Abstract
Post-translational modification of histones is critical for gene expression, mitosis, cell growth, apoptosis, and cancer development. Thus, finding protein kinases that are responsible for the phosphorylation of histones at critical sites is considered an important step in understanding the process of histone modification. The serine/threonine kinase Cot is a member of the mitogen-activated protein kinase (MAPK) kinase kinase family. We show here that Cot can phosphorylate histone H3 at Ser-10 in vivo and in vitro, and that the phosphorylation of histone H3 at Ser-10 is required for Cot-induced cell transformation. We found that activated Cot is recruited to the c-fos promoter resulting in increased activator protein-1 (AP-1) transactivation. The formation of the Cot-c-fos promoter complex was also apparent when histone H3 was phosphorylated at Ser-10. Furthermore, the use of dominant negative mutants of histone H3 revealed that Cot was required for phosphorylation of histone H3 at Ser-10 to induce neoplastic cell transformation. These results revealed an important function of Cot as a newly discovered histone H3 kinase. Moreover, the transforming ability of Cot results from the coordinated activation of histone H3, which ultimately converges on the regulation of the transcriptional activity of the c-fos promoter, followed by AP-1 transactivation activity.
Keywords: c-fos promoter, AP-1
Post-translational modification of histones is an important element in the regulation of gene expression, and histone H3 phosphorylation at Ser-10 has traditionally been regarded as a marker for mitosis (1, 2). Increased phosphorylation of histone H3 at Ser-10 has been observed in mitogen-stimulated and oncogene-transformed mouse fibroblasts (3). The normal checkpoints regulating mitosis are ignored or overridden by the cancer cell. We previously reported that phosphorylation of histone H3 at Ser-10 is essential for neoplastic cell transformation (4). Thus, it is important to identify the responsible kinases and the circumstances under which histone H3 at Ser-10 becomes phosphorylated in order to understand tumorigenesis.
The serine/threonine protein kinase Cot was originally identified as a carboxyl-truncated protein encoded by the cot oncogene, which was isolated by cellular transformation assays on transfection of a human thyroid carcinoma cell line DNA into hamster cells (5). Similarly, the rat cot homologue, designated tpl-2, was identified as a target for provirus insertion in Moloney murine leukemia virus-induced rat T cell lymphomas, which resulted in enhanced expression of a carboxyl-terminally truncated kinase (6, 7). The cot gene appears to be highly expressed in a number of tissues, including the spleen, thymus, liver, and lung (8), and is also expressed at lower levels in many other tissues and cell lines (8, 9). Furthermore, mitogenic stimuli such as concanavalin A (8), inflammatory mediators such as interleukin-1 (IL-1) (10), and tumor promoters such as okadaic acid (10) were all shown to potently induce the expression of cot transcripts in a variety of cell types. However, the molecular mechanisms responsible for Cot’s oncogenic potential are still poorly defined. Cot activates the MAPK and SAPK pathways (11, 12) and induces IL-2 and TNF-α expression in T cell lines (13, 14) by activating the transcription factors NFAT and NF-κB (15, 16). The MAPK and SAPK pathways both play an important role in the transduction of signals generated by growth factors produced in mammary epithelial neoplasms (17, 18). NFAT and NF-κB are also key transcription factors for the activation of cytokines and growth factors (19, 20). Cot has been suggested to regulate neoplastic cell transformation through transcriptional transactivation. Indeed, Chiariello et al. (21) reported that the transforming ability of Cot results from the expression and activity of the product of the c-jun proto-oncogene. Although some signaling events of Cot have been elucidated, the signaling pathway of Cot related to the post-translational modification of chromatin is unknown.
Members of the AP-1 (activator protein-1) family of transcription factors are frequently regulated at the transcriptional and post-transcriptional levels by MAPKs. AP-1 complexes have been shown to be necessary for cell cycle progression in several cell systems (22, 23) and also for cell transformation induced by a variety of oncogenes, including src, ras, and raf (24, 25). Members of the AP-1 family of transcription factors are usually classified into two subfamilies, namely, the Jun (c-Jun, JunB, and JunD) (26–28) and the Fos (c-Fos, FosB, Fra-1, and Fra-2) (29–32) families. Homodimerization of Jun proteins of the two subfamilies (33) or other transcriptional factors, including the ATF2, CREB, NFAT, or SMAD proteins (34–36), confers on the complexes the ability to recognize specific DNA sequences known as tetradecanoyl phorbol acetate-responsive elements or AP-1 sites. In addition, phosphorylation of histone H3 plays a critical role in mitosis, chromatin remodeling, and condensation and occurs concurrently with transcriptional activation of the immediate-early response genes (e.g., c-jun, c-fos, and c-myc) (37). Most recently our group reported that the epidermal growth factor (EGF) -induced phosphorylation of histone H3 at Ser-10 up-regulates c-fos and c-jun transcriptional activity (4).
In the present study we showed that Cot could bind with and phosphorylate histone H3 at Ser-10, and thereby was sufficient to induce the transcriptional activity of the c-fos promoter. In turn, histone H3 was required for Cot-induced cell transformation. In this regard, we found that ultraviolet B (UVB) induced Cot was recruited at the c-fos promoter and induced c-fos transcriptional activity, a mechanism that appeared to increase AP-1 transactivation activity and transformation. Thus, these findings indicated that Cot represents a new example of a serine/threonine kinase that induces cell transformation as a result of its interaction with histone H3 and induction of the c-fos promoter.
MATERIALS AND METHODS
Reagents and antibodies
Chemical reagents, including Tris, NaCl, and SDS for molecular biology and buffer preparation, were purchased from Sigma-Aldrich (St. Louis, MO, USA). EGF, PD 98059, SB 202190, and the Cot kinase inhibitor (4-(3-chlor-4-fluorophenyl-amino)-6-(pyridin-3-yl-methylamino)-3-cyano-1, 7-naphthylridine) were purchased from Calbiochem-Novabiochem (San Diego, CA, USA). Restriction enzymes and some modifying enzymes were obtained from New England BioLabs, Inc. (Beverly, MA, USA). Cell culture medium and other supplements were purchased from Invitrogen (Carlsbad, CA, USA). The DNA ligation kit (version 2.0) was from TAKARA Bio Inc. (Otsu, Shiga, Japan). [γ-32P]ATP and [35S]methionine were purchased from Amersham Biosciences (Piscataway, NJ, USA). Antibodies for immunoblotting and immunocytochemial analysis were purchased from Cell Signaling Technology, Inc. (Beverly, MA, USA), Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA), or Upstate Biotechnology, Inc. (Charlottesville, VA, USA). NE-PER, nuclear, and cytoplasmic extraction reagent were purchased from Pierce (Rockford, IL, USA). The Check-Mate mammalian two-hybrid system, including expression vectors and the reporter luciferase vector, was obtained from Promega Corp. (Madison, WI, USA). The NE-PER Nuclear and Cytoplasmic Extraction Reagents for fractionation of cells were purchased from Pierce (Rockford, IL, USA).
Cell culture conditions and transfection
Human embryonic kidney 293 (HEK293), human cervix adenocarcinoma (HeLa), and mouse embryo fibroblast (NIH3T3) cells were purchased from American Type Culture Collection (ATCC). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) or 10% bovine calf serum (BCS). DNA transfection of cells was performed using Fugene 6 (Roche, Palo Alto, CA, USA).
Construction of mammalian expression and small interfering RNA vectors
For the mammalian two-hybrid (M2H) system, the cDNAs of 60 human kinases were amplified by polymerase chain reaction (PCR), and each was introduced into the pBIND two-hybrid system vector. The cDNA encoding histone H3.3 (a generous gift from Aichi Cancer Center Research Institute, Nagoya, Japan) was recombined into the BamH I/KpnI site of the pACT vector. The point mutation of histone H3 at Ser-10 (S10A), Ser-28 (S28A), or Ser-10/Ser-28 (S10/28A) was generated by using the Quick Change II site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA) and introduced into the pcDNA3.1/V5-His vector (pV5-H3-S10A, -S28A, -S10/28A), respectively. The pRK-myc-Cot plasmid was provided by Warner C. Greene (University of California, San Francisco, CA, USA) and subcloned into BamHI/EcoRI site of pcDNA4-hisMaxA. To construct the siRNA-Cot, pSilencer 3.0-H1 (Ambion, Austin, TX, USA) was digested with XbaI and BbsI. The annealed synthetic primers were then introduced following the recommended protocols (Ambion, Austin, TX, USA): 1) sense siRNAs: GATCCACTGA TCCCAGTAGA TCAAT-TCAAG AGATTGATCT ACTGGGATCA GTTTTTTTGG AAA; 2) antisense siRNAs: AGCTTTTCCA AAAAAACTGA TCCCAGTAGA TCAATCTCTT GAATTGATCT ACTGGGATCA GTG. The human c-fos promoter was a gift from Akihiko Yoshimura (Kyushu University, Fukuoka, Japan) and the AP-1 luciferase reporter plasmid (−73/+63 collagenase-luciferase) was constructed as reported (38).
