Abstract
A bioinformatic search was carried for plant homologues of human serine-threonine protein kinases involved in regulation of cell division and microtubule protein phosphorylation (SLK, PAK6, PAK7, MARK1, MAST2, TTBK1, TTBK2, AURKA, PLK1, PLK4 and PASK). A number of SLK, MAST2 and AURKA plant homologues were identified. The closest identified homologue of human AURKA kinase was a protein of unknown function, A7PY12/GSVIVT00026259001 from Vitis vinifera (herein named as "STALK", Serine-Threonine Aurora-Like Kinase). Analysis of STALK's three-dimensional structure confirmed its relationship to the subgroup of AURKA-like protein kinases.
Background
Microtubules are essential and universal cytoskeleton component of all eukaryotic cells [1-3]. Due to their dynamic behaviour, microtubules participate in many cellular processes, including cell division, shape formation, intracellular trafficking and organelle positioning. Their protofilaments are composed of tubulin heterodimers, a highly conserved microtubule protein [4,5]. Their multiple isotypes and isoforms arise from variable gene expression as well as post-translational modifications, most studied of which are phosphorylation, acetylation, tyrosination/detyrosination, polyglutamylation, polyglycylation, and palmitoylation [3,6,7]. Among these modifications, phosphorylation represents a special case because of its wide occurrence [8] and ability to regulate structure/function of around 30% of all proteins in eukaryotes [9,10].
The genome of Arabidopsis thaliana contains more then 1000 protein kinase genes [8]. Analysis of plant gene similarity to the phosphate-binding regions of animal tyrosine kinases showed that genes for Zap70 tyrosine kinase family are the most similar to its potential plant homologues [11,12]. tBLASTn http://blast.ncbi.nlm.nih.gov/ scanning of Arabidopsis genome against mouse Zap70 catalytic domain detected 503 consensus regions corresponding to 494 protein kinases [12]. It means that only ~50% of Arabidopsis kinases are homologous to animal kinases. These genes are distributed between different chromosomes: 150 - on I, 71 - on II, 94 - on III, 65 - on IV and 114 - on chromosome V. Analysis in SMART http://smart.embl-heidelberg.de/ confirms the correspondence of their products to the models of catalytic domains of S_TKc, STYKc, TyrKc, Pkinase, Pkinase_Tyr, S_TKc, D1phk (SCOP: 88854), D1pme (SCOP: 88854), D1qpca (SCOP: 88854), D1fgka (SCOP: 88854), D1ir3a (SCOP: 88854), D1b6cb (SCOP: 88854), 1A06, 1BYG, 1CM8, 1E1V, 1F3M, 1FGI, 1FOT, 1FPU, 1FVR, 1G3N, 1GOL, 1IA8, 1IAN, 1JPA, 1KOA, 1PME, 1QCF, 1QL6, 1QPD, 2PTK, 2SRC, 3LCK and d1b6cb kinases [12]. We previously demonstrated that microtubule tubulin in plants, like in animals, is highly phosphorylated by different serine/threonine [13] and tyrosine kinases [14,15]. It is interesting that among potential products of the genes we identified proteins corresponding to HMMs and patterns of catalytic domains of atypical Tyr-specific kinases (Pkinase_Tyr), dual specific (STYKc) and canonical animal tyrosine kinases (TyrKc) [11,12].
Phosphorylation of other proteins forming plant microtubules and involved in cell division, may also be mediated by kinases having well characterized homologues in the animal kingdom [15]. Several microtubule proteins show homology between animals and plants, e.g., microtubule-associated proteins type I (MAP1) [16]. Therefore, these proteins may be expected to have similar phosphorylation sites and to be phosphorylated by corresponding conserved protein kinases.
Microtubule-related proteins are both species- and tissue-specific. Not surprisingly, these proteins differ significantly between plants and animals [16-18]. Accordingly, the total microtubule-related kinase subset (termed "kinome") is distinct in animals and plants. However, microtubule proteins and associated kinases in plant cells have not been studied in as much details as in their human/animal counterparts. Further insight requires identifying of plant kinases and corresponding genes.
All protein kinases are characterized by a conserved catalytic (kinase) domain of 250-300 residues which, in turn, contains unique sub-domain motifs related to serine/threonine, tyrosine or dual kinases [19,20]. These features enable search and identification by database scanning using BLAST family tools [21]. It is this catalytic domain with the characteristic structure/function relationship that aids in identification of new kinases by in silico methods. The homology of these domains has been previously used to identify several protein kinases from multicellular organisms [22-27].
At present, several eukaryotic genomes have been completely sequenced and partially annotated. Specific examples of plant genomic information include plant genomic data in NCBI http://www.ncbi.nlm.nih.gov/, and, additionally, the project Genoscope http://www.genoscope.cns.fr which provides information and in silico search engines for the following plants: Arabidopsis thaliana, Ectocarpus siliculosus, Medicago truncatula, Oryza sativa, Alnus glutinosa, Aphanomyces euteiches, Casuarina glauca, Citrus clementina, Eucalyptus, Juglans regia, Phaseolus vulgaris, Pinus pinaster, Populus trichocarpa × deltoides, Quercus, Saccharum ssp., Theobrama cacao, Triticum ssp., Vitis vinifera.
The goal of the present work was to identify plant homologues of human kinases [28] that could participate in phosphorylation of microtubule proteins and/or regulation of cell division. The investigated kinase families included AGC (containing kinases A, G and C families), CAMK (calcium/calmodulin-dependent), CK1 (casein kinase 1) and STE (homologues of yeast Sterile 7, Sterile 11 and Sterile 20 kinases) protein kinases. For the bioinformatic search and reconstruction of plant microtubule kinome, we used sequences of the following human protein kinases: SLK, PAK6, PAK7 (from the superfamily STE); MARK1, PASK (from the superfamily CAMK); MAST2, AURKA (from the superfamily AGC); TTBK1, TTBK2 (from the superfamily CK1); kinases Plk1 and Plk4 (not belonging to any specific group) [28].
Results and discussion
Using public databases and published research papers [28-47], we have selected a range of human kinases involved in microtubule proteins phosphorylation and cell division regulation (Table 1). Depending on the complexity level of protein kinases domain organization, either full sequences (in case of AURKA) or catalytic domain sequence (SLK, PAK7, MARK1, SLK, MAST2 TTBK1, PLK1, PLK4 and PASK, as seen on Figure 1) were used for SIB BLASTp scanning. As a result of the UniProt database scanning, we identified plant homologues of human kinases from the families SLK, MAST2 and AURKA.
Table 1.
Human kinases taking part in microtubule protein phosphorylation and cell division regulation [28-30]; in italics are kinases plant homologues of which were identified in the present study
| Protein kinase | GenBank (mRNA) | Gene | Locus | UniProt |
|---|---|---|---|---|
| SLK (KIAA0204, LOSK, MGC133067, STK2, bA16H23.1, se20-9) [32,33,47] | NM_014720 | NC_000010.10 | Chr.10; Loc.10q25.1 | Q9H2G2 |
| PAK7 (RP5-1119D9.3, KIAA1264, MGC26232, PAK5) [35,39] | NM_177990 | NC_000020.10 | Chr.20; Loc.20p12 | Q9P286 |
| PAK6 (PAK5) [35,39] | NM_020168 | NC_000015.9 | Chr.15; Loc.15q14 | Q9NQU5 |
| MARK1 (KIAA1477, MARK, MGC126512, MGC126513) [34] | NM_018650 | NC_000001.10 | Chr.1; Loc.1q41 | Q9P0L2 |
| MAST2 (MTSSK, MAST205, FLJ39200, KIAA0807, RP4-533D7.1) [45] | NM_015112 | NC_000001.10 | Chr.1; Loc.1p34.1 | Q6P0Q8 |
| TTBK1 (BDTK, KIAA1855, RP3-330M21.4) [42] | NM_032538 | NC_000006.11 | Chr.6; Loc.6p21.1 | Q5TCY1 |
| TTBK2 (KIAA0847, SCA11, TTBK) [37] | NM_173500 | NC_000015.9 | Chr.15; Loc.15q15.2 | Q6IQ55 |
| AURKA (AIK, ARK1, AURA, AURORA2, BTAK, MGC34538, STK15, STK6, STK7) [38,41,43,46] | NM_198436 | NC_000020.10 | Chr.20; Loc.20q13.2-q13.3 | O14965 |
| PLK1 (PLK, STPK13) [40,41,43,44] | NM_005030 | NC_000016.9 | Chr.16; Loc.16p12.1 | P53350 |
| PLK4 (SAK, STK18) [31,41] | NM_014264 | NC_000004.11 | Chr.4; Loc.4q28 | O00444 |
| PASK (DKFZp434O051, DKFZp686P2031, KIAA0135, PASKIN, STK37) [36] | NM_015148 | NC_000002.11 | Chr.2; Loc.2q37.3 | Q96RG2 |
Figure 1.
