Abstract
KCNE1 associates with the pore-forming α-subunit KCNQ1 to generate the slow (IKs) current in cardiac myocytes. Mutations in either KCNQ1 or KCNE1 can alter the biophysical properties of IKs and mutations in KCNE1 underlie cases of long QT syndrome type 5 (LQT5). We previously investigated a mutation in KCNE1, T58P/L59P, which causes severe attenuation of IKs. However, how T58P/L59P acts to disrupt IKs has not been determined. In this study, we investigate and compare the effects of T58P/L59P with three other LQT5 mutations (G52R, S74L, and R98W) on the biophysical properties of the current, trafficking of KCNQ1, and assembly of the IKs channel. G52R and T58P/L59P produce currents that lack the kinetic behavior of IKs. In contrast, S74L and R98W both produce IKs-like currents but with rightward shifted voltage dependence of activation. All of the LQT5 mutants express protein robustly, and T58P/L59P and R98W cause modest, but significant, defects in the trafficking of KCNQ1. Despite defects in trafficking, in the presence of KCNQ1, T58P/L59P and the other LQT5 mutants are present at the plasma membrane. Interestingly, in comparison to KCNE1 and the other LQT5 mutants, T58P/L59P associates only weakly with KCNQ1. In conclusion, we identify the disease mechanisms for each mutation and reveal that T58P/L59P causes disease through a novel mechanism that involves defective IKs complex assembly.
Keywords: K channel, KCNQ1, KCNE1, arrhythmia
long qt syndrome (lqts) is characterized by a prolongation of the QTc interval on the electrocardiogram. LQTS causes sudden death in affected individuals due to the development of a characteristic ventricular tachycardia known as torsade de pointes and subsequently fatal ventricular fibrillation. LQTS can be acquired in origin, most commonly due to drugs, or more rarely occur as a part of an inherited syndrome. Progress has been made in understanding the molecular basis of the inherited forms of LQTS and mutations in genes encoding ion channels and their ancillary proteins have been identified. Mutations in the cardiac Na+ channel and two K+ channels, and their related proteins, that form the rapid (IKr) and slow (IKs) currents have been shown to be the commonest cause of hereditary LQTS (19, 20).
IKs is composed of the pore-forming KCNQ1 α-subunit and the auxiliary β-subunit KCNE1. The IKs channel complex consists of a tetramer of KCNQ1 α-subunits and probably two KCNE1 β-subunits (1, 5, 24). In the absence of KCNE1, KCNQ1 produces smaller and more rapidly activating K+ selective currents that also inactivate upon prolonged depolarization. When the two subunits are coexpressed, currents are significantly enhanced, activation and deactivation kinetics are markedly slowed, inactivation is lost, and the voltage dependence of activation is shifted rightward to more depolarized potentials (1, 24). Mutations in both KCNQ1 and KCNE1 cause LQTS and account for types 1 and 5, respectively (LQT1 and LQT5). These mutations lead to two distinct clinical syndromes, the autosomal dominant Romano-Ward syndrome (RWS) and the rarer autosomal recessive Jervell-Lange Nielsen syndrome (JLNS). Individuals with JLNS also suffer from profound hearing loss that is not found in individuals with RWS (19, 20). Mutations in KCNE1 have been documented to alter the biophysical properties of IKs in a variety of ways. Usually, LQT5 mutations cause alterations that result in a reduction of IKs current density. This reduction in current density acts to prolong repolarization and therefore promote the onset of ventricular tachycardia (19). For example, D76N, a mutation found in patients with RWS, acts in a dominant-negative manner to shift the voltage dependence of activation toward positive potentials and therefore decrease current density (28). In general, studies have concentrated on the actions of LQT5 mutations on these biophysical properties and have not assessed whether other mechanisms can contribute to disease pathogenesis. In contrast to LQT1, where it is clear that defects in trafficking and IKs channel assembly influence disease pathogenesis (8, 26, 33), whether defects in trafficking and assembly are important in LQT5 disease pathogenesis has not been extensively investigated. To date, only one LQT5 mutation, L51H, has been identified that can cause defective trafficking of the IKs channel (2, 14), and whether defective IKs channel assembly can affect disease pathogenesis has not been established.
We previously investigated an LQT5 mutation found in JLNS, T58P/L59P, which acts to cause a severe attenuation of IKs (10, 31). However, the mechanism by which this mutation acts to disrupt IKs has not been determined. In this study, by comparing the effects of this mutant, with three other LQT5 mutants (G52R, S74L, and R98W), on the biophysical properties, trafficking of KCNQ1, and assembly of the IKs channel, we determine the disease mechanism for T58P/L59P.
MATERIALS AND METHODS
Molecular Biology
KCNQ1, KCNQ1-green fluorescent protein (GFP), and DsRed2-endoplasmic reticulum (ER) are as previously described (33). KCNE1 was cloned into pcDNA3.1/Zeo(+) on BamHI/EcoRI ends. Mutations were introduced into KCNE1 using site-directed mutagenesis (QuikChange, Stratagene). Myc-KCNQ1 was a kind gift from Dan Roden (12). S373P-KCNQ1-GFP was generated by splicing by overlap extension PCR as described in Ref. 33. 3XFlag-KCNE1 and LQT5 mutants were generated following the method described in Ref. 18. Briefly, KCNE1 and LQT5 mutants were amplified, from the untagged versions, using primers that introduced EcoRI and BamHI at the 5′- and 3′-ends, respectively, and then digested and ligated into the 3XFLAG cytomegalovirus (CMV)-10 vector (Sigma) that introduces NH2-terminal flag tags. 3XFlag-KCNE1Δ79−129, 3XFlag-G52RΔ79−129, and 3XFlag-T58P/L59PΔ79−129 were generated on the basis of the method described in Ref. 29. By using the same forward primer used to generate 3XFlag-KCNE1 and a reverse (antisense) primer that introduced a stop codon after amino acid 78, and a BamHI restriction site, the 1–78 amino acid region of KCNE1, G52R, and T58P/L59P was amplified from the untagged versions. The amplification products were then digested and ligated into the 3XFLAG CMV-10 vector (Sigma). All constructs were verified by automated sequencing.
Cell Culture
Chinese Hamster Ovary-K1 (CHO-K1) cells were obtained from the American Type Culture Collection and cultured in F-12 Ham nutrient mixture (Gibco-Invitrogen) supplemented with 10% fetal bovine serum and penicillin-streptomycin (Gibco-Invitrogen). Human embryonic kidney (HEK)-293 cells were cultured in Dulbecco's modified Eagle's medium (Gibco-Invitrogen) supplemented with 10% fetal bovine serum and penicillin-streptomycin (Gibco-Invitrogen). Unless otherwise stated, 500 ng of cDNA, per vector, was transfected using Lipofectamine (Invitrogen), and cells were seeded onto glass coverslips 5–6 h after transfection.
