Abstract
DNA nanoparticles of approximately 250nm were produced by rolling circle replication of circular oligonucleotide templates which results in highly condensed DNA particulates presenting concatemeric sequence repeats. Using templates containing randomized sequences, high diversity libraries of particles were produced. A biopanning method that iteratively screens for binding and uses PCR to recover selected particles was developed. The initial application of this technique was the selection of particles that bound to human dendritic cells (DCs). Following 9 rounds of selection the population of particles was enriched for particles that bound DCs, and individual binding clones were isolated and confirmed by flow cytometry and microscopy. This process, which we have termed DeNAno, represents a novel library technology akin to aptamer and phage display, but unique in that the selected moiety is a multivalent nanoparticle whose activity is intrinsic to its sequence. Cell targeted DNA nanoparticles may have applications in cell imaging, cell sorting, and cancer therapy.
The paradigm of nanotechnology for applications in the medical field has been oriented around the framework of bottom-up construction. Generally, a scaffold of polymer or metal serves as a basis for the addition of functional moieties to lend the nanomaterial the desired capabilities such as selective targeting, transport of therapeutic and imaging agents, and immune evasion1. When biopolymers such as DNA are used, they are often rationally designed to form a predetermined structure 2. However, this approach has overlooked a powerful tool of molecular biology: the simple creation and efficient combing of libraries with diversity of 109 or more 3–5. Small nucleic acid aptamer sequences have been identified with binding 6, 7 and enzymatic8 properties, but their use in nanoparticle based applications has mostly involved grafting them onto other materials9. In this study, we have fused the concepts of diverse library selection methods with nanoparticles by creating libraries of DNA nanoparticles by rolling circle replication of randomized circular templates and selecting for particles that bind to a target cell type.
Rolling circle replication of a circular oligonucleotide template using a strand displacing DNA polymerase produces a continuous single strand of DNA that is the concatemeric complement of the template. The single strand condenses into a discrete particle 10–12 that can be visualized by fluorescent microscopy and flow cytometry if fluorescently labeled (Fig. 1). The processivity of the strand displacing enzyme most commonly used, phi29 DNA polymerase, is ~60kb so that a particle produced from a 100–200 oligonucleotide template will consist of several hundred complementary copies. The size of the particles is a function of the reaction kinetics and can be controlled by stopping the reaction with saturating amounts of EDTA and/or heat in activation of the polymerase. Dynamic light scattering (DLS) estimates that particles produced from reactions of 10–60 minutes have hydrodynamic radii between 217–338nm with polydispersity indices of .228–.333 (Fig. 1). These measurements are in good agreement with a freely joined chain model of polymer condensation which estimates a 60kb ssDNA strand to have a hydrodynamic radius of 379nm13. Because of their large size and chaotic single stranded structure, the particles will not migrate in an agarose gel.
Figure 1.