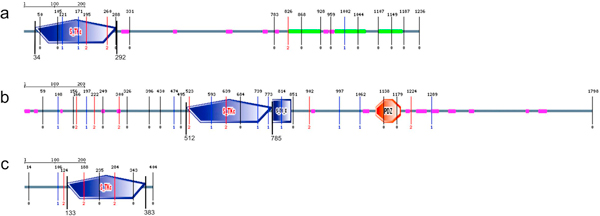
SLK_HUMAN (a), MAST_HUMAN (b) and STK6_HUMAN (c) kinase domain architecture. S_TKc - catalytic domain of serine/threonine-kinases; S_TK_X - auxiliary S_TKc domain; DUF1908 - domain of unknown function (DUF1908); PDZ (also referred as DHR (Dlg homology region) or GLGF (relatively well conserved tetrapeptide in these domains) - domain found in PSD-95, Dlg and ZO-1/2[74]. These domains help anchor transmembrane proteins to the cytoskeleton and hold together signaling complexes [72].
Plant homologues of SLK_HUMAN (Q9H2G2)
Uniprot scanning against SLK_HUMAN catalytic domain has revealed 11 plant homologues to STE20-like human kinase (Table 2) from A. thaliana (Q9LQA1/AT1G69210, O24527/At1g69220), Hordeum vulgare var. distichum (Q9ARL7/GenBank: AY013246.1), O. sativa ssp. japonica (Q10CN6/Os03g0755000), O. sativa ssp. indica (B8AK85/OsI_13553), Physcomitrella patens ssp. patens (A9RVK0/PHYPADRAFT_119967), Populus balsamifera ssp. trichocarpa (B9HXI5/POPTRDRAFT_226120), Ricinus communis (B9REC4/RCOM_1620250), Solanum chacoense (B3GK00/MAP4K1) [48 Champion et al., 2004a], Sorghum bicolor (Q8LKU7/Sb01g007720) and V. vinifera (A7P2E2/GSVIVT00030023001). Consensus region identity to Homo sapiens kinase SLK (Q9H2G2) catalytic region reached 45-46% with similarity of 65-66% (Table 2, Figure 2). Analysis of full sequences of potential plant homologues in SMART confirmed their relationship to serine/threonine-specific protein kinases. All identified plant homologues were found in TrEMBL database as deposited proteins with unknown function. As an exception, putative STE20 protein O24527/At1g69220 (TrEMBL database), from A. thaliana, is annotated as potential homologue of protein kinase SIK1.
Table 2.
Plant homologues for animal kinases taking part in microtubule protein phosphorylation and cell division regulation
| Protein kinase from Homo sapiens | Potential plant homologues | Species | Database | Length of consensus | Score | Expect (E-value) |
Identity (%) |
Similarity (%) |
Gaps (%) |
|---|---|---|---|---|---|---|---|---|---|
| SLK | B9REC4_RICCO | Ricinus communis | tr | 260 | 245 | 3e-63 | 46 | 66 | 2 |
| A9RVK0_PHYPA | Physcomitrella patens ssp. patens | tr | 260 | 243 | 1e-62 | 46 | 65 | 2 | |
| A7P2E2_VITVI | Vitis vinifera | tr | 260 | 243 | 1e-62 | 46 | 65 | 2 | |
| B9HXI5_POPTR | Populus balsamifera ssp. trichocarpa | tr | 261 | 239 | 2e-61 | 46 | 65 | 2 | |
| Q9LQA1_ARATH | A. thaliana | tr | 260 | 242 | 2e-62 | 46 | 66 | 2 | |
| O24527_ARATH | - | tr | 260 | 242 | 2e-62 | 46 | 66 | 2 | |
| Q10CN6_ORYSJ | Oryza sativa ssp. japonica | tr | 260 | 241 | 4e-62 | 45 | 65 | 2 | |
| B8AK85_ORYSI | Oryza sativa ssp. indica | tr | 260 | 241 | 4e-62 | 45 | 65 | 2 | |
| Q8LKU7_SORBI | Sorghum bicolor | tr | 260 | 241 | 5e-62 | 45 | 65 | 2 | |
| Q9ARL7_HORVD | Hordeum vulgare var. distichum | tr | 260 | 239 | 1e-61 | 45 | 65 | 2 | |
| B3GK00_SOLCH | Solanum chacoense | tr | 260 | 238 | 3e-61 | 45 | 66 | 2 | |
| MAST2 | A7QWR7_VITVI | Vitis vinifera | tr | 359 | 314 | 5e-83 | 47 | 63 | 4 |
| A7PHB5_VITVI* | - | tr | 346 | 313 | 7e-83 | 46 | 64 | 1 | |
| A7NTE9_VITVI* | - | tr | 327 | 295 | 3e-77 | 46 | 63 | 4 | |
| A5BWH0_VITVI* | - | tr | 338 | 241 | 4e-61 | 39 | 56 | 9 | |
| Q9LE81_ARATH | A. thaliana | tr | 350 | 310 | 9e-82 | 46 | 63 | 4 | |
| Q94F38_ARATH* | - | tr | 343 | 301 | 4e-79 | 46 | 62 | 3 | |
| Q8GZ40_ARATH* | - | tr | 360 | 300 | 6e-79 | 44 | 63 | 4 | |
| Q0WLU7_ARATH | - | tr | 343 | 294 | 4e-77 | 45 | 63 | 4 | |
| Q10E10_ORYSJ | O. sativa ssp. japonica | tr | 384 | 309 | 2e-81 | 44 | 58 | 7 | |
| Q2QM12_ORYSJ | - | tr | 359 | 305 | 3e-80 | 46 | 61 | 6 | |
| A2XLA4_ORYSI | O. sativa ssp.indica | tr | 384 | 309 | 2e-81 | 44 | 58 | 7 | |
| A2ZMW0_ORYSI | - | tr | 359 | 279 | 2e-72 | 44 | 59 | 9 | |
| Q32YB5_MEDTR | Medicago truncatula | tr | 364 | 307 | 6e-81 | 45 | 62 | 6 | |
| A9TQ65_PHYPA* | Physcomitrella patens ssp. patens | tr | 347 | 286 | 1e-74 | 43 | 60 | 5 | |
| A9TWY7_PHYPA | - | tr | 327 | 285 | 2e-74 | 43 | 61 | 6 | |
| A9TUB0_PHYPA* | - | tr | 326 | 285 | 3e-74 | 44 | 61 | 5 | |
| A9T694_PHYPA* | - | tr | 364 | 284 | 4e-74 | 42 | 56 | 9 | |
| AURKA | Q5SNH4_ORYSJ | O. sativa ssp. japonica | tr | 285 | 369 | e-100 | 64 | 77 | 2 |
| Q4R1K7_ORYSJ | - | tr | 264 | 359 | 2e-97 | 66 | 80 | 1 | |
| A2WLL4_ORYSI | O. sativa ssp.indica | tr | 285 | 369 | e-100 | 64 | 77 | 2 | |
| A9PFI9_POPTR | Populus balsamifera ssp. trichocarpa | tr | 285 | 368 | e-100 | 62 | 78 | 1 | |
| A7P4F7_VITVI | V. vinifera | tr | 285 | 368 | e-100 | 63 | 77 | 1 | |
| A5BPE0_VITVI | - | tr | 283 | 367 | e-100 | 63 | 77 | 1 | |
| A7PY12_VITVI | - | tr | 279 | 339 | 3e-91 | 56 | 76 | 1 | |
| B4F8A1_MAIZE | Zea mays | tr | 292 | 368 | e-100 | 62 | 76 | 2 | |
| AUR2_ARATH | A. thaliana | sp | 272 | 365 | 3e-99 | 65 | 78 | 1 | |
| AUR2_ARATH sof.2 | - | sp | 268 | 365 | 4e-99 | 65 | 79 | 1 | |
| AUR1_ARATH | - | sp | 284 | 363 | 2e-98 | 60 | 76 | 1 | |
| AUR3_ARATH | - | sp | 280 | 337 | 1e-90 | 57 | 76 | 1 | |
* - presence of auxiliary catalytic S_TK_X domain
Figure 2.

