative Platination of SBBD and BBD (Figure 2c)
Twenty micromolar (0.2 nmol) of BBD RNA, each individual SBBD strand, or the SBBD hybrid was annealed and rested on ice for 30 min. RNAs were then incubated in the presence of absence of 200 μM cisplatin (added as a 1 mM solution with 8 mM NaCl) in 5 mm triethanolamine (TEA) for 12–16 h at 37 °C. Reaction mixtures were analyzed on 20% dPAGE and visualized by staining with methylene blue.
Isolation of 5′ End-Labeled, Cross-Linked RNAs
BBD: 5′ End-labeled BBD in the presence of 0.1 μM unlabeled BBD was annealed and reacted with aquated cisplatin in 100 mM NaNO3, 1 mM Mg(NO3)2, and 5 mM 3-(N-morpholino)propanesulfonic acid (MOPS) (pH 6.8) at 37 °C for 23 h. Reaction products were isolated via excision from 18% dPAGE. RNA was eluted overnight into deionized water and desalted using in-house prepared G-25 sephadex spin columns (BioRad). SBBD: One 5′ end-labeled strand was annealed in 10 μM of the unlabeled complement RNA in 12.5 mM NH4NO3 and reacted with 100 μM aquated cisplatin for 23 h. The cross-linked product was excised from 20% PAGE and eluted overnight into deionized water. Following speedvac concentration, SBBD cross-links were desalted using G-25 sephadex spin columns (GE Healthcare).
Hydrolysis Mapping of Cross-Linked RNAs
Trace 5′ end-labeled, cross-linked RNAs were dried to completion in the presence of 0.2 pmol unlabeled RNA corresponding to the 5′ end-labeled strand. Samples were then resuspended in 50 mM Na2CO3/NaHCO3 (pH 9.5) and 1 mM EDTA and reacted at 90 °C for times ranging from 5 to 25 min. The reaction was quenched by the addition of 8 M urea, 10 mM sodium citrate (pH 3.5), and 0.005% (w/v) xylene cyanol loading buffer and held on dry ice until electrophoresis. The results were analyzed on 15 or 20% dPAGE (BBD and SBBD, respectively) then visualized via phosphorimaging.
Reference lanes of 5′ end-labeled un-cross-linked RNAs were generated from hydrolysis as above and by partial nuclease digestion by RNase T1 (Ambion) or U2 (Pierce/Thermo Scientific). Briefly, 5′ end-labeled RNA with 0.2 μM of the corresponding unlabeled RNA in 8 M urea, 10 mM sodium citrate (pH 3.5), and 0.005% (w/v) xylene cyanol was reacted for at 50 °C for 12–15 min with 1U T1 RNase or 0.2U U2 RNase. Samples were then held on dry ice until electrophoresis.
Kinetic Analysis
Prior to kinetic analysis, 0.1 μM oligonucleotide with trace 5′ end-labeled material was annealed in buffered solution by heating to 90 °C for 90 s followed by gradual cooling to room temperature or, for the case of the SBBD hybrid, resting on ice for 30 min. Buffers included either 100 mM NaNO3, 1 mM Mg(NO3)2, and 5 mM triethanolamine (TEA) (pH 7.8) or 100 mM NaNO3, 1 mM Mg(NO3)2, and 5 mM (MOPS) (pH 6.8) depending on desired pH. Freshly aquated cisplatin was added to final concentrations of 13, 25, or 50 μM, and the reactions were incubated at 37 °C for times ranging from 1 min to 35 h. Aliquots were removed and stopped by ethanol precipitation or dilution with formamide and freezing. These aliquots were applied to 18–19% dPAGE and visualized by autoradiography. Each kinetic experiment was repeated at least three times. Molecular dynamics ImageQuant software version 5.0 was used to quantify the reaction mixtures from each kinetics experiment. Rate constants were generated from data analysis using SigmaPlot version 8.0.
