Abstract
The classical nonhomologous end-joining (C-NHEJ) DNA double-strand break (DSB) repair pathway employs the Ku70/80 complex (Ku) for DSB recognition and the XRCC4/DNA ligase 4 (Lig4) complex for ligation. During IgH class switch recombination (CSR) in B lymphocytes, switch (S) region DSBs are joined by C-NHEJ to form junctions either with short microhomologies (MHs; “MH-mediated” joins) or no homologies (“direct” joins). In the absence of XRCC4 or Lig4, substantial CSR occurs via “alternative” end-joining (A-EJ) that generates largely MH-mediated joins. Because upstream C-NHEJ components remain in XRCC4- or Lig4-deficient B cells, residual CSR might be catalyzed by C-NHEJ using a different ligase. To address this, we have assayed for CSR in B cells deficient for Ku70, Ku80, or both Ku70 and Lig4. Ku70- or Ku80-deficient B cells have reduced, but still substantial, CSR. Strikingly, B cells deficient for both Ku plus Lig4 undergo CSR similarly to Ku-deficient B cells, firmly demonstrating that an A-EJ pathway distinct from C-NHEJ can catalyze CSR end-joining. Ku-deficient or Ku- plus Lig4-deficient B cells are also biased toward MH-mediated CSR joins; but, in contrast to XRCC4- or Lig4-deficient B cells, generate substantial numbers of direct CSR joins. Our findings suggest that more than one form of A-EJ can function in CSR.
There are two well-characterized mammalian DSB repair pathways. Homologous recombination accurately repairs post-replicative DSBs via a long, homologous template from a sister chromatid, whereas nonhomologous end joining (C-NHEJ) fuses DSB ends that lack substantial junctional homology (Bassing and Alt, 2004). Thus, C-NHEJ is particularly important during the G1 cell cycle phase when homologous templates from sister chromatids are not available (Lieber et al., 2008). Studies of the repair of RAG endonuclease-generated DSBs, in the context of lymphocyte-specific V(D)J recombination, were critical for elucidation of C-NHEJ. In this context, V(D)J recombination is abrogated in the absence of any of the four evolutionarily conserved “core” C-NHEJ factors, including Ku70, Ku80, XRCC4, and Lig4 (Taccioli et al., 1994; Li et al., 1995; Gu et al., 1997; Frank et al., 1998). Ku70 and Ku80 form the “Ku” DNA end-binding complex which functions as the DSB recognition component of C-NHEJ, whereas the XRCC4/Lig4 complex is specific for C-NHEJ ligation. DNA-dependent protein kinase catalytic subunit (DNA-PKcs) and the Artemis endonuclease are nonevolutionarily conserved C-NHEJ factors. Ku and DNA-PKcs form the DNA-PK holoenzyme, which, upon Ku binding to DSBs, phosphorylates Artemis, which can then process a subset of DSBs, including the hairpin coding ends generated during V(D)J recombination (Lieber et al., 2008). C-NHEJ also plays a key role in general DSB repair, as indicated by the impaired DSB repair, increased radiosensitivity and marked genomic instability of C-NHEJ–deficient cells (Rooney et al., 2004). Yet, studies of C-NHEJ deficient mammalian cells have revealed a still poorly characterized, but surprisingly robust, alternative end-joining (A-EJ) mechanism.
Early evidence for A-EJ came from linear plasmid rejoining assays using Ku-, Xrcc4-, or Lig4-deficient cell lines (Boulton and Jackson, 1996; Kabotyanski et al., 1998; Wang et al., 2003), and interest in A-EJ was stimulated by findings that it fuses chromosomal DSBs to generate oncogenic translocations in lymphomas from Xrcc4- or Lig4-deficient mice that were also deficient for p53 (Roth, 2002; Zhu et al., 2002). More recently, A-EJ was found to join ISceI endonuclease-generated DSBs in substrates chromosomally integrated into C-NHEJ-deficient cells (Guirouilh-Barbat et al., 2004, 2007) and to join physiologically relevant Ig heavy chain (IgH) class switch recombination (CSR)-associated DSBs in C-NHEJ deficient mouse B cells (Soulas-Sprauel et al., 2007; Yan et al., 2007; Han and Yu, 2008). Moreover, the absolute dependence of V(D)J recombination on core C-NHEJ factors was found to result from RAG endonuclease channeling the reaction into C-NHEJ and excluding A-EJ (Corneo et al., 2007; Deriano et al., 2009). Thus, A-EJ clearly appears to be a relevant chromosomal end joining mechanism. Yet, A-EJ and its components remain largely uncharacterized, and A-EJ might represent more than one pathway. With respect to components, the Xrcc1/Ligase 3 base excision repair ligation complex has been implicated in extra-chromosomal A-EJ, but potential roles in chromosomal A-EJ are unknown (Wang et al., 2003; Audebert et al., 2004). In addition, very recent studies have implicated the MRN complex in both C-NHEJ and A-EJ (Deng et al., 2009; Deriano et al., 2009; Dinkelmann et al., 2009; Rass et al., 2009; Xie et al., 2009), potentially via an end-processing function in A-EJ by itself and/or indirectly via the DSB response (Zha et al., 2009).