Catalytic domain alignment of human SLK_HUMAN (Q9H2G2) and putative plant SLK-like proteins. NP_BIND (ATP) - nucleotide phosphate binding region; Binding site (ATP) - ATP binding site; ACT_SITE - proton acceptor; consensus conserved motif in catalytic loop region of the subdomain VIb (animals (red): H-R-D-[LI]-K-[GA]-x-N and A. thaliana (blue): H-[RC]-D-[ILV]-K-x-x-N); consensus conserved motif in activation loop of the subdomain VIII (in animals (red): G-T-P-[YF]-[WY]-M-A-P-E and in A. thaliana (blue): G-[TS]-x-x-[WYF]-[ML]-[AS]-P-E)
To identify the closest homologues among the potential plant SLK-like kinases, we performed multiple alignments of catalytic domains with that of human SLK and built a phylogenetic tree using the neighbour joining method. According to the cladistic data, a common clade with human SLK is formed by proteins Q9LQA1/At1g69210 and O24527/At1g69220 from A. thaliana as well as A9RVK0 from P. patens ssp. patens (Figure 3). Following that, the closest plant homologue of human SLK was A9RVK0 from P. patens ssp. patens with the identity of 46% and similarity of 65%. Inside the group, the newly found plant homologues demonstrated a high degree of sequence identity with conserved functionally important positions specific to SLK kinases. Also, the analysis of the known conserved residues, implicated in substrate recognition in animal and A. thaliana STE-kinases [49 Champion et al., 2004b], confirmed the existence of the consensus motifs in catalytic loop region of the subdomain VIb (animals: H-R-D-[LI]-K-[GA]-x-N and A. thaliana: H-[RC]-D-[ILV]-K-x-x-N) and activation loop of the subdomain VIII (animals: G-T-P-[YF]-[WY]-M-A-P-E and A.thaliana: G-[TS]-x-x-[WYF]-[ML]-[AS]-P-E) in all identified plant homologues(Figure 2).
Figure 3.

Phylogenetic tree constructed for SLK kinase of H. sapiens and its plant homologues.
Plant homologues of MAST2_HUMAN (Q6P0Q8)
As with SLK above, we searched the UniProt database for potential plant homologues of MAST2_HUMAN with the catalytic domain sequence identified in SMART tool http://smart.embl-heidelberg.de/[50].
Swiss-Prot/TrEMBL scanning has revealed 17 plant MAST2_HUMAN homologues: 4 from V. vinifera, 4 from A. thaliana, 2 from O. sativa ssp. japonica, 2 from O. sativa ssp. indica, 1 from Medicago truncatula and 4 from P. patens ssp. patens (Table 2). Animal MAST2 kinases contain a typical auxiliary C-terminal S_TK_X domain (sTKc or sTKx), termed C-terminal kinase domain [51], which, together with the catalytic domain, is responsible for interactions with microtubules [45]. Results of the domain architecture analysis of full sequences of potential plant homologues of human MAST2 kinase demonstrated that only 8 out of 17 found sequences have the C-terminal S_TK_X domain (SMART SM00133): A7PHB5/GSVIVT00017880001, A7NTE9/GSVIVT00014640001, A5BWH0/VITISV_001730 (all from V. vinifera); Q94F38/At1g48490, Q8GZ40/At3g17850 (A. thaliana) and A9TQ65/PHYPADRAFT_148894, A9TUB0/PHYPADRAFT_150879, A9T694/PHYPADRAFT_140919 (P. patens ssp. patens) (Table 2). Consequently, the maximum consensus identity for the query catalytic domain in BLASTp was observed with a protein of unknown function, A7QWR7/GSVIVT00008775001, from grape (identity-47%, similarity-63%) (Figure 4, Table 2). Also, the analysis of the known conserved residues, implicated in substrate recognition in animal and A. thaliana AGC-kinases[49]., confirmed the existence of the consensus motifs in catalytic loop region of the subdomain VIb (animals: [HY]-R-D-[LI]-K-[PL]-[ED]-N and A. thaliana: [HY]-[RY]-D-[LI]-K-P-[ED]-N) and activation loop of the subdomain VIII (animals: G-T-P-[EA]-Y-[IM]-A-P-E and A.thaliana: G-T-x-D-Y-L-A-P-E) in all identified plant MAST-homologues(Figure 2).
Figure 4.
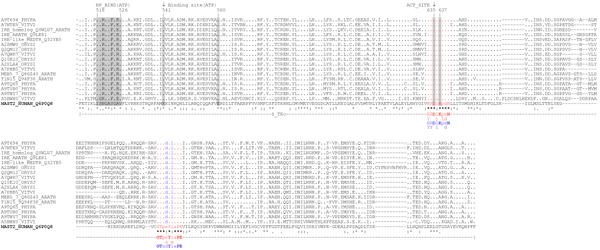
Catalytic domain alignment of MAST_HUMAN (Q6P0Q8) with its plant homologues. NP_BIND (ATP) - nucleotide phosphate binding region; Binding site (ATP) - ATP binding site; ACT_SITE - proton acceptor; consensus conserved for AGC kinases motif in catalytic loop region of the subdomain VIb (animals (red): [HY]-R-D-[LI]-K-[PL]-[ED]-N and A. thaliana (blue): [HY]-[RY]-D-[LI]-K-P-[ED]-N); consensus conserved for AGC kinases motif in activation loop of the subdomain VIII (in animals (red): G-T-P-[EA]-Y-[IM]-A-P-E and in A. thaliana (blue): G-T-x-D-Y-L-A-P-E)
Phylogenetic analysis using Neighbour-Joining algorithm based on catalytic domain similarity demonstrated that a common clade with human MAST2 is formed by the following plant homologues: Q0WLU7/At1g45160, Q9LE81/At5g62310 (A. thaliana), A7NTE9_VITVI*/GSVIVT00014640001, A5BWH0_VITVI*/VITISV_001730 (V. vinifera) and Q32YB5/GenBank: AY770392.1 (M. truncatula) (Figure 5).
Figure 5.