Isolation of histone proteins
To isolate histone proteins, cells (2×107–5×107) were homogenized in 1 ml of nuclear preparation buffer (10 mM Tris-HCl pH 7.6, 150 mM NaCl, 1.5 mM MgCl2, 0.65% Nonidet P-40, and 1 mM phenylmethylsulfonyl fluoride [PMSF]) in the presence of protein phosphatase inhibitors (10 mM NaF, 1 mM sodium orthovanadate, and 25 mM β-glycerophosphate). Nuclei were recovered by centrifugation at 1500 g for 10 min. All centrifugations were carried out at 4°C. Nuclei were resuspended in 0.3 ml of resuspension buffer (10 mM Tris-HCl pH 7.6, 3 mM MgCl2, 10 mM NaCl, 1 mM PMSF, and protein phosphatase inhibitors). Nuclei were extracted with 0.4 N H2SO4 to isolate total histones. The samples were precipitated with trichloroacetic acid (TCA), then resuspended in double distilled H2O.
Immunoblotting
The proteins were resolved by sodium dodecyl (lauryl) sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene fluoride membranes. The membranes were blocked and hybridized with the appropriate primary antibody overnight at 4°C. Histone H3 and phospho-histone H3 (Ser-10) were detected with the respective specific antibodies. Protein bands were visualized by the chemiluminescence detection kit (ECL, Amersham Biosciences) after hybridization with the horseradish peroxidase-conjugated secondary antibody from rabbit or mouse.
In vitro binding assay and GST protein expression
For expression of the Xpress epitope-tagged Cot, DYRK3, deletion mutant Cot, the appropriate plasmids (pcDNA4/Xpress-Cot, deletion mutants Cot) were in vitro translated with l-[35S]methionine using the TNTQuick coupled transcription/translation system (Promega). For the glutathione-S-transferase (GST) pulldown assay, 5 μg GST fusion proteins were collected on glutathione-Sepharose beads (Amersham Biosciences) and incubated for 4 h at 4°C with [35S]-labeled Cot. The bound proteins were denatured in sample buffer and separated by 10–20% SDS-PAGE, and expression was detected by autoradiography (KODAK, New Haven, CT, USA).
Phosphorylation assay for histone H3 in vitro
Phosphorylation of histone H3 by Cot in vitro was carried out as described previously (39). In brief, the myc-Cot protein was immunoprecipitated from transiently transfected HEK293 cells, combined with 1 μg of bacterial expressed histone H3 in 50 μl kinase buffer (20 mM Tris pH 7.4, 20 mM β-glycerophosphate, 0.1 mM Na3VO4, 10 mM MgCl2, 50 mM NaCl, 1 mM dithiothreitol, 50 mM ATP, 1 mM NaF, and 10 μCi [γ-32P]) for 1 h at 30°C. The samples were separated by 15% SDS-PAGE and gels were dried. The [γ-32P] ATP-labeled histone H3 was visualized by autoradiography or by using a phospho-specific antibody against histone H3 (Ser-10).
Mammalian two-hybrid assay
The DNAs, pACT-histone H3, pBIND-kinases, and pG5-luciferase were combined in the same molar ratio and the total amount of DNA was not more than 100 ng/well. The transfection was performed using the Fugene 6 reagent as described by the manufacturer’s recommended protocols. The cells were disrupted by the addition of 200 μl of cell lysis buffer directly into each well of the 48-well plate, then aliquots of 100 μl were added to individual wells of a 96-well luminescence plate. Luminescence activity was measured automatically by computer program (MTX Lab, INC, Vienna, VA, USA). The relative luciferase activity was calculated and normalized based on the pG5-luciferase basal control. To assess transfection efficiency and protein amount, the Renilla luciferase activity assay or Lowry protein assay was used.
Reporter gene assays
The reporter gene assay for firefly luciferase activity was performed using lysates from transfected cells. In addition, the reporter gene vector phRL-SV40 (Promega) was co-transfected into each cell line and the Renilla luciferase activity generated by this vector was used to normalize the results for transfection efficiency. Cell lysates were prepared by first washing the transfected HEK293 cells (grown in 60 mm diameter dishes) once in phosphate-buffered saline (PBS) at 37°C. After removing the PBS completely, 500 μl of passive lysis buffer (PLB, Promega Dual Lucif-erase Reporter Assay System) was added, then cells were incubated for 1 h with gentle shaking. The supernatant fraction was used to measure firefly and Renilla luciferase activities. Cell lysates (20 μl each) were mixed with 100 μl of luciferase assay II reagent and firefly luciferase light emission was measured by a Luminoskan Ascent plate reader (Thermo Electron Corp., Helsinki, Finland). Subsequently, 100 μl of Renilla luciferase substrate (Promega) was added in order to normalize the firefly luciferase data. The results are expressed as relative c-fos or AP-1 activity (fold) and are presented as luciferase activity relative to the c-fos- or AP-1-only transfected control cells.
Immunofluorescence assay
For translocation of endogenous Cot, the cells were fixed in 4% paraformaldehyde and Cot was detected with a monoclonal Cot antibody and a Texas Red-conjugated secondary antibody. Phosphorylation of histone H3 (Ser-10) was detected with monoclonal anti-phospho-histone H3-FITC (Ser-10). Nuclei were stained with 4, 6-diamidino-2-phenylindole. Cells were incubated for 24 h, starved in serum-free media for an additional 24 h, then were or were not irradiated with UVB (4 kJ/m2) and harvested after 30 min additional incubation. Samples were analyzed using a fluorescence microscope system (Leica) and the Image-Pro software program v.4.
Chromatin immunoprecipitation assay
Nuclear factors associated with chromatin in HEK293 cells were cross-linked to DNA by using formaldehyde (1%). Cells were harvested, and cross-linked chromatin was sheared by sonication. DNA fragments were < 1 kb and averaged 450 bp as verified by agarose gel electrophoresis. Immunoprecipitation was performed with 100 μg (DNA content) of chromatin extracts diluted in ChIP dilution buffer (1.1% Triton X-100, 0.01% SDS, 1.2 mM EDTA, 16.7 mM Tris-HCl pH 8.1, and 167 mM NaCl). Samples were precleared with Salmon Sperm DNA/Protein A agarose beads (Upstate) for 30 min, then incubated overnight (16 h) with 4 μg of anti-histone H3, anti-phospho-histone H3 (Ser-10), or anti-myc, respectively, at 4°C. DNA present in the immunoprecipitated chromatin was isolated after reversed cross-link and proteinase K digestion, then the specific region of the c-fos promoter was confirmed by PCR amplification using the following primer sets: 5′-CCCGACCTCGGGAACAAGGG-3′ (− 491 forward) and 5′-ATGAGGGGTTTCGGGGATGG-3′ (−233 reverse).
Anchorage-independent cell transformation assay (soft agar assay)
EGF-induced cell transformation was investigated in mock, psi-H3, or pV5-H3 stably transfected cells. In brief, cells (8×103/ml) were exposed to EGF (0.1–10 ng/ml) in 1 ml of 0.3% basal medium Eagle (BME) agar containing 10% FBS. The cultures were maintained at 37°C in a 5% CO2 incubator for 10 days, and the cell colonies were scored using a microscope and the Image-Pro PLUS computer software program (Media Cybernetics, Silver Spring, MD, USA).
Focus-forming assay
Transformation of NIH3T3 cells was performed following standard protocols (40). Cells were plated in 100 mm dishes at a density of 1 × 104 cells and, after incubation for 3 wk, were transiently transfected with 0.1 μg of the H-RasG12V, 2.5 μg pV5, 2.5 μg pRK-myc-Cot, and/or 2.5 μg pV5-H3 plasmid. Cells were kept in MEM with 5% BCS and media were changed every 3 days for a period of 3 wk. Foci were enumerated by staining the monolayer with methanol for fixation and 0.4% crystal violet for visualization. Data shown represent data obtained from three independent plates for each transfection.