CSR provides a useful model for studies of chromosomal A-EJ. CSR in activated mature B lymphocytes exchanges the Cμ IgH constant region (CH) exons for one of several sets of CH exons (e.g., Cγ, Cα, and Cε) that lie 100 to 200kb downstream (Chaudhuri et al., 2007). Long, repetitive switch (S) regions lie just upstream of each set of CH exons. Activation-induced cytidine deaminase (AID) initiates CSR by generating lesions that lead to multiple DSBs in the donor S region flanking Cμ (Sμ) and in a downstream acceptor S region. CSR is completed when a DSB in Sμ is end-joined to a DSB in a downstream S region. Several different studies found that CSR was abrogated by Ku70- and/or Ku80-deficiency (Casellas et al., 1998; Manis et al., 1998). To resolve these findings with the relatively robust CSR via A-EJ in XRCC4- and Lig4-deficient B cells (Yan et al., 2007), one might speculate that Ku functions in both C-NHEJ and A-EJ during CSR. However, given that CSR requires B cell proliferation (Rush et al., 2005), an alternative, not mutually exclusive, possibility is that the previously reported severe proliferation defects of Ku-deficient B cells led indirectly to impaired CSR (Manis et al., 1998; Reina San-Martin et al., 2003; Chaudhuri and Alt, 2004; Chaudhuri et al., 2007). In this context, substantial end joining of chromosomal ISceI-initiated DSBs occurs in Ku-deficient cells (Guirouilh-Barbat et al., 2004, 2007).
C-NHEJ can join both DSB ends that lack junctional MH to form “direct” joins and DSBs ends with very short stretches of junctional homology to form microhomology (MH)-mediated joins. Correspondingly, both V(D)J recombination and CSR junctions generated in C-NHEJ-proficient B cells show substantial numbers of direct joins (up to 50%), with the remainder mostly containing short (1-2 bp) junctional MHs (Gilfillan et al., 1993; Komori et al., 1993). However, essentially all CSR junctions in XRCC4-deficient B cells contain MH which are, on average, longer than those observed in normal CSR junctions (Yan et al., 2007). Studies of XRCC4- or Lig4-deficient cells also revealed increased usage of MH in plasmid circularization junctions (Kabotyanski et al., 1998), ISceI DSB junctions (Guirouilh-Barbat et al., 2007), oncogenic translocation junctions (Zhu et al., 2002; Wang et al., 2008, 2009), and V(D)J junctions (Corneo et al., 2007). Such findings have led to the description of A-EJ as MH-mediated end joining (McVey and Lee, 2008). Yet, outside of CSR, end-joining events in the absence of XRCC4 do not exclusively use MH (Kabotyanski et al., 1998; Corneo et al., 2007; Guirouilh-Barbat et al., 2007). Moreover, in Ku80-deficient cells, A-EJ of ISceI-initiated DSBs generates a substantial proportion of direct junctions (Guirouilh-Barbat et al., 2004, 2007). Conceivably, the relative predominance of MH-mediated junctions in various A-EJ studies might reflect differences in substrates used, which in turn might influence pathway choice and junctions by providing differing degrees of MH (Yan et al., 2007). Alternatively, but not mutually exclusively, there may be different A-EJ pathways with distinct MH requirements and differential dependence on Ku versus XRCC4/Lig4. In the latter context, the relatively robust CSR observed in XRCC4 and Lig4 deficient B cells (Yan et al., 2007, Corneo et al., 2007) has been proposed to potentially represent a less efficient form of C-NHEJ that uses all of the C-NHEJ upstream components but a different DNA ligase (Lieber et al., 2008).