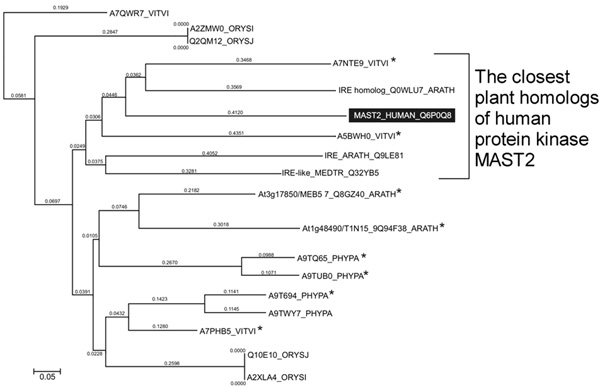
Phylogenetic tree built for H. sapiens MAST2 kinase and its plant homologues; * - plant homologues containing typical for animal MAST2 kinases auxiliary S_TK_X catalytic S_TK_X domain.
Therefore, the most distinguished plant homologues of the MAST2 family are A7NTE9_VITVI*/GSVIVT00014640001 (423 residues; chromosome 18), A5BWH0_VITVI*/VITISV_001730 (550 residues; chromosome 5) from V. vinifera. In summary, based on the analysis of human MAST2 and its potential plant homologues for such functional elements as auxiliary S_TK_X domain, Asn635 in putative active centre, ATP-binding motif (NP_BIND ATP) and Lys541, allowed us to conclude that A7NTE9 (ORF Name GSVIVT00014640001) from grape (Figure 6) is the most promising for future research including spatial structure reconstruction and molecular genetic methods.
Figure 6.
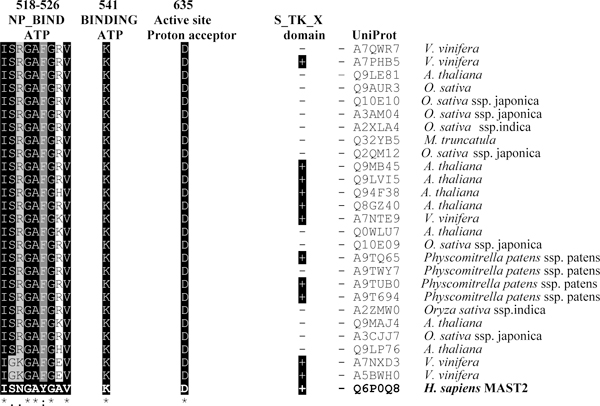
Similarity of H. sapiens MAST2 and potential plant homologues for auxiliary S_TK_X domain, conserved active site Asp-635 and ATP-binding motifs (NP_BIND ATP and Lys-541).
Plant homologues of human Aurora A kinase (STK6_HUMAN, O14965)
Protein kinase Aurora A belongs to the serine/threonine kinase family AGC [28] and plays a fundamental role in mitosis regulation [30,31]. In the case of Aurora A kinase, we used the full sequence of human STK6 (AURKA; O14965) as a 'query' sequence for the BLASTp search. As a result of database scanning, we have identified 12 potential plant homologues of the STK6 kinase (Table 2) characterized by a similar domain structure and high sequence similarity of kinase domains and N- and C-terminals. Sequence similarity has reached 76-80% with the identity of 56-66%.(Table 2)
In an earlier paper on plant homologues of Aurora kinases [52], the authors described a high level of identity for Aurora catalytic domains from Arabidopsis (64-95%). We now report additional homologues belonging to O. sativa ssp. japonica (Q5SNH4/Os01g0191800, Q4R1K7/OsAUR1), O. sativa ssp. indica (A2WLL4/OsI_00731), P. balsamifera ssp. trichocarpa (A9PFI9/POPTRDRAFT_819526), V. vinifera (A7P4F7/GSVIVT00032134001, A5BPE0/VITISV_023325, A7PY12/GSVIVT00026259001), Zea mays (B4F8A1/LOC100191291) and A. thaliana (AUR2 Isof. 1 and AUR2 Isof. 2 (At2g25880), AUR1/At4g32830, AUR3/At2g45490). Together with a low gap percentage (1-2%), such a high identity reflects not only sequence homology but also homology of structure and function (Figure 7). As evident from the annotations in the Swiss-Prot database kinases AUR1/At4g32830, AUR2 Isof. 1 and AUR2 Isof. 2 (At2g25880) and AUR3/At2g45490 from A. thaliana could be functional homologues. Sequence Q4R1K7/OsAUR1 from O. sativa ssp. japonica in the TrEMBL database [53] is also annotated as a potential plant Aurora kinase.
Figure 7.
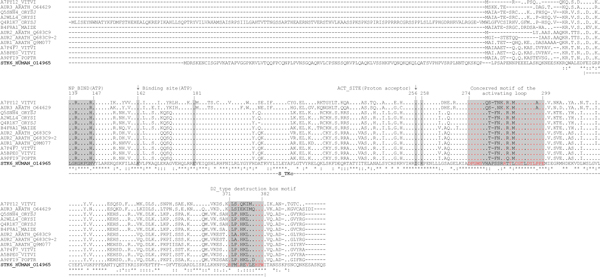
Alignment of human Aurora A and its plant homologues. NP_BIND (ATP) - nucleotide phosphate binding site; Binding site (ATP) - ATP binding site; ACT_SITE - proton acceptor site
As a result of highly conserved positions of the functionally important residues and motifs, together with homology of a primary sequence and domains (analysis using domain profiles and hidden Markov models) all S_TKc kinases were identified as plant structural and functional homologues of human Aurora A. Based on the cladistic analysis (Figure 8), the closest homologue of STK6_HUMAN was a protein of unknown function, A7PY12/GSVIVT00026259001 from V. vinifera (identity-76%, similarity-56%, E-value = 3e-91).
Figure 8.
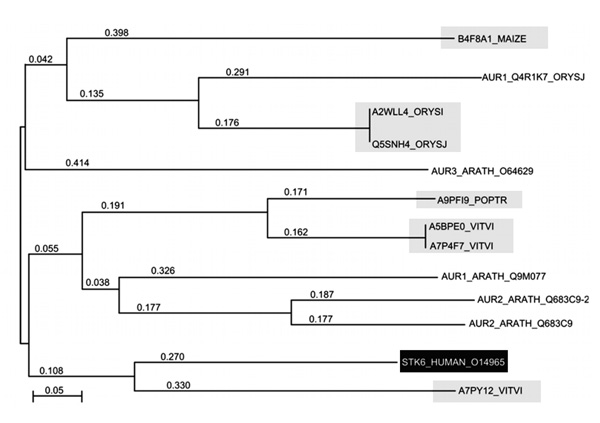
Phylogenetic tree of STK6_HUMAN (Aurora A) kinase and its plant homologues demonstrating their phylogenetic relationships. Highlighted are the proteins identified in the paper.
Taking into account the critical role of Aurora kinases in microtubule protein [46] and cell division regulation [30,41,53], the plant homologue A7PY12/GSVIVT00026259001 from V. vinifera, referred herein as STALK (S_T AURKA LIKE KINASE), has been chosen for in silico spatial structure prediction (Figure 9). Sequence identity and similarity between STALK and Aurora are 60.1% and 81.7%, respectively (Table 2). In addition, identical and similar residues of STALK are distributed evenly along the whole protein chain (Figure 7), resulting in high reliability of the predicted spatial structure.
Figure 9.