RESULTS
Cot interacts with and phosphorylates histone H3
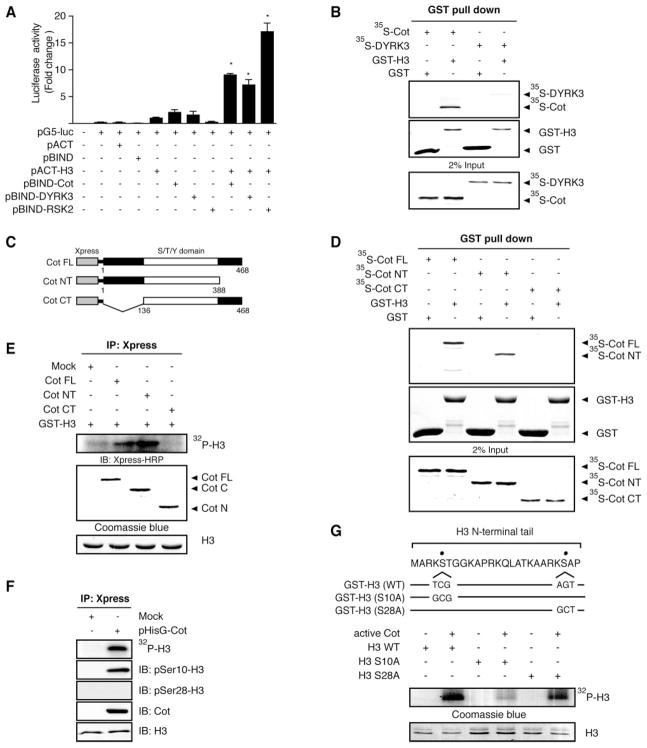
Several lines of evidence have recently shown that mitotic phosphorylation of histone H3 at Ser-10 is responsible for chromosome instability, and thus histone H3 was suggested to play a role in carcinogenesis (41). Therefore, we investigated potential protein binding partners of histone H3 by screening with the M2H system (Promega). Among the 50 protein kinases screened, the Cot oncoprotein was found to interact with histone H3 in vitro. In this system, histone H3 was cloned into the pACT expression vector (pACT-H3) and the Cot kinase was cloned into the pBIND expression vector (pBIND-Cot) in combination with the pG5 luciferase reporter gene. An interaction between histone H3 and Cot brings together the Gal4 binding domain (from pBIND) and the VP16 transactivation domain (from pACT) of the fusion proteins and activates the luciferase reporter gene in NIH3T3 cells. From results of the M2H assay, we confirmed that histone H3 interacted with Cot (Fig. 1A, lane 9) as well as DYRK3 or RSK2 (Fig. 1A, lanes 10 and 11, respectively), which served as positive controls. Data indicated that an ~10-fold increase in luciferase activity was observed in Cot and histone H3 cotransfected cells compared with cells transfected with only pACT-H3. We next examined the in vitro interaction between Cot and purified GST-histone H3. First, the cDNA coding sequence of Cot was subcloned into the pcDNA4His/Max vector to generate the Xpress epitope-tagged Cot, and the fusion protein was in vitro translated using the TNT Quick coupled transcription/translation system (Promega). Affinity-purified GST-histone H3 immobilized to GST beads was incubated with [35S] methionine-labeled Cot. The bound proteins eluted from the beads were separated by SDS-PAGE, then detected by autoradiography. Results (Fig. 1B) showed that Cot efficiently interacted with histone H3 by in vitro GST pulldown assay. To identify the region of Cot that is responsible for its interaction with histone H3, WT full-length Cot (Cot FL) and C-terminal deletion (Cot NT) and N-terminal deletion (Cot CT) mutants (Fig. 1C) were in vitro translated using the TNT Quick coupled transcription/translation system, and the interaction with GST-histone H3 was determined by GST pulldown assay. The results suggested that the N-terminal of Cot was required for the interaction with histone H3 (Fig. 1D). To further determine whether histone H3 was a substrate of Cot, we next performed an in vitro kinase assay with histone H3 using HEK293 cells overexpressing Cot. In this experiment, wild-type-Cot, N-terminal, and C-terminal deletion mutant cells were subjected to immunoprecipitation using anti-Xpress against an Xpress epitope-tagged Cot. Cot kinase activity with histone H3 as substrate was measured at 30°C for 1 h. Results confirmed that phosphorylation of histone H3 was markedly increased by kinase activity of Cot WT or the C-terminal deletion mutant, but not the N-terminal deletion mutant (Fig. 1E). We next explored whether Cot phosphorylation of histone H3 occurred at Ser-10 or Ser-28 in vitro. Cot phosphorylated histone H3 only at Ser-10 but not at Ser-28 in vitro (Fig. 1F). To confirm that Cot specifically phosphorylated histone H3 at Ser-10, but not Ser-28, we replaced Ser-10 of histone H3 with alanine (S10A), Ser-28 with alanine (S28A), or both Ser-10/28 with alanine (Ser-10/28), then subcloned these constructs into the pcDNA3.1/V5-His vector (Fig. 1G, top panel). Compared with histone H3 WT, the mutation of histone H3 (Ser-10) suppressed the ability of Cot to phosphorylate histone H3 whereas the mutation of histone H3 (Ser-28) had no effect, confirming that Ser-10 of histone H3 is the specific Cot phosphorylation site (Fig. 1G).
Figure 1.
Cot binds with histone H3. A) Direct interaction of Cot with histone H3 as determined by the M2H assay. For a negative control, pACT or pBIND plasmids were transfected along with the pG5-luc reporter plasmid into NIH3T3 cells (18,000 cells/ml). For a positive control, histone pACT-H3 and pBIND-RSK2 or pBIND-DYRK3 were transfected with the pG5-luc reporter plasmid into NIH3T3 cells. Histone pACT-H3 and/or pBIND-Cot were cotransfected with the pG5-luc plasmid to confirm the interaction of Cot with histone H3. After a 36 h incubation, firefly luciferase activity was determined in the cell lysates and normalized against Renilla luciferase activity. All experiments were performed at least twice with triplicate samples and are depicted as the means ± se. An asterisk (*) indicates a significant increase in activity compared with negative control, pACT-H3 only (P<0.05). The data were recorded as a relative fold change in luciferase activity as measured by a Luminoskan Ascent plate reader (Thermo Electron Corp., Helsinki, Finland). B) In vitro binding with an 35S-labeled Cot protein and a GST-histone H3 fusion protein. The cDNA of Cot or DYRK3 was translated in vitro, then the 35S-Cot or 35S-DYRK3 (positive control) proteins were mixed with GST-histone H3 and a pulldown assay was performed. Proteins were visualized by autoradiography. Input represents 2% of the material used for the in vitro binding assays. C) Schematic diagram of full-length Xpress-Cot (1–468, Cot FL), the N-terminal (1–368) fragment of Xpress-Cot (Cot NT), or the C-terminal (136–468) fragment of Xpress-Cot (Cot CT). D) In vitro interactions of the 35S-labeled full-length Cot (Cot FL), the N-terminal fragment of Xpress-Cot (Cot NT) or the C-terminal fragment of Xpress-Cot (Cot CT) with GST only or GST-H3. E) In vitro phosphorylation of GST-H3 by Cot FL, Cot NT, or Cot CT, which were immunoprecipitated with anti-Xpress, from Cot-overexpressing HEK293 cells. F) In vitro phosphorylation of histone H3 by Cot (top panel) purified from Cot-overexpressing HEK293 cells was detected by immunoblotting with anti-phospho-histone H3 (Ser-10) or anti-phospho-histone H3 (Ser-28). Overexpression of Cot was detected with anti-Xpress in cells transiently transfected with Xpress-Cot (fourth panel). Total histone H3 levels are shown as loading controls (bottom panel). “Mock” denotes a pcDNA4/hisMaxA-transfected control cell line. IB, immunoblotting; IP, immunoprecipitation. G) Schematic diagrams of histone H3 mutation of Ser-10 to alanine (S10A) or mutation of Ser-28 to alanine (S28A) (upper panel). GST pulldown proteins histone H3 wild-type (WT) or histone H3 mutants, S10A or S28A, were incubated with active Cot (1 μg) for 60 min at 30°C for an in vitro kinase assay and the 32P-labeled histone H3 was visualized by autoradiography. For visualizing equal loading of protein, total histone H3 was detected by Coomassie blue (bottom panel). The figures are representative of at least three separate experiments that yielded similar results.