To unequivocally determine whether CSR can be catalyzed by an A-EJ pathway that is distinct from C-NHEJ, we have now assayed for CSR in B cells that lack Ku70, Ku80 or both Ku70 and Lig4.
RESULTS
Ku-independent IgH class switching
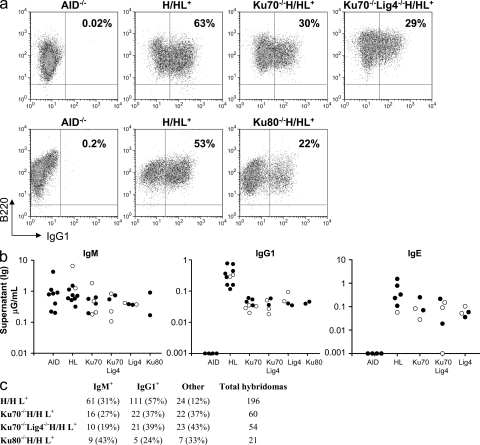
Several independent studies from more than a decade ago reported that Ku70- or Ku80-deficient B cells had very severe CSR defects (Manis et al., 1998; Casellas et al., 1998), leading to the assumption that, as in V(D)J recombination, CSR end-joining is catalyzed strictly by C-NHEJ (Honjo et al., 2004). However, normal CSR requires B cell proliferation, and previously studied Ku70- or Ku80-deficient B cells had severe proliferation defects, with many dying when activated for CSR (Casellas et al., 1998; Manis et al., 1998; Reina San-Martin et al., 2003). Therefore, we have suggested that although Ku70 and Ku80 are required for normal CSR, their overall contribution via C-NHEJ remained to be determined (Manis et al., 2002; Chaudhuri and Alt, 2004; Chaudhuri et al., 2007). Our current protocols for activating B cells stimulate much higher CSR levels in WT B cells than those used in the original Ku70−/− and Ku80−/− B cell studies; e.g., in the earlier studies of Ku-deficient cells, only ∼10% of WT B cells underwent CSR to IgG1 (Casellas et al., 1998; Manis et al., 1998), whereas under current stimulation conditions (different purification of cells, sources of cytokines, and activators, etc.) we observe CSR to IgG1 in 60% or more of WT B cells (Cheng et al., 2009; see Fig. 2). Therefore, we have reexamined the requirement for Ku70 and Ku80 in CSR. As Ku70 and Ku80 are required for V(D)J recombination and generation of mature B cells, we generated Ku70−/− or Ku80−/− mice that harbored preassembled (“knock-in”) IgH (B1-8-HC) and IgL (3–83k-LC) variable region exons (Pelanda et al., 1997; Sonoda et al., 1997). Various studies have shown that the B1-8-HC V(D)J knock-in allele undergoes normal CSR (Casellas et al., 1998; Manis et al., 1998; Yan et al., 2007). Because of the complex breeding strategies necessary to generate these mice, we generated Ku70−/− and Ku80−/− strains that were either homozygous or heterozygous for the B1-8-HC knock-in allele (referred to, respectively, as H/HL and H/+L). The breakdown of results with these genotypes is indicated in each figure or in supplementary figures and tables. Because H/HL and H/+L lines gave similar results for most experiments, we generically refer to both genotypes as HL mice, except where otherwise noted. We verified homozygous disruption of the Ku70 and Ku80 locus and the absence of the Ku70 protein in the Ku70−/−HL mice and cells analyzed (Fig. 1 a and not depicted).
Figure 2.