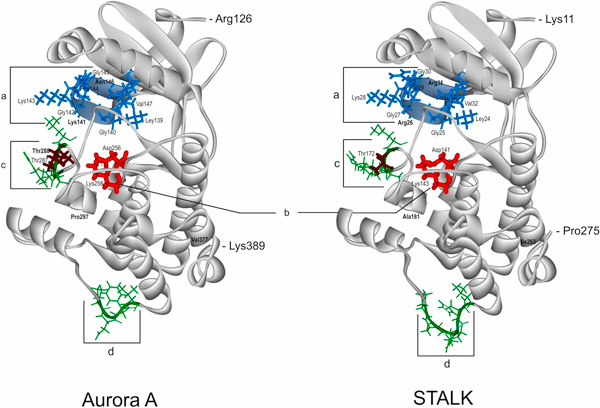
Comparison of the catalytic domain spatial structures of the human protein kinase Aurora A (AURKA, STK6, PDB: 2J4Z) and the protein of unknown function STALK (S_T AURKA LIKE KINASE, UniProt: A7PY12) from V. vinifera. "a" (marked by blue) ATP-binding regions in Aurora A and STALK; "b" (marked by red) is active site; "c", "d" (marked by green) are the most spatially variable regions between the two proteins; phosphorylated Thr residues (287, 288) in the Aurora A are marked by brown. In bold are marked the only discrepancies between the corresponding functionally important residues in Aurora A versus STALK: Asn146↔Arg31, Lys141↔Arg26 in "a"; Thr288↔Thr172 in variable region "c"; Pro297↔Ala181 in the DFGWSxxxxxxxRxTxCGTxDYLPPE motif of the activating loop; Val377↔Ile263 in the D2_type destruction box - Rxx(L/I)xxVxxHPW
Differences in spatial orientation between the STALK residues in the active centre (Asp141, Lys143) and the ATP-binding site (Leu24, Gly25, Arg26, Gly27, Lys28, Phe29, Gly30, Arg31, Val32, Lys47) and Aurora A do not exceed 0.3 Å, while the corresponding value for the whole STALK domain is 0.43 Å. This finding confirms more conserved spatial structure of these areas in relationship to the whole protein. Together with the high sequence homology, it further confirms similar properties and catalytic functions of the template and target proteins. The largest difference of the 3D structures between STALK and Aurora is observed in STALK's positions 169-173 (corresponding partly to the activation loop in Aurora A containing pThr288 which is crucial for Aurora A activation [54]) and 233-238 (marked "c" and "d", respectively, on Figure 9). In addition, Thr171 of the STALK's first variable range is identical to Thr288 of Aurora (Figure 9).
The activation loop of animal Aurora kinases contains a conserved motif (DFGWSxxxxxxxRxTxCGTxDYLPPE) carrying Thr288 phosphorylated by the cAMP-dependent kinase (Figure 10) [54]. Also, the C-terminal part of the Aurora kinases catalytic domain contains a so-called D2-type destruction box motif [Rxx(L/I)xxVxxHPW] which is thought to play a role in proteasome-dependent protein degradation [53].
Figure 10.
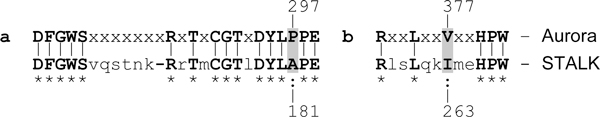
Conserved motifs typical for Aurora animal kinases found in STALK protein from V. vinifera: a - activation loop conserved motif and b - D2-type destruction box. Marked are the residues different in both proteins.
In the STALK sequence we have identified, with near 100% identity, both motifs essential for animal Auroras (Figure 10). This confirms functional homology of the STALK (A7PY12/GSVIVT00026259001) protein and human STK6 kinase.
Thus, the results of the search for plant homologues of human kinases SLK, PAK6, PAK7 MARK1, MAST2, TTBK1, TTBK2, AURKA, PLK1, PLK4 and PASK clearly demonstrated the existence of corresponding plant kinases SLK, MAST2 and AURKA. Plants do not seem to contain homologues of kinases PAK6, PAK7, MARK1, TTBK1, TTBK2, PLK1, PLK4 and PASK. This observation could be explained by significant differences in microtubule proteins between plants and animals. Despite the high similarity of animal and plant tubulins and the presence of plant homologues for some other proteins like MAP1 [56], there are differences in a number of microtubule proteins. Plants, for example, do not possess the type of MAP that contains tau repeats (MAP2) [16,56], which explains absence of some protein kinases phosphorylating given substrates in plants. At the same time, kinases involved in pathways common in animal and plant cells and phosphorylated conserved sites in microtubule proteins (tubulin, MAP1, motor proteins, etc.), exhibit considerable similarity.
An example of the corresponding homologue is Aurora A, a kinase responsible for daughter centrosome formation, asymmetric cell division and mitotic spindle formation [46]. It is believed that the signal from the cyclin-dependent kinase Cdk1 is relayed to the "second" level kinases, one of which is Aurora A [38,46]. In human and animal cells alike, AURKA (Aurora kinase A) is involved in the initiation of mitosis [30]. Activation of AURKA occurs by means of AURKA - BORA (protein BORA, FLJ22624; RP11-342J4.2; C13orf34) complex formation [29,38,43,57]. A mechanism was suggested recently, according to which BORA and AURKA together control transition from phase G2 of cell cycle to phase M [44]. According to the proposed mechanism, Cdk1 causes BORA to exit from the nucleus and binds reversibly with Plk1. This changes Plk1 conformation so that its Thr210 in the catalytic domain becomes accessible for phosphorylation by AURKA with subsequent Plk1 activation [44]. This leads to further Cdk1 activation, degradation of Bora and, eventually, to start of mitosis [44,46] (Figure 11).
Figure 11.
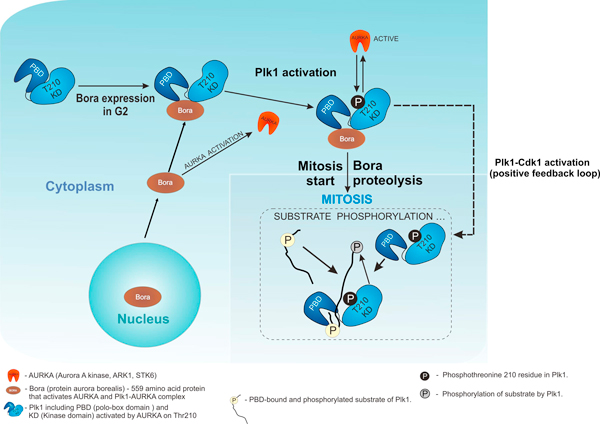
Interplay of Plk1, AURKA and BORA in cell cycle based on literature data: adapted from [43,46]and is based on references [29,38,43,44,46,57].
STALK, the closest plant homologue for human Aurora kinase, may have a similar mode of interaction in plant cells.
In relation to the potential plant homologues of kinase SLK, some questions remain. Human SLK kinase is highly expressed in various cell types and is involved in the regulation of microtubule radial orientation [44]. By analogy to the Xenopus laevis polo-like kinase xPlk1 [58], it may be hypothesized that human protein kinase SLK regulates activity of Plk1, the homologue of the X. laevis Plx1 [32]. However, so far the data is inconclusive.
In summary, we have identified plant homologues of human kinases SLK, MAST2 and Aurora A. MAST2 plant homologues are identified here for the first time. BLASTp scanning did not produce any plant homologues of human PAK6, PAK7, MARK1, TTBK1, TTBK2, PLK1, PLK4 and PASK. We hope that new bioinformatic and instrumental genomic and proteomic data will reveal additional plant kinases involved in microtubule protein phosphorylation and cell division regulation.
Methods
Target sequences of human protein kinases SLK, PAK7, PAK6, MARK1, MAST2, TTBK1, TTBK2, AURKA, PLK1, PLK4 and PASK were obtained from UniProtKB http://www.uniprot.org/ and NCBI GenBank http://www.ncbi.nlm.nih.gov/Genbank/[59] based on the information presented in respective publications. The target sequences were used to search for plant homologues against the UniProt database (Swiss-Prot and TrEMBL) [60]. We performed a BLASTp query (SIB BLAST Network Service) with the protein weight matrix BLOSUM62, with the following settings: (the E threshold = 10 (number of expected matches in a random database), "Filter the sequence for low-complexity regions" and "Gapped alignment" http://www.clustal.org[21].