UVB-induced Cot kinase activity mediates histone H3 phosphorylation at Ser-10
UVB irradiation markedly induces phosphorylation of histone H3 at Ser-10 and Ser-28. Studies have shown that a MEK1 or a p38 chemical inhibitor as well as dominant negative mutant ERK or dominant negative mutant p38 cells (39) could suppress phosphorylation. In the present study, we first used the M2H assay to determine whether UVB stimulation could affect the interaction between Cot and histone H3. NIH3T3 cells were seeded in 48-well plates in 10% FBS/DMEM and cultured until they reached 70% confluence. The cells were transfected with pACT-H3 and pBIND-Cot, then treated or not treated with UVB (4 kJ/m2). The results confirmed that after treatment with UVB, the interaction of histone H3 and Cot was greater compared with unstimulated cells (Fig. 2A). These results suggested that the interaction between Cot and histone H3 was enhanced by UVB and that Cot could mediate the UVB-induced phosphorylation of histone H3 at Ser-10 in vivo. To further confirm that UVB-induced Cot regulated the phosphorylation of histone H3 at Ser-10 in vivo, we examined UVB-induced phosphorylation of Cot in HEK293 cells transiently expressing myc-tagged Cot. These results indicated that UVB induced phosphorylation of Cot at serine residues, but not at tyrosine or threonine residues (Fig. 2B). Furthermore, the UVB-induced phosphorylation of Cot was recognized by anti-phospho-Cot (Ser-400), concomitant with the phosphorylation of histone H3 at Ser-10 (Fig. 2C). Next we performed an immunoprecipitation kinase assay in vivo with GST-histone H3 using Cot-overexpressing HEK293 cells transfected with myc-Cot. At 24 h after transfection of myc-Cot, cells were starved for an additional 24 h, then treated or not treated with UVB (4 kJ/m2) and harvested at various times after UVB (5, 15, or 30 min). UVB-induced Cot was immunoprecipitated from cell lysates using anti-myc. UVB-induced Cot kinase activity with histone H3 as a substrate was measured at 30°C and results confirmed that Cot mediated the phosphorylation of histone H3 induced by UVB (Fig. 2D). Hence, we sought to determine whether UVB-induced Cot is recruited with histones. Chromatin immunoprecipitation results indicated that Cot is recruited to the chromatin proteins, especially histone H3, by treatment with UVB (Fig. 2E). The phosphorylation of histone H3 at Ser-10 was enhanced in Cot-overexpressing HEK293 cells treated or not treated with UVB compared with mock cells with or without UVB (Fig. 2F). Enhanced phosphorylation of histone H3 under serum-deprived conditions might have been the result of phosphorylation of ERK, but not JNK or p38, induced by Cot. We confirmed that signaling through Cot also appeared to activate the well-established pathway of ERK after stimulation by UVB (Fig. 2F). Therefore, we determined whether PD 98059, a specific inhibitor of the activation and phosphorylation of ERK by MEK, and/or SB 202190, a specific inhibitor of p38, affected UVB-induced phosphorylation of histone H3 at Ser-10 in Cot overexpressing HEK293 cells. Our results clearly showed that UVB-induced phosphorylation of histone H3 at Ser-10 in control cells was blocked by these inhibitors, the 25 μM PD 98059 together with 1 μM SB 20290 (Fig. 2G, left, top panel). UVB-induced phosphorylation of histone H3 at Ser-10 was not significantly affected by PD98059 in this cell line, because UVB most likely induced the signaling pathway of JNK or p38 rather than ERK. Surprisingly, these inhibitors had no effect on histone H3 phosphorylation at Ser-10 in Cot overexpressing HEK293 cells (Fig. 2G, right, top panel). However, the inhibitors effectively suppressed ERK and p38 phosphorylation in Cot overexpressing HEK293 cells (Fig. 2G, right, 3rd and 4th panels), indicating that Cot-mediated phosphorylation of histone H3 at Ser-10 occurred independent of the ERKs and p38 MAP kinase pathways. 4-(3-Chlor-4-fluorophenylamino)-6-(pyridin-3-yl-methylamino)-3-cyano-(1, 7)-naphthylridine is a very strong inhibitor of Cot and may be a compound with significant therapeutic potential for treating rheumatoid arthritis and other inflammatory diseases (42). This ATP competitive inhibitor of Cot almost completely blocked the UVB-induced phosphorylation of histone H3 at Ser-10 and directly inhibited phosphorylation of Cot at Ser-400 (Fig. 2H). We also used the siRNA method to knock down the endogenous Cot expression level (Fig. 2I), then determined the effects on UVB-induced phosphorylation of histone H3 at Ser-10. The pBsi-Cot construct for targeting the specific sequence of Cot was transiently transfected into HEK293 cells. At 48 h after transfection, total protein was isolated from pBsi-sc (control) or pBsi-Cot transfected cells, and the levels of Cot protein were compared with control cells. The expression of pBsi-Cot specifically knocked down the Cot protein level (Fig. 2I, 3rd panel). Further results indicated that UVB-induced phosphorylation of histone H3 at Ser-10 was almost blocked in the pBsi-Cot transfected cells, but not in pBsi-sc transfected cells (Fig. 2I, 1st panel). Taken together, our results strongly indicated that UVB-induced Cot plays an important role in mediating phosphorylation of histone H3 at Ser-10 in vivo.
Figure 2.
Cot mediates UVB-induced phosphorylation of histone H3 at Ser-10. A) UVB stimulation causes an enhanced association between histone H3 and Cot. HEK293 cells were cotransfected with the pG5-luc plasmid and pACT-H3 and/or pBIND-Cot, then incubated for 2 h in fresh serum-deprived DMEM. The cells were either exposed or not exposed to 4 kJ/m2 UVB and incubated for an additional 12 h at 37°C, then luciferease activity was measured. B) Time course of UVB-induced Cot phosphorylation of serine residues. After cells were starved in serum-free media, the time course was investigated by exposing HEK293 cells, transiently transfected with myc-Cot, to UVB (4 kJ/m2). After UVB, cells were incubated for the times indicated, then harvested. The phosphorylation of Cot at serine, threonine, or tyrosine residues was detected by immunoblotting with anti-phospho-serine, anti-phospho-threonine, or anti-phospho-tyrosine, respectively. C) Time course of phosphorylation of histone H3 at Ser-10 and phosphorylation of Cot at Ser-400 was investigated by exposing HEK293 cells to UVB (4 kJ/m2). The phosphorylation of histone H3 (Ser-10) or Cot (Ser-400) was detected by immunoblotting with anti-phospho-histone H3 (Ser-10) or anti-phospho-Cot (Ser-400), respectively. D) Time-dependent effect of UVB-induced Cot activation on the phosphorylation of histone H3. Transiently transfected Cot in HEK293 cells is activated in response to UVB (4 kJ/m2) irradiation. After UVB, cells were incubated for the indicated times and kinase activity (KA) was measured with histone H3 as substrate. Phosphorylated histone H3 was visualized by autoradiography. Immunoprecipitated (IP) myc-Cot was determined by immunoblot using anti-myc. E) The binding of Cot and chromatin-immunoprecipitated histone H3 induced by UVB was evaluated. Myc-Cot was transiently transfected into HEK293 cells and activated by UVB (4 kJ/m2). Chromatin immunoprecipitation (ChIP) was performed to precipitate histone H3, then the myc-Cot proteins were detected by immunoblotting with anti-myc. F) The effect of Cot overexpression on the phosphorylation of histone H3 (Ser-10), ERK1/2, p38, JNK, or MSK1 (Ser-360) in cells treated or not treated with UVB (4 kJ/m2). G) Effect of PD 98059 and/or SB 202190 on UVB-induced phosphorylation of histone H3 at Ser-10 in control and Cot overexpressing cells. Mock vector (left panels) and Cot (right panels) transfected HEK293 cells were M starved for 24 h by incubating in serum-deprived DMEM at 37°C in a 5% CO2 atmosphere. After pretreatment with 25 μ PD 98059 and/or 1 μM SB 202190 for 1 h, cells were exposed to UVB (4 kJ/m2) and incubated for an additional 30 min. Phosphorylation of histone H3 at Ser-10, ERK1/2, or p38 was determined by immunoblotting with anti-phospho-histone H3 (Ser-10), anti-phospho-ERK1/2, or anti-phospho-p38, respectively. Overexpression of Cot was detected with anti-myc and total histone H3 and β-actin were used as internal controls to monitor equal protein loading. H) Effect of a Cot inhibitor on UVB-induced phosphorylation of histone H3 at Ser-10. Cells were starved for 24 h by incubation in serum-deprived DMEM at 37°C in a 5% CO2 atmosphere. After pretreatment with the Cot inhibitor for 2 h, cells were exposed to UVB (4 kJ/m2) and incubated for an additional 30 min. Phosphorylation of histone H3 and Cot was determined by immunoblotting with anti-phospho-histone H3 (Ser-10) and anti-phospho-Cot (Ser-400). Total histone H3 and β-actin were used as internal controls to monitor equal protein loading. I) Effect of Cot knockdown on the UVB-induced phosphorylation of endogenous histone H3 (Ser-10). Control siRNA (pBsi-sc) and Cot knockdown (pBsi-Cot) cells were starved for 24 h by incubating in serum-deprived DMEM at 37°C in a 5% CO2 atmosphere. Cells were then exposed to UVB (4 kJ/m2) and incubated for an additional 30 min. After removal of media and harvesting, phosphorylation of histone H3 at Ser-10 was determined by immunoblotting with anti-phospho-histone H3 (Ser-10). Endogeneous Cot expression was measured by immunoblotting with anti-Cot, and total histone H3 and β-actin were used as internal controls to monitor equal protein loading.
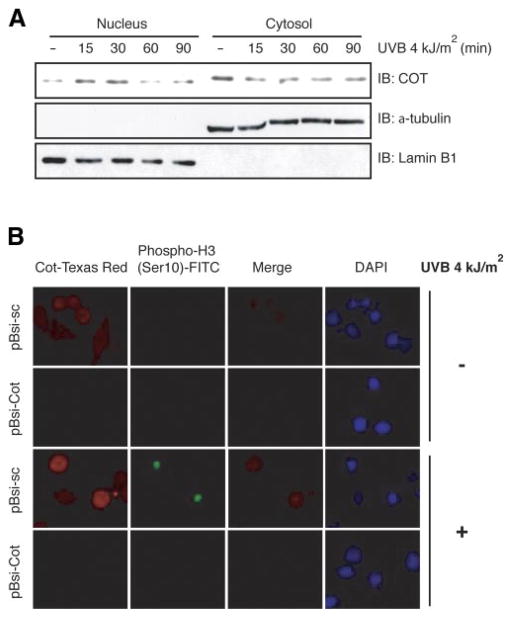
Nuclear localization of Cot induced by UVB
For Cot to phosphorylate the chromatin protein histone H3, translocation of Cot to the nucleus must occur. To address whether Cot translocation into the nucleus occurs and is affected by UVB, we investigated the cellular localization of endogenous Cot and phosphorylation of histone H3 at Ser-10 induced or not induced with UVB irradiation. Cot was located in the cytoplasm of nonirradiated HEK293 cells, but at 15–30 min after UVB exposure, the Cot protein was translocated into the nucleus (Fig. 3A). In the immunofluorescence assay, we also found that phosphorylation of histone H3 was increased in response to UVB irradiation (Fig. 3B, 3rd panel in phospho-histone H3 [Ser-10]-FITC lane) but was suppressed by knockdown of Cot expression (Fig. 3B, 4th panel in phospho-histone H3 [Ser-10]-FITC lane). These results indicated that Cot accumulates in the nucleus to regulate histone H3 after stimulation of cells with UVB irradiation.