MALDI-MS
Isolation of platinated oligonucleotides for MALDI-MS analysis: 30 μM of an oligonucleotide was annealed in 100 mM NaNO3, 1 mM Mg(NO3)2, and 5 mM MOPS (pH 6.8) and reacted with either 90 μM aquated cisplatin for LSBBD or 150 μM for RNA HP and DNA HP. Samples were incubated at 37 °C for 5 h, at which time they were applied to 19% dPAGE. Products were stained with methylene blue and excised. Oligonucleotides were recovered via electroelution using a Schleicher and Schuell Elutrap electro-separation system, concentrated, and desalted. Bulk time-course reactions for MALDI-MS analysis: 0.6 nmol of an oligonucleotide was reacted under identical conditions to those used in kinetics experiments (see above). Reactions were incubated for 1 or 23 h and stopped by ethanol precipitation, dried, and desalted using G-25 sephadex spin columns.
Oligonucleotide samples (~50–100 pmol) were additionally desalted on C18 ZipTips (Millipore) following the manufacturer’s protocol for RNA. RNA was eluted in a matrix solution containing 41 mg/mL 3-hydroxypiccolinic acid and 4.5 mg/mL diammonium citrate then applied to the sample plate. MALDI-MS analysis was performed on a Waters QTof Premier mass spectrometer in positive-ion mode using V-mode optics.
SBBD Thermal Denaturation
Thermal denaturation of SBBD, in a “kinetics” buffer of 100 mM NaNO3, 1 mM Mg(NO3)2, and 5 mM TEA (pH 7.8), was monitored on a Varian Cary 300 Bio UV–visible spectrophotometer with multicell holder and temperature controller. These data were fit with the accompanying software, giving a Tm of 59 °C and a calculated Kd at 37 °C of 6.8 × 10−9 M. On the basis of this Kd, approximately 60% of the SBBD hybrid would be formed during the kinetic analyses, which used a concentration of 0.1 μM for each SBBD strand.
Supplementary Material
Acknowledgments
We thank Dr. Janell Schaak and Sarah Tate for their initial work on the BBD subdomain. We thank Dr. Steve Golledge and the Center for Advanced Materials Characterization in Oregon for support of MALDI-MS facilities, which were obtained under NSF-CRIF CHE-0639170. Funding from the NIH (GM058096), the Robert A. Welch Foundation, and the University of Oregon is gratefully acknowledged.
Footnotes
Supporting Information Available: Alkali hydrolysis mapping of SBBD1 and SBBD2 in platinated SBBD-XL products, dPAGE radiograms and resultant kinetic data for the SBBD construct, kinetic data for BBD platination under varying platinum concentrations depicting first-order kinetics, and a table of platination rates for oligonucleotide constructs. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Jung Y, Lippard SJ. Chem Rev. 2007;107:1387–1407. doi: 10.1021/cr068207j. [DOI] [PubMed] [Google Scholar]
- 2.Wang D, Lippard SJ. Nat Rev Drug Discovery. 2005;4:307–320. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- 3.Reedijk J. Proc Natl Acad Sci USA. 2003;100:3611–3616. doi: 10.1073/pnas.0737293100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akaboshi M, Kawai K, Maki H, Akuta K, Ujeno Y, Miyahara T. Jpn J Cancer Res. 1992;83:522–526. doi: 10.1111/j.1349-7006.1992.tb01959.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pascoe JM, Roberts JJ. Biochem Pharmacol. 1974;23:1345–1357. doi: 10.1016/0006-2952(74)90354-2. [DOI] [PubMed] [Google Scholar]
