Abstract
Carcinogenesis is a multistage process, involving oncogene activation and tumor suppressor gene inactivation as well as complex interactions between tumor and host tissues, leading ultimately to an aggressive metastatic phenotype. Among many genetic lesions, mutational inactivation of p53 tumor suppressor, the “guardian of the genome,” is the most frequent event found in 50% of human cancers. p53 plays a critical role in tumor suppression mainly by inducing growth arrest, apoptosis, and senescence, as well as by blocking angiogenesis. In addition, p53 generally confers the cancer cell sensitivity to chemoradiation. Thus, p53 becomes the most appealing target for mechanism-driven anticancer drug discovery. This review will focus on the approaches currently undertaken to target p53 and its regulators with an overall goal either to activate p53 in cancer cells for killing or to inactivate p53 temporarily in normal cells for chemoradiation protection. The compounds that activate wild type (wt) p53 would have an application for the treatment of wt p53-containing human cancer. Likewise, the compounds that change p53 conformation from mutant to wt p53 (p53 reactivation) or that kill the cancer cells with mutant p53 using a synthetic lethal mechanism can be used to selectively treat human cancer harboring a mutant p53. The inhibitors of wt p53 can be used on a temporary basis to reduce the normal cell toxicity derived from p53 activation. Thus, successful development of these three classes of p53 modulators, to be used alone or in combination with chemoradiation, will revolutionize current anticancer therapies and benefit cancer patients.
Introduction
Cancer is usually associated with aberrant cell cycle progression and defective apoptosis induction due to the activation of proto-oncogenes and/or inactivation of tumor suppressor genes [1]. The evolving molecular events often provide the intervening candidate targets for the development of cancer therapy. One of the most promising targets is p53, a well-established and frequently mutated tumor suppressor in human cancer. Since its first discovery in 1979 as an oncogene [2,3], and particularly after its rediscovery as a tumor suppressor gene in 1989 [4,5], p53 has been the hot spot gene for cancer biologists seeking to elucidate the mechanisms of tumor formation and to validate it as a potential cancer therapy target [6–8].
It is well known now that p53 acts biochemically as a transcription factor and biologically as a powerful tumor suppressor. Under normal, unstressed conditions, p53 protein remains undetectable due to its short half-life. The p53 instability is primarily controlled by its negative regulator Mdm2, which, as an E3 ubiquitin ligase, targets p53 for proteasome-mediated degradation [9,10]. Other E3 ubiquitin ligases, which are also implicated in p53 degradation, are Pirh2 and COP1 [11,12]. Another source of p53 instability comes from its own physical property with a melting temperature slightly above body temperature [13]. p53 responds to a wide variety of cellular stresses including genotoxic damages, oncogene activation, and hypoxia [14,15] and is activated on posttranslational modifications by phosphorylation, acetylation, ubiquitination, and methylation [16–18]. Activated p53 then performs its two well-known biological functions: inducing apoptosis or inducing growth arrest [15,19]. The p53-induced apoptosis is mediated by the mitochondrial pathway through transcription-dependent or transcription-31independent mechanisms and by the death receptor pathway through transcriptional activation of FAS and KILLER/DR5 [8,19,20]. p53 also transcriptionally represses cell survival genes such as Bcl-2, survivin, IGFR, Mcl-1, and PIK3CA [21–24] through multiple mechanisms [25]. Conversely, p53-induced growth arrest is mainly mediated through up-regulation of p21, Gadd45, 14-3-3σ, and PTGFβ among others, through a direct DNA binding and transactivation [8,26]. Other p53-involved anticancer mechanisms include induction of cellular senescence [27,28], inhibition of angiogenesis [29,30], and regulation of autophagy [31]. Although the major function of p53 is the “killer,” p53 is also implicated in some cases as a “healer” to enhance the cell survival [21,32].
Given the central role of p53 in cancer prevention and suppression and in chemosensitization or radiosensitization, p53 has to be abrogated during carcinogenesis for most cancers to arise. Indeed, p53 is inactivated by point mutations in more than 50% of human cancers (see http://www.iarc.fr/p53) with a majority of mutations occurring in the DNA binding domain, which either change wt p53 conformation (conformation mutants, e.g., 175H, 249S, 281G) or abolish its DNA contact (contact mutants, e.g., 248W, 273H) [33]. Furthermore, in cancer carrying a wt p53, p53 is often nonfunctional as a result of either being degraded by overexpressed Mdm2 [9,10] or being excluded from the nucleus where p53 acts as a transcriptional factor [19,34,35]. In this review, we aimed to discuss various approaches 1) to activate wt p53, 2) to reactivate mutant p53 or selectively kill cancer cells with mutant p53, and 3) to temporarily inhibit wt p53 for normal cell protection (see Figure 1 and Table 1). Successful clinical development of these three classes of novel compounds would eventually revolutionize the current cancer therapies to benefit a majority of cancer patients.
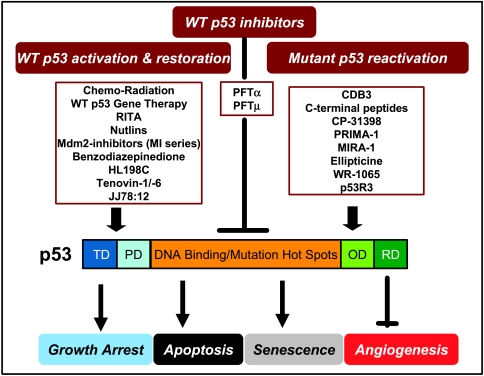
Figure 1.
Current approaches for p53 targeting: p53, the “guardian of the genome,” consists of 393 amino acids with four distinct functional domains. The transactivation domain (TD) and proline-rich domain (PD) is located at the N-terminus, the DNA binding andmutation hot spots domain at the central of themolecule, whereas the oligomerization domain (OD) and regulatory domain (RD) at the C-terminus. On activation, p53 plays a pivotal role in tumor suppression by inducing growth arrest, apoptosis, and senescence, as well as by blocking angiogenesis. Wild-type p53 also confers the sensitivity of cancer cells to chemoradiation. Thus, p53 becomes an appealing therapeutic target for anticancer drug discovery. As illustrated in the figure, three classes of p53 targeting compounds have been identified and characterized. The first class are the compounds that activate or restore wild-type p53 function and can be used in human cancers harboring a wt p53. The second class of compounds reactivates and rescues the mutant p53 with an application in human cancers carrying a p53 mutation. The third class is capable of inhibiting wt p53 and can be used during chemoradiation to block p53 activation in normal cells, thus reducing cytotoxicity.
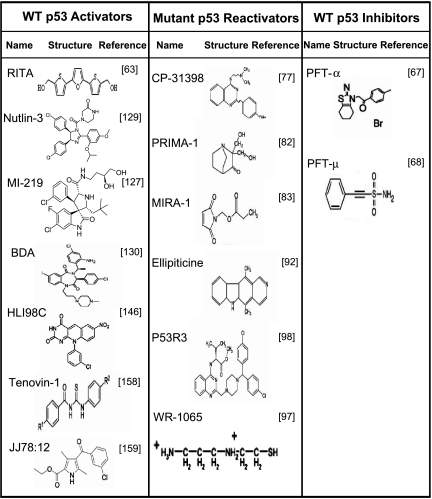
Table 1.
Structure of Small-Molecule p53 Activators, Reactivators, and Inhibitors.
Targeting p53 Itself
Targeting wt p53 — To Activate
The approaches include the use of chemoradiation to activate endogenous wt p53, of gene therapy to introduce wt p53 or modified adenovirus to kill cancer cells with mutant p53, and of synthetic peptides or nongenotoxic small molecules to activate endogenous wt p53.
Chemoradiation
Conventional anticancer therapies target p53 because almost all genotoxic anticancer drugs as well as ionizing radiation (IR) cause the substantial DNA damage, which triggers p53 activation and stabilization [36]. Early preclinical studies using both in vitro cell and in vivo tumor models showed that cells or tumors with a wt p53 are more sensitive to chemoradiation [37,38]. The early clinical studies further showed that mutant p53 confers chemoresistance in patients with ovarian cancer [39,40], breast cancer [41,42], gastric and colorectal cancer [43], and hematological malignancies [42]. Extensive follow-up studies in both preclinical and clinical settings showed that in general, cancer cells with a wt p53 are more sensitive to chemoradiation [44–47], but there are quite few exceptions. For example, breast cancer patients with a transcription-deficient mutant p53 had a better response to chemotherapy than patients with a wt p53 [48]. The same was true for MCF7 breast cancer cells and HCT116 colon cancer cells with p53 either abrogated or deleted, as a result of increased cellular vulnerability to G2 checkpoint abrogators [49,50]. In multiple head and neck cancer cell lines, the absence of p53 appeared to be associated with radiosensitivity [51]. Furthermore, the p53 status determined the cellular response to chemotherapy in an anticancer drug-dependent manner. Colon cancer cells with the p53 gene deleted was found to be more sensitive to the DNA-damaging agent, doxorubicin, but was much more resistant to the antimetabolite 5-fluorouracil [52]. Taken together, these data indicate that because of the nature of tumor heterogeneity, the cellular response to chemoradiation is not solely determined by wt p53. However, understanding such responses in conjunction with p53 status would help the rational design of anticancer therapies to maximize their efficacy.
Gene therapy
To reintroduce wild-type p53
Because p53 function is lost in many cancers, it is logical to restore p53 function by reintroducing wt p53. One common gene therapy approach is the use of viruses to deliver p53. An early study, using retrovirus-mediated gene transfer of wt p53 into human lung cancer cells, showed the inhibition of tumor growth both in vitro and in vivo [53]. Gene therapy using human wt p53, delivered by replication-defective adenovirus (Ad-p53) for better transduction efficiency and lower toxicity, has been extensively studied in the preclinical and clinical settings with an impressive anticancer activity, resulting from p53-induced growth arrest and apoptosis [54]. The Ad-p53, under the brand name of Gendicine, or Advexin, has been currently in clinical use in China since 2003 [55] or in phase 1 to 3 clinical trial in the United States, respectively [7]. The results showed that Gendicine/Advexin is well tolerated in patients and efficacious in the treatment of numerous cancers, particularly head and neck cancer and lung cancer, as a single agent or in combination with chemotherapy or radiation therapy [7,56,57].
To eliminate mutant p53-containing cancer cells by adenovirus
Another p53-related gene therapy is the use of an E1B-deleted adenovirus, designated as ONYX-015, which selectively replicates in p53-deficient cancer cells and subsequently lyse the cells [58]. Preclinical studies showed that ONYX-015 has antitumor activity both in vitro and in vivo, particularly in combination with chemotherapy or radiation therapy [59,60]. Clinical trials revealed that ONYX-015 had a marginal antitumor activity when administrated alone, but a significant effect when combined with standard chemotherapies in a number of human cancers [56,61,62].
Small molecules
Reactivation of p53 and induction of tumor cell apoptosis (RITA) was identified through a cell proliferation assay using a pair of isogenic cancer cell lines differing in p53 status [63]. Biochemically, RITA bound to p53 at the N-terminal domain with a high affinity. Although an initial study showed that RITA could block p53-Mdm2 binding [63], a subsequent in vitro study using nuclear magnetic resonance suggested that it might not disrupt p53-Mdm2 binding [64]. Biologically, RITA suppressed tumor cell growth both in vitro and in vivo by inducing massive apoptosis in a p53-dependent manner in multiple cancer cell models [63]. A recent mechanistic study revealed that RITA, through activating p53, abrogates key oncogenic pathways. Specifically, RITA-activated p53 causes the transcriptional repression of antiapoptotic proteins, including Mcl-1, Bcl-2, survivin, and MAP4, downregulates oncogenic proteins, c-Myc, cyclin E, and β-catenin, and blocks the AKT pathway at multiple levels [65]. Thus, RITA can be further developed as a single agent or used in combination with chemoradiation for effective cancer therapy through p53-mediated abrogation of cancer cell survival and oncogenic pathways.