Electrophysiology
Currents produced by KCNQ1 were recorded from transfected CHO-K1 cells, 2 days after transfection. Whole cell voltage-clamp recording was carried out at room temperature using an Axopatch 200B amplifier (Axon Instruments). Transfected cells were identified by epifluorescence. Pipette solution contained (in mM) 150 KCl, 5 EGTA, 10 HEPES, 2 MgCL2, 1 CaCl2, and 5 (Na)2ATP (pH 7.2 with KOH). Extracellular solution contained (in mM) 150 NaCl, 5 KCl, 10 HEPES, 2 MgCL2, and 1 CaCl2 (pH 7.4 with NaOH). Currents were recorded by holding the cell at a voltage of −80 mV followed by stepped depolarizations from −80 to +80 mV for 6 s in 10-mV increments, followed by a repolarizing pulse back to −20 mV for 3 s (to measure tail currents) and back to −80 mV. Series resistance was at least 70% compensated using the amplifier circuitry. Pipette resistance, when filled with intracellular solution, was ∼1.5–2.5 MΩ, and pipette capacitance was reduced by coating the tip with a parafilm-oil suspension. Data were analyzed using Clampfit and Microcal Origin software (10, 33). Current-voltage relationships were determined by normalizing the maximal current densities at the end of each pulse potential to cell capacitance (nA/pF). The voltage dependence of channel activation (V0.5, indicates the potential at which the activation is half-maximal) was determined by fitting the normalized amplitude of the peak tail currents (y/ymax) versus test potential (Vt) with a Boltzmann function: y/ymax = 1/{1 + exp[(V0.5 − Vt)/k]}, where k indicates the slope factor. Voltage-dependent activation (activation τ) time constants, at +40 mV, were determined by fitting the currents induced by the voltage protocol to a single exponential function. Time to peak values, instead of voltage-dependent activation time constants, were generated for currents that lacked a slow component of activation because these currents could not be fitted to a single exponential function or easily be described by an exponential rise. Therefore, to fit an activation time for these currents, we simply determined the time taken to reach the maximal current at +40 mV. To determine channel deactivation time constants (deactivation τ) the cells were repolarized to −20 mV for 3 s following a 6-s pulse to +40 mV, and the resulting tail currents were fit to a single exponential function.
Microscopy
Imaging and analysis for colocalization studies were performed as previously described (33). Briefly, live cells were imaged using a Bio-Rad Radiance 2100 laser-scanning confocal microscope. GFP was excited using an argon 488-nm laser, and emission recorded using a HQ515/15 filter. DsRed2 was excited using a helium/neon laser at 543 nm, and images were collected with an HQ590/15 filter set. Colocalization analysis was performed using Laserpix software (Bio-Rad). All images were acquired sequentially. The colocalization coefficient is the sum of the green pixel intensities (GFP) that have a red component (ER) divided by the sum of all the green pixel intensities in the image. All images visualized for immunofluorescence experiments were also taken using the same microscopy system, and FITC was excited using an argon 488-nm laser and emission was recorded using a HQ515/15 filter. Low-magnification images were acquired using a ×20 objective, and individual cells were visualized using a ×60 oil-immersion objective. Images were pseudocolored, filtered, and converted to 24-bit RGB files for display (TIFF).
Biochemistry
Coimmunoprecipitation.
For coimmunoprecipitation (Co-IP) experiments, HEK-293 cells were grown to ∼70% confluence in 100-mm dishes. In general, HEK-293 cells were used, instead of CHO-K1 cells, for Co-IP studies because they proved easier to transfect on the large scale required and, once transfected, provided more robust and equal expression of each construct. HEK-293 cells were transiently transfected using a calcium phosphate transfection technique (32) with the α-subunit (KCNQ1) (12.5 μg) and/or the β-subunit (KCNE1) (12.5 μg). Empty vector DNA was cotransfected to keep the total amount of DNA transfected (25 μg) constant. Forty-eight hours after transfection, the cells were washed with PBS+ (PBS + 0.1 mM CaCl2 and 1 mM MgCL2) and scraped into 500 μl of ice-cold immunoprecipitation buffer [TDET buffer: 1% Triton X-100, 0.4% sodium deoxycholate, 5 mM EDTA, and 25 mM Tris (pH 7.4)] containing protease inhibitors (Roche). The cell lysate was then incubated on ice for 30 min with brief but repeated vortexing. After incubation, lysates were cleared by centrifugation at 16,000 g for 10 min at 4°C. Following centrifugation, the supernatant was placed into a fresh Eppendorf tube and the pellet was discarded. From the cleared lysate, 50 μl was removed and combined with 25 μl of reducing 3× SDS-PAGE loading buffer (15) (R-STB) to provide a total cell lysate sample. To preclear the samples, 10 μl of protein G Sepharose (Zymed) was added and the samples were incubated for 2 h at 4°C with end-over-end rotation. After rotation, the samples were centrifuged at 376 g for 3 min to pellet the Sepharose beads and the supernatant was removed and transferred to a fresh tube. To the supernatants, 2 μg of immunoprecipitation antibody [α-GFP mouse monoclonal (no. 1181446001, Roche) or α-KCNQ1 NH2-terminal mouse monoclonal (no. N37/10, NeuroMab)] was then added and the samples were incubated overnight at 4°C with end-over-end rotation. To precipitate the antibody, 25 μl of protein G Sepharose were added and incubated for 2 h at 4°C with end-over-end rotation. The beads were then harvested by centrifugation at 376 g for 3 min and the supernatant was discarded. After centrifugation, the beads were washed six times with 750 μl ice-cold immunoprecipitation buffer. Once washed, the remaining wash buffer was removed and precipitated proteins were eluted from the beads by incubating with 35 μl R-STB for 10 min at 95°C. For details of the biochemical methods used for the coimmunoprecipitation studies performed in CHO-K1 cell, please see supplemental data (supplemental material for this article is available online at the American Journal of Physiology-Cell Physiology website).
SDS-PAGE and Western blot analysis.
For the Co-IP studies, 8 μl of the immunoprecipitate and 20 μl of the lysate were separated on Laemmli (15) gels for KCNQ1-GFP or KCNQ1 and on tricine gels for KCNE1 (25). Gels were transferred to a polyvinylidene difluoride membrane and blocked in PBS (pH 7.4) containing 5% dried milk powder. After blocking, membranes were incubated with either anti-FLAG [mouse monoclonal (M2), 1:3,000 (no. F3165, Sigma)], anti-KCNE1 [rabbit polyclonal, 1:500 (no. APC-008, Alomone)], anti-GFP [mouse monoclonal, 1:2,000 (no. 11814460001, Roche)], or anti-KCNQ1 [rabbit polyclonal, 1:5,000 (no. APC-022, Alomone)] antibodies for 2 h. Membranes were then washed with PBS three times for 5 min. After washing, the primary antibody was detected by adding a horseradish peroxidase (HRP)-conjugated goat anti-mouse Fcγ chain-specific secondary antibody [1:10,000 (no. 115-035-164, Jackson)] or a HRP-conjugated goat anti-rabbit antibody [1:5,000 (no. sc-2054, Santa Cruz)] for 1 h. The membranes were then washed with PBS three times for 5 min and developed using the ECL Western blotting chemiluminescent reagent kit (Amersham) as per the manufacturer's instructions.
Immunofluorescence.
CHO-K1 cells were transfected, using Lipofectamine (Invitrogen), when at 70% confluence on 25-mm glass coverslips in six-well plates. Cells were transfected with 0.75 μg myc-KCNQ1 in the presence or absence of 1 μg KCNE1/LQT5 mutant. Empty pcDNA3.1/Zeo(+) plasmid DNA (Invitrogen) was cotransfected in samples containing only myc-KCNQ1 or KCNE1/LQT5 mutant to keep the total amount of DNA transfected constant. Forty-eight hours after transfection, cells were washed, two times for 5 min, in PBS+ and then fixed for 20 min at 4°C in 4% paraformaldehyde that had been freshly prepared in PBS+. After fixation, cells were washed three times for 5 min in PBS+ and blocked for 20 min at room temperature (RT), with gentle rocking, in either permeabilizing buffer [PBS+ containing 1% IgG-free BSA (Sigma) and 0.2% Triton X-100 (Sigma)] for permeabilized samples or blocking buffer [PBS+ containing 1% IgG-free BSA (Sigma)] for unpermeabilized samples. After blocking, cells were washed two times for 5 min in PBS+ and then incubated with 5 μg/ml of fluorescein (FITC)-conjugated mouse monoclonal anti-flag antibody (M2) (no. F4049, Sigma) in either permeabilizing or blocking buffer for 1.5 h at RT, in the dark, with gentle rocking. Cells were then washed with PBS+ three times for 5 min and were either visualized directly or dried in the dark.