- 6.Harder HC, Rosenberg B. Int J Cancer. 1970;6:207–216. doi: 10.1002/ijc.2910060207. [DOI] [PubMed] [Google Scholar]
- 7.Jung Y, Lippard SJ. J Biol Chem. 2006;281:1361–1370. doi: 10.1074/jbc.M509688200. [DOI] [PubMed] [Google Scholar]
- 8.Damsma GE, Alt A, Brueckner F, Carell T, Cramer P. Nat Struct Mol Biol. 2007;14:1127–1133. doi: 10.1038/nsmb1314. [DOI] [PubMed] [Google Scholar]
- 9.Schmittgen TD, Ju JF, Danenberg KD, Danenberg PV. Int J Oncol. 2003;23:785–789. [PubMed] [Google Scholar]
- 10.Rosenberg JM, Sato PH. Mol Pharmacol. 1993;43:491–497. [PubMed] [Google Scholar]
- 11.Rosenberg J, Sato P. Mol Pharmacol. 1988;33:611–616. [PubMed] [Google Scholar]
- 12.Hagerlof M, Papsai P, Chow CS, Elmroth SKC. J Biol Inorg Chem. 2006;11:974–990. doi: 10.1007/s00775-006-0157-y. [DOI] [PubMed] [Google Scholar]
- 13.Papsai P, Snygg AS, Aldag J, Elmroth SKC. Dalton Trans. 2008:5225–5234. doi: 10.1039/b719542g. [DOI] [PubMed] [Google Scholar]
- 14.Papsai P, Aldag J, Persson T, Elmroth SKC. Dalton Trans. 2006:3515–3517. doi: 10.1039/b603833f. [DOI] [PubMed] [Google Scholar]
- 15.Hagerlof M, Papsai P, Hedman HK, Jungwirth U, Jenei V, Elmroth SKC. J Biol Inorg Chem. 2008;13:385–399. doi: 10.1007/s00775-007-0327-6. [DOI] [PubMed] [Google Scholar]
- 16.Danenberg PV, Shea LCC, Danenberg KD, Horikoshi T. Nucleic Acids Res. 1991;19:3123–3128. doi: 10.1093/nar/19.11.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rijal K, Chow CS. Chem Commun. 2009:107–109. doi: 10.1039/b816633a. [DOI] [PubMed] [Google Scholar]
- 18.DeRose VJ, Burns S, Kim N-K, Vogt M. Comprehensive Coordination Chemistry II. Elsevier; St. Louis, MO: 2003. pp. 787–813. [Google Scholar]
- 19.Pyle AM. J Biol Inorg Chem. 2002;7:679–690. doi: 10.1007/s00775-002-0387-6. [DOI] [PubMed] [Google Scholar]
- 20.Klein DJ, Moore PB, Steitz TA. RNA. 2004;10:1366–1379. doi: 10.1261/rna.7390804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wahl MC, Will CL, Luhrmann R. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 22.Smith DJ, Query CC, Konarska MM. Mol Cell. 2008;30:657–666. doi: 10.1016/j.molcel.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wachtel C, Manley JL. Mol Biosyst. 2009;5:311–316. doi: 10.1039/b820828j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valadkhan S, Manley JL. Nature. 2001;413:701–707. doi: 10.1038/35099500. [DOI] [PubMed] [Google Scholar]
- 25.Hagerlof M, Papsai P, Chow CS, Elmroth SKC. J Biol Inorg Chem. 2006;11:974–990. doi: 10.1007/s00775-006-0157-y. [DOI] [PubMed] [Google Scholar]
- 26.Ourliac-Garnier I, Bombard S. J Inorg Biochem. 2007;101:514–524. doi: 10.1016/j.jinorgbio.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 27.Sigurdsson ST, Eckstein F. Anal Biochem. 1996;235:241–242. doi: 10.1006/abio.1996.0120. [DOI] [PubMed] [Google Scholar]
- 28.Huggins W, Shapkina T, Wollenzien P. RNA. 2007;13:2000–2011. doi: 10.1261/rna.656907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Behlen LS, Sampson JR, Uhlenbeck OC. Nucleic Acids Res. 1992;20:4055–4059. doi: 10.1093/nar/20.15.4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Butcher SE, Burke JM. Biochemistry. 1994;33:992–999. doi: 10.1021/bi00170a018. [DOI] [PubMed] [Google Scholar]