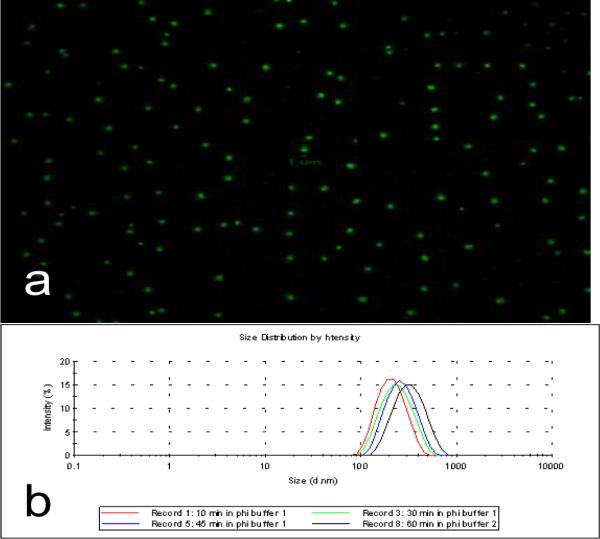
Production and basic characterization of DNA nanoparticles. a) DNA nanoparticles are produced by circularizing a 100nM concentration of a 94 base ssDNA template with T4 Ligase and a 300nM concentration of a 31 base templating primer. Polymerization was done with phi29 DNA polymerase at 30°C for 30 minutes and terminated with EDTA. Discrete particles are stained with SYBR Green and viewed under a 100X oil objective. b) Nanoparticles created for various reaction times are measured with Dynamic Light Scattering to validate size and demonstrate positive correlation of hydrodynamic radius with reaction time
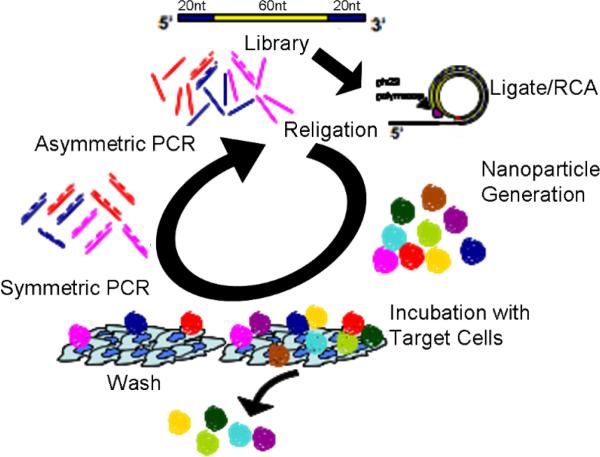
The library screening process consists of three major steps which are performed iteratively: particle synthesis, selection, and amplification. A random library template sequence (5'-Phos-GCGCGGTACATTTGCTGGACTA-N60-TGGAGGTTGGGGATTTGATGTTG 3') (Integrated DNA Technologies, Coralville, IA) was circularized with a template sequence (TCC AGC AAA TGT ACC GCG CCA ACA TCA AAT CCC CAA CCT) using T4 DNA ligase (New England BioLabs, Ipswich, MA) and polymerized with phi29 DNA polymerase (NEB) for 30 minutes at 30°C and terminated by addition of 50mM EDTA. The initial library particle synthesis reaction produced over 1010 unique nanoparticles and was used to begin a selection directed against primary human dendritic cells with an eye towards vaccine or cancer immunotherapy14 applications. Bound particles were amplified by PCR using primers that bound to the sequences flanking the random region. Because each particle contains several hundred copies of the sequence unit, PCR amplification from a single particle is robust. To regenerate the library, the desired single strand template was enriched after symmetric PCR by adding a 20 fold excess of the desired strand's phosphorylated primer v6F (5'-Phos-GCG CGG TAC ATT TGC TGG ACT A). The regenerated single strands were then circularized to form a pool of template circles for the next round of particle synthesis and selection (Fig.2).
Figure 2.
DNA nanoparticle iterative selection scheme. ssDNA libraries are ligated with T4 ligase and polymerized with phi29 DNA polymerase. 3'–5' exonuclease activity of phi29 DNA polymerase ensures nanoparticle purity from extraneous DNA. Immature DCs were cultured in RPMI 1640 medium supplemented with 2 mM L-glutamine, 50 mM 2-mercaptoethanol, 10 mM HEPES, penicillin (100 U/mL), streptomycin (100mg/mL), 5% human AB serum, 1000 U GM-CSF/mL and 200U IL-4/mL and harvested in days 5–7. Cell incubation and washing followed by QPCR (200nM primers, 95°C 2min, cycle 95°C 30sec, 61°C 1min, 72°C 20sec to completion. 5μL of resultant reaction was added to 45 μL fresh PCR buffer with 400nM phosphorlyated template primer v6F. 10 additional cycles of PCR generate an excess of the desired single strand. DNA was purified with a QIAquick Nucleotide Removal Kit (Qiagen, Valencia, CA), eluted into T4 DNA Ligase Buffer and recircularized to begin the next round. Nine rounds were produced after which sequences were cloned using a pGEM-T cloning kit (Promega, Madison, WI).