Data Analysis
For the electrophysiological data, statistical comparisons were made using a two-tailed Student's t-test. For the ER retention data, statistical comparisons were made using a one-way ANOVA with Bonferroni post hoc test for multiple comparisons. For the Co-IP data, statistical comparisons were made using a one-sample t-test. Data were considered to be statistically significant when P < 0.05. Western blot band intensity was determined and quantified using ImageJ software (National Institutes of Health, Bethesda, MD). To determine the relative level of KCNE1/mutant coprecipitation with KCNQ1, we used the following method: Initially, the band intensities seen for KCNE1 or the LQT5 mutants in the lysate or Co-IP blots were normalized to the levels of expression/precipitation seen for KCNE1, respectively. The normalized Co-IP band intensities were then divided by the normalized band intensities seen for KCNE1 or LQT5 mutants in the lysate. To determine the relative amount, in comparison to KCNE1, of association with KCNQ1, the Co-IP/lysate ratios were then divided by the normalized levels of immunoprecipitated KCNQ1 (the level of immunoprecipitated KCNQ1 was normalized to the level seen for immunoprecipitation of KCNQ1 + KCNE1).
RESULTS
We generated and compared the effects of KCNE1 mutants, described by others and us in LQT5, in mammalian cell expression systems (CHO-K1 and HEK-293 cells). Specifically, we made a detailed study of T58P/L59P which occurs in JLNS (10, 31). We also examined G52R (RWS), S74L (RWS), and R98W (RWS) for comparison (16, 22, 28). We chose to compare the effects of T58P/L59P with G52R, S74L, and R98W because, like T58P/L59P, their effects on trafficking or assembly of the IKs complex have not been determined.
Effects of LQT5 Mutations on Channel Function
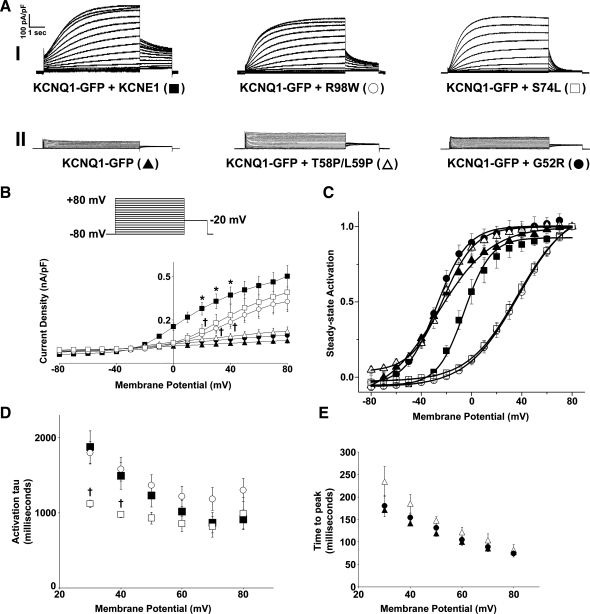
The mutants were coexpressed with KCNQ1-GFP, and the currents produced were compared with those produced by expression of KCNQ1-GFP alone or coexpression of KCNQ1-GFP and KCNE1 (Fig. 1). As previously reported, in comparison to the currents produced by expression of KCNQ1-GFP alone, the coexpression of KCNE1 with KCNQ1-GFP acted to significantly (P < 0.05) increase current density, cause a shift in the voltage dependence of activation to more positive potentials, and slow activation and deactivation kinetics (Fig. 1, A–E, and Table 1) (1, 24, 33). We have previously shown that the addition of GFP to the COOH terminus of KCNQ1 does not affect the biophysical properties of the currents produced by KCNQ1 (33). In comparison to KCNE1, all four LQT5 mutants produced abnormal currents, and these effects could be split into two types of behavior. S74L and R98W produced currents with the slow activation profile characteristic of IKs (Fig. 1AI). However, in comparison to those produced by KCNQ1-GFP and KCNE1, these currents had reduced current density (P < 0.05) and shifts in the voltage dependences of activation to more positive test potentials (P < 0.05) (Fig. 1AI, B, and C and Table 1). Although both S74L and R98W acted to significantly (P < 0.05) shift the voltage dependence of activation to a similar degree, R98W appeared to reduce current density more strongly (Fig. 1B and Table 1). In contrast to the effects of S74L and R98W, G52R and T58P/L59P produced currents that lacked the slow activation profile characteristic of IKs (Fig. 1AII). In fact, the currents produced by G52R and T58P/L59P were not statistically (P > 0.05) different in terms of current density or the voltage dependence of activation from those produced by expression of KCNQ1-GFP in the absence of KCNE1 (Fig. 1A, B, and C and Table 1). In comparison to KCNE1, the LQT5 mutants also affected the kinetics of channel activation and deactivation. S74L but not R98W acted to modestly but significantly (P < 0.05) increase the rate of activation in comparison to KCNE1 (Fig. 1D and Table 1). It was not possible to fit a single exponential to the activation of the currents that lacked slow activation. Therefore, for these currents we determined the time taken to reach maximal peak current (time to peak) instead. The time taken to reach maximal peak current for G52R or T58P/L59P was not significantly (P > 0.05) different from that seen for KCNQ1-GFP when expressed without KCNE1 (Fig. 1E and Table 1). The activation time constants for KCNE1, S74L, or R98W were significantly (P < 0.05) different from the time taken to reach maximal peak current for KCNQ1-GFP when expressed without KCNE1 (Table 1). In terms of the effects of the LQT5 mutants on channel deactivation, both S74L and R98W acted to significantly (P < 0.05) accelerate the rate of channel deactivation in comparison to KCNE1 (Table 1).
Fig. 1.
Differential effects of long QT syndrome type 5 (LQT5) mutants on the slow K+ current (IKs) channel function. A: representative traces of KCNQ1-green fluorescent protein (GFP) alone or in the presence of KCNE1 or the LQT5 mutants G52R, T58P/L59P, S74L, and R98W. Currents were normalized to cell capacitance, and the traces have been split into two categories: those that exhibit IKs characteristics (I) and those that do not possess the characteristics of IKs (II). The voltage protocol used is shown in the inset. B: mean current-voltage relationships. Current-voltage relationships were determined by normalizing the maximal current densities at the end of each pulse potential to cell capacitance (nA/pF). C: normalized voltage-dependent activation curves (steady-state activation) of KCNQ1-GFP expressed alone or in the presence of KCNE1 or the LQT5 mutants. The activation curves were determined by fitting the normalized amplitude of the peak tail currents versus test potential with a Boltzmann function (solid lines). D and E: mean activation time constants (activation τ) and time to peak values. These values were determined as described in materials and methods. Data are presented as means ± SE (n = 5–6 cells). *P < 0.05, significantly different from KCNQ1-GFP. †P < 0.05, significantly different from KCNQ1-GFP + KCNE1.
Table 1.