Figure 3.
Nuclear localization of Cot and phosphorylation of histone H3 induced by UVB. A) UVB-induced translocation of endogenous Cot protein from cytoplasm to nucleus. Nuclear localization of Cot induced by treatment with UVB (4 kJ/m2) was assessed in HEK293 cells. The cells were seeded and cultured for 24 h in 10% FBS/DMEM in a 37°C, 5% CO2 incubator. The cells were then starved in serum-deprived DMEM for 24 h, exposed or not exposed to UVB (4 kJ/m2), and harvested after incubation for 15, 30, 60, or 90 min. The distribution of endogenous Cot protein in the cytoplasmic or nuclear fractions was assessed by immunoblotting using anti-Cot. For visualizing equal loading of protein and confirming localization, α-tubulin (cytosolic marker) or lamin B1 (nuclear marker) was detected with a specific antibody against each. B) Effect of knockdown of the endogenous Cot protein on UVB-induced phosphorylation of histone H3 (Ser-10). The phosphorylation of histone H3 (Ser-10) induced by UVB (4 kJ/m2 and harvested at 30 min) was visualized by an immunofluorescence assay using anti-phospho-histone H3 (Ser-10)-FITC in control siRNA- and Cot siRNA-transfected HeLa cells as described in Materials and Methods.
Cot is recruited to histone H3, which is associated with the c-fos promoter
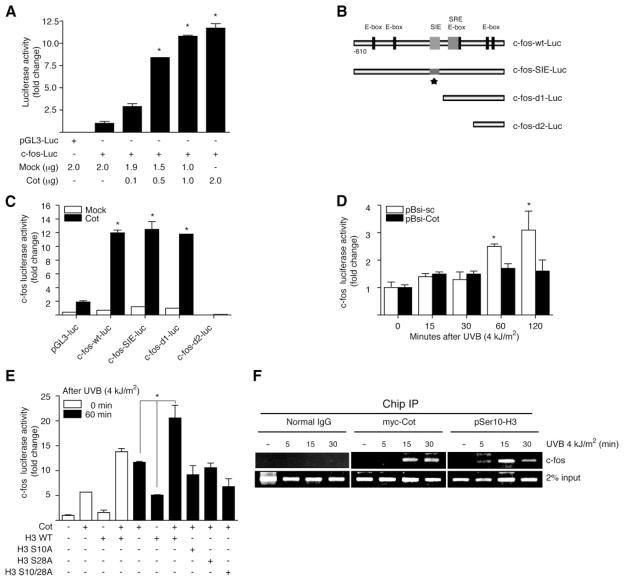
Indeed, only a limited number of genes are activated by mitogen stimulation in mammalian cells. The phosphorylated histone H3 has been used in chromatin immunoprecipitation (ChIP) assays to show that phosphorylated (Ser-10) histone H3 is associated with c-fos and c-myc genes following EGF treatment (3). The c-Fos proteins are nuclear proto-oncoproteins whose expression is stimulated by a variety of growth-promoting agents and activated oncogenes (43). We next wanted to investigate whether Cot might activate the c-fos promoter to induce cell transformation. The activity of the c-fos promoter is tightly regulated on key regulatory elements, including the four elements of the E-box or the SIE or SRE region. The transcriptional activity of c-fos was significantly increased by the transient expression of Cot in HEK293 cells (Fig. 4A). To further confirm that c-fos transcriptional activation was cooperatively induced by Cot and histone H3, a reporter gene assay using various WT and mutant c-fos promoter luciferase constructs (Fig. 4B) was performed. Using mutated or truncated forms of the c-fos promoter, we found that the SRE/E-box region is critical for activation by Cot (Fig. 4C). We next assessed UVB-induced transcriptional activation of c-fos in control siRNA or Cot siRNA-transfected HEK293 cells. Transcriptional activation of c-fos in HEK293 cells was increased 30–60 min after irradiation with UVB (4 kJ/m2), but the knockdown of Cot expression significantly suppressed UVB-induced c-fos activation compared with control siRNA cells (Fig. 4D). To further investigate the effect of Cot-induced histone H3 phosphorylation on c-fos activation, we transfected HEK293 cells harboring WT Cot with WT or mutants (S10A, S28A, S10/28A) of histone H3. Serum-starved HEK293 cells were irradiated with UVB (4 kJ/m2) and harvested after 60 min. Results indicated that the transcriptional activation of c-fos in cells transfected with Cot and WT histone H3 was dramatically induced compared with activation in cells transfected with only Cot or histone H3 (Fig. 4E) or the histone H3 mutants (S10A, S28A, S10/28A). Unexpectedly, the Cot-induced transcriptional activation of c-fos was also inhibited in the histone H3 mutant (S28A) cells, suggesting that Cot enhanced the ERK signaling pathway, then had an indirect effect on the phosphorylation of histone H3 at Ser-28. Next, to examine whether myc-Cot and phosphorylated H3 (Ser-10) are associated with the c-fos promoter, we used anti-myc and anti-phospho-histone H3 (Ser-10) in ChIP assays. UVB-stimulated cells were cross-linked with formaldehyde and sonicated chromatin from these cells was immunoprecipitated with anti-myc and anti-phospho-histone H3 (Ser-10). The amount of c-fos promoter DNA present in exogenous or immunoprecipitated chromatin fractions was then determined by PCR. The control for DNA input was included in each PCR reaction. Immunoprecipitation with anti-myc or anti-phospho-histone H3 (Ser-10) of equivalent amounts of chromatin from UVB-stimulated cells showed a dramatic increase in the association of myc-Cot or phosphorylated histone H3 (Ser-10) with the c-fos promoter after UVB stimulation (Fig. 4F). Thus, these data indicated that UVB-induced Cot is recruited to the c-fos transcription complex to mediate c-fos gene expression concomitant with the phosphorylation of histone H3 (Ser-10). These results also illustrated that Cot is essential for UVB-induced c-fos transcriptional activation, and the targeted area in the c-fos promoter is the SRE region. Furthermore, these observations suggested that phosphorylation of histone H3 plays an important role in Cot-induced transcriptional activity of c-fos.
Figure 4.
Cot and histone H3 cooperatively up-regulate the transcriptional activity of the c-fos promoter. A) Cot up-regulates c-fos promoter activity. HEK293 cells were cotransfected with a plasmid mixture containing the c-fos-luciferase reporter gene (0.5 μg) and the myc-Cot (0.1, 0.5, 1.0, 2.0 μg) or mock (1.0–2.0 μg) and the phRL-SV40 gene (50 ng). At 36 h after transfection, firefly luciferase activity was determined in cell lysates and normalized against Renilla luciferase activity. The asterisk (*) indicates a significant increase in c-fos luciferase activity in cells transfected with Cot compared to cells transfected with the mock plasmid (P<0.05). B) A diagram of the c-fos-luciferase reporter gene containing five E-boxes, the SIE region, and SRE region. The asterisk (*) indicates mutations in the SIE region; two deletion mutants, d1 (SIE deletion) and d2 (SIE/SRE deletion), are also shown. C) The region of the c-fos promoter induced by Cot is the SRE (serum-responsive element)/E-Box region. HEK293 cells were cotransfected with a plasmid mixture containing the c-fos-luciferase reporter gene and phRL-SV40 gene for wild-type (WT), SIE mutant, d1 or d2 mutant with mock or pRK-myc-Cot. At 36 h after transfection, the firefly luciferase activity was determined in cell lysates and normalized against Renilla luciferase activity. The asterisk (*) indicates a significant increase in c-fos luciferase activity in cells transfected with various forms of the c-fos-luc promoter and Cot compared to cells transfected with pGL3-luc and Cot (P<0.05). D) Comparison of c-fos promoter activity in control and Cot knockdown HEK293 cells. Control siRNA (pBsi-sc) or Cot-specific siRNA (pBsi-Cot) were cotransfected with a plasmid mixture containing the c-fos luciferase reporter gene and the phRL-SV40 gene. At 24 h post-transfection, cells were starved for 24 h by incubating them in serum-deprived DMEM at 37°C in a 5% CO2 atmosphere, then exposed or not exposed to UVB (4 kJ/m2) and harvested after incubation for 15, 30, 60, or 120 min. The firefly luciferase activity was determined in cell lysates and normalized against Renilla luciferase activity. An asterisk (*) indicates a significant increase in c-fos luciferase activity in cells treated with UVB compared with control cells (P<0.05). E) Comparison of c-fos promoter activity in HEK293 cells expressing WT, S10A, S28A, or S10/28A histone H3 and treated or not treated with UVB (4 kJ/m2). pV5-H3 WT, S10A, S28A, or S10/28A were cotransfected with a plasmid mixture containing the c-fos luciferase reporter gene and the phRL-SV40 gene with or without the Cot gene. At 24 h post-transfection, cells were starved for an additional 24 h by incubating them in serum-deprived MEM at 37°C in a 5% CO2 atmosphere, then exposed or not exposed to UVB (4 kJ/m2) and harvested after incubation for 60 min. Firefly luciferase activity was determined in cell lysates and normalized against Renilla luciferase activity. The asterisk (*) indicates a significant increase in UVB-induced c-fos luciferase activity in cells transfected with Cot and histone H3 WT compared to cells transfected with only Cot or histone H3 WT. A, C–E) All data are presented as means ± sd of 2 independent experiments with triplicate assays. F) Analysis of histone H3 phosphorylation (Ser-10) in chromatin at the c-fos promoter. HEK293 cells were transfected with myc-Cot. At 24 h after transfection, cells were starved in serum-free DMEM, then treated or not treated (control) with UVB (4 kJ/m2) and harvested at 5, 15, or 30 min later. ChIP assays were performed using either normal mouse IgG, anti-myc, or anti-phospho-histone H3 (Ser-10). PCR amplification of the c-fos promoter was performed using template DNA isolated from chromatin immunoprecipitated by normal mouse IgG, anti-myc or anti-phospho-histone H3 (Ser-10). Final PCR products of the c-fos promoter were obtained after completion of 35 cycles using either input DNA or ChIP DNA as templates. Bottom histogram represents the relative c-fos levels normalized to 2% input.