Targeting wt p53 — To Inhibit
Chemotherapy or radiation therapy kills cancer cells, but at the same time causes many adverse effects because of normal cell toxicity, resulting at least in part from p53 activation and apoptosis induction in normal proliferating cells/tissues, such as bone marrow, lymphoid organs, hair follicles, and epithelium lining of the small intestine. Temporary blockage of p53 activation in normal cells during the treatment of p53-deficient tumors should reduce these adverse effects [66]. Because p53 induces apoptosis through transcription-dependent and transcription-independent, but mitochondria-dependent mechanisms [20], two classes of small molecules have been identified, which target either p53 transcriptional activity or p53-mitochondrial binding activity, respectively. The first class of compound, designated as Pifithrin (PFT)-α, reversibly inhibits p53-dependent transcriptional activation and apoptosis and protects mice from the lethal dose of IR without promoting tumor formation [67]. The second class of compound, designated as PFT-µ, inhibits p53 binding to mitochondria by reducing p53-binding affinity to Bcl-xL and Bcl-2 without affecting p53 transactivation. Similar to PFT-α, PFT-µ also protects primary mouse thymocytes from p53-mediated apoptosis on exposure to radiation and protects mice from radiation-induced lethal hematopoietic syndrome [68]. Thus, these two classes of compounds could be further developed for clinical use in combination of chemoradiation during the treatment of cancers, particularly those with p53 mutation or deletion for normal cell protection.
Targeting Mutant p53 - To Rescue
Because p53 mutation occurs in ∼50% of human cancers, an effective cancer therapy would be the reactivation of p53 mutants. The rescue strategies vary dependent on the mutation types. In tumors harboring a DNA-contact mutant, the attempts were to introduce the functional groups that create new contacts or to stabilize the scaffolding positioning in the remaining contact sites [69,70]. The functions of conformation mutants can be restored by specific small peptides or small molecules that aid the proper folding of the unfolded p53 conformation [70].
Synthetic peptides
CDB3
CDB3 is a short synthetic nine-residue peptide (REDE-DEIEW), derived from p53-binding protein 2 (53BP2 or ASPP), a known p53-binding protein that interacts with the p53 core domain and upregulates p53-dependent transactivation [71]. It was identified through a rational approach searching for small molecules that bind to p53 core domain and stabilize p53 [72]. The notion behind this approach is that a peptide binding with higher affinity to a properly folded state than to an unfolded form of the mutant will shift the equilibrium toward the active wild-type conformation. The in vitro studies showed that CDB3 indeed restored the sequence-specific DNA binding to various p53 mutants by stabilizing them in a bioactive conformation [72]. CDB3 also restored the transcriptional activity of two hot spot p53 mutants, R273H and R175H, for transactivation of two p53 target genes, Mdm2, p21 [73]. Furthermore, CDB3 induced the accumulation of wt p53 and sensitized cancer cells to radiation [73]. Although as a small peptide, CDB3 will likely have poor bioavailability, it can serve as a lead for the further development of p53-reactivating small molecules.
p53 C-terminal peptide
In addition to CDB3, several other studies demonstrated that short synthetic peptides derived from the p53 C-terminal region (peptide 46) can reactivate mutant p53 through stabilization of the core domain folding and/or establishment of novel DNA contacts [74,75]. Although the exact mechanism of action is still unclear, these peptides were indeed shown to rescue the function of endogenous mutant p53 proteins, leading to growth inhibition, apoptosis, and suppression of solid tumor growth in an in vivo animal tumor model [76].
Small molecules
CP-31398
CP-31398 was the first small molecule that has activity to change p53 conformation from mutant to wild type. It was identified through a chemical library screening based on an in vitro biochemical assay that detects wt or mutant p53 conformation through two specific antibodies [77]. CP-31398 was found to stabilize the core domain and enhance transcriptional activity of p53 in living cells [77], but the detailed mechanism of action of CP-31398 remains elusive. Nuclear magnetic resonance studies failed to detect any binding of CP-31398 to the p53 core domain [78]. CP-31398 had no effect on p53-Mdm2 binding, did not cause DNA damage response, or induce p53 phosphorylation at Ser 15 or 20, but rather it increased wt p53 levels by reducing p53 ubiquitination [79]. Biologically, CP-31398 induced growth arrest and apoptosis in a number of human cancer cell lines both in vitro and in vivo [77,80] and inhibited UVB-induced skin carcinogenesis [81]. Furthermore, CP-31398 seems to have other targets in addition to p53 because it could induce cell death in both p53-dependent and -independent manners [70].
PRIMA-1 and MIRA-1
p53 reactivation and induction of massive apoptosis (PRIMA-1) and mutant p53 reactivation and induction of rapid apoptosis (MIRA-1) are two classes of compounds with unique chemical structures that were identified through a cell-based screening for compounds that suppress the growth of tumor cells expressing an exogenous mutant p53 [82,83]. The compounds were found to restore the sequence-specific DNA binding and change the mutant p53 conformation to wild type, leading to transactivation of p53 target genes [82,83]. In contrast to both CP-31398 and CDB3, PRIMA-1 does not activate wild-type p53. A recent mechanistic study revealed that PRIMA-1 is converted to a decomposition product that forms covalent thiol adducts on mutant p53 to restore its tumor suppressor activity [84]. Biologically, both compounds showed antitumor activity in multiple cancer cell lines and in vivo xenograft tumors harboring a mutant p53 without apparent normal tissue toxicity [82–84]. Furthermore, PRIMA-1 was found to be active against p53-null cancer cell lines through a mechanism involving the JNK pathway and heat shock protein 90 [85,86]. Finally, PRIMA-1 or its analog, PRIMA-1(met), sensitized lung cancer cells or prostate cancer cells to adriamycin or radiation, respectively [87,88] and inhibited in vivo xenograft tumor growth [89].
Ellipticine
Ellipticine and its derivatives were initially identified in the drug sensitivity profiling of the NCI-60 cancer cell line panel to be more effective against tumor cells with a mutant p53 [90]. An ellipticine derivative, 9-hydroxyellipticine was found to increase the transcription of p21 and Bax and to induce G1 arrest in a mutant p53-dependent manner [91]. The detailed follow-up work showed that ellipticine changed the p53 conformation from mutant to wild type, restored the sequence-specific DNA binding and transactivation of p53-driven luciferase reporter, and activated mutant p53 to induce p53 target genes, p21 and Mdm2 in mouse xenograft tumor tissues [92]. Furthermore, ellipticine was recently identified to increase nuclear localization of both wt and mutant p53 in a manner independent of DNA damage [93]. Because the ellipticine series of compounds has many off-target activities including promoting p27 degradation [94], it is unlikely to be further developed for the clinic use.
WR-1065
WR-1065 (aminothiol) is an active metabolite of the cytoprotector amifostine and acts as a classic scavenger of reactive oxygen species [95]. WR-1065 was found to activate both wt and mutant p53 and increase the expression of p53 target genes in a manner independent of DNA damage [96,97].
P53R3
p53R3 is a recently identified small molecule that restores the sequence-specific DNA binding of p53 mutants (R175H and R273H) after screening a small library of compounds using an in vitro gel shift assay [98]. The compound was found to enhance the recruitment of both wt and mutant p53 to target promoters and to induce the expression of a number of p53 target genes. Biologically, the compound induces mutant p53-dependent growth arrest and sensitizes TRAIL-induced cell death in multiple glioma cell lines [98].
Targeting Mutant p53 - To Kill
Synthetic lethality for p53 mutation
Synthetic lethality is a situation where a cancer-associated mutation itself is nonlethal but renders cancer cells susceptible to a second hit that becomes lethal on inactivation [99,100]. In the case of p53, which is mutated in 50% of human cancer, application of a synthetic lethality strategy to identify chemical compounds that selectively kill human cancer cells harboring a mutant p53 will be of significant importance for the discovery of a novel class of anticancer drugs (Figure 2). Synthetic lethality has been shown in a number of human cancer cells with deletion of PTEN or DPC4, mutation of K-Ras or B-Raf, and overexpression of c-Myc [101–105], demonstrating the feasibility of using synthetic lethality strategy for anticancer drug discovery. The p53 synthetic lethal drugs, if identified and developed, can be used 1) for cancer treatment to selectively kill mutant p53-containing cancer cells and 2) for chemoprevention to eliminate mutant p53-containing cancer-prone cells, at the early stage of carcinogenesis. Furthermore, p53 synthetic lethal drugs should have, in theory, minimal adverse effects, because normal cells do not contain a p53 mutation.
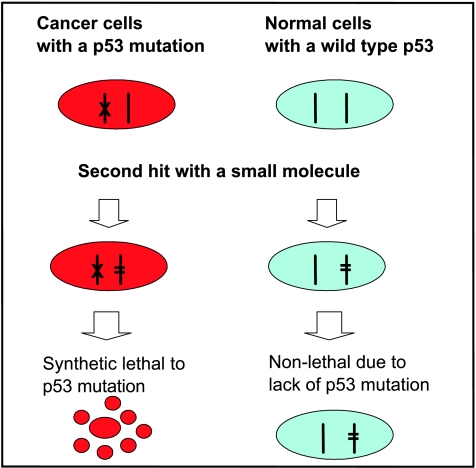
Figure 2.
Synthetic lethality for p53 mutation: Synthetic lethality refers to the situation in which the cancer-associated mutation itself is nonlethal but renders cancer cells susceptible to the second hit, which results in lethal phenotype. Because p53 is most frequently mutated in more than half of human cancer cells, it is feasible in theory to use this strategy to identify drug candidates that preferentially kill cancer cells with a p53 mutation. The p53 synthetic lethal drugs, if identified and developed, should have a minimal toxicity to normal cells and can be used for cancer chemoprevention and treatment of mutant p53-containing cancers.
A “synthetic lethal-like” screening was conducted in a panel of 60 NCI cancer cell lines in an attempt to identify compounds that selectively inhibited the growth of cancer cells devoid of p53 [106]. Few classes of compounds were identified, and the only “popular” hit was paclitaxel, an inhibitor of microtubule polymerization [106]. It was speculated that microtubule-associated protein 4, a p53 transcriptional repressed target, may mediate the sensitivity of cells lacking p53 activity to paclitaxel-induced cell killing [107]. Another drug that selectively inhibited p53-deficient tumor cell growth is metformin, a diabetic drug, with the mechanism associated with activation of AMP kinase and inhibition of oxidative phosphorylation, which created an environment more vulnerable to mutant p53 cells [108].
Small molecules that abrogate the G2/M checkpoint control can be considered to act through the synthetic lethal mechanism against p53 mutation. The hypothesis is based on the fact that p53-deficient cells have abrogated G1 checkpoint control (lack of p53-mediated p21 induction in response to DNA damage), and thus, further abrogation of G2 checkpoint control will selectively kill p53-deficient cancer cells through the induction of mitotic catastrophe. This turns out to be the case. UCN01, a potent G2 checkpoint abrogator sensitized p53 mutant cancer cells to IR by abrogating G2 checkpoint control, whereas cancer cells with wt p53 were much more resistant [109]. Similarly, our previous work showed that PD0166285, a potent Wee-1 kinase inhibitor that abrogated G2 checkpoint control, selectively sensitized p53 mutant cancer cells to radiation [110]. A recent study from Vogelstein's group further supported this notion of synthetic lethality. Transcription profiles of four paired colon cancer cell lines, isogenic for p53 deletion or mutation, revealed a consistent up-regulation of polo-like kinase 1 (PLK-1), a well-known protein that regulates G2/M checkpoint control, in p53-deficient lines after exposure to IR. Consistently, p53-deficient cells are much more sensitive to PLK-1 inhibitors when combined with p53 activators, such as radiation and Nutlin-3. Furthermore, in vivo xenograft tumor studies showed that a PLK-1 inhibitor used as a single agent caused significant regression of p53-null tumors [111]. These studies highlight the feasibility of using synthetic lethal mechanism for effective cancer therapy.