Analysis of the effects of the LQT5 mutants on IKs channel function
| Channel | Current Density, nA/pF (+40 mV) | V0.5, mV | Slope Factor, mV | Activation τ, ms (Time to Peak) | Deactivation τ, ms |
|---|---|---|---|---|---|
| KCNQ1-GFP | 0.038 ± 0.012† | −26.73 ± 2.52† | 21.36 ± 1.97† | 142 ± 6† | ND |
| KCNQ1-GFP + KCNE1 | 0.372 ± 0.057* | −5.83 ± 1.77* | 10.58 ± 1.06* | 1,491 ± 179* | 1,402.16 ± 64.69 |
| KCNQ1-GFP + G52R | 0.066 ± 0.021† | −27.62 ± 1.59† | 12.35 ± 1.54* | 154 ± 9† | ND |
| KCNQ1-GFP + T58P/L59P | 0.087 ± 0.021† | −22.67 ± 1.58† | 12.88 ± 1.39* | 185 ± 21† | ND |
| KCNQ1-GFP + S74L | 0.232 ± 0.076§ | 36.40 ± 2.24*† | 19.64 ± 1.12† | 978 ± 41*†‡ | 415.94 ± 26.27†‡ |
| KCNQ1-GFP + R98W | 0.184 ± 0.042*† | 37.03 ± 2.43*† | 20.60 ± 1.47† | 1,580 ± 159* | 850.55 ± 69.32† |
Values are means ± SE (n = 5–6 cells). LQT5, long QT syndrome type 5; IKs, slow component of the delayed rectifier K+ current; V0.5, potential at which the activation is half-maximal; GFP, green fluorescent protein.
P < 0.05, significantly different from KCNQ1-GFP;
P < 0.05, significantly different from KCNQ1-GFP + KCNE1;
P < 0.05, significantly different from KCNQ1-GFP + R98W;
P = 0.06, different from KCNQ1-GFP. ND, not determined.
KCNE1 (LQT5) Mutants Can Increase ER Retention of KCNQ1
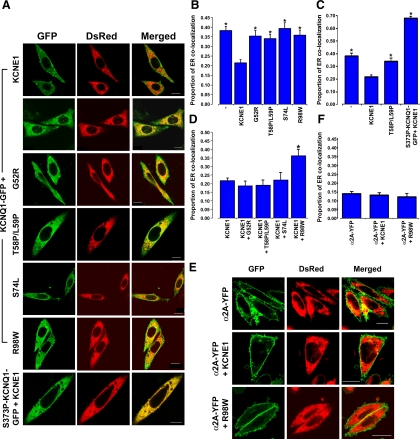
Using an assay based on microscopy (see materials and methods), all the LQT5 mutants, when expressed with KCNQ1-GFP (mimicking the JLNS phenotype, two mutant alleles), caused a small but statistically significant (P < 0.05) increase in the level of ER retention of KCNQ1-GFP compared with that seen in response to coexpression of KCNQ1-GFP with KCNE1 (Fig. 2, A and B). The level of ER retention seen, in comparison to that seen for the LQT1 KCNQ1 mutant S373P-KCNQ1-GFP (11), was modest and similar to that seen when KCNQ1-GFP is expressed in the absence of KCNE1 (Fig. 2C). Three of the LQT5 mutants described here (G52R, S74L, and R98W) were identified from individuals that have RWS. Therefore, we assessed which mutants were able to have a dominant-negative effect on trafficking. When the mutants were expressed with KCNE1 in a 1:1 manner, in an effort to mimic the phenotype seen in RWS, (one mutant, one wild-type allele) only R98W was able to significantly (P < 0.05) increase the level of KCNQ1-GFP retention in the ER (Fig. 2D). These effects were specific for KCNQ1-GFP because coexpression of KCNE1 or R98W had no effect on the surface delivery of the α2A-adrenergic receptor (α2A) (Fig. 2, E and F). We have previously shown that the behavior of α2A-yellow fluorescent protein is comparable to an untagged version of the protein (21).
Fig. 2.
Effects of KCNE1 and LQT5 mutants on the trafficking of KCNQ1-GFP. A: confocal images of Chinese hamster ovary (CHO)-K1 cells transfected with either S373P-KCNQ1-GFP in the presence of KCNE1 and the endoplasmic reticulum (ER) marker DsRed2-ER or KCNQ1-GFP and DsRed2-ER alone or in the presence of KCNE1 or the LQT5 mutants. Images are shown for GFP alone, Ds-Red2, and the merged image. Colocalization between GFP and DsRed2 appears as yellow. Scale bar indicates 10 μm. B: mean data showing the proportion of KCNQ1-GFP ER colocalization in the absence or presence of KCNE1 or the LQT5 mutants G52R, T58P/L59P, S74L, and R98W. Data are presented as means ± SE (n = 36–66 cells). *P < 0.05 compared with control. Minus sign (−) indicates expression of KCNQ1-GFP alone. C: comparison of the effects on trafficking of KCNQ1-GFP, of the LQT5 mutant T58P/L59P with those produced by the KCNQ1 (LQT1) mutation S373P. Data are presented as means ± SE (n = 36–66 cells). *P < 0.05 compared with control. Minus sign (−) indicates expression of KCNQ1-GFP alone. D: dominant-negative effects of coexpression of KCNE1 and the LQT5 mutants on the trafficking of KCNQ1-GFP. KCNE1 and the LQT5 mutants are expressed at a 1:1 ratio. Data are presented as means ± SE (n = 13–66 cells). *P < 0.05 compared with control. E: confocal images of CHO-K1 cells transfected with a yellow fluorescent protein (YFP)-tagged α2A-adrenergic receptor (α2A-YFP) in the presence of the ER marker DsRed2-ER and the presence or absence of KCNE1 or the LQT5 mutant R98W. α2A-YFP was imaged using conditions optimized for GFP visualization as described in materials and methods. Scale bar indicates 10 μm. F: mean data showing the proportion of α2A-YFP ER colocalization in the presence or absence of KCNE1 or the LQT5 mutant R98W. Data are presented as means ± SE (n = 23–28 cells).