The domain organization of identified plant homologues was analysed with Simple Modular Architecture Research Tool (SMART - http://smart.embl-heidelberg.de/) [50], Pfam http://pfam.sanger.ac.uk/[61], PROSITE http://www.expasy.ch/prosite/[62] and data from the Swiss-Prot database. Protein kinase relation to specific kinase family was confirmed on the basis of SMART scanning results. Anonymous SMART scanning (RAW-format) was performed with activation of all analytical modules. We also took into account the results of pairwise alignments of query and subject sequences as well as the results of the SMART analysis regarding domain composition and organization (including HMMER results). Homologues were selected based on the meaning of the sequence identities (%), positives % and E-value [63]. Multiple amino acid alignments were generated with ClustalX (2.0.5) http://www.clustal.org using the BLOSUM protein weight matrix [64].
Phylogenetic trees of plant homologues and human SLK, MAST2 and STK6 (AURKA, Aurora A) were generated based on multiple alignment results for the protein kinase domain (SLK, MAST2) [19,20] or the full sequence (STK6), using the neighbour-joining (NJ) algorithm [65,66]. The boundaries of the protein kinase domain were determined using SMART profiles. Dendrogram analysis and visualization was performed using the TreeView X Ver.5 http://taxonomy.zoology.gla.ac.uk/rod/treeview.html[67] and MEGA http://www.megasoftware.net/[68] programs. Conserved functionally important residues and motifs in plant homologues were determined based on database annotation for template sequences of corresponding human protein kinases http://www.expasy.org.
For the three-dimensional (3D) structure reconstruction of A7PY12/GSVIVT00026259001 from V. vinifera we used method homologous (profile) modelling [69]. As a matrix we used a 3D structure of AURORA2 kinase in a complex with the inhibitor PHA-6806260 [70] from the Protein Data Bank (PDB) (PDB ID 2J4Z) [71]. Pro-model geometry optimisation was performed by energy minimization using the L-BFGS method [72]. Visualization of obtained three-dimensional model was done using program DS Visualizer 2.0 (Accelrys Software Inc. - http://accelrys.com/). Differences between structures of the STALK and AURORA were estimated by calculating RMSD (root median square deviation) using the Swiss-Pdb Viewer 4.0 software [73].
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
PAK made a major contribution to conception and design, acquisition of data, analysis and interpretation of bioinformatic data: blast query, analysis of domain organization, alignments and sequences analysis, cladistic analysis. ESN was involved in revising the manuscript critically for important intellectual content. AIY took part in discussion and interpretation of data (blast query, sequence alignments, cladistic data). VGM made contributions to conception and design, acquisition of data, analysis and interpretation of data. AYN performed acquisition of spatial structure data, analysis and interpretation, molecular modelling and was involved in drafting of the part of manuscript dedicated to spatial structure analysis. NYS participated in the discussion of sequence alignments data. YBB took part in discussion of data, contribution to conception, design and interpretation of data and coordination of whole project. All authors read and approved the final manuscript.
Contributor Information
Pavel A Karpov, Email: karpov.p.a@gmail.com.
Elena S Nadezhdina, Email: elena.nadezhdina@gmail.com.
Alla I Yemets, Email: alyemets@univ.kiev.ua.
Vadym G Matusov, Email: matusovvadim@gmail.com.
Alexey Yu Nyporko, Email: dfnalex@gmail.com.
Nadezhda Yu Shashina, Email: clewer@rambler.ru.
Yaroslav B Blume, Email: cellbio@cellbio.freenet.viaduk.net.
Acknowledgements
This work is supported by bilateral grant No. 08-04-90454 of the National Academy of Sciences of Ukraine (NAS of Ukraine) and Russian Foundation for Basic Research (RFBR) 2008-2009. The authors thank Dr Konstantin Linnik for critical reading of the manuscript.
This article has been published as part of BMC Genomics Volume 11 Supplement 1, 2010: International Workshop on Computational Systems Biology Approaches to Analysis of Genome Complexity and Regulatory Gene Networks. The full contents of the supplement are available online at http://www.biomedcentral.com/1471-2164/11?issue=S1.
References
- Rodionov V, Nadezhdina E, Borisy G. Centrosomal control of microtubule dynamics. Proc Natl Acad Sci USA. 1999;96:115–120. doi: 10.1073/pnas.96.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos LA. In: Nature Encyclopedia of Life Sciences. Zheng Y, Tickle C, Jansson R, Kehrer-Sawatzki H, Cooper DN, Melino G, Delves P, Battista J, Levitan I, Roberts K, Bynum WF, Harper H, editor. Chichester, John Wiley & Sons, Ltd; 2004. Tubulin and microtubules; pp. 1–12.http://www2.mrc-lmb.cam.ac.uk/archive/papers/2004-029.pdf [Google Scholar]
- McKean PG, Vaughan S, Gull K. The extended tubulin superfamily. J Cell Sci. 2001;114:2723–2733. doi: 10.1242/jcs.114.15.2723. [DOI] [PubMed] [Google Scholar]
- Demchuk ON, Blume YaB. Phylogenetic tree of bacterial and eukaryotic FtsZ-proteins based on the homology of their primary sequences. Tsitol Genet. 2005;39(4):3–12. [PubMed] [Google Scholar]
- Little M, Seehaus T. Comparative analysis of tubulin sequences. Comp Biochem Physiol B. 1988;90:655–670. doi: 10.1016/0305-0491(88)90320-3. [DOI] [PubMed] [Google Scholar]
- Blume YaB, Smertenko AP, Ostapets NN, Viklický V, Draber P. Post-translational modifications of plant tubulin. Cell Biol Int. 1997;21:918–920. [Google Scholar]
- Smertenko AP, Blume YaB, Viklicky V, Opatrny Z, Dráber P. Posttranslational modifications and multiple isoforms of tubulin in Nicotiana tabacum cells. Planta. 1997;201:349–358. doi: 10.1007/s004250050077. [DOI] [PubMed] [Google Scholar]
- Wang H, Chevalier D, Larue C, Cho SK, Walkera JC. In: The protein phosphatases and protein kinases of Arabidopsis thaliana. Somerville CR, Meyerowitz EM, editor. The Arabidopsis Book, Rockville, MD; 2007. pp. 1–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves AJ, Wandosell F, Avila J. Phosphorylation of tubulin enhances its interaction with membranes. Nature. 1986;323:827–828. doi: 10.1038/323827a0. [DOI] [PubMed] [Google Scholar]
- Robinson DR, Wu YM, Lin SF. The protein tyrosine kinase family of the human genome. Oncogene. 2000;19:5548–5557. doi: 10.1038/sj.onc.1203957. [DOI] [PubMed] [Google Scholar]
- Karpov PA, Yemets AI, Blume YB. In: Plant GEM 2008 (Plant Genome European Meetings). 24-27 September 2008, Albena, Bulgaria. Atanassov AI, Christov NK, Rusanov K, Todorovska EG Sofia, editor. 2008. Kinome of the Arabidopsis thaliana chromosome I based on the database search and bioinformatics analysis; p. 78. Abstract 2.5. [Google Scholar]
- Karpov PA, Blume YB. In: Plant GEM 2008 (Plant Genome European Meetings). 24-27 September 2008, Albena, Bulgaria. Atanassov AI, Christov NK, Rusanov K, Todorovska EG Sofia, editor. 2008. Search of Arabidopsis thaliana homologies of animal protein-tyrosine kinases based on tBLASTn scaning of genome; p. 80. Abstract 2.6. [Google Scholar]
- Blume YaB, Lloyd CW, Yemets AI. In: The Plant Cytoskeleton: a Key Tool for Agro-Biotechnology. Blume YaB, Baird WV, Yemets AI, Breviario D, editor. Dordrecht, Springer; 2008. Plant tubulin phosphorylation and its role in cell cycle progression; pp. 145–155. [Google Scholar]
- Blume Y, Yemets A, Sulimenko V, Sulimenko T, Chan J, Lloyd C, Dráber P. Tyrosine phosphorylation of plant tubulin. Planta. 2008;229:143–150. doi: 10.1007/s00425-008-0816-z. [DOI] [PubMed] [Google Scholar]
- Yemets A, Sheremet Y, Vissenberg K, Van Orden J, Verbelen JP, Blume YB. Effects of tyrosine kinase and phosphatase inhibitors on microtubules in Arabidopsis root cells. Cell Biol Int. 2008;32:630–637. doi: 10.1016/j.cellbi.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Karpov PA, Blume YB. In: The Plant Cytoskeleton: a Key Tool for Agro-Biotechnology. Blume YaB, Baird WV, Yemets AI, Breviario D, editor. Dordrecht, Springer; 2008. Bioinformatic search for plant homologues of animal structural MAPs in the Arabidopsis thaliana genome; pp. 373–397. [Google Scholar]
- Hamada T. Microtubule-associated proteins in higher plants. J Plant Res. 2007;20:79–98. doi: 10.1007/s10265-006-0057-9. [DOI] [PubMed] [Google Scholar]
- Sedbrook JC. MAPs in plant cells: delineating microtubule growth dynamics and organization. Cur Opin Plant Biol. 2004;7:632–640. doi: 10.1016/j.pbi.2004.09.017. [DOI] [PubMed] [Google Scholar]
- Zhang C, Brankle S, Mallery E, Szymanski D. Composition and function of the Arabidopsis WAVE complex during epidermal morphogenesis. 16th Internatianal Conference on Arabidopsis Research. 15-19 June 2005, Madison. 2005. p. 131.