We have recently attempted to identify novel small molecules that act through synthetic lethal mechanism to selectively kill cancer cells with a mutant p53 mutation. We conducted this p53 synthetic lethal screening using a well-characterized p53 temperature-sensitive model in which p53-null H1299 cells were transfected with a temperature-sensitive mutant p53A138V (H1299-p53ts). The p53 in these cells adopts a mutant conformation when grown at 39°C and a wild type conformation at 32°C. Temperature shifting from 39 to 32°C restores the wild type p53 conformation that induces growth arrest but not apoptosis [112,113]. Screening of both chemical and natural product libraries was conducted at both temperatures, and the compounds that selectively killed cells at 39°C (mutant p53 status), but not at 32°C (wt p53 status), were subjected to the secondary counterscreening. In the counterscreening, we used H1299-neo vector control cells at both temperatures to filter out potential false-positives derived merely from temperature shifting. Candidates that showed selective killing of H1299-p53ts at 39°C only were identified, and they are being further characterized (unpublished data). A disadvantage of this type of screening is the lack of mechanism of drug action, which will need follow-up study to identify the “second” target that is synthetically lethal to p53 mutation. The ideal situation would be to conduct simultaneously the same screening using a small interfering RNA (siRNA) library, leading to identification of the “second” target.
Elimination of cancer cells with the gain-of-function p53 mutants
During the validation of our candidate hits, identified from this p53 synthetic lethal screening, we serendipitously found that cardiac glycoside drugs, such as digoxin and ouabain, reduced the p53 levels in a time- and dose-dependent manner in a subset of cancer cell lines but not in immortalized normal cell lines. The drug sensitivity to p53 reduction is cancer cell line-dependent but independent of p53 status of a wild type or mutants. A mechanistic study revealed that the drug-induced p53 decrease neither occurs at the messenger RNA level nor is due to enhanced degradation, which is a common mechanism for p53 regulation. Rather, it occurs at the protein level as a result of reduced de novo synthesis of p53 protein. The drug-induced p53 reduction can be rescued by the inhibitors of Src and MEK, suggesting an involvement of Src/mitogen-activated protein kinase signaling pathways, initiated on the drug binding to Na+/K+-ATPase [114]. Given the fact that glycoside drugs are being used in clinic for the treatment of congestive heart failure [115] and that cardiac glycosides are inactive against wt p53 in normal cells, but potently active in the elimination of mutant p53 in some cancer cells, the drugs may have utility in the treatment of human cancers harboring a gain-of-function p53 mutant [116,117].
Targeting p53 Regulators - To Activate p53
Mdm-2 (Murine Double Minute-2, Hdm2 in Human)
Another effective approach to activate wt p53 is to inhibit its negative regulators. The best-known naturally occurring p53 inhibitor is its downstream target, Mdm2 [118]. Mdm2 encodes a 90-kDa protein that was first identified as the gene responsible for the spontaneous transformation of immortalized murine BALB/c 3T3 cells [119]. It contains a p53 binding domain at the N-terminus and a RING domain at the C-terminus. Mdm2 inhibits p53 through two well-characterized mechanisms: 1) Mdm2 binds to p53 at the N-terminus and blocks p53 transactivation activity and 2) Mdm2 acts as an E3 ubiquitin ligase to promote p53 degradation. In addition, Mdm2 also exports p53 out of nucleus where p53 acts as a transcription factor [34,120]. Mdm2 is overexpressed in approximately 7% of human cancers with a much higher incidence in soft tissue tumors, such as osteosarcoma [121]. Both in vitro and in vivo studies indicated that oncogenic activity of Mdm2 is mainly attributable to its binding and degrading p53 [122,123]. Thus, inhibition of Mdm2 in this subset of human cancer should reactivate p53 to induce cell killing. Indeed, this notion was supported by many proof-of-concept studies, including 1) the blockage of the interaction between Mdm2 and p53 with synthetic peptides or monoclonal antibodies [124] and 2) reduction of Mdm2 levels with antisense oligonucleotides or siRNA [125]. So far, two classes of small molecules have been identified, which either disrupt Mdm2-p53 binding or inhibit Mdm2 E3 ubiquitin ligase activity to reactivate wt p53.
Small Molecules that Disrupt Mdm2-p53 Interaction
Historically, it has been difficult to develop small-molecule inhibitors that disrupt large protein-protein interactions. However, the crystal structure of the Mdm2-p53 peptide binding revealed that the binding relies on the contact of the p53 peptide side chains of Phe19, Trp23, and Leu26 with the N-terminus of Mdm2 (amino acids 17–125) in a deep hydrophobic pocket [126], which made it possible for small molecules to disrupt binding. Several classes of structurally distinctive compounds have been reported to disrupt Mdm2-p53 binding [127,128]. These compounds include Nutlins, benzodiazepinediones (BDAs), and an Mdm2 inhibitor (MI) series of spiro-oxindoles derivatives, including MI-63, MI-219, and MI-43 [127,129–133].
Nutlins
The nutlin series was the first class of small molecules, identified through a screening of a diverse library that disrupted Mdm2-p53 peptide binding. This series of compounds effectively abrogates Mdm2-p53 interaction through a high-affinity binding to Mdm2 [129] with a high selectivity for Mdm2 over Mdmx, a homologue of Mdm2 [127]. Nutlin-3, an analog of the series, induced p53 levels, activated p53 transcription activity, and has a broad activity against cancer cells harboring a wt p53 both in vitro and in vivo [129]. Examples include colon and breast cancer cell lines and osteosarcoma cells [129], lymphoblastic leukemia [134], and retinoblastoma [135]. In combination with chemoradiation, Nutlin-3 showed synergistic activity against prostate cancer [136], lung cancer [137], lymphocytic leukemia [138,139], and neuroblastoma [140]. Nutlin-3 also showed some normal cell protective activity against chemotherapy by inducing cell cycle arrest [141,142]. Furthermore, Nutlins showed a direct antiangiogenic and antiosteoclastic activity, which may have an application for tumors harboring a mutant p53 [143].
Benzodiazepinedione
Benzodiazepinedione (BDA) and its derivatives were identified through a chemical library screening using a miniaturized thermal denaturation-based assay. X-ray crystal structure confirmed that the BDA interacts with the p53-binding pocket of Mdm2 [130]. The compounds increased the p53 transcriptional activity, inhibited the proliferation of cancer cells in a wt p53-dependent manner, and synergized with doxorubicin to inhibit tumor cell growth both in vitro and in vivo [130,133].
MI series
The MI series of spiro-oxindole compounds, including MI-219, MI-63, and MI-43, were discovered through structure-based design by Ding et al. [131] by mimicking all four Mdm2-contacting residues (Phe19, Trp 23, Leu 22, and Leu 26) on p53. The MI series of compounds, with MI-219 being the most potent, bind to Mdm2 with a high affinity. The drug-induced disruption of Mdm2-p53 binding caused p53 accumulation leading to up-regulation of many p53 target genes and to induction of apoptosis in a number of human cancer cell lines derived from breast, colon and prostate cancers in vitro in a wt p53-dependent manner. MI-219 as a single agent also caused a complete inhibition of xenograft tumor growth in vivo at a dose that has no obvious toxicity to animals [127,144,145].
We tested the efficacy of MI-43 against human lung cancer cells and found that the compound preferentially inhibited the growth of wt p53-containing cells over p53-null cells. MI-43 induced the growth arrest at both G1 and G2 phases of the cell cycle, at the low concentration as a result of p21 induction and apoptosis at the high concentration due to Puma/Noxa induction. Importantly, MI-43 was much less toxic to normal lung cells than cancer cells. Finally, when used in combination, MI-43 sensitized chemo resistant A549 cells to etoposide-induced apoptosis [132].
Mdm2 E3 ubiquitin ligase inhibitors
HLI98
The HLI98 series of compounds was identified through the high-throughput screening of a chemical library using an in vitro Mdm2 autoubiquitination assay [146]. The follow-up experiment showed that HLI98C, an analog, indeed inhibited Mdm2 E3 ligase activity. In the cell-based assays, the compound stabilized p53 and Mdm2 and activated p53-dependent transcription and apoptosis. The compound was much more efficacious against cancer cells with wt p53, although it demonstrated some p53-independent cytotoxicity [146]. Although the evidence of in vivo antitumor activity for HLI98C was lacking [146], this proof-of-concept study did indicate that an Mdm2 E3 ligase inhibitor, which has selective activity against wt p53-containing cancer cells, can be identified.
Two concerns should be borne in mind in the clinical development of Mdm2-based inhibitors for anticancer therapy. The first is the therapeutic window because the compounds would also induce p53 in normal cells to cause some adverse effects. Second is the Mdm2 itself, which is accumulated after compound treatment as a result of p53 activation. In addition to targeting p53, Mdm2 interacts with a number of other proteins, such as p73, p63, E2F1, and HIF1α [147–149]. A recent study also showed that Mdm2 caused the accumulation of XIAP to inhibit apoptosis [150]. These p53-independent but Mdm2-dependent effects would eventually affect the efficacy of this class of compounds.
SirT1/SirT2
SirT1 and SirT2 are two members of the NAD+-dependent class III histone deacetylases with a total of seven members in humans [151,152]. SirT1 and SirT2 catalyze the reaction between an acetylated lysine with NAD+, leading to the production of deacetylated lysine, 2′-O-acetyl-ADP-ribose, and nicotinamide [153]. It is well established that acetylation of p53 at lysine 382 enhances p53 DNA binding activity [154]. Thus, SirT1-catalyzed deacetylation of p53 at lysine 382 would deactivate and destabilize p53 [155–157]. These studies established that SirT1 is yet another negative regulator of p53.
Tenovin-1 and Tenovin-6
This series of compounds was identified in a cell-based screening for compounds that activate p53, using a p53-driven transactivation assay [158]. Tenovins stabilized wt p53, induced p53-dependent cell cycle arrest and apoptosis, and suppressed xenograft tumor growth in vivo as a single agent [158]. Mechanistically, Tenovins are nongenotoxic agents and do not induce DNA damage. Rather, they inhibited NAD-dependent deacetylase SirT1/T2, thus restabilizing and reactivating p53. Tenovins, therefore, are a novel class of p53-activating agents that can be further developed for clinical use as well as for the study of sirtuin function as biological tools.
JJ78:12
This series of compounds was also identified by Lain et al. [158] in the same reporter assay used for p53 activator screening. The JJ78:12 series contains much more potent p53 activators with a nanomolar dose range and showed clear anticancer activity both in vitro and in vivo. Mechanistically, this series of compounds caused tubulin depolymerization and may not be further developed for clinical use because of their high cytotoxicity, resulting from tubulin poison [159]. Interestingly, our effort in screening for compounds that change p53 conformation from mutant to wt using a luciferase reporter-based H1299-p53 temperature-sensitive model [112,113] led to the identification of a series of microtubulin poison compounds, including nocodazole, albendazole, parbendazole, and mebendazole (unpublished data), suggesting a nonspecific or indirect nature of p53 activation by this class of compounds.
Targeting Other p53 Family Members
p53 has two family members, p73 and p63 with significant sequence homology among them [160,161]. Like p53, both p73 and p63 bind to the p53-specific DNA binding motif, transactivate p53 downstream target genes, and suppress tumor cell growth by inducing growth arrest and apoptosis [160,161]. The tumor suppression activity of p73 and p63 was further demonstrated in a mouse knockout study, which showed various tumor predispositions on heterozygous deletion of either gene [162]. Unlike p53, p73 or p63 is rarely mutated in human cancers [163] but could be inactivated by binding to a subset of p53 mutants [164]. Targeting p73 and p63 has been proposed for cancer therapy [165,166] with approaches including gene therapy, small peptides, and small molecules.