Analyzing the Effects of LQT5 Mutations on KCNQ1-KCNE1 Complex Formation
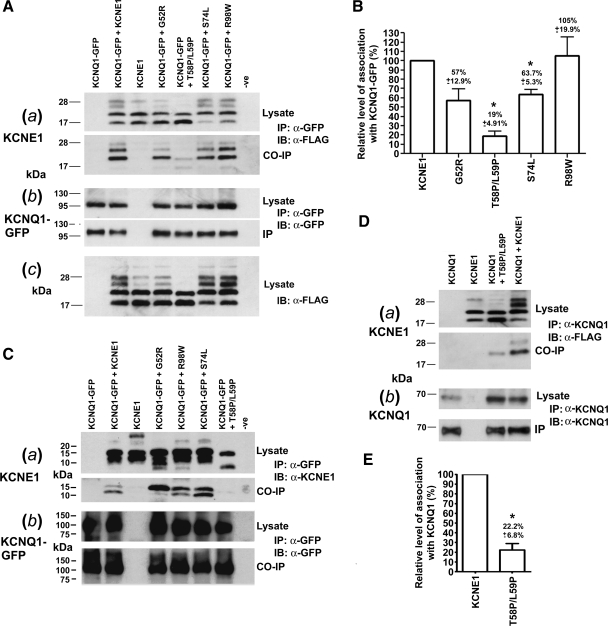
The currents produced by expression of T58P/L59P or G52R with KCNQ1-GFP were indistinguishable from those produced when KCNQ1-GFP is expressed without KCNE1 (Fig. 1). A trivial explanation for this would be that these mutations lead to the expression of unstable proteins that are poorly expressed. However, it was clear from immunoblotting that these mutants were expressed at comparable levels, in HEK-293 cells (Fig. 3A) and CHO-K1 cells (Fig. 3C), to KCNE1. We did, however, notice that the gel mobility of T58P/L59P is faster than for KCNE1 and the other LQT5 mutants and that T58P/L59P appears to lack the higher-molecular-weight forms (Fig. 3Ac) that have been attributed to complex glycosylation (4, 7). The inability of T58P/L59P and G52R to modify the current produced by KCNQ1 led us to hypothesize that these particular mutations may act to disrupt the association between KCNQ1 and KCNE1. To investigate this, we used coimmunoprecipitation to assess whether the mutants associate with KCNQ1. Primarily, we used 3XFlag-tagged versions of KCNE1 and mutants instead of the untagged versions because this tag provides an epitope that can be easily identified by immunoblotting. This detection system is also more sensitive than blotting using an antibody directed against the COOH terminus of untagged KCNE1 (Supplemental Fig. 1). The currents generated by 3XFlag-KCNE1 in conjunction with KCNQ1-GFP are similar to the untagged version (Supplemental Fig. 2) (18), and the flag-tag addition did not affect expression of KCNE1 or the LQT5 mutants in CHO-K1 or HEK-293 cells (Supplemental Fig. 2D and Fig. 3A). 3XFlag-tagged-KCNE1 and the LQT5 mutants coimmunoprecipitated with KCNQ1-GFP (Fig. 3A). However, the amount of T58P/L59P that coimmunoprecipitated with KCNQ1-GFP was considerably lower (Fig. 3A). When quantified, the level of T58P/L59P that interacted with KCNQ1-GFP was significantly (P < 0.05) reduced to 19 ± 4.9% of the level seen for KCNE1 (Fig. 3B). S74L also acted to significantly (P < 0.05) reduce interaction with KCNQ1-GFP, although not as dramatically, to 63.7 ± 5.3% of the level seen for KCNE1 (Fig. 3B). G52R also appeared to cause a reduction in interaction (57 ± 12.9%), but this did not reach significance (P = 0.074) (Fig. 3B). A similar pattern was also seen, although not quantified, using untagged KCNE1 and LQT5 mutants in CHO-K1 cells (Fig. 3C). Furthermore, in CHO-K1 cells we could not even detect coimmunoprecipitated untagged T58P/L59P. The inability to detect T58P/L59P was probably, as described earlier, due to the reduced sensitivity of the antibody detection system used (Supplemental Fig. 1). We also saw a similar reduction in interaction for T58P/L59P when immunoprecipitated with untagged KCNQ1 instead of KCNQ1-GFP (Fig. 3D). In this system, T58P/L59P acted to significantly (P < 0.05) reduce interaction with untagged KCNQ1 to 22.2 ± 6.8% of the level seen for KCNE1 (Fig. 3, D and E).
Fig. 3.
LQT5 mutations can affect the ability of KCNE1 to associate with KCNQ1. A: coimmunoprecipitation (Co-IP) of 3XFlag-tagged KCNE1 and LQT5 mutants with KCNQ1-GFP. Aa, top: 3XFlag-tagged KCNE1 and LQT5 mutant expression in human embryonic kidney (HEK)-293 cell lysates. Aa, bottom: 3XFlag-tagged KCNE1 or LQT5 mutants that coimmunoprecipitated with KCNQ1-GFP. Ab, top: KCNQ1-GFP expression in HEK-293 cell lysate. Ab, bottom: KCNQ1-GFP immunoprecipitation. The Co-IP study shown in Aa and Ab is representative of three experiments. Ac: T58P/L59P does not display the higher forms seen for KCNE1 and the other LQT5 mutants in the presence of KCNQ1-GFP. The gel order is the same as shown in Aa. B: the LQT5 mutations T58P/L59P and S74L, but not G52R or R98W, act to reduce the level of KCNE1 that coimmunoprecipitates with KCNQ1-GFP. The relative level of association of the LQT5 mutants in comparison to KCNE1 was determined from three separate experiments. Data are displayed as a percentage of the level seen for KCNE1 and are presented as means ± SE. *Significantly (P < 0.05) different from the value seen for control (KCNQ1-GFP + 3XFlag-KCNE1). C: coimmunoprecipitation of untagged KCNE1 and the LQT5 mutants with KCNQ1-GFP from CHO-K1 cells. Ca, top: untagged KCNE1 and LQT5 mutant expression in CHO-K1 cell lysate. Ca, bottom: KCNE1 or LQT5 mutants that coimmunoprecipitated with KCNQ1-GFP. Cb, top: KCNQ1-GFP expression in CHO-K1 cell lysate. Cb, bottom: KCNQ1-GFP immunoprecipitation. The Co-IP study shown in Ca and Cb is representative of two experiments. D: coimmunoprecipitation of 3XFlag-tagged KCNE1 and T58P/L59P with untagged KCNQ1 from HEK-293 cells. Da, top: 3XFlag-tagged KCNE1 and T58P/L59P expression in HEK-293 cells. Da, bottom: 3XFlag-tagged KCNE1 or T58P/L59P that coimmunoprecipitated with untagged KCNQ1. Db, top: KCNQ1 expression in HEK-293 cell lysate. Db, bottom: KCNQ1 immunoprecipitation. The Co-IP study shown in Da and Db is representative of three experiments. E: T58P/L59P acts to reduce the level of KCNE1 that coimmunoprecipitates with untagged KCNQ1. The relative level of association of 3X Flag-tagged T58P/L59P in comparison to KCNE1 was determined from three separate experiments. Data are displayed as a percentage of the level seen for KCNE1 and are presented as means ± SE. *Significantly (P < 0.05) different from the value seen for control (KCNQ1 +3XFlag-KCNE1). Co-IP, coimmunoprecipitation; IP, immunoprecipitation; IB, immunoblot. −ve, blank transfection control.
Are KCNE1 Mutants Present at the Cell Surface?
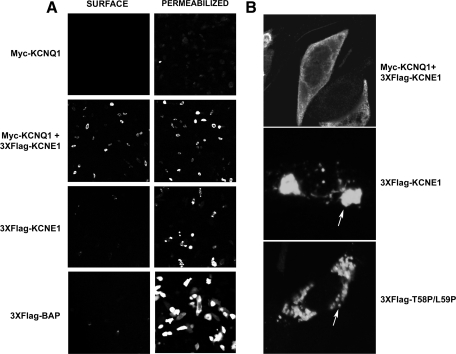
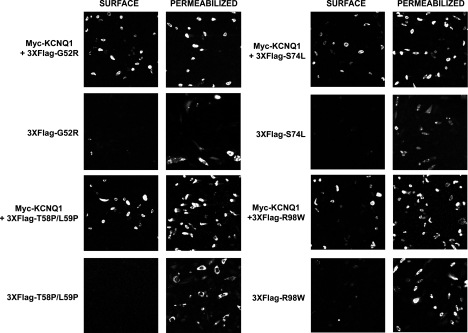
KCNE1 requires coassembly with KCNQ1 to reach the cell surface in CHO-K1 cells. In the absence of KCNQ1, KCNE1 is retained in the early stages of the secretory pathway (4). Given the differences in the strength of interaction between KCNE1 and the LQT5 mutants with KCNQ1, we wanted to see what effect these mutations had on transport of KCNE1 to the cell membrane. The 3XFlag tag addition at the NH2 terminus of KCNE1 and the LQT5 mutants, previously described, provides an extracellular epitope that can be identified by immunofluorescence. To validate the system, we used 3XFlag-BAP, a bacterial alkaline phosphatase fusion protein that produces a Flag-tagged cytosolic protein, as a control. As expected, staining for 3XFlag-BAP can be detected in permeabilized but not unpermeabilized cells (Fig. 4A). As previously reported (4), 3XFlag-KCNE1 in the absence of myc-KCNQ1 did not reach the cell surface but a strong level of FLAG signal was detected in permeabilized cells (Fig. 4A). In comparison, in the presence of myc-KCNQ1, 3XFlag-KCNE1 could be detected at the cell surface (Fig. 4A). It was also clear that the intracellular staining profile of 3XFlag-KCNE1 was different in the presence of myc-KCNQ1. In the absence of myc-KCNQ1, staining could be observed in either punctate spots or large aggregates (Fig. 4B). Though we have not performed a formal analysis, they bear resemblance to Russell bodies, an intermediary in protein degradation (13, 27). In contrast, in the presence of myc-KCNQ1, staining is observed in a more diffuse pattern (Fig. 4B). Interestingly, when the mutants were tested for cell surface expression in the presence of myc-KCNQ1, all could be detected at the cell surface (Fig. 5). In the absence of myc-KCNQ1, the mutant forms of KCNE1 did not reach the cell surface and clustered into similar punctate spots or aggregates within the cell (Figs. 4B and 5).