- Hanks SK, Quinn AM. Protein kinase catalytic domain sequence database: identification of conserved features of primary structure and classification of family members. Methods Enzymol. 1991;200:38–62. doi: 10.1016/0076-6879(91)00126-h. full_text. [DOI] [PubMed] [Google Scholar]
- Korf I, Yandell M, Bedell J. BLAST. Sebastopol: O'Reilly & Associates, Inc; 2003. [Google Scholar]
- Lucini C, Castaldo L, Lamanna C, Maruccio L, Vega JA, Gargiulo G. Neuronal and non-neuronal Trk neurotrophin receptor-like proteins in Eisenia foetida (Annelida oligochaeta) Neurosci Lett. 1999;261:163–166. doi: 10.1016/S0304-3940(99)00011-7. [DOI] [PubMed] [Google Scholar]
- Miller MA, Malik IA, Shenk MA, Steele RE. The Src/Csk regulatory circuit arose early in metazoan evolution. Oncogene. 2000;19:3925–3930. doi: 10.1038/sj.onc.1203714. [DOI] [PubMed] [Google Scholar]
- Müller P, Aichinger C, Feldbrügge M, Kahmann R. The MAP kinase kpp2 regulates mating and pathogenic development in Ustilago maydis. Mol Microbiol. 1999;34:1007–1017. doi: 10.1046/j.1365-2958.1999.01661.x. [DOI] [PubMed] [Google Scholar]
- Rikke BA, Murakami S, Johnson TE. Paralogy and orthology of tyrosine kinases that can extend the life span of Caenorhabditis elegans. Mol Biol Evol. 2000;17:671–683. doi: 10.1093/oxfordjournals.molbev.a026346. [DOI] [PubMed] [Google Scholar]
- Sakuma M, Onodera H, Suyemitsu T, Yamasu K. The protein tyrosine kinases of the sea urchin Anthocidaris crassispina. Zool Sci. 1997;14:941–946. doi: 10.2108/zsj.14.941. [DOI] [PubMed] [Google Scholar]
- Suga H, Hoshiyama D, Kuraku S, Katoh K, Kubokawa K, Miyata T. Protein tyrosine kinase cDNAs from amphioxus, hagfish, and lamprey: isoform duplications around the divergence of cyclostomes and gnathostomes. J Mol Evol. 1999;49:601–608. doi: 10.1007/PL00006581. [DOI] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Bailly E, Doree M, Nurse P, Bornens M. p34cdc2 is located in both nucleus and cytoplasm; part is centrosomally associated at G2/M and enters vesicles at anaphase. EMBO J. 1989;8:3985–3995. doi: 10.1002/j.1460-2075.1989.tb08581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Giet R, Sinka R, Mazumdar A, Lock WG, Balloux F, Zafiropoulos PJ, Yamaguchi S, Winter S, Carthew RW, Cooper M, Jones D, Frenz L, Glover DM. Genome-wide survey of protein kinases required for cell cycle progression. Nature. 2004;432:980–987. doi: 10.1038/nature03160. [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Rodrigues-Martins A, Carpenter L, Riparbelli M, Lehmann L, Gatt MK, Carmo N, Balloux F, Callaini G, Glover DM. SAK/PLK4 is required for centriole duplication and flagella development. Curr Biol. 2005;15:2199–2207. doi: 10.1016/j.cub.2005.11.042. [DOI] [PubMed] [Google Scholar]
- Burakov AV, Zhapparova ON, Kovalenko OV, Zinovkina LA, Potekhina ES, Shanina NA, Weiss DG, Kuznetsov SA, Nadezhdina ES. Ste20-related protein kinase LOSK (SLK) controls microtubule radial array in interphase. Mol Biol Cell. 2008;19:1952–1961. doi: 10.1091/mbc.E06-12-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaar Z, O'reilly P, Gelman I, Sabourin LA. v-Src-dependent down-regulation of the Ste20-like kinase SLK by casein kinase II. J Biol Chem. 2006;281:28193–28199. doi: 10.1074/jbc.M605665200. [DOI] [PubMed] [Google Scholar]
- Drewes G, Ebneth A, Preuss U, Mandelkow E-M, Mandelkow E. MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell. 1997;89:297–308. doi: 10.1016/S0092-8674(00)80208-1. [DOI] [PubMed] [Google Scholar]
- Eswaran J, Lee WH, Debreczeni JE, Filippakopoulos P, Turnbull A, Fedorov O, Deacon SW, Peterson JR, Knapp S. Crystal structures of the p21-activated kinases PAK4, PAK5, and PAK6 reveal catalytic domain plasticity of active group II PAKs. Structure. 2007;15:201–213. doi: 10.1016/j.str.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer T, Spielmann P, Stengel P, Stier B, Katschinski DM, Desbaillets I, Gassmann M, Wenger RH. Mammalian PASKIN, a PAS-serine/threonine kinase related to bacterial oxygen sensors. Biochem Biophys Res Commun. 2001;288:757–764. doi: 10.1006/bbrc.2001.5840. [DOI] [PubMed] [Google Scholar]
- Houlden H, Johnson J, Gardner-Thorpe C, Lashley T, Hernandez D, Worth P, Singleton AB, Hilton DA, Holton J, Revesz T, Davis MB, Giunti P, Wood NW. Mutations in TTBK2, encoding a kinase implicated in tau phosphorylation, segregate with spinocerebellar ataxia type 11. Nat Genet. 2007;39:1434–1436. doi: 10.1038/ng.2007.43. [DOI] [PubMed] [Google Scholar]
- Hutterer A, Berdnik D, Wirtz-Peitz F, Žigman M, Schleiffer A, Knoblich JA. Mitotic activation of the kinase Aurora-A requires its binding partner Bora. Dev Cell. 2006;11:147–157. doi: 10.1016/j.devcel.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Matenia D, Griesshaber B, Li XY, Thiessen A, Johne C, Jiao J, Mandelkow E, Mandelkow EM. PAK5 kinase is an inhibitor of MARK/Par-1, which leads to stable microtubules and dynamic actin. Mol Biol Cell. 2005;16:4410–4422. doi: 10.1091/mbc.E05-01-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef R, Gruneberg U, Kopajtich R, Li X, Nigg EA, Sillje H, Barr FA. Choice of Plk1 docking partners during mitosis and cytokinesis is controlled by the activation state of Cdk1. Nat Cell Biol. 2007;9:436–444. doi: 10.1038/ncb1557. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Mitotic kinases as regulators of cell division and its checkpoints. Nature Rev Mol Cell Biol. 2001;2:21–32. doi: 10.1038/35048096. [DOI] [PubMed] [Google Scholar]
- Sato S, Cerny RL, Buescher JL, Ikezu T. A Tau-tubulin kinase 1 (TTBK1), a neuron-specific tau kinase candidate, is involved in tau phosphorylation and aggregation. J Neurochem. 2006;98:1573–1584. doi: 10.1111/j.1471-4159.2006.04059.x. [DOI] [PubMed] [Google Scholar]