Gene Therapy
Like the gene therapy approach using wt p53, the same approach has been extended to its family members, p73 and p63. The adenovirus-mediated transduction of p63 and p73 into tumor cells was found to be an alternatively efficient gene therapy approach both in vivo and in vitro [167,168], especially when tumors are resistant to p53-mediated gene therapy [168,169]. Ad-p73 demonstrated a significant cytotoxicity against multiple cancer cell lines tested by inducing both growth arrest and apoptosis [169]. Ad-p73 also sensitized p53 mutant-containing cells to adriamycin with a higher efficiency than Ad-p53 [169]. The apoptosis-inducing effects of Ad-p63γ were also found to be greater than those of wild-type p53 in osteosarcoma cells with Mdm2 amplification [168].
Small Peptides
Short-interfering mutant p53 peptides
One activity of gain-of-function p53 mutants is to physically interact with, sequester, and inactivate p73 [170,171]. Short-interfering mutant p53 (SIMP) peptides were designed based on mutant p53/p73 binding regions. Indeed, SIMP effectively disrupted the protein complex of mutant p53/p73 and restored the p73 function. Biologically, SIMP sensitized mutant p53-containing tumor cells to adriamycin and cisplatin. Notably, the effects of SIMPs are mutant p53-specific and had no effect on cancer cells either with a wt p53 or with a p53-null [172].
37AA
This peptide consists of 37 amino acids from p53 after a noncontiguous fusion of evolutionarily conserved boxes II and III on the DNA binding domain of p53. The 37AA induced cell death through activation of p53/p73 target genes in p53-null cells. Further studies revealed that 37AA was able to bind to iASPP, an inhibitory member of the ASPP family, resulting in the release TA-p73 (a p73 isoform) from iASSP/TA-p73 complex. Anticancer activity of 37AA was further demonstrated in a colon cancer xenograft model, in which 37AA was driven by an expression vector and delivered in dendrimer-based nanoparticles [165,173].
Small Molecules
Wang et al. [174] recently identified a number of smallmolecules in a screening for activating p53 target genes and apoptosis in p53 mutant-containing cells. The follow-up studies using an siRNA-silencing approach confirmed that some of these compounds mediated their activity through transactivating p73. Although the detailed mechanism is unknown, these compounds had anticancer activity when assayed in vivo using the p53-null HCT116 xenograft tumor model [174].
Reactivate transcriptional activity
Reactivate transcriptional activity (RETRA) was identified in a cell-based screening for compounds that reactivate the transcriptional activity of p53 in mutant p53-containing cells [175]. Follow-up studies showed that RETRA activates a number of p53 target genes and selectively inhibited the growth of mutant p53-containing cancer cells both in vitro and in mouse xenografts. Mechanistic studies in mutant p53-containing cells revealed that the compound increased the p73 levels and released p73 from complexing with mutant p53. Importantly, RETRA is active against mutant p53-containing cancer cells without affecting normal cells. Taken together, these studies demonstrated that reactivation of p73 from p53-null or p53 mutant cells is a promising approach for effective cancer therapy.
Conclusions and Future Perspectives
Cancer is a complex disease with multiple genetic and epigenetic alterations. Genetic alterations in any given cancer, even those originating from the same tissue/organ, could have a dramatic difference. Conversely, cancers derived from different tissues/organs may have similar alterations in a given signaling pathway. Thus, an effective personalized cancer therapy requires a thorough understanding of genetic and epigenetic alterations of each individual cancer, followed by rational design of combinational therapies targeting these altered molecules and pathways. These drugs, if further developed from p53 lead compounds described in this review (Table 1), will revolutionize the current cancer therapies by targeting p53 and its regulators on an individual tumor basis. For example, for wt p53-containing tumors, chemoradiation can be used in combination with p53-activating drugs. Similarly, chemoradiation in combination with p53-reactivating drugs or drugs acting through a synthetic lethal mechanism could increase the efficacy against tumors with a mutant p53. The synergistic effect on cancer cell killing of p53 drugs-chemoradiation combination allows a lower dose regimen of routine chemoradiation to reduce normal cell/tissue toxicity. Another approach to reduce normal cell/tissue toxicity is to use p53 inhibitors on a temporary basis during chemoradiation therapy. Furthermore, p53 drugs can be used in combination with other to-be-developed mechanism-driven drugs to achieve a synergistic effect by targeting the same signaling pathway both upstream and downstream. One example will be the combination of Nutlins or MI-219, which activates p53, but with an adverse effect of Mdm2 accumulation [127,132,144], with Smac mimetic drugs, which disrupt XIAP-caspases inhibitory binding to release activated caspases [176–178]. A recent report showed that Mdm2 causes the accumulation of XIAP [150]. This mechanism-driven combination would lead to apoptosis induced by p53 being fully executed with activated caspases. Furthermore, as we gain a better understanding of p53 signaling pathways, additional p53-related targets, upstream and downstream of p53, can be identified and validated for future discovery of novel compounds that target p53 signaling pathways [26,179].
Finally, as the old proverb goes, “prevention is the best medicine.” Chemoprevention is a widely accepted concept, aiming to kill cancer-prone cells at the early stage of carcinogenesis to prevent tumor formation in the first place. Nontoxic natural products from vegetables, teas, and fruits as chemoprevention agents have been extensively studied, and some promising components have been advanced to clinical trials [180–183]. Although our current effort in discovery of p53 drugs is mainly focused on cancer therapy, one promising future direction will be to identify and fully develop natural products, that act through a synthetic lethal mechanism to kill cancer-prone cells with p53 mutations, as chemoprevention drugs. Thus, p53 provides us many opportunities in our fight against cancer, a deadly disease to humankind.
Acknowledgments
The authors thank Min Zheng, Wei Zheng, and Jie Yang for their effort in the screening of synthetic lethal compounds targeting mutant p53, described in this review.
Footnotes
This work was supported by the National Cancer Institute grants (CA111554 and CA118762) and Department of Defense concept award (W81XWH-08-1-0539) to Y.S.
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Linzer DI, Levine AJ. Characterization of a 54K Dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell. 1979;17(1):43–52. doi: 10.1016/0092-8674(79)90293-9. [DOI] [PubMed] [Google Scholar]
- 3.Lane DP, Crawford LV. Tantigen is bound to a host protein in SV40-transformed cells. Nature. 1979;278(5701):261–263. doi: 10.1038/278261a0. [DOI] [PubMed] [Google Scholar]
- 4.Baker SJ, Fearon ER, Nigro JM, Hamilton SR, Preisinger AC, Jessup JM, van Tuinen P, Ledbetter DH, Barker DF, Nakamura Y, et al. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science. 1989;244(4901):217–221. doi: 10.1126/science.2649981. [DOI] [PubMed] [Google Scholar]
- 5.Finlay CA, Hinds PW, Levine AJ. The p53 proto-oncogene can act as a suppressor of transformation. Cell. 1989;57(7):1083–1093. doi: 10.1016/0092-8674(89)90045-7. [DOI] [PubMed] [Google Scholar]
- 6.Koshland DE., Jr Molecule of the year. Science. 1993;262(5142):1953. doi: 10.1126/science.8266084. [DOI] [PubMed] [Google Scholar]
- 7.Vazquez A, Bond EE, Levine AJ, Bond GL. The genetics of the p53 pathway, apoptosis and cancer therapy. Nat Rev Drug Discov. 2008;7(12):979–987. doi: 10.1038/nrd2656. [DOI] [PubMed] [Google Scholar]
- 8.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408(6810):307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 9.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387(6630):296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 10.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387(6630):299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 11.Dornan D, Wertz I, Shimizu H, Arnott D, Frantz GD, Dowd P, O'Rourke K, Koeppen H, Dixit VM. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature. 2004;429(6987):86–92. doi: 10.1038/nature02514. [DOI] [PubMed] [Google Scholar]
- 12.Leng RP, Lin Y, Ma W, Wu H, Lemmers B, Chung S, Parant JM, Lozano G, Hakem R, Benchimol S. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell. 2003;112(6):779–791. doi: 10.1016/s0092-8674(03)00193-4. [DOI] [PubMed] [Google Scholar]
- 13.Bullock AN, Henckel J, Fersht AR. Quantitative analysis of residual folding and DNA binding in mutant p53 core domain: definition of mutant states for rescue in cancer therapy. Oncogene. 2000;19(10):1245–1256. doi: 10.1038/sj.onc.1203434. [DOI] [PubMed] [Google Scholar]
- 14.Giaccia AJ, Kastan MB. The complexity of p53 modulation: emerging patterns from divergent signals. Genes Dev. 1998;12(19):2973–2983. doi: 10.1101/gad.12.19.2973. [DOI] [PubMed] [Google Scholar]
- 15.Ko LJ, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10(9):1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 16.Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer. 2004;4(10):793–805. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- 17.Brooks CL, Gu W. Ubiquitination, phosphorylation and acetylation: the molecular basis for p53 regulation. Curr Opin Cell Biol. 2003;15(2):164–171. doi: 10.1016/s0955-0674(03)00003-6. [DOI] [PubMed] [Google Scholar]
- 18.Scoumanne A, Chen X. Protein methylation: a new mechanism of p53 tumor suppressor regulation. Histol Histopathol. 2008;23(9):1143–1149. doi: 10.14670/hh-23.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88(3):323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 20.Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458(7242):1127–1130. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oren M. Decision making by p53: life, death and cancer. Cell Death Differ. 2003;10(4):431–442. doi: 10.1038/sj.cdd.4401183. [DOI] [PubMed] [Google Scholar]