Fig. 4.
KCNE1 requires coexpression of KCNQ1 to reach the cell surface. A: analysis of the presence of cell surface expression of 3XFlag-tagged KCNE1 in the presence or absence of myc-KCNQ1, in CHO-K1 cells, by immunofluorescence . 3XFlag-tagged KCNE1 and LQT5 mutants were detected using a fluorescein (FITC)-conjugated mouse monoclonal antibody that recognizes the FLAG epitope. Images displaying representative immunostaining from unpermeabilized (surface staining) and permeabilized (intracellular staining) cells are shown. The 3XFlag-tagged cytosolic protein BAP (bacterial alkaline phosphatase) is shown as a control to highlight the presence of intracellular but lack of surface staining. B: the presence of KCNQ1 changes the intracellular distribution of KCNE1. Imaging the cellular distribution of 3XFlag-KCNE1 in the presence or absence of myc-KCNQ1, and 3XFlag-T58P/L59P in the absence of myc-KCNQ1, in permeabilized CHO-K1 cells. Arrows indicate the presence of punctate spots or aggregates of 3XFlag-tagged KCNE1 and T58P/L59P when expressed in the absence of myc-KCNQ1. Images are representative of a range of cells that were analyzed over separate experiments.
Fig. 5.
The LQT5 mutants require coexpression of KCNQ1 to reach the cell surface. Analysis of the presence of cell surface expression of 3XFlag-tagged LQT5 mutants in the presence or absence of myc-KCNQ1, in CHO-K1 cells, by immunofluorescence. 3XFlag-tagged KCNE1 and LQT5 mutants were detected using a fluorescein (FITC)-conjugated mouse monoclonal antibody that recognizes the FLAG epitope. Images displaying representative immunostaining from unpermeabilized (surface staining) and permeabilized (intracellular staining) cells are shown.
Removal of the COOH Terminus of KCNE1 Reduces KCNQ1/KCNE1 Interaction
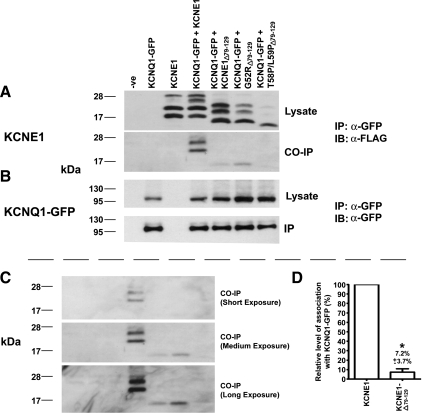
Although the interaction between KCNE1 and KCNQ1 is weakened by T58P/L59P, the complex still assembles and reaches the cell surface. The reduction but not abolition in the level of interaction between T58P/L59P and KCNQ1 suggests that KCNE1 can interact with KCNQ1 through other regions. Tapper and George (29) using electrophysiological methods have identified that two regions of KCNE1 are critical for the KCNQ1/KCNE1 interaction. The regions identified in their study are the transmembrane region (residues 43–66) and the COOH terminus (67–129) (29). Since T58P/L59P resides within the transmembrane domain, we analyzed what effect the removal of the COOH terminus of KCNE1, G52R, and T58P/L59P would have on the interaction with KCNQ1. The removal of the COOH terminus of KCNE1 (residues 79–129) resulted in a significant (P < 0.05) reduction in the strength of the association with KCNQ1-GFP (Fig. 6A) to 7.2 ± 3.7% of the level seen for KCNE1 (Fig. 6D). In a similar manner to 3XFlag-KCNE1Δ79−129, 3XFlag-G52RΔ79−129 was also still able to interact with KCNQ1-GFP (Fig. 6, A and C). In contrast, an interaction between 3XFlag-T58P/L59PΔ79−129 and KCNQ1-GFP could not be detected, even after prolonged exposures (Fig. 6, A and C).
Fig. 6.
Removal of the COOH terminus, residues 79–129, of KCNE1 reduces the level of coimmunoprecipitation with KCNQ1-GFP. Coimmunoprecipitation of 3XFlag-tagged-KCNE1, 3XFlag-tagged-KCNE1 and the LQT5 mutants, G52R and T58P/L59P, with the COOH terminus removed (3XFlag-KCNE1Δ79−129, 3XFlag-G52RΔ79−129, and 3XFlag-T58P/L59PΔ79−129, respectively) with KCNQ1-GFP. A, top: 3XFlag-tagged-KCNE1, ΔC-term79−129-KCNE1, and ΔC-term79−129-LQT5 mutant expression in HEK-293 cell lysate. A, bottom: 3XFlag-tagged-KCNE1, ΔC-term79−129-KCNE1, and ΔC-term79−129-LQT5 mutants that coimmunoprecipitated with KCNQ1-GFP. B, top: KCNQ1-GFP expression in HEK-293 cell lysate. B, bottom: KCNQ1-GFP immunoprecipitation. The Co-IP study shown in A and B is representative of three experiments. C: panels displaying varying lengths of exposure of the Co-IP panel displayed in A. The gel order is the same as that shown for A and B. D: deletion of the COOH terminus of KCNE1 reduces the relative level of association with KCNQ1-GFP. The relative level of association of 3XFlag-KCNE1Δ79−129 in comparison to KCNE1 was determined from three separate experiments. Data are displayed as a percentage of the level seen for KCNE1 and are presented as means ± SE. *Significantly (P < 0.05) different from the value seen for control (KCNQ1-GFP + 3XFlag-KCNE1).
DISCUSSION
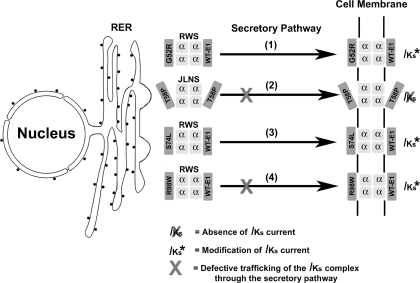
The aims of this study were to determine the disease mechanism by which the LQT5 mutation, T58P/L59P, acts to cause severe attenuation of IKs and to determine whether defective trafficking and assembly of KCNQ1/KCNE1 can influence disease pathogenesis in LQT5. For G52R, an inability to modulate channel gating when present in correctly trafficked and assembled complexes at the membrane results in a severe attenuation of IKs. Ma et al. (16) previously reported that G52R can act in a dominant-negative fashion to cause a 50% reduction in wild-type current amplitude. On the basis of the data in this study we can now report that this ability to reduce wild-type current density does not involve defects in trafficking or assembly and is solely due to an inability to modulate the gating properties of KCNQ1 (Fig. 7). Although T58P/L59P is also unable to modulate channel gating, the mechanism by which this occurs is different from that for G52R. T58P/L59P causes a small defect in trafficking and severely weakens the interaction of KCNE1 with KCNQ1. This defect in assembly leads to an inability to modulate channel gating and the production of currents that lack the slow activation profile (Fig. 7). The effects of S74L and R98W on the biophysical properties of the current are, as previously described, similar (22, 28). Both cause shifts in the voltage dependence of activation toward positive potentials, result in reductions in current density, and act to accelerate channel deactivation (22, 28). However, the disease mechanisms are slightly different. R98W but not S74L causes a small defect in trafficking (Fig. 7), and, in contrast, S74L but not R98W affects the assembly of the IKs complex (Fig. 7). Interestingly, R98W tended to decrease current density more strongly than S74L, and it is possible that the defects in trafficking caused by R98W may account for the ability of this mutant to reduce current amplitude more strongly.