A dominant negative mutant form of histone H3 inhibits Cot-induced transformation of NIH3T3 cells
Cellular transformation is the best-characterized biological effect resulting from Cot overexpression and/or carboxyl-terminal truncation (6, 8, 10, 44, 45). The AP-1 transcription factor is a dimeric complex that comprises members of the Jun, Fos activating transcription factor and musculoaponeurotic fibrosarcoma protein families (46). The regulation of cell proliferation by AP-1 might be of crucial importance for the multistage development of tumors (47). We thus investigated whether the AP-1 transcription factor might participate in Cot-induced cell transformation. When Cot was transiently overexpressed in HEK293 cells, AP-1 luciferase activity was increased in a dose-dependent manner, similar to that observed for c-fos activity (Fig. 5A). We next assessed UVB-induced transactivation activity of AP-1 in control siRNA or Cot siRNA-transfected HEK293 cells. Transactivation activity of AP-1 in control cells was significantly increased 30–60 min after irradiation with UVB (4 kJ/m2), but was suppressed in Cot siRNA-transfected cells (Fig. 5B). To investigate the effect of Cot-induced histone H3 phosphorylation on the transactivation activity of AP-1, we transfected Cot with the WT or various mutants (S10A, S28A, S10/28A) of histone H3 into HEK293 cells. After irradiation of serum-starved HEK293 cells with UVB (4 kJ/m2), the transactivation activity of AP-1 in cells transfected with Cot and WT histone H3 was dramatically increased compared with cells transfected with only Cot or histone H3 (Fig. 5C). In contrast, mutants of histone H3, especially H3S10A, suppressed Cot-induced AP-1 transactivation (Fig. 5C). To investigate whether Cot-induced AP-1 transactivation results in increased neoplastic cell transformation, we confirmed the effect of a Cot inhibitor on the epidermal growth factor (EGF) -induced cell transformation. EGF is a well-known tumor promotion agent used to study malignant cell transformation in cell and animal models of cancer (48). EGF also induces activation of AP-1 (38) and phosphorylation of histone H3 at Ser-10, which is mediated by RSK2 (49). The Cot kinase inhibitor almost completely blocked the EGF-induced cell transformation (Fig. 5D, E). We next decided to assess the effect of WT or mutant histone H3 on Cot-induced cell transformation. Indeed, whereas transfection of the cot proto-oncogene with WT histone H3 into NIH3T3 cells readily induced the appearance of foci after 3 wk of culture, cotransfection with mutants of histone H3 (S10A, S28A, or S10/28A) caused a marked inhibition of Cot transforming potential (Fig. 5F). Taken together, these results suggested that phosphorylation of histone H3 is necessary for the transforming ability of Cot.
Figure 5.

Dominant negative mutants of histone H3 inhibit AP-1 activity and foci formation mediated by Cot. A) Cot up-regulates AP-1 activity. HEK293 cells were cotransfected with a plasmid mixture containing the AP-1-luciferase reporter gene (0.5 μg) and myc-Cot (0.1, 0.5, 1.0, 2.0 μg) or mock (1.0–2.0 μg) plus the phRL-SV40 gene (50 ng). At 36 h after transfection, firefly luciferase activity was determined in cell lysates and normalized against Renilla luciferase activity. The asterisk (*) indicates a significant increase in AP-1 luciferase activity in cells transfected with Cot compared to cells transfected with mock (P<0.05). B) A comparison of AP-1 activity in control and Cot knockdown HEK293 cells. Control siRNA (pBsi-sc) or Cot-specific siRNA (pBsi-Cot) was cotransfected with a plasmid mixture containing the AP-1-luciferase reporter gene and the phRL-SV40 gene. At 24 h post-transfection, cells were starved for an additional 24 h by incubating them in serum-deprived DMEM at 37°C in a 5% CO2 atmosphere, then exposed or not exposed to UVB (4 kJ/m 2) and harvested after additional incubation for 15, 30, 60, or 120 min. The asterisk (*) indicates a significant increase in AP-1 luciferase activity in cells treated with UVB compared with untreated control cells (P<0.05). (C) Comparison of WT and S10A, S28A, and S10/28A mutants of histone H3 on AP-1 activity in HEK293 cells treated or not treated with UVB (4 kJ/m2). pV5-H3 WT, S10A, S28A, and S10/28A were cotransfected with a plasmid mixture containing the AP-1 luciferase reporter gene and the phRL-SV40 gene with or without the Cot gene. At 24 h post-transfection, cells were starved for an additional 24 h by incubating them in serum-deprived MEM at 37°C in a 5% CO2 atmosphere, then exposed or not exposed to UVB (4 kJ/m2) and harvested after incubation for 60 min. The asterisk (*) indicates a significant increase in UVB-induced AP-1 luciferase activity in cells transfected with Cot and histone H3 WT compared with cells transfected with only Cot or histone H3 WT. A–C) All data are presented as means ± sd of 2 independent experiments with triplicate assays. D, E) A Cot inhibitor suppresses EGF-induced cell transformation. JB6 Cl41 cells (8×103 cells/ml) were subjected to a soft agar assay in the presence of EGF (1 ng/ml) with or without a Cot inhibitor (1, 10, or 20 μM) in 1 ml of 0.3 basal medium Eagle agar containing 10% FBS. The cultures were maintained at 37°C in a 5% CO2 atmosphere for 10 days. The average colony number was calculated and photographed from 3 separate experiments. Significant differences were evaluated using the Student’s t test. The asterisk (*P<0.005) indicates a significant decrease in EGF-induced cell transformation in Cot inhibitor-treated cells compared with control cells. F) In the focus-forming assay, the foci were markedly increased in cells transfected with Cot and histone H3 WT, but not with Cot or any of the histone H3 mutants (S10A, S28A, S10/28A). NIH3T3 cells were transfected with the myc-Cot (0.2 μg) plasmid with or without histone H3 WT or mutant histone H3 S10A, H3 S28A, or H3 S10/28A (0.2 μg), respectively. Cells were cultured for 3 wk in 5% calf serum, fixed, then stained as described in Materials and Methods. Representative plates from three experiments are shown.
DISCUSSION
Cell transformation occurs because of the loss of negative growth regulators or genes responsible for the maintenance of genome integrity, or it may occur because of the amplification, overexpression, or mutational activation of an oncogene (50, 51). The cot proto-oncogene encodes a serine/threonine protein kinase that is activated by provirus insertion in Moloney leukemia virus (MoMuLV) -induced T cell lymphomas and MMTV-induced mammary carcinomas (8, 45). Earlier studies have shown that overexpression of Cot activates the ERKs, JNKs, and p38 MAPK pathways and NFAT and NF-κB transcription factors, induces IL-2 expression in Jurkat cells, promotes cell cycle progression, and is highly oncogenic in mice (6, 11, 12, 15). Although the chromatin remodeling events take place during the induction of oncogenic signal transduction of Cot proteins, little is known about chromatin modification of Cot as a biological mechanism of Cot-induced cell transformation. In this study we have shown that Cot is a newly discovered histone H3 kinase that phosphorylates histone H3 at Ser-10 induced by UVB.