High IgG1 and IgE CSR levels in Ku70−/−, Ku70−/−Lig4−/−, Ku80−/−, and Lig4−/− B cells. (a) Representative FACS panel for IgG1 CSR in B cells stimulated with αCD40 plus IL-4 and assayed at day 3.5. Data shown are representative of >10 experiments, with one or more mouse of each genotype per experiment. Table S1 contains a summary of all FACS experiments. (b) ELISA quantification of IgM, IgG1, and IgE secretion (μG/ml) in B cells stimulated as in a. Black circles, H/H (the knock-in prerearranged heavy chain receptor is present on both alleles); white circles, H/+ (knock-in heavy chain on one allele). All experiments completed are shown, with one mouse per genotype per experiment and each circle represents one mouse. Table S2 contains a detailed summary of all ELISA experiments. (c) Frequency of IgG1-secreting hybridomas generated from αCD40/IL-4–stimulated B cells. Hybridomas for one mouse per genotype are shown. The number and percentage of hybridomas is indicated for each isotype assayed. “Other” denotes hybridomas negative for both IgM and IgG1 secretion, some of which may be IgE+ because IgE ELISA was not performed for the fusions shown. Data shown are representative of three independent hybridoma fusion experiments (one mouse each) for WT-HL and Ku70−/−HL and two independent hybridoma fusions (one mouse each) for the Ku70−/−Lig4−/−HL mice. Table S3 contains a summary of all hybridoma data, with multiple mice of each genotype.
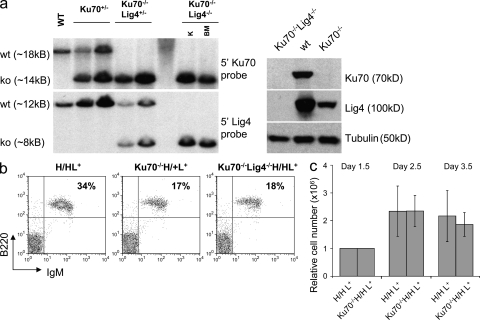
Figure 1.
Ku70−/−HL and Ku70−/−Lig4−/−HL mice posses mature splenic B cells which proliferate in response to stimulation by αCD40/IL-4. (a) Disruption of the Ku70 and Lig4 genomic loci and absence of Ku70 and Lig4 proteins in Ku70−/−Lig4−/− mice. (left) Southern blotting: BamHI-digested genomic DNA was probed with Ku70- or Lig4-specific probes. (right) Thymus protein extracts were assayed for the presence of Ku70 or Lig4 proteins. K, kidney protein extracts; BM, bone marrow protein extracts. Both the Southern and the Western blotting were repeated two times on independent samples. (b) WT-HL, Ku70−/−HL and Ku70−/−Lig4−/−HL mice have mature splenic B cells. Whole cell suspensions from spleens of WT-HL, Ku70−/−HL, and Ku70−/−Lig4−/−HL were used to quantify the percentage of B220+IgM+ mature B cells. Data shown are representative of more than 10 experiments, with at least one mouse of each genotype per experiment. (c) Ku70−/−HL mature B cells proliferate at levels comparable to WT-HL after αCD40/IL-4 stimulation. Splenic B cells were enriched with B220 magnetic beads and counted by Trypan blue staining starting at day 1.5 from the beginning of the culture, to allow for the disappearance of non–B cells. Data shown are based on two independent stimulations and a total of 4 WT-HL and 5 Ku70−/−HL mice were used. Error bars are based on the relative cell number for each mouse.
Ku70−/−HL and Ku80−/−HL mice generate substantial numbers of mature splenic B cells, which express surface Ig that derives from their IgH and Igκ knock-in alleles (Fig. 1 b). To test CSR potential to IgG1 and IgE, we purified Ku70−/−HL and Ku80−/−HL splenic B cells and stimulated them for 4 d with αCD40 plus IL-4 or, for some experiments, with bacterial LPS plus IL-4. In contrast to earlier findings, neither the Ku70−/− nor Ku80−/− B cells showed a dramatic proliferation defect compared with control cells (Fig. 1 c and not depicted). Under these stimulation conditions, up to 50–60% of WT/HL B cells switched to IgG1 as measured by flow cytometry staining for surface IgG1 expression (Fig. 2 a, Fig. S1, and Table S1). Strikingly, however, the surface staining assay also revealed that αCD40 plus IL-4 or LPS plus IL-4–stimulated Ku70−/−HL B cells and Ku80−/−HL B cells underwent CSR to IgG1 at levels 20–50% of those of WT/HL B cells (Fig. 2 a, Fig. S1, and Table S1). We confirmed the unexpected CSR ability of Ku70−/− and Ku80−/− B cells by using numerous additional types of class switching assays, including measurement of secreted IgG1 by ELISA (Fig. 2 b and Table S2), as well as by the more quantitative clonal B cell ELISPOT (not depicted) or hybridoma IgG1 secretion analyses (Fig. 2 c and Table S3). Southern blotting also allowed us to confirm that CSR in Ku70−/−HL B cell hybridomas occurred at the DNA level via Sμ to Sγ1 recombination (Fig. S2). IgH class switching to IgE cannot be assayed quantitatively via surface staining because of binding of secreted IgE to Fcε receptors on B cells. However, ELISA demonstrated that Ku70−/−HL B cells switched to IgE at ∼25% of WT/HL levels (Fig. 2 b). Moreover, hybridoma analyses revealed that ∼50% of WT/HL B cells switched to IgE under our current stimulation conditions and that Ku70−/−HL B cell hybridomas switched to IgE at ∼30% of WT/HL levels, which is consistent with the ELISA data (Table S3). Note that because the WT hybridomas were derived from the H/HL+ genotype, ∼15% were IgE/IgG1 double producers caused by productive CSR to IgG1 on one allele and to IgE on the other (see Table S3 legend). Finally, the hybridoma analyses also allowed us to confirm IgE CSR at the DNA level via Southern blotting (Fig. S2). Thus, these studies implicate a relatively robust Ku-independent form of A-EJ that supports CSR to IgG1 and IgE.