Fig. 7.
Disease mechanisms in LQT5. In 1, G52R assembles and traffics correctly to the membrane. Once there, G52R fails to modulate gating and in a dominant-negative manner acts to reduce wild-type current by ∼50% (16). In 2, T58P/L59P reduces the strength of interaction with KCNQ1 and causes a slight disruption in trafficking. This weakened complex reaches the cell surface and once there produces a current that lacks the characteristic slow activation of IKs and is identical to that seen when KCNQ1 is expressed alone. In 3, S74L affects the strength of complex assembly but still traffics correctly to the membrane. Once there, S74L shifts the voltage dependence of activation to potentials that are more positive and reduces current density. In 4, R98W causes defects in trafficking and alters the characteristics of IKs by shifting the voltage dependence of activation to positive potentials and reducing current density. T58P, LQT5 mutation T58P/L59P; WT-E1, KCNE1; RER, rough endoplasmic reticulum; RWS, Romano-Ward syndrome; JLNS, Jervell-Lange Nielson syndrome.
In this study, we identify that T58P/L59P and R98W can cause defects in trafficking. It must, however, be noted that these changes are small in comparison to those seen in LQT1 and that the level of ER retention does not exceed that seen when KCNQ1-GFP is expressed alone. In other studies, only one other LQT5 mutant, L51H, a mutation found in JLNS, has previously been shown to affect trafficking of the IKs complex. L51H reduces surface expression of KCNQ1 and does not reach the cell surface, even in the presence of KCNQ1, because it is sequestered in the ER (2, 14). Overall, on the basis of the effects of the mutations described here and elsewhere (2, 14), it is clear that defective trafficking can influence disease pathogenesis in LQT5. However, it does not appear to be as common or severe as for LQT1 (33).
Two of the mutations we studied, T58P/L59P and S74L, acted to impair channel assembly. Interestingly, Tapper and George (29, 30) have identified that the transmembrane (residues 43 to 66) region of KCNE1 is essential for gating modulation and association with KCNQ1. Furthermore, the KCNE1 residue threonine-58 and the two surrounding amino acids are critically involved in gating modulation (17, 23). It is perhaps therefore not surprising that the introduction of two proline residues (T58P/L59P) into this α-helical region severely disrupts secondary structure and interaction at this site. It is also interesting to note the differences between the effects of G52R and T58P/L59P on gating modulation and assembly. Both G52R and T58P/L59P are unable to modulate channel gating but only T58P/L59P weakens channel assembly significantly (P < 0.05). This difference either indicates that different areas of the transmembrane region are involved in interaction and assembly or indicates that the ability to modulate gating is more sensitive to changes. It also highlights that disrupting an interaction/assembly site within the transmembrane domain is sufficient but not necessary to disrupt gating. Although to a lesser degree than T58P/L59P, the COOH-terminal mutation S74L was also able to significantly (P < 0.05) weaken the interaction with KCNQ1. It has recently been described that functional interactions between the COOH terminus of KCNE1 and the coiled-coil helix C domain of KCNQ1 are important for IKs channel assembly and that these interactions can be disrupted by a different COOH-terminal LQT5 mutation, D76N (9). It is therefore possible that other LQT5 mutations found in the COOH terminus of KCNE1, excluding R98W, could also affect assembly, and this warrants further investigation.
Although T58P/L59P severely weakened interaction with KCNQ1-GFP, it was, in the presence of KCNQ1, able to traffic to the membrane. Since KCNE1 cannot traffic to the membrane without assembling with KCNQ1 (4), this finding indicates that, although reduced, the level of interaction is still sufficient to allow T58P/L59P passage to the cell surface. As the COOH terminus of KCNE1 has recently been shown to functionally interact with KCNQ1 (6, 9), it is possible that T58P/L59P interacts through this region. This would likely produce an interaction that is weaker than that for KCNE1 but would still allow loose association and trafficking with KCNQ1. We found that removal of the COOH terminus of KCNE1 (residues 79–129) weakens the interaction with KCNQ1, and this finding is consistent with recent studies (6, 9). However, when the COOH terminus of T58P/L59P (residues 79–129) is removed, interaction with KCNQ1 is lost. Overall, these data are in agreement with a model in which T58P/L59P impairs assembly with KCNQ1 and causes a severe attenuation of IKs, by disrupting an interaction site located in the transmembrane region of KCNE1 that is essential for gating control. These data are also consistent with the presence of two interaction sites in the transmembrane and COOH-terminal regions of KCNE1.
One drawback of our study is that we were not able to investigate these phenomena in a more native setting, i.e., cardiac myocytes. The mutant T58P/L59P occurs in JLNS, and these patients have two mutant alleles. Therefore, transfecting the mutant KCNE1 into native myocytes where KCNQ1/KCNE1 are natively expressed only mimics the situation in the carriers and not those with the condition. The second alternative is to make a knock-in mouse. This is a time-consuming and expensive undertaking beyond the scope of our current study. In addition, it may not be an optimal strategy because the currents responsible for the repolarization of the mouse ventricular action potential are different from those in humans. This has even prompted some investigators to develop transgenic rabbit models overexpressing mutants involved in autosomal dominant LQT1 and LQT2 (3).
In summary, we have analyzed in detail the disease mechanisms of four LQT5 mutations and have identified that defects in trafficking and channel assembly can influence disease pathogenesis in LQT5. In addition, by comparing these disease mechanisms, we identify that T58P/L59P causes disease through a novel mechanism that results in a loss of the normal regulation of channel gating due to compromised interaction with KCNQ1.
GRANTS
This work was supported by the British Heart Foundation.
DISCLOSURES
No conflicts of interest are declared by the author(s).