The best-characterized link between signal transduction and histone modification has been observed in mammalian cells exposed to mitogens or various stresses. Many cellular processes fall under the tight regulation of the Ras/MAPK pathway, and its persistent activation has been reported to result in the chromatin remodeling and altered gene expression observed in cancer (52, 53). Growth factors such as EGF, phorbol esters including 12-O-tetradecanoylphorbol-13-acetate (TPA), or UVB activate the Ras/MAPK pathway, which results in the phosphorylation of downstream targets, including various transcription factors and nucleosomal proteins. One downstream event is the phosphorylation of the basic NH2-terminal tail of histone H3 at Ser-10 and Ser-28 (54). Post-translational modification of histone H3 has been shown to play significant roles in chromosome condensation during mitosis in many organisms, and the phosphorylation of histone H3 has been directly associated with immediate-early gene induction in mouse fibroblasts (3, 55, 56). We recently reported a direct link between the phosphorylation of histone H3 at Ser-10 and neoplastic cell transformation mediated through the activation of AP-1 factors, including the transcriptional activation of c-jun and c-fos (4). UVB stimulation of Cot- and histone H3-overexpressing HEK293 cells resulted in an increased activation of AP-1 due to a stimulation of c-fos transcriptional activation compared with control cells. These results also showed that the oncogene cot could increase foci formation cooperatively with WT histone H3, but not with mutant histone H3 (S10A, S28A, and S10/28A), suggesting that Cot-induced cell transformation resulted from the phosphorylation of histone H3.
An interesting idea is whether suppressing Cot activity might prove to be a valid strategy for the treatment of diseases in which it is found to be aberrantly active. Whether inhibition of Cot activity could enhance the effectiveness of current cancer therapies should also be considered. In a recent publication, PHA-680632, a novel aurora kinase inhibitor, was shown to have potent anticancer activity, resulting in inhibition of the phosphorylation of histone H3 at Ser-10 (57). Aurora kinases have been implicated in cancer and tumorigenesis (58). Our results suggest that Cot inactivation resulting in the inhibition of histone H3 activity and c-fos promoter activity might also be a valid strategy for cancer therapy.
Acknowledgments
This work was supported in part by the Hormel Foundation and National Institutes of Health grants CA77646, CA81064, CA111356, and CA120388.
References
- 1.Bradbury EM. Reversible histone modifications and the chromosome cell cycle. Bioessays. 1992;14:9–16. doi: 10.1002/bies.950140103. [DOI] [PubMed] [Google Scholar]
- 2.Gurley LR, D’Anna JA, Barham SS, Deaven LL, Tobey RA. Histone phosphorylation and chromatin structure during mitosis in Chinese hamster cells. Eur J Biochem. 1978;84:1–15. doi: 10.1111/j.1432-1033.1978.tb12135.x. [DOI] [PubMed] [Google Scholar]
- 3.Chadee DN, Hendzel MJ, Tylipski CP, Allis CD, Bazett-Jones DP, Wright JA, Davie JR. Increased Ser-10 phosphorylation of histone H3 in mitogen-stimulated and oncogene-transformed mouse fibroblasts. J Biol Chem. 1999;274:24914–24920. doi: 10.1074/jbc.274.35.24914. [DOI] [PubMed] [Google Scholar]
- 4.Choi HS, Choi BY, Cho YY, Mizuno H, Kang BS, Bode AM, Dong Z. Phosphorylation of histone H3 at serine 10 is indispensable for neoplastic cell transformation. Cancer Res. 2005;65:5818–5827. doi: 10.1158/0008-5472.CAN-05-0197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyoshi J, Higashi T, Mukai H, Ohuchi T, Kakunaga T. Structure and transforming potential of the human cot oncogene encoding a putative protein kinase. Mol Cell Biol. 1991;11:4088–4096. doi: 10.1128/mcb.11.8.4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ceci JD, Patriotis CP, Tsatsanis C, Makris AM, Kovatch R, Swing DA, Jenkins NA, Tsichlis PN, Copeland NG. Tpl-2 is an oncogenic kinase that is activated by carboxy-terminal truncation. Genes Dev. 1997;11:688–700. doi: 10.1101/gad.11.6.688. [DOI] [PubMed] [Google Scholar]
- 7.Makris A, Patriotis C, Bear SE, Tsichlis PN. Genomic organization and expression of Tpl-2 in normal cells and Moloney murine leukemia virus-induced rat T-cell lymphomas: activation by provirus insertion. J Virol. 1993;67:4283–4289. doi: 10.1128/jvi.67.7.4283-4289.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patriotis C, Makris A, Bear SE, Tsichlis PN. Tumor progression locus 2 (Tpl-2) encodes a protein kinase involved in the progression of rodent T-cell lymphomas and in T-cell activation. Proc Natl Acad Sci U S A. 1993;90:2251–2255. doi: 10.1073/pnas.90.6.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohara R, Miyoshi J, Aoki M, Toyoshima K. The murine cot proto-oncogene: genome structure and tissue-spe-cific expression. Jpn J Cancer Res. 1993;84:518–525. doi: 10.1111/j.1349-7006.1993.tb00170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan AM, Chedid M, McGovern ES, Popescu NC, Miki T, Aaronson SA. Expression cDNA cloning of a serine kinase transforming gene. Oncogene. 1993;8:1329–1333. [PubMed] [Google Scholar]
- 11.Salmeron A, Ahmad TB, Carlile GW, Pappin D, Narsimhan RP, Ley SC. Activation of MEK-1 and SEK-1 by Tpl-2 proto-oncoprotein, a novel MAP kinase kinase kinase. EMBO J. 1996;15:817–826. [PMC free article] [PubMed] [Google Scholar]
- 12.Patriotis C, Makris A, Chernoff J, Tsichlis PN. Tpl-2 acts in concert with Ras and Raf-1 to activate mitogen-activated protein kinase. Proc Natl Acad Sci U S A. 1994;91:9755–9759. doi: 10.1073/pnas.91.21.9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsatsanis C, Patriotis C, Bear SE, Tsichlis PN. The Tpl-2 protooncoprotein activates the nuclear factor of activated T cells and induces interleukin 2 expression in T cell lines. Proc Natl Acad Sci U S A. 1998;95:3827–3832. doi: 10.1073/pnas.95.7.3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ballester A, Velasco A, Tobena R, Alemany S. Cot kinase activates tumor necrosis factor-alpha gene expression in a cyclosporin A-resistant manner. J Biol Chem. 1998;273:14099–14106. doi: 10.1074/jbc.273.23.14099. [DOI] [PubMed] [Google Scholar]
- 15.Tsatsanis C, Patriotis C, Tsichlis PN. Tpl-2 induces IL-2 expression in T-cell lines by triggering multiple signaling pathways that activate NFAT and NF-kappaB. Oncogene. 1998;17:2609–2618. doi: 10.1038/sj.onc.1202460. [DOI] [PubMed] [Google Scholar]
- 16.Belich MP, Salmeron A, Johnston LH, Ley SC. TPL-2 kinase regulates the proteolysis of the NF-kappaB-inhibitory protein NF-kappaB1 p105. Nature. 1999;397:363–368. doi: 10.1038/16946. [DOI] [PubMed] [Google Scholar]
- 17.Cowley S, Paterson H, Kemp P, Marshall CJ. Activation of MAP kinase kinase is necessary and sufficient for PC12 differentiation and for transformation of NIH 3T3 cells. Cell. 1994;77:841–852. doi: 10.1016/0092-8674(94)90133-3. [DOI] [PubMed] [Google Scholar]
- 18.Johnson R, Spiegelman B, Hanahan D, Wisdom R. Cellular transformation and malignancy induced by ras require c-jun. Mol Cell Biol. 1996;16:4504–4511. doi: 10.1128/mcb.16.8.4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rao A, Luo C, Hogan PG. Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 20.Royds JA, Dower SK, Qwarnstrom EE, Lewis CE. Response of tumour cells to hypoxia: role of p53 and NFκB. Mol Pathol. 1998;51:55–61. doi: 10.1136/mp.51.2.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiariello M, Marinissen MJ, Gutkind JS. Multiple mitogen-activated protein kinase signaling pathways connect the cot oncoprotein to the c-jun promoter and to cellular transformation. Mol Cell Biol. 2000;20:1747–1758. doi: 10.1128/mcb.20.5.1747-1758.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kovary K, Bravo R. The jun and fos protein families are both required for cell cycle progression in fibroblasts. Mol Cell Biol. 1991;11:4466–4472. doi: 10.1128/mcb.11.9.4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishikura K, Murray JM. Antisense RNA of proto-oncogene c-fos blocks renewed growth of quiescent 3T3 cells. Mol Cell Biol. 1987;7:639–649. doi: 10.1128/mcb.7.2.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okuno H, Suzuki T, Yoshida T, Hashimoto Y, Curran T, Iba H. Inhibition of jun transformation by a mutated fos gene: design of an anti-oncogene. Oncogene. 1991;6:1491–1497. [PubMed] [Google Scholar]