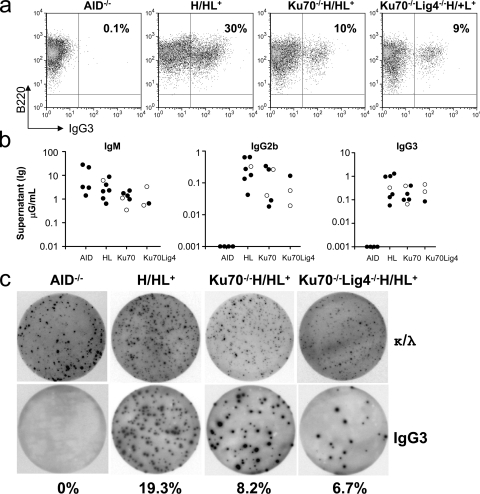
DNA-PKcs–deficient B cells appear more impaired for CSR to other IgH isotypes than for CSR to IgG1 (Manis et al., 2002; Rooney et al., 2005; Callén et al., 2009). Therefore, because Ku functions in part via its role in the DNA–PK complex, we assessed the ability of Ku70−/−HL or Ku80−/−HL B cells to undergo CSR to additional IgH isotypes. Stimulation of splenic B cells with LPS alone induces CSR to IgG2b and IgG3. However, we have recently shown that DNA-PKcs– or Artemis-deficient B cells activated for CSR are much more prone to cell death upon LPS stimulation compared with stimulation with αCD40 plus IL-4 because of a p53-dependent response to unrepaired DNA DSBs (Franco et al., 2008). LPS plus anti–IgD-dextran stimulates class switching to IgG2b and IgG3 more robustly than treatment with LPS alone (Cheng et al., 2009). Therefore, to test CSR to these two IgH isotypes, we stimulated purified Ku70−/−HL splenic B cells for 4 d with LPS plus anti–IgD-dextran. Based on the flow cytometry assay for surface IgG3 expression, this stimulation led to IgG3 switching in up to 40% of the WT/HL B cells (Fig. 3 a; Table S1). Strikingly, Ku70−/−HL B cells underwent class switching to IgG3 on average at nearly 30% of WT levels under these conditions (Fig. 3 a and Table S1). LPS plus anti–IgD-dextran treatment of Ku70−/−HL B cells also induced switching to IgG3 at a substantial fraction of WT/HL levels, based on ELISA and ELISPOT analyses (Fig. 3, b and c, and Table S2) and to IgG2b based on ELISA analyses (Fig. 3 b and Table S2). A more limited set of studies also demonstrated that LPS-stimulated Ku80−/−HL B cells switch to IgG3 or IgG2b, as assayed by flow cytometry (Table S1), ELISPOT (not depicted), or ELISA (Table S2). Finally, we used stimulation with LPS–IL-4–IL-5 and TGFβ for class switching to IgA and found that Ku70−/−HL B cells also switched to IgA at levels that were a substantial fraction of those observed for WT/HL B cells (Fig. S3).
Figure 3.