Supplementary Material
REFERENCES
- 1.Barhanin J, Lesage F, Guillemare E, Fink M, Lazdunski M, Romey G. K(V)LQT1 and lsK (minK) proteins associate to form the I(Ks) cardiac potassium current. Nature 384: 78–80, 1996 [DOI] [PubMed] [Google Scholar]
- 2.Bianchi L, Shen ZJ, Dennis AT, Priori SG, Napolitano C, Ronchetti E, Bryskin R, Schwartz PJ, Brown AM. Cellular dysfunction of LQT5-minK mutants: abnormalities of I-Ks, I-Kr and trafficking in long QT syndrome. Hum Mol Genet 8: 1499–1507, 1999 [DOI] [PubMed] [Google Scholar]
- 3.Brunner M, Peng X, Liu GX, Ren XQ, Ziv O, Choi BR, Mathur R, Hajjiri M, Odening KE, Steinberg E, Folco EJ, Pringa E, Centracchio J, Macharzina RR, Donahay T, Schofield L, Rana N, Kirk M, Mitchell GF, Poppas A, Zehender M, Koren G. Mechanisms of cardiac arrhythmias and sudden death in transgenic rabbits with long QT syndrome. J Clin Invest 118: 2246–2259, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chandrasekhar KD, Bas T, Kobertz WR. KCNE1 subunits require co-assembly with K+ channels for efficient trafficking and cell surface expression. J Biol Chem 281: 40015–40023, 2006 [DOI] [PubMed] [Google Scholar]
- 5.Chen H, Kim LA, Rajan S, Xu S, Goldstein SA. Charybdotoxin binding in the I(Ks) pore demonstrates two MinK subunits in each channel complex. Neuron 40: 15–23, 2003 [DOI] [PubMed] [Google Scholar]
- 6.Chen J, Zheng R, Melman YF, McDonald TV. Functional interactions between KCNE1 C-terminus and the KCNQ1 channel. PLoS ONE 4: 5143, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freeman LC, Lippold JJ, Mitchell KE. Glycosylation influences gating and pH sensitivity of I(sK). J Membr Biol 177: 65–79, 2000 [DOI] [PubMed] [Google Scholar]
- 8.Ghosh S, Nunziato DA, Pitt GS. KCNQ1 assembly and function is blocked by long-QT syndrome mutations that disrupt interaction with calmodulin. Circ Res 98: 1048–1054, 2006 [DOI] [PubMed] [Google Scholar]
- 9.Haitin Y, Wiener R, Shaham D, Peretz A, Cohen EB, Shamgar L, Pongs O, Hirsch JA, Attali B. Intracellular domains interactions and gated motions of I(KS) potassium channel subunits. EMBO J 28: 1994–2005, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang LQ, Bitner-Glindzicz M, Tranebjaerg L, Tinker A. A spectrum of functional effects for disease causing mutations in the Jervell and Lange-Nielsen syndrome. Cardiovasc Res 51: 670–680, 2001 [DOI] [PubMed] [Google Scholar]
- 11.Jongbloed RJ, Wilde AA, Geelen JL, Doevendans P, Schaap C, van Langen I, van Tintelen JP, Cobben JM, Beaufort-Krol GC, Geraedts JP, Smeets HJ. Novel KCNQ1 and HERG missense mutations in Dutch long-QT families. Hum Mutat 13: 301–310, 1999 [DOI] [PubMed] [Google Scholar]
- 12.Kanki H, Kupershmidt S, Yang T, Wells S, Roden DM. A structural requirement for processing the cardiac K+ channel KCNQ1. J Biol Chem 279: 33976–33983, 2004 [DOI] [PubMed] [Google Scholar]
- 13.Kopito RR, Sitia R. Aggresomes and Russell bodies. Symptoms of cellular indigestion? EMBO Rep 1: 225–231, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krumerman A, Gao XH, Bian JS, Melman YF, Kagan A, McDonald TV. An LQT mutant minK alters KvLQT1 trafficking. Am J Physiol Cell Physiol 286: C1453–C1463, 2004 [DOI] [PubMed] [Google Scholar]
- 15.Laemmli UK. Cleavage of structural proteins during assembly of head of bacteriophage T4. Nature 227: 680–685, 1970 [DOI] [PubMed] [Google Scholar]
- 16.Ma L, Lin C, Teng S, Chai Y, Bahring R, Vardanyan V, Li L, Pongs O, Hui R. Characterization of a novel Long QT syndrome mutation G52R-KCNE1 in a Chinese family. Cardiovasc Res 59: 612–619, 2003 [DOI] [PubMed] [Google Scholar]
- 17.Melman YF, Krumerman A, McDonald TV. A single transmembrane site in the KCNE-encoded proteins controls the specificity of KvLQT1 channel gating. J Biol Chem 277: 25187–25194, 2002 [DOI] [PubMed] [Google Scholar]
- 18.Melman YF, Um SY, Krumerman A, Kagan A, McDonald TV. KCNE1 binds to the KCNQ1 pore to regulate potassium channel activity. Neuron 42: 927–937, 2004 [DOI] [PubMed] [Google Scholar]
- 19.Moss AJ, Kass RS. Long QT syndrome: from channels to cardiac arrhythmias. J Clin Invest 115: 2018–2024, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nerbonne JM, Kass RS. Molecular physiology of cardiac repolarization. Physiol Rev 85: 1205–1253, 2005 [DOI] [PubMed] [Google Scholar]
- 21.Nobles M, Benians A, Tinker A. Heterotrimeric G-proteins precouple with G-protein coupled receptors in living cells. Proc Natl Acad Sci USA 102: 18706–18711, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohno S, Zankov DP, Yoshida H, Tsuji K, Makiyama T, Itoh H, Akao M, Hancox JC, Kita T, Horie M. N- and C-terminal KCNE1 mutations cause distinct phenotypes of long QT syndrome. Heart Rhythm 4: 332–340, 2007 [DOI] [PubMed] [Google Scholar]
- 23.Panaghie G, Tai KK, Abbott GW. Interaction of KCNE subunits with the KCNQ1 K+ channel pore. J Physiol 570: 455–467, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanguinetti MC, Curran ME, Zou A, Shen J, Spector PS, Atkinson DL, Keating MT. Coassembly of K(V)LQT1 and minK (IsK) proteins to form cardiac I(Ks) potassium channel. Nature 384: 80–83, 1996 [DOI] [PubMed] [Google Scholar]
- 25.Schagger H. Tricine-SDS-PAGE. Nat Protoc 1: 16–22, 2006 [DOI] [PubMed] [Google Scholar]
- 26.Schmitt N, Schwarz M, Peretz A, Abitbol I, Attali B, Pongs O. A recessive C-terminal Jervell and Lange-Nielsen mutation of the KCNQ1 channel impairs subunit assembly. EMBO J 19: 332–340, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sitia R, Braakman I. Quality control in the endoplasmic reticulum protein factory. Nature 426: 891–894, 2003 [DOI] [PubMed] [Google Scholar]
- 28.Splawski I, TristaniFirouzi M, Lehmann MH, Sanguinetti MC, Keating MT. Mutations in the hminK gene cause long QT syndrome and suppress I-Ks function. Nat Genet 17: 338–340, 1997 [DOI] [PubMed] [Google Scholar]
- 29.Tapper AR, George AL., Jr MinK subdomains that mediate modulation of and association with KvLQT1. J Gen Physiol 116: 379–390, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tapper AR, George AL., Jr Location and orientation of minK within the I(Ks) potassium channel complex. J Biol Chem 276: 38249–38254, 2001 [DOI] [PubMed] [Google Scholar]
- 31.Tyson J, Tranebjaerg L, Bellman S, Wren C, Taylor JF, Bathen J, Aslaksen B, Sorland SJ, Lund O, Malcolm S, Pembrey M, Bhattacharya S, Bitner-Glindzicz M. IsK and KvLQT1: mutation in either of the two subunits of the slow component of the delayed rectifier potassium channel can cause Jervell and Lange-Nielsen syndrome. Hum Mol Genet 6: 2179–2185, 1997 [DOI] [PubMed] [Google Scholar]
- 32.Urabe M, Kume A, Tobita K, Ozawa K. DNA/calcium phosphate precipitates mixed with medium are stable and maintain high transfection efficiency. Anal Biochem 278: 91–92, 2000 [DOI] [PubMed] [Google Scholar]
- 33.Wilson AJ, Quinn KV, Graves FM, Bitner-Glindzicz M, Tinker A. Abnormal KCNQ1 trafficking influences disease pathogenesis in hereditary long QT syndromes (LQT1). Cardiovasc Res 67: 476–486, 2005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.