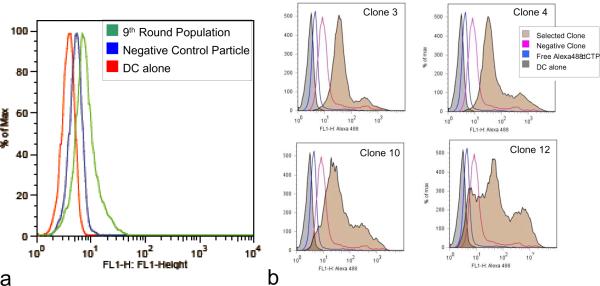
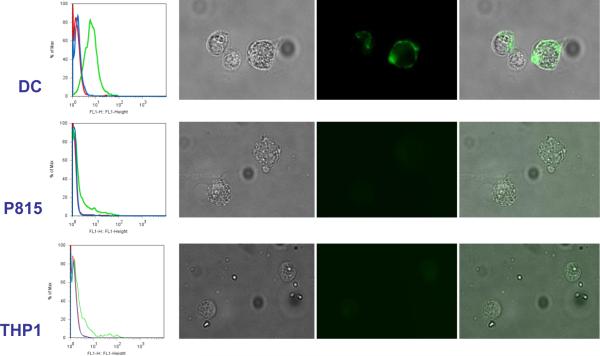
After nine rounds of selection the pool of selected sequences served as templates for the generation of fluorescent DNA nanoparticles by replacing 10% of total dCTPs with ChromaTide® Alexa Fluor® 488-7-OBEA-dCTP (Invitrogen, Carlsbad, CA) in the polymerization reaction and incubating for 30 minutes at 30°C followed by inactivation by EDTA. These fluorescent nanoparticles were used in all analyses of binding by flow cytometry and microscopy. An increase in total population fluorescence was observed compared to a negative DNA nanoparticle control, suggesting that cell binding particles had become enriched (Fig. 3). Individual population members were cloned, sequenced, regenerated as fluorescent particles, and similarly tested for binding by flow cytometry. Several clones were found to bind to DCs more than an irrelevant particle control with some of them demonstrating similar binding patterns. The multivalent binding nature of these nanoparticles may lend them the ability to bind a pattern of surface markers on a cell surface rather than a single target. In the four clones tested in Figure 3, there is definitive homology in the binding characteristics of Clones 3 and 4 that differs significantly from Clones 10 and 12. It is possible that subpopulations of nanoparticles have been selected that bind to unique but distinctive cell surface patterns. It is also interesting to note that even among clones that exhibited similar binding patterns by flow, there was no obvious primary sequence homology. The shape space of such long concatemers is enormous and it likely that even divergent primary sequences may accommodate similar cell surface targets. Consequently, a single clone (Clone 3) (5'GCGCGGTACATTTGCTGGACTATGCATGTTCGTAGTTATATAGGGGGATTGTTTGATAGTCGGAACCGCTGTGCTCAAAGTTTGGAGGTTGGGGATTTGATGTTG 3') was pursued for additional validation (primer sites indicated in bold). Particles with the sequence of Clone 3 were independently generated from a synthetic oligonucleotide template for all subsequent experiments. A control particle made from the reverse complement of the Clone 3 template was also produced. While the selection scheme used did not include a subtractive or counter-selective step to exclude generic cell binding, the selected DNA nanoparticles bound only to DCs and not to human THP1 (acute monocytic leukemia) and mouse P815(mastocytoma) cell lines (Fig. 4). Both flow cytometry and fluorescent microscopy supported the conclusion that the selected particles bind to DCs specifically while the reverse complement control particle did not. Other cell types tested including K562 (chronic myelogenous leukemia) and primary CLL cells (chronic lymphocytic leukemia) also showed no difference between control and selected nanoparticles (data not shown). It is of note that the nature of the fluorescent staining appears much more homogeneous on the cell surface in Figure 4 than would have been predicted from the discrete particle nature seen in Figure 1. This may be a product of the different fluorescent staining methods, OliGreen in Figure 1 and Alexa488-dCTP modified nucleotides in Figure 4, or it may be the result of the nanoparticle in effect spreading out along the cell surface if many possible interactions are possible that are stronger in sum than the net hybridization energy of the unit nanoparticle. Cell binding could be completely abrogated by incubation of the nanoparticles with oligonucleotides that hybridize to the selected random regions, though hybridizing a smaller oligonucleotide to the flanking sequence did not affect the DC binding (data not shown).