- Seki OA, Coppinger JA, Jang C-Y, Yates JR, Fang G. Bora and the kinase Aurora A cooperatively activate the kinase Plk1 and control mitotic entry. Science. 2008;320:1655–1658. doi: 10.1126/science.1157425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki A, Coppinger JA, Du H, Jang CY, Yates JR, Fang G. Plk1- and beta-TrCP-dependent degradation of Bora controls mitotic progression. J Cell Biol. 2008;181:65–78. doi: 10.1083/jcb.200712027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walden PD, Cowan NJ. A novel 205-kilodalton testis-specific serine/threonine protein kinase associated with microtubules of the spermatid manchette. Mol Cell Biol. 1993;13:7625–7635. doi: 10.1128/mcb.13.12.7625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese C, O'Brien LL. What's so Bor(a)ing about Aurora-A activation? Dev Cell. 2006;11:133–134. doi: 10.1016/j.devcel.2006.07.011. [DOI] [PubMed] [Google Scholar]
- Potekhina ES, Zinovkina LA, Nadezhdina ES. Enzymatic activity of protein kinase LOSK: possible regulatory role of the structural domain. Biochemistry (Mosc) 2003;68:188–195. doi: 10.1023/A:1022649428881. [DOI] [PubMed] [Google Scholar]
- Champion A, Picaud A, Picaud A, Henry Y. Reassessing the MAP3K and MAP4K relationships. TRENDS in Plant Science. 2004;9(3):123–129. doi: 10.1016/j.tplants.2004.01.005. [DOI] [PubMed] [Google Scholar]
- Champion A, Kreis M, Mockaitis K, Picaud A, Henry Y. Arabidopsis kinome: after the casting. Funct Integr Genomics. 2004;4:163–187. doi: 10.1007/s10142-003-0096-4. [DOI] [PubMed] [Google Scholar]
- Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006. pp. D257–260. [DOI] [PMC free article] [PubMed]
- Sun L, Gu S, Li X, Sun Y, Zheng D, Yu K, Ji C, Tang R, Xie Y, Mao Y. Identification of a novel human MAST4 gene, a new member of the microtubule associated serine-threonine kinase family. Mol Biol (Mosk) 2006;40:808–815. [PubMed] [Google Scholar]
- Demidov D, VanDamme D, Geelen D, Blattner FR, Houben A. Identification and dynamics of two classes of Aurora-like kinases in Arabidopsis and other plants. Plant Cell. 2005;17:836–848. doi: 10.1105/tpc.104.029710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe A, Matsunaga S, Nakagawa K, Kurihara D, Yoneda A, Hasezawa S, Uchiyama S, Fukui K. Characterization of plant Aurora kinases during mitosis. Plant Mol Biol. 2005;58:1–13. doi: 10.1007/s11103-005-3454-x. [DOI] [PubMed] [Google Scholar]
- Walter AO, Seghezzi W, Korver W, Sheung J, Lees E. The mitotic serine/threonine kinase Aurora2/AIK is regulated by phosphorylation and degradation. Oncogene. 2000;19:4906–4916. doi: 10.1038/sj.onc.1203847. [DOI] [PubMed] [Google Scholar]
- Arlot-Bonnemains Y, Klotzbucher A, Giet R, Uzbekov R, Bihan R, Prigent C. Identification of a functional destruction box in the Xenopus laevis aurora-A kinase pEg2. FEBS Lett. 2001;508:149–152. doi: 10.1016/S0014-5793(01)03048-4. [DOI] [PubMed] [Google Scholar]
- Guo L, Ho CM, Kong Z, Lee YR, Qian Q, Liu B. Evaluating the microtubule cytoskeleton and its interacting proteins in monocots by mining the rice genome. Ann Bot. 2009;103:387–402. doi: 10.1093/aob/mcn248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riabowol K, Draetta G, Brizuela L, Vandre D, Beach D. The cdc2 kinase is a nuclear protein that is essential for mitosis in mammalian cells. Cell. 1989;57:393–401. doi: 10.1016/0092-8674(89)90914-8. [DOI] [PubMed] [Google Scholar]
- Qian YW, Erikson E, Maller JL. Purification and cloning of a protein kinase that phosphorylates and activates the polo-like kinase Plx1. Science. 1998;282:1701–1704. doi: 10.1126/science.282.5394.1701. [DOI] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2005. pp. D34–38. [DOI] [PMC free article] [PubMed]
- UniProt Consortium. The universal protein resource (UniProt) Nucleic Acids Res. 2008. pp. D190–195. [DOI] [PMC free article] [PubMed]
- Finn RD, Tate J, Mistry J, Coggill PC, Sammut JS, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2008. pp. D281–288. [DOI] [PMC free article] [PubMed]
- Hulo N, Bairoch A, Bulliard V, Cerutti L, Cuche B, De Castro E, Lachaize C, Langendijk-Genevaux PS, Sigrist CJA. The 20 years of PROSITE. Nucleic Acids Res. 2008. pp. D245–249. [DOI] [PMC free article] [PubMed]
- Claverie J-M, Notredame C. Bioinformatics for dummies. New York: Wiley Publishing; 2007. [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Atteson K. In: Lecture Notes in Computer Science. Jiang T, Lee D, editor. Berlin, Springer-Verlag; 1997. The performance of neighbor-joining algorithms of phylogeny reconstruction; pp. 101–110. full_text. [Google Scholar]
- Nei M, Kumar S. Molecular evolution and phylogenetics. New York, Oxford University Press; 2000. [Google Scholar]
- Page RD. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Kumar S, Dudley J, Nei M, Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger E, Nabuurs SB, Vriend G. In: Structural Bioinformatics. Bourne PE, Weissig H, editor. Hoboken: John Wiley & Sons; 2003. Homology modeling; pp. 509–524. full_text. [Google Scholar]
- Fancelli D, Moll J, Varasi M, Bravo R, Artico R, Berta D, Bindi S, Cameron A, Candiani I, Cappella P, Carpinelli P, Croci W, Forte B, Giorgini ML, Klapwijk J, Marsiglio A, Pesenti E, Rocchetti M, Roletto F, Severino D, Soncini C, Storici P, Tonani R, Zugnoni P, Vianello P. 1,4,5,6-tetrahydropyrrolo[3,4-c]pyrazoles: identification of a potent Aurora kinase inhibitor with a favorable antitumor kinase inhibition profile. J Med Chem. 2006;49:7247–7251. doi: 10.1021/jm060897w. [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das B, Meirovitch H, Navon IM. Performance of hybrid methods for large-scale unconstrained optimization as applied to models of proteins. J Comput Chem. 2003;24:1222–12231. doi: 10.1002/jcc.10275. [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Ranganathan R, Ross EM. PDZ domain proteins: scaffolds for signaling complexes. Curr Biol. 1997;7:R770–773. doi: 10.1016/S0960-9822(06)00401-5. [DOI] [PubMed] [Google Scholar]