- 25.Suzuki T, Murakami M, Onai N, Fukuda E, Hashimoto Y, Sonobe MH, Kameda T, Ichinose M, Miki K, Iba H. Analysis of AP-1 function in cellular transformation pathways. J Virol. 1994;68:3527–3535. doi: 10.1128/jvi.68.6.3527-3535.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ball AR, Jr, Bos TJ, Loliger C, Nagata LP, Nishimura T, Su H, Tsuchie H, Vogt PK. Jun: oncogene and transcriptional regulator. Cold Spring Harb Symp Quant Biol. 1988;53:687–693. doi: 10.1101/sqb.1988.053.01.078. [DOI] [PubMed] [Google Scholar]
- 27.Ryder K, Lanahan A, Perez-Albuerne E, Nathans D. jun-D: a third member of the jun gene family. Proc Natl Acad Sci U S A. 1989;86:1500–1503. doi: 10.1073/pnas.86.5.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryder K, Lau LF, Nathans D. A gene activated by growth factors is related to the oncogene v-jun. Proc Natl Acad Sci U S A. 1988;85:1487–1491. doi: 10.1073/pnas.85.5.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cohen DR, Curran T. fra-1: a serum-inducible, cellular immediate-early gene that encodes a fos-related antigen. Mol Cell Biol. 1988;8:2063–2069. doi: 10.1128/mcb.8.5.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsui M, Tokuhara M, Konuma Y, Nomura N, Ishizaki R. Isolation of human fos-related genes and their expression during monocyte-macrophage differentiation. Oncogene. 1990;5:249–255. [PubMed] [Google Scholar]
- 31.Nishina H, Sato H, Suzuki T, Sato M, Iba H. Isolation and characterization of fra-2, an additional member of the fos gene family. Proc Natl Acad Sci U S A. 1990;87:3619–3623. doi: 10.1073/pnas.87.9.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zerial M, Toschi L, Ryseck RP, Schuermann M, Muller R, Bravo R. The product of a novel growth factor activated gene, fos B, interacts with JUN proteins enhancing their DNA binding activity. EMBO J. 1989;8:805–813. doi: 10.1002/j.1460-2075.1989.tb03441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ransone LJ, Verma IM. Nuclear proto-oncogenes fos and jun. Annu Rev Cell Biol. 1990;6:539–557. doi: 10.1146/annurev.cb.06.110190.002543. [DOI] [PubMed] [Google Scholar]
- 34.Benbrook DM, Jones NC. Heterodimer formation between CREB and JUN proteins. Oncogene. 1990;5:295–302. [PubMed] [Google Scholar]
- 35.Northrop JP, Ullman KS, Crabtree GR. Characterization of the nuclear and cytoplasmic components of the lymphoid-specific nuclear factor of activated T cells (NF-AT) complex. J Biol Chem. 1993;268:2917–2923. [PubMed] [Google Scholar]
- 36.Zhang Y, Feng XH, Derynck R. Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-beta-induced transcription. Nature. 1998;394:909–913. doi: 10.1038/29814. [DOI] [PubMed] [Google Scholar]
- 37.Mahadevan LC, Willis AC, Barratt MJ. Rapid histone H3 phosphorylation in response to growth factors, phorbol esters, okadaic acid, and protein synthesis inhibitors. Cell. 1991;65:775–783. doi: 10.1016/0092-8674(91)90385-c. [DOI] [PubMed] [Google Scholar]
- 38.Huang C, Ma WY, Dong Z. Requirement for phosphatidylinositol 3-kinase in epidermal growth factor-induced AP-1 transactivation and transformation in JB6 P+ cells. Mol Cell Biol. 1996;16:6427–6435. doi: 10.1128/mcb.16.11.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhong SP, Ma WY, Dong Z. ERKs and p38 kinases mediate ultraviolet B-induced phosphorylation of histone H3 at serine 10. J Biol Chem. 2000;275:20980–20984. doi: 10.1074/jbc.M909934199. [DOI] [PubMed] [Google Scholar]
- 40.Clark GJ, Cox AD, Graham SM, Der CJ. Biological assays for Ras transformation. Methods Enzymol. 1995;255:395–412. doi: 10.1016/s0076-6879(95)55042-9. [DOI] [PubMed] [Google Scholar]
- 41.Ota T, Suto S, Katayama H, Han ZB, Suzuki F, Maeda M, Tanino M, Terada Y, Tatsuka M. Increased mitotic phosphorylation of histone H3 attributable to AIM-1/Aurora-B overexpression contributes to chromosome number instability. Cancer Res. 2002;62:5168–5177. [PubMed] [Google Scholar]
- 42.Gavrin LK, Green N, Hu Y, Janz K, Kaila N, Li HQ, Tam SY, Thomason JR, Gopalsamy A, Ciszewski G, et al. Inhibition of Tpl2 kinase and TNF-alpha production with 1, 7-naphthyridine-3-carbonitriles: synthesis and structure-activity relationships. Bioorganic Med Chem Lett. 2005;15:5288–5292. doi: 10.1016/j.bmcl.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 43.Herschman HR. Primary response genes induced by growth factors and tumor promoters. Annu Rev Biochem. 1991;60:281–319. doi: 10.1146/annurev.bi.60.070191.001433. [DOI] [PubMed] [Google Scholar]
- 44.Aoki M, Akiyama T, Miyoshi J, Toyoshima K. Identification and characterization of protein products of the cot oncogene with serine kinase activity. Oncogene. 1991;6:1515–1519. [PubMed] [Google Scholar]
- 45.Erny KM, Peli J, Lambert JF, Muller V, Diggelmann H. Involvement of the Tpl-2/cot oncogene in MMTV tumorigenesis. Oncogene. 1996;13:2015–2020. [PubMed] [Google Scholar]
- 46.Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 47.Liu Y, Ludes-Meyers J, Zhang Y, Munoz-Medellin D, Kim HT, Lu C, Ge G, Schiff R, Hilsenbeck SG, Osborne CK, Brown PH. Inhibition of AP-1 transcription factor causes blockade of multiple signal transduction pathways and inhibits breast cancer growth. Oncogene. 2002;21:7680–7689. doi: 10.1038/sj.onc.1205883. [DOI] [PubMed] [Google Scholar]
- 48.Di Marco E, Pierce JH, Aaronson SA, Di Fiore PP. Mechanisms by which EGF receptor and TGF alpha contribute to malignant transformation. Nat Immun Cell Growth Regul. 1990;9:209–221. [PubMed] [Google Scholar]
- 49.Sassone-Corsi P, Mizzen CA, Cheung P, Crosio C, Monaco L, Jacquot S, Hanauer A, Allis CD. Requirement of Rsk-2 for epidermal growth factor-activated phosphorylation of histone H3. Science. 1999;285:886–891. doi: 10.1126/science.285.5429.886. [DOI] [PubMed] [Google Scholar]
- 50.Hunter T. Oncoprotein networks. Cell. 1997;88:333–346. doi: 10.1016/s0092-8674(00)81872-3. [DOI] [PubMed] [Google Scholar]
- 51.Kinzler KW, Vogelstein B. Cancer-susceptibility genes. Gatekeepers and caretakers. Nature 386. 1997;761:763. doi: 10.1038/386761a0. [DOI] [PubMed] [Google Scholar]
- 52.Davis RJ. The mitogen-activated protein kinase signal transduction pathway. J Biol Chem. 1993;268:14553–14556. [PubMed] [Google Scholar]
- 53.Davie JR, Samuel SK, Spencer VA, Holth LT, Chadee DN, Peltier CP, Sun JM, Chen HY, Wright JA. Organization of chromatin in cancer cells: role of signalling pathways. Biochem Cell Biol. 1999;77:265–275. [PubMed] [Google Scholar]
- 54.Dunn KL, Davie JR. Stimulation of the Ras-MAPK pathway leads to independent phosphorylation of histone H3 on serine 10 and 28. Oncogene. 2005;24:3492–3502. doi: 10.1038/sj.onc.1208521. [DOI] [PubMed] [Google Scholar]
- 55.Thomson S, Clayton AL, Hazzalin CA, Rose S, Barratt MJ, Mahadevan LC. The nucleosomal response associated with immediate-early gene induction is mediated via alternative MAP kinase cascades: MSK1 as a potential histone H3/HMG-14 kinase. EMBO J. 1999;18:4779–4793. doi: 10.1093/emboj/18.17.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Strelkov IS, Davie JR. Ser-10 phosphorylation of histone H3 and immediate early gene expression in oncogene-transformed mouse fibroblasts. Cancer Res. 2002;62:75–78. [PubMed] [Google Scholar]
- 57.Soncini C, Carpinelli P, Gianellini L, Fancelli D, Vianello P, Rusconi L, Storici P, Zugnoni P, Pesenti E, Croci V, et al. PHA-680632, a novel Aurora kinase inhibitor with potent antitumoral activity. Clin Cancer Res. 2006;12:4080–4089. doi: 10.1158/1078-0432.CCR-05-1964. [DOI] [PubMed] [Google Scholar]
- 58.Katayama H, Brinkley WR, Sen S. The Aurora kinases: role in cell transformation and tumorigenesis. Cancer Metastasis Rev. 2003;22:451–464. doi: 10.1023/a:1023789416385. [DOI] [PubMed] [Google Scholar]