Figure 3.
Selection of dendritic cell binding DNA nanoparticles. (a) Nine rounds of selection were performed, after which the selected population was labeled by incorporation of fluorescent nucleotides and the binding to dendritic cells evaluated by flow cytometry. A random clone from the library was used as a negative control. (b) From the ninth round of selection, individual population members were cloned, sequenced and regenerated with fluorescent nucleotides. The incorporation efficiency of Alexa488 OBEA-dCTPs by phi29 polymerase was calculated to be ~1.5%. Controls include an irrelevant DNA nanoparticle and the reaction mix containing the fluorescent dNTPs.
Figure 4.
DC specific binding by clone 3 DNA nanoparticle. Clone 3 particles were generated with incorporated fluorescent nucleotides and evaluated for binding to DCs as well as P815 and THP1 cell lines by flow cytometry and fluorescent microscopy. For each flow cytometry plot (shown on the left), the cells with the labeled clone 3 particles are shown in green, a control particle that is made from the complementary sequence of clone 3 is shown in blue, and the cells alone are indicated by the red curve. Microscopy images show bright field, fluorescent and overlays (from left to right) of fixed cells incubated on ice with the labeled clone 3 particles. Cells were washed 3 times before imaging. Fluorescent staining was seen only on the DC. The complementary control particles did not produce any fluorescent labeling (data not shown).
This suggests that the binding is a consequence of the single stranded nature of the particle, presumably due to specific secondary structure. It is important to note that the DC specificity that we observed was an inadvertent result that cannot be assumed in most positive selection mechanisms. Both the power and weakness of random library selections against complex targets such as cells is that the binding target need not be known in advance so there is no reason to believe that any selected ligand would bind a target unique to a particular cell type. However, subtractive or counter-selective screens against non-specific cell types can be used if necessary to enrich for cell specificity.
An important component of many biological nanoparticle applications for in vivo use is the ability to selectively target the desired cells or tissue. Monoclonal antibodies are the primary tool for biomolecular recognition both experimentally and in vivo. However, the general immunogenicity of non-human antibodies and the immune clearance of nanoparticle aggregated humanized antibodies raise concerns about this approach with nanoparticles. As a result, many nanoparticle applications have turned to molecular selection of aptamers and peptides for targeting ligands in place of antibodies. However, since each of these methods produces a small affinity ligand, the transition to a multivalent platform is commonly performed by the relatively crude method of simply attaching several monomers to a common surface, assuming the coupling can be performed without losing the binding activity of each monomer ligand. A potential problem with this approach is that weak non-specific binding can gain sufficient avidity to dilute the desired specificity. In contrast, because our DNA nanoparticles are composed of concatemeric repeats of a sequence they offer a native multivalent platform in a single particle that allows us to perform a selection on whole particles in the same context of ultimate usage.
DNA has a unique complement of overlapping biochemical, structural, and functional activities when compared to other polymers typically used in nanoparticle synthesis. DNA motifs can act as ligands to specific biomolecules, DNA can be immunogenic if it contains unmethylated CpG motifs 15, it can act as a scaffold for hybridizing other oligonucleotide conjugates, it can have enzymatic activity 16, it is easily chemically modified to allow small molecule or metal ion attachment and metals can be directly deposited onto DNA 17 for imagining, and it can carry DNA binding drugs. DNA has a long clinical history and a favorable toxicity and biodegradability profile18. Cell specific DNA nanoparticles are a potential affinity reagent for research work and are an attractive platform for targeted imaging or therapeutic applications.