- 22.Ho J, Benchimol S. Transcriptional repression mediated by the p53 tumour suppressor. Cell Death Differ. 2003;10(4):404–408. doi: 10.1038/sj.cdd.4401191. [DOI] [PubMed] [Google Scholar]
- 23.Astanehe A, Arenillas D, Wasserman WW, Leung PC, Dunn SE, Davies BR, Mills GB, Auersperg N. Mechanisms underlying p53 regulation of PIK3CA transcription in ovarian surface epithelium and in ovarian cancer. J Cell Sci. 2008;121(Pt 5):664–674. doi: 10.1242/jcs.013029. [DOI] [PubMed] [Google Scholar]
- 24.Pietrzak M, Puzianowska-Kuznicka M. p53-dependent repression of the human MCL-1 gene encoding an anti-apoptotic member of the BCL-2 family: the role of Sp1 and of basic transcription factor binding sites in the MCL-1 promoter. Biol Chem. 2008;389(4):383–393. doi: 10.1515/BC.2008.039. [DOI] [PubMed] [Google Scholar]
- 25.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9(5):402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 26.Sun Y. p53 and its downstream proteins as molecular targets of cancer. Mol Carcinog. 2006;45(6):409–415. doi: 10.1002/mc.20231. [DOI] [PubMed] [Google Scholar]
- 27.Campisi J, d'Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8(9):729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 28.Schmitt CA, Fridman JS, Yang M, Lee S, Baranov E, Hoffman RM, Lowe SW. A senescence program controlled by p53 and p16INK4a contributes to the outcome of cancer therapy. Cell. 2002;109(3):335–346. doi: 10.1016/s0092-8674(02)00734-1. [DOI] [PubMed] [Google Scholar]
- 29.Teodoro JG, Evans SK, Green MR. Inhibition of tumor angiogenesis by p53: a new role for the guardian of the genome. J Mol Med. 2007;85(11):1175–1186. doi: 10.1007/s00109-007-0221-2. [DOI] [PubMed] [Google Scholar]
- 30.Folkman J. Tumor suppression by p53 is mediated in part by the antiangiogenic activity of endostatin and tumstatin. Sci STKE. 2006;2006(354):pe35. doi: 10.1126/stke.3542006pe35. [DOI] [PubMed] [Google Scholar]
- 31.Levine B, Abrams J. p53: the Janus of autophagy? at Cell Biol. 2008;10(6):637–639. doi: 10.1038/ncb0608-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janicke RU, Sohn D, Schulze-Osthoff K. The dark side of a tumor suppressor: anti-apoptotic p53. Cell Death Differ. 2008;15(6):959–976. doi: 10.1038/cdd.2008.33. [DOI] [PubMed] [Google Scholar]
- 33.Hollstein M, Rice K, Greenblatt MS, Soussi T, Fuchs R, Sorlie T, Hovig E, Smith-Sorensen B, Montesano R, Harris CC. Database of p53 gene somatic mutations in human tumors and cell lines. Nucleic Acids Res. 1994;22(17):3551–3555. [PMC free article] [PubMed] [Google Scholar]
- 34.Freedman DA, Levine AJ. Regulation of the p53 protein by the MDM2 oncoprotein-thirty-eighth G.H.A. Clowes Memorial Award Lecture. Cancer Res. 1999;59(1):1–7. [PubMed] [Google Scholar]
- 35.Freedman DA, Wu L, Levine AJ. Functions of the MDM2 oncoprotein. Cell Mol Life Sci. 1999;55(1):96–107. doi: 10.1007/s000180050273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.El-Deiry WS. The role of p53 in chemosensitivity and radiosensitivity. Oncogene. 2003;22(47):7486–7495. doi: 10.1038/sj.onc.1206949. [DOI] [PubMed] [Google Scholar]
- 37.Lowe SW, Bodis S, McClatchey A, Remington L, Ruley HE, Fisher DE, Housman DE, Jacks T. p53 status and the efficacy of cancer therapy in vivo. Science. 1994;266(5186):807–810. doi: 10.1126/science.7973635. [DOI] [PubMed] [Google Scholar]
- 38.Lowe SW, Ruley HE, Jacks T, Housman DE. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell. 1993;74(6):957–967. doi: 10.1016/0092-8674(93)90719-7. [DOI] [PubMed] [Google Scholar]
- 39.Herod JJ, Eliopoulos AG, Warwick J, Niedobitek G, Young LS, Kerr DJ. The prognostic significance of Bcl-2 and p53 expression in ovarian carcinoma. Cancer Res. 1996;56(9):2178–2184. [PubMed] [Google Scholar]
- 40.Shelling AN. Role of p53 in drug resistance in ovarian cancer. Lancet. 1997;349(9054):744–745. doi: 10.1016/S0140-6736(05)60195-X. [DOI] [PubMed] [Google Scholar]
- 41.Borresen AL, Andersen TI, Eyfjord JE, Cornelis RS, Thorlacius S, Borg A, Johansson U, Theillet C, Scherneck S, Hartman S, et al. TP53 mutations and breast cancer prognosis: particularly poor survival rates for cases with mutations in the zinc-binding domains. Genes Chromosomes Cancer. 1995;14(1):71–75. doi: 10.1002/gcc.2870140113. [DOI] [PubMed] [Google Scholar]
- 42.Preudhomme C, Fenaux P. The clinical significance of mutations of the P53 tumour suppressor gene in haematological malignancies. Br J Haematol. 1997;98(3):502–511. doi: 10.1046/j.1365-2141.1997.2403057.x. [DOI] [PubMed] [Google Scholar]
- 43.Hamada M, Fujiwara T, Hizuta A, Gochi A, Naomoto Y, Takakura N, Takahashi K, Roth JA, Tanaka N, Orita K. The p53 gene is a potent determinant of chemosensitivity and radiosensitivity in gastric and colorectal cancers. J Cancer Res Clin Oncol. 1996;122(6):360–365. doi: 10.1007/BF01220804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Connor PM, Jackman J, Bae I, Myers TG, Fan S, Mutoh M, Scudiero DA, Monks A, Sausville EA, Weinstein JN, et al. Characterization of the p53 tumor suppressor pathway in cell lines of the National Cancer Institute anticancer drug screen and correlations with the growth-inhibitory potency of 123 anticancer agents. Cancer Res. 1997;57(19):4285–4300. [PubMed] [Google Scholar]
- 45.Lu C, El-Deiry WS. Targeting p53 for enhanced radio- and chemosensitivity. Apoptosis. 2009;14(4):597–606. doi: 10.1007/s10495-009-0330-1. [DOI] [PubMed] [Google Scholar]
- 46.Williams JR, Zhang Y, Zhou H, Gridley DS, Koch CJ, Russell J, Slater JS, Little JB. A quantitative overview of radiosensitivity of human tumor cells across histological type and TP53 status. Int J Radiat Biol. 2008;84(4):253–264. doi: 10.1080/09553000801953342. [DOI] [PubMed] [Google Scholar]
- 47.Gudkov AV, Komarova EA. The role of p53 in determining sensitivity to radiotherapy. Nat Rev Cancer. 2003;3(2):117–129. doi: 10.1038/nrc992. [DOI] [PubMed] [Google Scholar]
- 48.Bertheau P, Plassa F, Espie M, Turpin E, de Roquancourt A, arty M, Lerebours F, Beuzard Y, Janin A, de The H. Effect of mutated TP53 on response of advanced breast cancers to high-dose chemotherapy. Lancet. 2002;360(9336):852–854. doi: 10.1016/S0140-6736(02)09969-5. [DOI] [PubMed] [Google Scholar]
- 49.Fan S, Smith ML, Rivet DJ, II, Duba D, Zhan Q, Kohn KW, Fornace AJ, Jr, O'Connor PM. Disruption of p53 function sensitizes breast cancer MCF-7 cells to cisplatin and pentoxifylline. Cancer Res. 1995;55(8):1649–1654. [PubMed] [Google Scholar]
- 50.Gupta M, Fan S, Zhan Q, Kohn KW, O'Connor PM, Pommier Y. Inactivation of p53 increases the cytotoxicity of camptothecin in human colon HCT116 and breast MCF-7 cancer cells. Clin Cancer Res. 1997;3(9):1653–1660. [PubMed] [Google Scholar]
- 51.Servomaa K, Kiuru A, Grenman R, Pekkola-Heino K, Pulkkinen JO, Rytomaa T. p53 mutations associated with increased sensitivity to ionizing radiation in human head and neck cancer cell lines. Cell Prolif. 1996;29(5):219–230. doi: 10.1046/j.1365-2184.1996.01009.x. [DOI] [PubMed] [Google Scholar]
- 52.Bunz F, Hwang PM, Torrance C, Waldman T, Zhang Y, Dillehay L, Williams J, Lengauer C, Kinzler KW, Vogelstein B. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J Clin Invest. 1999;104(3):263–269. doi: 10.1172/JCI6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fujiwara T, Grimm EA, Mukhopadhyay T, Cai DW, Owen-Schaub LB, Roth JA. A retroviral wild-type p53 expression vector penetrates human lung cancer spheroids and inhibits growth by inducing apoptosis. Cancer Res. 1993;53(18):4129–4133. [PubMed] [Google Scholar]
- 54.Roth JA. Adenovirus p53 gene therapy. Expert Opin Biol Ther. 2006;6(1):55–61. doi: 10.1517/14712598.6.1.55. [DOI] [PubMed] [Google Scholar]
- 55.Pearson S, Jia H, Kandachi K. China approves first gene therapy. Nat Biotechnol. 2004;22(1):3–4. doi: 10.1038/nbt0104-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bouchet BP, de Fromentel CC, Puisieux A, Galmarini CM. p53 as a target for anti-cancer drug development. Crit Rev Oncol Hematol. 2006;58(3):190–207. doi: 10.1016/j.critrevonc.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 57.Peng Z. Current status of gendicine in China: recombinant human Ad-p53 agent for treatment of cancers. Hum Gene Ther. 2005;16(9):1016–1027. doi: 10.1089/hum.2005.16.1016. [DOI] [PubMed] [Google Scholar]
- 58.Bischoff JR, Kirn DH, Williams A, Heise C, Horn S, Muna M, Ng L, Nye JA, Sampson-Johannes A, Fattaey A, et al. An adenovirus mutant that replicates selectively in p53-deficient human tumor cells. Science. 1996;274(5286):373–376. doi: 10.1126/science.274.5286.373. [DOI] [PubMed] [Google Scholar]
- 59.Heise C, Sampson-Johannes A, Williams A, McCormick F, Von Hoff DD, Kirn DH. ONYX-015, an E1B gene-attenuated adenovirus, causes tumorspecific cytolysis and antitumoral efficacy that can be augmented by standard chemotherapeutic agents. Nat Med. 1997;3(6):639–645. doi: 10.1038/nm0697-639. [DOI] [PubMed] [Google Scholar]
- 60.Rogulski KR, Freytag SO, Zhang K, Gilbert JD, Paielli DL, Kim JH, Heise CC, Kirn DH. In vivo antitumor activity of ONYX-015 is influenced by p53 status and is augmented by radiotherapy. Cancer Res. 2000;60(5):1193–1196. [PubMed] [Google Scholar]
- 61.Heise C, Lemmon M, Kirn D. Efficacy with a replication-selective adenovirus plus cisplatin-based chemotherapy: dependence on sequencing but not p53 functional status or route of administration. Clin Cancer Res. 2000;6(12):4908–4914. [PubMed] [Google Scholar]
- 62.Khuri FR, Nemunaitis J, Ganly I, Arseneau J, Tannock IF, Romel L, Gore M, Ironside J, MacDougall RH, Heise C, et al. A controlled trial of intratumoral ONYX-015, a selectively-replicating adenovirus, in combination with cisplatin and 5-fluorouracil in patients with recurrent head and neck cancer. Nat Med. 2000;6(8):879–885. doi: 10.1038/78638. [DOI] [PubMed] [Google Scholar]
- 63.Issaeva N, Bozko P, Enge M, Protopopova M, Verhoef LG, Masucci M, Pramanik A, Selivanova G. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat Med. 2004;10(12):1321–1328. doi: 10.1038/nm1146. [DOI] [PubMed] [Google Scholar]
- 64.Krajewski M, Ozdowy P, D'Silva L, Rothweiler U, Holak TA. NMR indicates that the small molecule RITA does not block p53-MDM2 binding in vitro. Nat Med. 2005;11(11):1135–1136. doi: 10.1038/nm1105-1135. author reply 1136–1137. [DOI] [PubMed] [Google Scholar]