Ku70- and Ku70/Lig4-deficient cells undergo robust CSR to IgG3 and IgG2b. (a) Representative FACS panel for IgG3 CSR in B cells stimulated with LPS and anti–IgD-dextran and assayed at day 3.5. Data shown are representative of >10 experiments, with one or more mouse of each genotype per experiment. Table S1 contains a summary of all FACS experiments. (b) ELISA quantification of IgM, IgG3, and IgG2b secretion (micrograms/milliliter) in B cells stimulated as in a. Black circles, H/H (the knock-in prerearranged heavy chain receptor is present on both alleles); white circles: H/+ (knock-in heavy chain on one allele). Each circle represents one mouse. All experiments completed are shown, with one mouse per genotype per experiment. Table S2 contains a summary of all ELISA experiments. (c) Representative ELISPOT assay on B cells stimulated as in A. (top) Dots represent Igκ- and Igλ-secreting B cells. (bottom) Dots represent IgG3-secreting B cells. Percentages of B cells undergoing CSR to IgG3 are shown. Data shown are representative of three experiments, with one mouse of each genotype per experiment.
Class switching in B cells lacking both Ku70 and Lig4
Although Lig4 deficiency is embryonically lethal (Frank et al., 1998), Ku80−/−Lig4−/− mice (Karanjawala et al., 2002) and Ku70−/−/Lig4−/− DT40 avian cell lines (Adachi et al., 2001) are viable. In addition, we find that Ku70−/−Lig4−/− mice are also viable (see below). Thus, we addressed the question of whether CSR can occur in a fully C-NHEJ–independent fashion by asking whether we observe CSR in cells deficient for both Ku70 and Lig4 (the specific DSB recognition and ligation C-NHEJ components). For this purpose, we generated Ku70−/−Lig4−/− mice, which also harbored the preassembled IgH and IgK alleles (HL). Ku70−/−Lig4−/−HL mice develop normally, albeit being slightly smaller than their littermates. We confirmed the deletion of Ku70 and Lig4 genes and absence of both proteins in extracts from kidney, brain and thymus by Southern blotting and Western blotting, respectively (Fig. 1 a). Ku70−/−Lig4−/−HL mice also generate IgM+ splenic B cells (Fig. 1 b), but lack mature thymic or peripheral T cells because of their V(D)J recombination block (not depicted). To test for ability of purified Ku70−/−Lig4−/−HL splenic B cells to undergo class switching, we stimulated them for 4 d with αCD40 plus IL-4 and measured class switching to IgG1 by surface staining and ELISA (Fig. 2, a and b, Table S1, and Table S2). We also stimulated Ku70−/−Lig4−/−HL splenic B cells for 4 d with LPS plus anti–IgD-dextran and measured class switching to IgG3 and IgG2b by surface staining, ELISA, and ELISPOT (Fig. 3, a–c, Table S1, and Table S2). Based on all of these assays, Ku70−/−Lig4−/−HL B cells showed class switching to all tested IgH isotypes at levels comparable to Ku70−/− B cells. Therefore, we conclude that there is a repair pathway that catalyzes substantial IgH class switching in the complete absence of C-NHEJ.
CSR is less robust in the absence of Ku70 versus ligase 4
Most studies of CSR tend to measure this process at time points after stimulation (e.g., day 4) when WT CSR is nearly complete. However, measurements at such late time points might underestimate kinetic differences between mutant and WT B cells, by allowing the mutant B cells to “catch up” (Cheng et al., 2009). Therefore, to further characterize the Ku-independent A-EJ pathway during CSR, we performed time-course kinetics of class switching to IgG1 in αCD40 plus IL-4 stimulated HL, Ku70−/−HL and Ku70−/−Lig4−/−HL purified splenic B cells. For this purpose, we measured surface IgG1expression at days 2.5, 3.5, and 4.5, with the earliest time point corresponding to about the time that AID expression is readily detectable in stimulated B cells (Cheng et al., 2009). These studies showed that Ku70−/−HL and Ku70−/−Lig4−/−HL B cells underwent class switching to IgG1 at a significant fraction of the level observed in WT/HL B cells at all assayed time points (Fig. 4, a and b, Fig. S1, and Table S1). Thus, the Ku70- and Ku70/Lig4-independent pathway of IgH class switching to IgG1 also is relatively robust kinetically as compared with that of WT/HL B cells. However, the relative level of class switching to IgG1 for Ku70−/−B cells and Ku70−/−Lig4−/−HL B cells, as compared with that of WT/HL B cells, appeared slightly lower on average at day 2.5 versus later time points (Fig. 4 b and Table S1), suggesting that CSR in the absence of Ku70 might be slightly less robust kinetically than that of CSR in WT cells (Yan et al., 2007). In this regard, while CSR in B cells in which XRCC4 was conditionally deleted in the periphery appeared kinetically similar to that of WT B cells (Yan et al., 2007), CSR in a Lig4-deficient B cell line appeared kinetically reduced (Han and Yu, 2008).