Acknowledgements
This work was supported by a grant from the US National Institute of Health - U54CA119335. We thank Hsu-Hsiang Chang, Rebecca Saenz, and Diahnn Futalan for assistance preparing cells.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ferrari M. Cancer nanotechnology: opportunities and challenges. Nat Rev Cancer. 2005;5:161–171. doi: 10.1038/nrc1566. [DOI] [PubMed] [Google Scholar]
- 2.Zhang C, et al. Conformational flexibility facilitates self-assembly of complex DNA nanostructures. Proc Natl Acad Sci U S A. 2008;105:10665–10669. doi: 10.1073/pnas.0803841105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilson DS, Szostak JW. In vitro selection of functional nucleic acids. Annu. Rev. Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 4.Smith GP, Scott JK. Libraries of peptides and proteins displayed on filamentous phage. Methods Enzymol. 1993;217:228–257. doi: 10.1016/0076-6879(93)17065-d. [DOI] [PubMed] [Google Scholar]
- 5.Clackson T, Hoogenboom HR, Griffiths AD, Winter G. Making antibody fragments using phage display libraries. Nature. 1991;352:624–628. doi: 10.1038/352624a0. [DOI] [PubMed] [Google Scholar]
- 6.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 7.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP, Szostak JW. Isolation of new ribozymes from a large pool of random sequences. Science. 1993;261:1411–1418. doi: 10.1126/science.7690155. see comment. [DOI] [PubMed] [Google Scholar]
- 9.Huang CC, Huang YF, Cao Z, Tan W, Chang HT. Aptamer-modified gold nanoparticles for colorimetric determination of platelet-derived growth factors and their receptors. Anal. Chem. 2005;77:5735–5741. doi: 10.1021/ac050957q. [DOI] [PubMed] [Google Scholar]
- 10.Blab GA, Schmidt T, Nilsson M. Homogeneous detection of single rolling circle replication products. Anal. Chem. 2004;76:495–498. doi: 10.1021/ac034987+. [DOI] [PubMed] [Google Scholar]
- 11.Jarvius J, et al. Digital quantification using amplified single-molecule detection. Nat Methods. 2006;3:725–727. doi: 10.1038/nmeth916. [DOI] [PubMed] [Google Scholar]
- 12.Larsson C, et al. In situ genotyping individual DNA molecules by target-primed rolling-circle amplification of padlock probes. Nat Methods. 2004;1:227–232. doi: 10.1038/nmeth723. [DOI] [PubMed] [Google Scholar]
- 13.Austin R. Nanopores: The art of sucking spaghetti. Nat Mater. 2003;2:567–568. doi: 10.1038/nmat962. [DOI] [PubMed] [Google Scholar]
- 14.Fong L, Engleman EG. Dendritic cells in cancer immunotherapy. Annu Rev Immunol. 2000;18:245–273. doi: 10.1146/annurev.immunol.18.1.245. [DOI] [PubMed] [Google Scholar]
- 15.Klinman DM. Adjuvant activity of CpG oligodeoxynucleotides. Int. Rev. Immunol. 2006;25:135–154. doi: 10.1080/08830180600743057. [DOI] [PubMed] [Google Scholar]
- 16.Breaker RR, Joyce GF. A DNA enzyme that cleaves RNA. Chem. Biol. 1994;1:223–229. doi: 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 17.Berti L, Alessandrini A, Facci P. DNA-Templated Photoinduced Silver Deposition. J. Am. Chem. Soc. 2005;127:11216–11217. doi: 10.1021/ja052461w. [DOI] [PubMed] [Google Scholar]
- 18.Fichou Y, Ferec C. The potential of oligonucleotides for therapeutic applications. Trends Biotechnol. 2006;24:563–570. doi: 10.1016/j.tibtech.2006.10.003. [DOI] [PubMed] [Google Scholar]