- 65.Grinkevich VV, Nikulenkov F, Shi Y, Enge M, Bao W, Maljukova A, Gluch A, Kel A, Sangfelt O, Selivanova G. Ablation of key oncogenic pathways by RITA-reactivated p53 is required for efficient apoptosis. Cancer Cell. 2009;15(5):441–453. doi: 10.1016/j.ccr.2009.03.021. [DOI] [PubMed] [Google Scholar]
- 66.Gudkov AV, Komarova EA. Dangerous habits of a security guard: the two faces of p53 as a drug target. Hum Mol Genet. 2007;16(Spec No 1):R67–R72. doi: 10.1093/hmg/ddm052. [DOI] [PubMed] [Google Scholar]
- 67.Komarov PG, Komarova EA, Kondratov RV, Christov-Tselkov K, Coon JS, Chernov MV, Gudkov AV. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science. 1999;285(5434):1733–1737. doi: 10.1126/science.285.5434.1733. [DOI] [PubMed] [Google Scholar]
- 68.Strom E, Sathe S, Komarov PG, Chernova OB, Pavlovska I, Shyshynova I, Bosykh DA, Burdelya LG, Macklis RM, Skaliter R, et al. Small-molecule inhibitor of p53 binding to mitochondria protects mice from gamma radiation. Nat Chem Biol. 2006;2(9):474–479. doi: 10.1038/nchembio809. [DOI] [PubMed] [Google Scholar]
- 69.Brachmann RK, Yu K, Eby Y, Pavletich NP, Boeke JD. Genetic selection of intragenic suppressor mutations that reverse the effect of common p53 cancer mutations. EMBO J. 1998;17(7):1847–1859. doi: 10.1093/emboj/17.7.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Selivanova G, Wiman KG. Reactivation of mutant p53: molecular mechanisms and therapeutic potential. Oncogene. 2007;26(15):2243–2254. doi: 10.1038/sj.onc.1210295. [DOI] [PubMed] [Google Scholar]
- 71.Samuels-Lev Y, O'Connor DJ, Bergamaschi D, Trigiante G, Hsieh JK, Zhong S, Campargue I, Naumovski L, Crook T, Lu X. ASPP proteins specifically stimulate the apoptotic function of p53. Mol Cell. 2001;8(4):781–794. doi: 10.1016/s1097-2765(01)00367-7. [DOI] [PubMed] [Google Scholar]
- 72.Friedler A, Hansson LO, Veprintsev DB, Freund SM, Rippin TM, Nikolova PV, Proctor MR, Rudiger S, Fersht AR. A peptide that binds and stabilizes p53 core domain: chaperone strategy for rescue of oncogenic mutants. Proc Natl Acad Sci USA. 2002;99(2):937–942. doi: 10.1073/pnas.241629998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Issaeva N, Friedler A, Bozko P, Wiman KG, Fersht AR, Selivanova G. Rescue of mutants of the tumor suppressor p53 in cancer cells by a designed peptide. Proc Natl Acad Sci USA. 2003;100(23):13303–13307. doi: 10.1073/pnas.1835733100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Selivanova G, Iotsova V, Okan I, Fritsche M, Strom M, Groner B, Grafstrom RC, Wiman KG. Restoration of the growth suppression function of mutant p53 by a synthetic peptide derived fromthe p53 C-terminal domain. Nat Med. 1997;3(6):632–638. doi: 10.1038/nm0697-632. [DOI] [PubMed] [Google Scholar]
- 75.Kim AL, Raffo AJ, Brandt-Rauf PW, Pincus MR, Monaco R, Abarzua P, Fine RL. Conformational and molecular basis for induction of apoptosis by a p53 C-terminal peptide in human cancer cells. J Biol Chem. 1999;274(49):34924–34931. doi: 10.1074/jbc.274.49.34924. [DOI] [PubMed] [Google Scholar]
- 76.Snyder EL, Meade BR, Saenz CC, Dowdy SF. Treatment of terminal peritoneal carcinomatosis by a transducible p53-activating peptide. PLoS Biol. 2004;2(2):E36. doi: 10.1371/journal.pbio.0020036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Foster BA, Coffey HA, Morin MJ, Rastinejad F. Pharmacological rescue of mutant p53 conformation and function. Science. 1999;286(5449):2507–2510. doi: 10.1126/science.286.5449.2507. [DOI] [PubMed] [Google Scholar]
- 78.Rippin TM, Bykov VJ, Freund SM, Selivanova G, Wiman KG, Fersht AR. Characterization of the p53-rescue drug CP-31398 in vitro and in living cells. Oncogene. 2002;21(14):2119–2129. doi: 10.1038/sj.onc.1205362. [DOI] [PubMed] [Google Scholar]
- 79.Wang W, Takimoto R, Rastinejad F, El-Deiry WS. Stabilization of p53 by CP-31398 inhibits ubiquitination without altering phosphorylation at serine 15 or 20 or MDM2 binding. Mol Cell Biol. 2003;23(6):2171–2181. doi: 10.1128/MCB.23.6.2171-2181.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Takimoto R, Wang W, Dicker DT, Rastinejad F, Lyssikatos J, El-Deiry WS. The mutant p53-conformation modifying drug, CP-31398, can induce apoptosis of human cancer cells and can stabilize wild-type p53 protein. Cancer Biol Ther. 2002;1(1):47–55. doi: 10.4161/cbt.1.1.41. [DOI] [PubMed] [Google Scholar]
- 81.Tang X, Zhu Y, Han L, Kim AL, Kopelovich L, Bickers DR, Athar M. CP-31398 restores mutant p53 tumor suppressor function and inhibits UVB-induced skin carcinogenesis in mice. J Clin Invest. 2007;117(12):3753–3764. doi: 10.1172/JCI32481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bykov VJ, Issaeva N, Shilov A, Hultcrantz M, Pugacheva E, Chumakov P, Bergman J, Wiman KG, Selivanova G. Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat Med. 2002;8(3):282–288. doi: 10.1038/nm0302-282. [DOI] [PubMed] [Google Scholar]
- 83.Bykov VJ, Issaeva N, Zache N, Shilov A, Hultcrantz M, Bergman J, Selivanova G, Wiman KG. Reactivation of mutant p53 and induction of apoptosis in human tumor cells by maleimide analogs. J Biol Chem. 2005;280(34):30384–30391. doi: 10.1074/jbc.M501664200. [DOI] [PubMed] [Google Scholar]
- 84.Lambert JM, Gorzov P, Veprintsev DB, Soderqvist M, Segerback D, Bergman J, Fersht AR, Hainaut P, Wiman KG, Bykov VJ. PRIMA-1 reactivates mutant p53 by covalent binding to the core domain. Cancer Cell. 2009;15(5):376–388. doi: 10.1016/j.ccr.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 85.Li Y, Mao Y, Brandt-Rauf PW, Williams AC, Fine RL. Selective induction of apoptosis in mutant p53 premalignant and malignant cancer cells by PRIMA-1 through the c-Jun-NH2-kinase pathway. Mol Cancer Ther. 2005;4(6):901–909. doi: 10.1158/1535-7163.MCT-04-0206. [DOI] [PubMed] [Google Scholar]
- 86.Rehman A, Chahal MS, Tang X, Bruce JE, Pommier Y, Daoud SS. Proteomic identification of heat shock protein 90 as a candidate target for p53 mutation reactivation by PRIMA-1 in breast cancer cells. Breast Cancer Res. 2005;7(5):R765–R774. doi: 10.1186/bcr1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Magrini R, Russo D, Ottaggio L, Fronza G, Inga A, Menichini P. PRIMA-1 synergizes with adriamycin to induce cell death in non-small cell lung cancer cells. J Cell Biochem. 2008;104(6):2363–2373. doi: 10.1002/jcb.21794. [DOI] [PubMed] [Google Scholar]
- 88.Supiot S, Zhao H, Wiman K, Hill RP, Bristow RG. PRIMA-1(met) radiosensitizes prostate cancer cells independent of their MTp53-status. Radiother Oncol. 2008;86(3):407–411. doi: 10.1016/j.radonc.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 89.Bykov VJ, Zache N, Stridh H, Westman J, Bergman J, Selivanova G, Wiman KG. PRIMA-1(MET) synergizes with cisplatin to induce tumor cell apoptosis. Oncogene. 2005;24(21):3484–3491. doi: 10.1038/sj.onc.1208419. [DOI] [PubMed] [Google Scholar]
- 90.Shi LM, Myers TG, Fan Y, O'Connor PM, Paull KD, Friend SH, Weinstein JN. Mining the National Cancer Institute Anticancer Drug Discovery Database: cluster analysis of ellipticine analogs with p53-inverse and central nervous system-selective patterns of activity. Mol Pharmacol. 1998;53(2):241–251. doi: 10.1124/mol.53.2.241. [DOI] [PubMed] [Google Scholar]
- 91.Sugikawa E, Hosoi T, Yazaki N, Gamanuma M, Nakanishi N, Ohashi M. Mutant p53 mediated induction of cell cycle arrest and apoptosis at G1 phase by 9-hydroxyellipticine. Anticancer Res. 1999;19(4B):3099–3108. [PubMed] [Google Scholar]
- 92.Peng Y, Li C, Chen L, Sebti S, Chen J. Rescue of mutant p53 transcription function by ellipticine. Oncogene. 2003;22(29):4478–4487. doi: 10.1038/sj.onc.1206777. [DOI] [PubMed] [Google Scholar]
- 93.Xu GW, Mawji IA, Macrae CJ, Koch CA, Datti A, Wrana JL, Dennis JW, Schimmer AD. A high-content chemical screen identifies ellipticine as a modulator of p53 nuclear localization. Apoptosis. 2008;13(3):413–422. doi: 10.1007/s10495-007-0175-4. [DOI] [PubMed] [Google Scholar]
- 94.Pamarthy D, Tan M, Wu M, Chen J, Yang D, Wang S, Zhang H, Sun Y. p27 degradation by an ellipticinium series of compound via ubiquitin-proteasome pathway. Cancer Biol Ther. 2007;6(3):360–366. doi: 10.4161/cbt.6.3.3703. [DOI] [PubMed] [Google Scholar]
- 95.Grdina DJ, Shigematsu N, Dale P, Newton GL, Aguilera JA, Fahey RC. Thiol and disulfide metabolites of the radiation protector and potential chemopreventive agent WR-2721 are linked to both its anti-cytotoxic and anti-mutagenic mechanisms of action. Carcinogenesis. 1995;16(4):767–774. doi: 10.1093/carcin/16.4.767. [DOI] [PubMed] [Google Scholar]
- 96.North S, El-Ghissassi F, Pluquet O, Verhaegh G, Hainaut P. The cytoprotective aminothiol WR1065 activates p21waf-1 and down regulates cell cycle progression through a p53-dependent pathway. Oncogene. 2000;19(9):1206–1214. doi: 10.1038/sj.onc.1203413. [DOI] [PubMed] [Google Scholar]
- 97.North S, Pluquet O, Maurici D, El-Ghissassi F, Hainaut P. Restoration of wild-type conformation and activity of a temperature-sensitive mutant of p53 (p53(V272M)) by the cytoprotective aminothiolWR1065 in the esophageal cancer cell line TE-1. Mol Carcinog. 2002;33(3):181–188. doi: 10.1002/mc.10038. [DOI] [PubMed] [Google Scholar]
- 98.Weinmann L, Wischhusen J, Demma MJ, Naumann U, Roth P, Dasmahapatra B, Weller M. A novel p53 rescue compound induces p53-dependent growth arrest and sensitises glioma cells to Apo2L/TRAIL-induced apoptosis. Cell Death Differ. 2008;15(4):718–729. doi: 10.1038/sj.cdd.4402301. [DOI] [PubMed] [Google Scholar]
- 99.Kaelin WG. The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer. 2005;5(9):689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- 100.Mizuarai S, Irie H, Schmatz DM, Kotani H. Integrated genomic and pharmacological approaches to identify synthetic lethal genes as cancer therapeutic targets. Curr Mol Med. 2008;8(8):774–783. doi: 10.2174/156652408786733676. [DOI] [PubMed] [Google Scholar]
- 101.Neshat MS, Mellinghoff IK, Tran C, Stiles B, Thomas G, Petersen R, Frost P, Gibbons JJ, Wu H, Sawyers CL. Enhanced sensitivity of PTEN-deficient tumors to inhibition of FRAP/mTOR. Proc Natl Acad Sci USA. 2001;98(18):10314–10319. doi: 10.1073/pnas.171076798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Torrance CJ, Agrawal V, Vogelstein B, Kinzler KW. Use of isogenic human cancer cells for high-throughput screening and drug discovery. Nat Biotechnol. 2001;19(10):940–945. doi: 10.1038/nbt1001-940. [DOI] [PubMed] [Google Scholar]