- 31.Davies MS, Diakos CI, Messerle BA, Hambley TW. J Inorg Biochem. 2000;79:167–172. doi: 10.1016/s0162-0134(99)00180-4. [DOI] [PubMed] [Google Scholar]
- 32.Bancroft DP, Lepre CA, Lippard SJ. J Am Chem Soc. 1990;112:6860–6870. [Google Scholar]
- 33.Chifotides HT, Koomen JM, Kang MJ, Tichy SE, Dunbar KR, Russell DH. Inorg Chem. 2004;43:6177–6187. doi: 10.1021/ic040040u. [DOI] [PubMed] [Google Scholar]
- 34.Green R, Doudna JA. ACS Chem Biol. 2006;1:335–338. doi: 10.1021/cb600277m. [DOI] [PubMed] [Google Scholar]
- 35.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 36.Farazi TA, Juranek SA, Tuschl T. Development. 2008;135:1201–1214. doi: 10.1242/dev.005629. [DOI] [PubMed] [Google Scholar]
- 37.Serganov A, Patel DJ. Nat Rev Genet. 2007;8:776–790. doi: 10.1038/nrg2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Montange RK, Batey RT. Annu Rev Biophys. 2008;37:117–133. doi: 10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- 39.Bayne EH, Allshire RC. Trends Genet. 2005;21:370–373. doi: 10.1016/j.tig.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 40.Mandal M, Breaker RR. Nat Rev Mol Cell Biol. 2004;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- 41.Strobel SA, Cochrane JC. Curr Opin Chem Biol. 2007;11:636–643. doi: 10.1016/j.cbpa.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mansfield KD, Keene JD. Biol Cell. 2009;101:169–181. doi: 10.1042/BC20080055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grundy FJ, Henkin TM. Crit Rev Biochem Mol Biol. 2006;41:329–338. doi: 10.1080/10409230600914294. [DOI] [PubMed] [Google Scholar]
- 44.Lukong KE, Chang KW, Khandjian EW, Richard S. Trends Genet. 2008;24:416–425. doi: 10.1016/j.tig.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 45.Thomas JR, Hergenrother PJ. Chem Rev. 2008;108:1171–1224. doi: 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- 46.Hermann T, Tor Y. Expert Opin Ther Pat. 2005;15:49–62. [Google Scholar]
- 47.Legendre F, Kozelka J, Chottard JC. Inorg Chem. 1998;37:3964–3967. doi: 10.1021/ic980008y. [DOI] [PubMed] [Google Scholar]
- 48.Redon S, Bombard S, Elizondo-Riojas MA, Chottard JC. Nucleic Acids Res. 2003;31:1605–1613. doi: 10.1093/nar/gkg259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Monjardet-Bas W, Chottard JC, Kozelka J. Chem–Eur J. 2002;8:1144–1150. doi: 10.1002/1521-3765(20020301)8:5<1144::aid-chem1144>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 50.Danford AJ, Wang D, Wang Q, Tullius TD, Lippard SJ. Proc Natl Acad Sci USA. 2005;102:12311–12316. doi: 10.1073/pnas.0506025102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ober M, Lippard SJ. J Am Chem Soc. 2008;130:2851–2861. doi: 10.1021/ja710220x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang CX, Chang PV, Lippard SJ. J Am Chem Soc. 2004;126:6536–6537. doi: 10.1021/ja049533o. [DOI] [PubMed] [Google Scholar]
- 53.Garnier IO, Bombard S. J Inorg Biochem. 2007;101:514–524. doi: 10.1016/j.jinorgbio.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 54.Bombard S, Kozelka J, Favre A, Chottard JC. Eur J Biochem. 1998;252:25–35. doi: 10.1046/j.1432-1327.1998.2520025.x. [DOI] [PubMed] [Google Scholar]