Figure 4.
Time course for IgG1 CSR in Ku70−/−H/HL+, Ku70−/−Lig4−/−H/HL+, and WT-H/HL+ B cells stimulated with αCD40 plus IL-4. (a) Representative FACS plots for IgG1 CSR quantification in B cells from the indicated backgrounds stimulated with αCD40 plus IL-4. IgG1 FACS was recorded at day 2.5, 3.5, and 4.5. AID, AID−/− B cells used as negative controls for CSR. Percentage of B220+IgG1+ cells that have undergone CSR is shown for each genotype at each time point. Data shown are representative of three independent B cell stimulation experiments, with one mouse of each genotype per experiment. (b) Quantification of CSR levels at days 2.5, 3.5, and 4.5 from the beginning of the in vitro stimulation. Data are based on at least three mice per genotype used in three separate experiments. Table S1 contains the summary of all FACS experiments.
CSR junctional sequences in Ku70 or Ku70 plus Lig4-deficient B cells
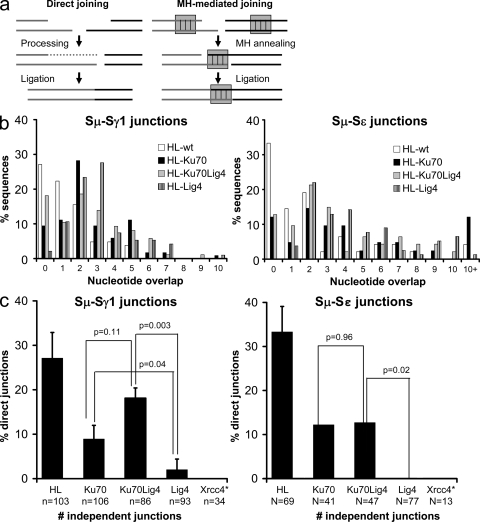
A substantial fraction of CSR junctions from WT B cells are direct, with most of the remainder displaying short MHs; whereas essentially all CSR junctions from XRCC4-deficient B cells display MHs that are on average longer than those from WT B cells (Yan et al., 2007). To further assess the relationship between the A-EJ pathway that generates CSR joins in Ku-deficient cells versus that which occurs in the absence of XRCC4 or Lig4, we sequenced Sμ/Sγ1 and Sμ/Sε junctions from Ku70−/−HL, Ku70−/−Lig4−/−HL, Lig4−/−HL, and WT/HL splenic B cells that were activated with αCD40 plus IL-4. As expected, a substantial percentage of Sμ/Sγ1 and Sμ/Sε junctions from WT/HL B cells were direct (27.1 and 33.3% direct, respectively; Fig. 5, b and c, and Table S4). On the other hand, Sμ/Sγ1 and Sμ/Sε junctions from Lig4−/−HL B cells were almost all MH-mediated (2 and 0% direct, respectively), consistent with the fact that XRCC4 and Lig4 act as a complex in C-NHEJ. Although Sμ/Sγ1 and Sμ/Sε junctions from Ku70−/−HL B cells also displayed increased usage and mean length of MH-mediated joins compared with those from WT/HL B cells (Fig. 5, b and c; Table S4), a substantial fraction were direct (8.9 and 12.2%, respectively; Fig. 5, b and c; Table S4). Similarly, a substantial fraction of Sμ/Sγ1 and Sμ/Sε junctions from Ku70−/−Lig4−/−HL B cells also were direct (18 and 13%, respectively; Fig. 5, b and c, and Table S4). Thus, CSR junctions in Ku-deficient B cells, whether or not Lig4 is present, show a substantial and significant increase in direct joins as compared with junctions generated in the absence of Lig4 (P = 0.04 for Ku70−/−HL; P = 0.003 for Ku70−/−Lig4−/−HL; Fig. 5 C and Table S4). Importantly, the fraction of direct joins is not significantly different between Ku70 versus Ku70 plus Lig4-deficient B cells (P = 0.11 for Sμ/Sγ1 and P = 0.96 for Sμ/Sε junctions; Fig. 5 C), which is consistent with the possibility that another ligase may be predominantly used for CSR end-joining in Ku70−/−HL B cells.