- 103.Wang H, Han H, Von Hoff DD. Identification of an agent selectively targeting DPC4 (deleted in pancreatic cancer locus 4)-deficient pancreatic cancer cells. Cancer Res. 2006;66(19):9722–9730. doi: 10.1158/0008-5472.CAN-05-4602. [DOI] [PubMed] [Google Scholar]
- 104.Wang Y, Engels IH, Knee DA, Nasoff M, Deveraux QL, Quon KC. Synthetic lethal targeting of MYC by activation of the DR5 death receptor pathway. Cancer Cell. 2004;5(5):501–512. doi: 10.1016/s1535-6108(04)00113-8. [DOI] [PubMed] [Google Scholar]
- 105.Guo W, Wu S, Liu J, Fang B. Identification of a small molecule with synthetic lethality for K-ras and protein kinase Cι. Cancer Res. 2008;68(18):7403–7408. doi: 10.1158/0008-5472.CAN-08-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Weinstein JN, Myers TG, O'Connor PM, Friend SH, Fornace AJ, Jr, Kohn KW, Fojo T, Bates SE, Rubinstein LV, Anderson NL, et al. An informationintensive approach to the molecular pharmacology of cancer. Science. 1997;275(5298):343–349. doi: 10.1126/science.275.5298.343. [DOI] [PubMed] [Google Scholar]
- 107.Zhang CC, Yang JM, Bash-Babula J, White E, Murphy M, Levine AJ, Hait WN. DNA damage increases sensitivity to vinca alkaloids and decreases sensitivity to taxanes through p53-dependent repression of microtubule-associated protein 4. Cancer Res. 1999;59(15):3663–3670. [PubMed] [Google Scholar]
- 108.Buzzai M, Jones RG, Amaravadi RK, Lum JJ, DeBerardinis RJ, Zhao F, Viollet B, Thompson CB. Systemic treatment with the antidiabetic drug metformin selectively impairs p53-deficient tumor cell growth. Cancer Res. 2007;67(14):6745–6752. doi: 10.1158/0008-5472.CAN-06-4447. [DOI] [PubMed] [Google Scholar]
- 109.Wang Q, Fan S, Eastman A, Worland PJ, Sausville EA, O'Connor PM. UCN-01: a potent abrogator of G2 checkpoint function in cancer cells with disrupted p53. J Natl Cancer Inst. 1996;88(14):956–965. doi: 10.1093/jnci/88.14.956. [DOI] [PubMed] [Google Scholar]
- 110.Wang Y, Li J, Booher RN, Kraker A, Lawrence T, Leopold WR, Sun Y. Radiosensitization of p53 mutant cells by PD0166285, a novel G(2) checkpoint abrogator. Cancer Res. 2001;61(22):8211–8217. [PubMed] [Google Scholar]
- 111.Sur S, Pagliarini R, Bunz F, Rago C, Diaz LA, Jr, Kinzler KW, Vogelstein B, Papadopoulos N. A panel of isogenic human cancer cells suggests a therapeutic approach for cancers with inactivated p53. Proc Natl Acad Sci USA. 2009;106(10):3964–3969. doi: 10.1073/pnas.0813333106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pochampally R, Fodera B, Chen L, Lu W, Chen J. Activation of an MDM2-specific caspase by p53 in the absence of apoptosis. J Biol Chem. 1999;274(21):15271–15277. doi: 10.1074/jbc.274.21.15271. [DOI] [PubMed] [Google Scholar]
- 113.Robinson M, Jiang P, Cui J, Li J, Wang Y, Swaroop M, Madore S, Lawrence TS, Sun Y. Global Genechip profiling to identify genes responsive to p53-induced growth arrest and apoptosis in human lung carcinomas. Cancer Biol Ther. 2003;2:406–415. doi: 10.4161/cbt.2.4.437. [DOI] [PubMed] [Google Scholar]
- 114.Wang Z, Zheng M, Li Z, Li R, Jia L, Xiong X, Southall N, Wang S, Xia M, Austin CP, et al. Cardiac glycosides inhibit p53 protein synthesis by a mechanism relieved by Src or MAPK inhibitors. Cancer Res. 2009;69:6556–6564. doi: 10.1158/0008-5472.CAN-09-0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schoner W, Scheiner-Bobis G. Endogenous and exogenous cardiac glycosides: their roles in hypertension, salt metabolism, and cell growth. Am J Physiol Cell Physiol. 2007;293(2):C509–C536. doi: 10.1152/ajpcell.00098.2007. [DOI] [PubMed] [Google Scholar]
- 116.Di Agostino S, Strano S, Emiliozzi V, Zerbini V, Mottolese M, Sacchi A, Blandino G, Piaggio G. Gain of function of mutant p53: the mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer Cell. 2006;10(3):191–202. doi: 10.1016/j.ccr.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 117.Song H, Hollstein M, Xu Y. p53 gain-of-function cancer mutants induce genetic instability by inactivating ATM. Nat Cell Biol. 2007;9(5):573–580. doi: 10.1038/ncb1571. [DOI] [PubMed] [Google Scholar]
- 118.Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7(7a):1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 119.Cahilly-Snyder L, Yang-Feng T, Francke U, George DL. Molecular analysis and chromosomal mapping of amplified genes isolated froma transformed mouse 3T3 cell line. Somat Cell Mol Genet. 1987;13(3):235–244. doi: 10.1007/BF01535205. [DOI] [PubMed] [Google Scholar]
- 120.Zhang Y, Xiong Y. Control of p53 ubiquitination and nuclear export by MDM2 and ARF. Cell Growth Differ. 2001;12(4):175–186. [PubMed] [Google Scholar]
- 121.Momand J, Jung D, Wilczynski S, Niland J. The MDM2 gene amplification database. Nucleic Acids Res. 1998;26(15):3453–3459. doi: 10.1093/nar/26.15.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378(6553):203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 123.deRozieres S, Maya R, Oren M, Lozano G. The loss of mdm2 induces p53-mediated apoptosis. Oncogene. 2000;19(13):1691–1697. doi: 10.1038/sj.onc.1203468. [DOI] [PubMed] [Google Scholar]
- 124.Wasylyk C, Salvi R, Argentini M, Dureuil C, Delumeau I, Abecassis J, Debussche L, Wasylyk B. p53 mediated death of cells overexpressing MDM2 by an inhibitor of MDM2 interaction with p53. Oncogene. 1999;18(11):1921–1934. doi: 10.1038/sj.onc.1202528. [DOI] [PubMed] [Google Scholar]
- 125.Wang H, Nan L, Yu D, Lindsey JR, Agrawal S, Zhang R. Anti-tumor efficacy of a novel antisense anti-MDM2 mixed-backbone oligonucleotide in human colon cancer models: p53-dependent and p53-independent mechanisms. Mol Med. 2002;8(4):185–199. [PMC free article] [PubMed] [Google Scholar]
- 126.Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274(5289):948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 127.Shangary S, Qin D, McEachern D, Liu M, Miller RS, Qiu S, Nikolovska-Coleska Z, Ding K, Wang G, Chen J, et al. Temporal activation of p53 by a specific MDM2 inhibitor is selectively toxic to tumors and leads to complete tumor growth inhibition. Proc Natl Acad Sci USA. 2008;105(10):3933–3938. doi: 10.1073/pnas.0708917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bassett EA, Wang W, Rastinejad F, El-Deiry WS. Structural and functional basis for therapeutic modulation of p53 signaling. Clin Cancer Res. 2008;14(20):6376–6386. doi: 10.1158/1078-0432.CCR-08-1526. [DOI] [PubMed] [Google Scholar]
- 129.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303(5659):844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 130.Grasberger BL, Lu T, Schubert C, Parks DJ, Carver TE, Koblish HK, Cummings MD, LaFrance LV, Milkiewicz KL, Calvo RR, et al. Discovery and cocrystal structure of benzodiazepinedione HDM2 antagonists that activate p53 in cells. J Med Chem. 2005;48(4):909–912. doi: 10.1021/jm049137g. [DOI] [PubMed] [Google Scholar]
- 131.Ding K, Lu Y, Nikolovska-Coleska Z, Wang G, Qiu S, Shangary S, Gao W, Qin D, Stuckey J, Krajewski K, et al. Structure-based design of spiro-oxindoles as potent, specific small-molecule inhibitors of the MDM2-p53 interaction. J Med Chem. 2006;49(12):3432–3435. doi: 10.1021/jm051122a. [DOI] [PubMed] [Google Scholar]
- 132.Sun SH, Zheng M, Ding K, Wang S, Sun Y. A small molecule that disruptsMdm2-p53 binding activates p53, induces apoptosis, and sensitizes lung cancer cells to chemotherapy. Cancer Biol Ther. 2008;7(6):845–852. doi: 10.4161/cbt.7.6.5841. [DOI] [PubMed] [Google Scholar]
- 133.Koblish HK, Zhao S, Franks CF, Donatelli RR, Tominovich RM, LaFrance LV, Leonard KA, Gushue JM, Parks DJ, Calvo RR, et al. Benzodiazepinedione inhibitors of the Hdm2:p53 complex suppress human tumor cell proliferation in vitro and sensitize tumors to doxorubicin in vivo. Mol Cancer Ther. 2006;5(1):160–169. doi: 10.1158/1535-7163.MCT-05-0199. [DOI] [PubMed] [Google Scholar]
- 134.Gu L, Zhu N, Findley HW, Zhou M. MDM2 antagonist nutlin-3 is a potent inducer of apoptosis in pediatric acute lymphoblastic leukemia cells with wild-type p53 and overexpression of MDM2. Leukemia. 2008;22(4):730–739. doi: 10.1038/leu.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Laurie NA, Donovan SL, Shih CS, Zhang J, Mills N, Fuller C, Teunisse A, Lam S, Ramos Y, Mohan A, et al. Inactivation of the p53 pathway in retinoblastoma. Nature. 2006;444(7115):61–66. doi: 10.1038/nature05194. [DOI] [PubMed] [Google Scholar]
- 136.Supiot S, Hill RP, Bristow RG. Nutlin-3 radiosensitizes hypoxic prostate cancer cells independent of p53. Mol Cancer Ther. 2008;7(4):993–999. doi: 10.1158/1535-7163.MCT-07-0442. [DOI] [PubMed] [Google Scholar]
- 137.Cao C, Shinohara ET, Subhawong TK, Geng L, Woon Kim K, Albert JM, Hallahan DE, Lu B. Radiosensitization of lung cancer by nutlin, an inhibitor of murine double minute 2. Mol Cancer Ther. 2006;5(2):411–417. doi: 10.1158/1535-7163.MCT-05-0356. [DOI] [PubMed] [Google Scholar]
- 138.Coll-Mulet L, Iglesias-Serret D, Santidrian AF, Cosialls AM, de Frias M, Castano E, Campas C, Barragan M, de Sevilla AF, Domingo A, et al. MDM2 antagonists activate p53 and synergize with genotoxic drugs in B-cell chronic lymphocytic leukemia cells. Blood. 2006;107(10):4109–4114. doi: 10.1182/blood-2005-08-3273. [DOI] [PubMed] [Google Scholar]
- 139.Kojima K, Konopleva M, McQueen T, O'Brien S, Plunkett W, Andreeff M. Mdm2 inhibitor Nutlin-3a induces p53-mediated apoptosis by transcription-dependent and transcription-independent mechanisms and may overcome Atm-mediated resistance to fludarabine in chronic lymphocytic leukemia. Blood. 2006;108(3):993–1000. doi: 10.1182/blood-2005-12-5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Barbieri E, Mehta P, Chen Z, Zhang L, Slack A, Berg S, Shohet JM. MDM2 inhibition sensitizes neuroblastoma to chemotherapy-induced apoptotic cell death. Mol Cancer Ther. 2006;5(9):2358–2365. doi: 10.1158/1535-7163.MCT-06-0305. [DOI] [PubMed] [Google Scholar]
- 141.Jiang M, Pabla N, Murphy RF, Yang T, Yin XM, Degenhardt K, White E, Dong Z. Nutlin-3 protects kidney cells during cisplatin therapy by suppressing Bax/Bak activation. J Biol Chem. 2007;282(4):2636–2645. doi: 10.1074/jbc.M606928200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kranz D, Dobbelstein M. Nongenotoxic p53 activation protects cells against S-phase.specific chemotherapy. Cancer Res. 2006;66(21):10274–10280. doi: 10.1158/0008-5472.CAN-06-1527. [DOI] [PubMed] [Google Scholar]