- 55.Boer J, Blount KF, Luedtke NW, Elson-Schwab L, Tor Y. Angew Chem, Int Ed. 2005;44:927–932. doi: 10.1002/anie.200461182. [DOI] [PubMed] [Google Scholar]
- 56.Yu YT, Maroney PA, Darzynkiewicz E, Nilsen TW. RNA. 1995;1:46–54. [PMC free article] [PubMed] [Google Scholar]
- 57.Fabrizio P, Abelson J. Nucleic Acids Res. 1992;20:3659–3664. doi: 10.1093/nar/20.14.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Manning GS. Q Rev Biophys. 1978;11:179–246. doi: 10.1017/s0033583500002031. [DOI] [PubMed] [Google Scholar]
- 59.Hambley TW. J Chem Soc, Dalton Trans. 2001:2711–2718. [Google Scholar]
- 60.Kozelka J, Legendre F, Reeder F, Chottard JC. Coord Chem Rev. 1999;192:61–82. [Google Scholar]
- 61.Legendre F, Bas V, Kozelka J, Chottard JC. Chem–Eur J. 2000;6:2002–2010. doi: 10.1002/1521-3765(20000602)6:11<2002::aid-chem2002>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 62.Davies MS, Berners-Price SJ, Hambley TW. Inorg Chem. 2000;39:5603–5613. doi: 10.1021/ic000847w. [DOI] [PubMed] [Google Scholar]
- 63.Davies MS, Berners-Price SJ, Hambley TW. J Am Chem Soc. 1998;120:11380–11390. [Google Scholar]
- 64.Zou Y, Van Houten B, Farrell N. Biochemistry. 1994;33:5404–5410. doi: 10.1021/bi00184a007. [DOI] [PubMed] [Google Scholar]
- 65.Redon S, Bombard S, Elizondo-Riojas MA, Chottard JC. Biochemistry. 2001;40:8463–8470. doi: 10.1021/bi001565a. [DOI] [PubMed] [Google Scholar]
- 66.Villanueva JM, Jia X, Yohannes PG, Doetsch PW, Marzilli LG. Inorg Chem. 1999;38:6069–6080. doi: 10.1021/ic990603f. [DOI] [PubMed] [Google Scholar]
- 67.Brabec V, Vrana O, Boudny V. Prog Biophys Mol Biol. 1996;65:PB113. [Google Scholar]
- 68.Snygg AS, Brindell M, Stochel G, Elmroth SKC. Dalton Trans. 2005:1221–1227. doi: 10.1039/b418966c. [DOI] [PubMed] [Google Scholar]
- 69.Monjardet-Bas V, Elizondo-Riojas MA, Chottard JC, Kozelka J. Angew Chem, Int Ed. 2002;41:2998–3001. doi: 10.1002/1521-3773(20020816)41:16<2998::AID-ANIE2998>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 70.Baik MH, Friesner RA, Lippard SJ. J Am Chem Soc. 2003;125:14082–14092. doi: 10.1021/ja036960d. [DOI] [PubMed] [Google Scholar]
- 71.Mantri Y, Lippard SJ, Baik MH. J Am Chem Soc. 2007;129:5023–5030. doi: 10.1021/ja067631z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang E, van Nimwegen E, Zavolan M, Rajewsky N, Schroeder M, Magnasco M, Darnell JE. Genome Res. 2003;13:1863–1872. doi: 10.1101/gr.1272403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.(a) Measured turnover rates range from 0.034 to 0.0048 μmol/kg · day for rRNA and 0.46–0.88 μmol/kg · day for tRNA. On the basis of the calculation put forth by Petersen et al., this roughly corresponds to 12–29 days for rRNA and 17–29 days for tRNA in average adults. Schoch G, Topp H, Held A, Hellerschoch G, Ballauff A, Manz F. Eur J Clin Nutr. 1990;44:647–658.Nakano K, Nakao T, Schram KH, Hammargren WM, McClure TD, Katz M, Petersen E. Clin Chim Acta. 1993;218:169–183. doi: 10.1016/0009-8981(93)90181-3.Marway JS, Anderson GJ, Miell JP, Ross R, Grimble GK, Bonner AB, Gibbons WA, Peters TJ, Preedy VR. Clin Chim Acta. 1996;252:123–135. doi: 10.1016/0009-8981(96)06300-0.Sander G, Topp H, Hellerschoch G, Wieland J, Schoch G. Clin Sci. 1986;71:367–374. doi: 10.1042/cs0710367.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.