- 143.Secchiero P, di Iasio MG, Gonelli A, Zauli G. The MDM2 inhibitor Nutlins as an innovative therapeutic tool for the treatment of haematological malignancies. Curr Pharm Des. 2008;14(21):2100–2110. doi: 10.2174/138161208785294663. [DOI] [PubMed] [Google Scholar]
- 144.Shangary S, Ding K, Qiu S, Nikolovska-Coleska Z, Bauer JA, Liu M, Wang G, Lu Y, McEachern D, Bernard D, et al. Reactivation of p53 by a specific MDM2 antagonist (MI-43) leads to p21-mediated cell cycle arrest and selective cell death in colon cancer. Mol Cancer Ther. 2008;7(6):1533–1542. doi: 10.1158/1535-7163.MCT-08-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Shangary S, Wang S. Targeting the MDM2-p53 interaction for cancer therapy. Clin Cancer Res. 2008;14(17):5318–5324. doi: 10.1158/1078-0432.CCR-07-5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yang Y, Ludwig RL, Jensen JP, Pierre SA, Medaglia MV, Davydov IV, Safiran YJ, Oberoi P, Kenten JH, Phillips AC, et al. Small molecule inhibitors of HDM2 ubiquitin ligase activity stabilize and activate p53 in cells. Cancer Cell. 2005;7(6):547–559. doi: 10.1016/j.ccr.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 147.Lau LM, Nugent JK, Zhao X, Irwin MS. HDM2 antagonist Nutlin-3 disrupts p73-HDM2 binding and enhances p73 function. Oncogene. 2008;27(7):997–1003. doi: 10.1038/sj.onc.1210707. [DOI] [PubMed] [Google Scholar]
- 148.Ambrosini G, Sambol EB, Carvajal D, Vassilev LT, Singer S, Schwartz GK. Mouse double minute antagonist Nutlin-3a enhances chemotherapy-induced apoptosis in cancer cells with mutant p53 by activating E2F1. Oncogene. 2007;26(24):3473–3481. doi: 10.1038/sj.onc.1210136. [DOI] [PubMed] [Google Scholar]
- 149.LaRusch GA, Jackson MW, Dunbar JD, Warren RS, Donner DB, Mayo LD. Nutlin3 blocks vascular endothelial growth factor induction by preventing the interaction between hypoxia inducible factor 1α and Hdm2. Cancer Res. 2007;67(2):450–454. doi: 10.1158/0008-5472.CAN-06-2710. [DOI] [PubMed] [Google Scholar]
- 150.Gu L, Zhu N, Zhang H, Durden DL, Feng Y, Zhou M. Regulation of XIAP translation and induction by MDM2 following irradiation. Cancer Cell. 2009;15(5):363–375. doi: 10.1016/j.ccr.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Haigis MC, Guarente LP. Mammalian sirtuins—emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20(21):2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 152.Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J. 2007;404(1):1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Jackson MD, Denu JM. Structural identification of 2′ and 3′-O-acetyl-ADP-ribose as novel metabolites derived from the Sir2 family of β-NAD+-dependent histone/protein deacetylases. J Biol Chem. 2002;277(21):18535–18544. doi: 10.1074/jbc.M200671200. [DOI] [PubMed] [Google Scholar]
- 154.Luo J, Li M, Tang Y, Laszkowska M, Roeder RG, Gu W. Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo. Proc Natl Acad Sci USA. 2004;101(8):2259–2264. doi: 10.1073/pnas.0308762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Langley E, Pearson M, Faretta M, Bauer UM, Frye RA, Minucci S, Pelicci PG, Kouzarides T. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 2002;21(10):2383–2396. doi: 10.1093/emboj/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2α promotes cell survival under stress. Cell. 2001;107(2):137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 157.Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107(2):149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 158.Lain S, Hollick JJ, Campbell J, Staples OD, Higgins M, Aoubala M, McCarthy A, Appleyard V, Murray KE, Baker L, et al. Discovery, in vivo activity, and mechanism of action of a small-molecule p53 activator. Cancer Cell. 2008;13(5):454–463. doi: 10.1016/j.ccr.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Staples OD, Hollick JJ, Campbell J, Higgins M, McCarthy AR, Appleyard V, Murray KE, Baker L, Thompson A, Ronseaux S, et al. Characterization, chemical optimization and anti-tumour activity of a tubulin poison identified by a p53-based phenotypic screen. Cell Cycle. 2008;7(21):3417–3427. doi: 10.4161/cc.7.21.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Jost CA, Marin MC, Kaelin WG., Jr p73 is a simian [correction of human] p53-related protein that can induce apoptosis. Nature. 1997;389(6647):191–194. doi: 10.1038/38298. [see comments] [published erratum appears in Nature 1999 Jun 24;399(6738):817] [DOI] [PubMed] [Google Scholar]
- 161.Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2(3):305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- 162.Flores ER, Sengupta S, Miller JB, Newman JJ, Bronson R, Crowley D, Yang A, McKeon F, Jacks T. Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell. 2005;7(4):363–373. doi: 10.1016/j.ccr.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 163.Deyoung MP, Ellisen LW. p63 and p73 in human cancer: defining the network. Oncogene. 2007;26(36):5169–5183. doi: 10.1038/sj.onc.1210337. [DOI] [PubMed] [Google Scholar]
- 164.Li Y, Prives C. Are interactions with p63 and p73 involved in mutant p53 gain of oncogenic function? Oncogene. 2007;26(15):2220–2225. doi: 10.1038/sj.onc.1210311. [DOI] [PubMed] [Google Scholar]
- 165.Bell HS, Ryan KM. Targeting the p53 family for cancer therapy: “big brother‘ joins the fight. Cell Cycle. 2007;6(16):1995–2000. doi: 10.4161/cc.6.16.4614. [DOI] [PubMed] [Google Scholar]
- 166.Vilgelm A, El-Rifai W, Zaika A. Therapeutic prospects for p73 and p63: rising from the shadow of p53. Drug Resist Updat. 2008;11(4–5):152–163. doi: 10.1016/j.drup.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sasaki Y, Morimoto I, Ishida S, Yamashita T, Imai K, Tokino T. Adenovirus-mediated transfer of the p53 family genes, p73 and p51/p63 induces cell cycle arrest and apoptosis in colorectal cancer cell lines: potential application to gene therapy of colorectal cancer. Gene Ther. 2001;8(18):1401–1408. doi: 10.1038/sj.gt.3301538. [DOI] [PubMed] [Google Scholar]
- 168.Oshima Y, Sasaki Y, Negishi H, Idogawa M, Toyota M, Yamashita T, Wada T, Nagoya S, Kawaguchi S, Yamashita T, et al. Antitumor effect of adenovirusmediated p53 family gene transfer on osteosarcoma cell lines. Cancer Biol Ther. 2007;6(7) doi: 10.4161/cbt.6.7.4320. [DOI] [PubMed] [Google Scholar]
- 169.Das S, Nama S, Antony S, Somasundaram K. p73 β-expressing recombinant adenovirus: a potential anticancer agent. Cancer Gene Ther. 2005;12(4):417–426. doi: 10.1038/sj.cgt.7700803. [DOI] [PubMed] [Google Scholar]
- 170.Strano S, Munarriz E, Rossi M, Cristofanelli B, Shaul Y, Castagnoli L, Levine AJ, Sacchi A, Cesareni G, Oren M, et al. Physical and functional interaction between p53 mutants and different isoforms of p73. J Biol Chem. 2000;275(38):29503–29512. doi: 10.1074/jbc.M003360200. [DOI] [PubMed] [Google Scholar]
- 171.Strano S, Fontemaggi G, Costanzo A, Rizzo MG, Monti O, Baccarini A, Del Sal G, Levrero M, Sacchi A, Oren M, et al. Physical interaction with human tumor-derived p53 mutants inhibits p63 activities. J Biol Chem. 2002;277(21):18817–18826. doi: 10.1074/jbc.M201405200. [DOI] [PubMed] [Google Scholar]
- 172.Di Agostino S, Cortese G, Monti O, Dell'Orso S, Sacchi A, Eisenstein M, Citro G, Strano S, Blandino G. The disruption of the protein complex mutant p53/p73 increases selectively the response of tumor cells to anticancer drugs. Cell Cycle. 2008;7(21):3440–3447. doi: 10.4161/cc.7.21.6995. [DOI] [PubMed] [Google Scholar]
- 173.Bell HS, Dufes C, O'Prey J, Crighton D, Bergamaschi D, Lu X, Schatzlein AG, Vousden KH, Ryan KM. A p53-derived apoptotic peptide derepresses p73 to cause tumor regression in vivo. J Clin Invest. 2007;117(4):1008–1018. doi: 10.1172/JCI28920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wang W, Kim SH, El-Deiry WS. Small-molecule modulators of p53 family signaling and antitumor effects in p53-deficient human colon tumor xenografts. Proc Natl Acad Sci USA. 2006;103(29):11003–11008. doi: 10.1073/pnas.0604507103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kravchenko JE, Ilyinskaya GV, Komarov PG, Agapova LS, Kochetkov DV, Strom E, Frolova EI, Kovriga I, Gudkov AV, Feinstein E, et al. Small-molecule RETRA suppresses mutant p53-bearing cancer cells through a p73-dependent salvage pathway. Proc Natl Acad Sci USA. 2008;105(17):6302–6307. doi: 10.1073/pnas.0802091105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Bockbrader KM, Tan M, Sun Y. A small molecule Smac-mimic compound induces apoptosis and sensitizes TRAIL- and etoposide-induced apoptosis in breast cancer cells. Oncogene. 2005;24(49):7381–7388. doi: 10.1038/sj.onc.1208888. [DOI] [PubMed] [Google Scholar]
- 177.Li L, Thomas RM, Suzuki H, De Brabander JK, Wang X, Harran PG. A small molecule Smac mimic potentiates TRAIL- and TNFα-mediated cell death. Science. 2004;305(5689):1471–1474. doi: 10.1126/science.1098231. [DOI] [PubMed] [Google Scholar]
- 178.Lu J, Bai L, Sun H, Nikolovska-Coleska Z, McEachern D, Qiu S, Miller RS, Yi H, Shangary S, Sun Y, et al. SM-164: a novel, bivalent Smac mimetic that induces apoptosis and tumor regression by concurrent removal of the blockade of cIAP-1/2 and XIAP. Cancer Res. 2008;68(22):9384–9393. doi: 10.1158/0008-5472.CAN-08-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Hupp TR, Lane DP, Ball KL. Strategies for manipulating the p53 pathway in the treatment of human cancer. Biochem J. 2000;352(Pt 1):1–17. [PMC free article] [PubMed] [Google Scholar]
- 180.Bode AM, Dong Z. Cancer prevention research—then and now. Nat Rev Cancer. 2009;9(7):508–516. doi: 10.1038/nrc2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Clement Y. Can green tea do that? A literature review of the clinical evidence. Prev Med. 2009;49:83–87. doi: 10.1016/j.ypmed.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 182.Khan N, Afaq F, Mukhtar H. Cancer chemoprevention through dietary antioxidants: progress and promise. Antioxid Redox Signal. 2008;10(3):475–510. doi: 10.1089/ars.2007.1740. [DOI] [PubMed] [Google Scholar]
- 183.Nagini S. Cancer chemoprevention by garlic and its organosulfur compounds—panacea or promise? Anticancer Agents Med Chem. 2008;8(3):313–321. doi: 10.2174/187152008783961879. [DOI] [PubMed] [Google Scholar]