Figure 5.
The Ku-dependent A-EJ pathway uses a distinct mechanism for S region joining. (a) Classical NHEJ performs both direct (left) and MH-mediated (right) end-joining. Direct joining occurs when DNA ends are joined either immediately, or after processing (removal of overhangs or fill-in by polymerases). MH-mediated joining (right) is facilitated by base-pairing interactions between short stretches of nucleotides at or near the ends of the DSB. (b) MH length in Sμ-Sγ1 (left) and Sμ-Sε (right) junctions from Ku70−/−HL, Ku70−/−Lig4−/−HL, Lig4−/−HL and WT/HL B cells stimulated in culture with αCD40 plus IL-4. Ku70-, Ku70/Lig4-, and Lig4-deficient cells have more Sμ-Sγ1 (left) and Sμ-Sε (right) joins with longer MH than WT. Data shown are based on 86 or more independent sequences per genotype for Sμ-Sγ1 junctions and 41 or more independent sequences for Sμ-Sε junctions. At least three mice per genotype in three independent experiments were used for Sμ-Sγ1 junctions and one Ku70−/−HL mouse, two Ku70−/−Lig4−/−HL mice, three Lig4−/−HL, and three WT-HL mice in three independent experiments were used for Sμ-Sε junctions. A detailed summary of all S region junction isolation experiments and mice is presented in Table S4. (C) Percentage of direct Sμ-Sγ1 (left) and Sμ-Sε (right) joins in Ku70−/−HL, Ku70−/−Lig4−/−HL, Lig4−/−HL, XRCC4−/−HL and WT/HL B cells. Ku70 and Ku70Lig4-deficient cells have significantly more direct Sμ-Sγ1 and Sμ-Sε joins than Lig4 germline-deficient cells. *, XRCC4 junctions were acquired and published previously (Yan et al., 2007) and are shown here for comparison. Two-tailed Student’s t test was used for p-value. Data shown are based on 86 or more independent sequences per genotype for Sμ-Sγ1 junctions and 41 or more independent sequences for Sμ-Sε junctions. At least three mice per genotype in three independent experiments were used for Sμ-Sγ1 junctions, and one Ku70−/−HL mouse, two Ku70−/−Lig4−/−HL mice, three Lig4−/−HL, and three WT-HL mice in three independent experiments were used for Sμ-Sε junctions. N, number of independent sequences.
DISCUSSION
Recent work has established the role of A-EJ in mammalian DNA repair and made this pathway a major current focus of the DNA repair field. Yet, the nature of A-EJ has remained enigmatic. In particular, other than recent studies that implicate the MRN complex in chromosomal A-EJ (Deng et al., 2009; Dinkelmann et al., 2009; Rass et al., 2009; Xie et al., 2009), little has been forthcoming regarding the identity of chromosomal A-EJ components. In addition, although A-EJ is often discussed as a single pathway, it has remained possible that there are multiple A-EJ pathways that use at least some different components. Our current studies provide several new insights into A-EJ. In particular, we now demonstrate that, in the absence of C-NHEJ, two forms of A-EJ, which appear to be potentially distinct based on both different preferences for junctional MHs and differential reliance on Ku, catalyze the physiological process of CSR. One form of A-EJ, that operates in the absence of XRCC4 or Lig4, utilizes Ku and either Lig1 or Lig3 and, at least in the context of CSR, is almost totally reliant on junctional MHs. Given its apparent dependence on Ku, this form of A-EJ may, as has been suggested (Lieber et al., 2008), reflect the use of the upstream components of C-NHEJ with a different ligase (Lieber et al., 2008); although it remains possible that there are other differences from C-NHEJ. Our current findings further demonstrate a second form of A-EJ during CSR that functions in the simultaneous absence of both the DSB recognition component (Ku70) and the specific ligation component (Lig4) of C-NHEJ. This second form of A-EJ, while also biased toward CSR junctional MHs, generates substantial levels of CSR direct joins. We conclude that this second (Ku plus Lig4-independent) form of A-EJ is totally distinct from C-NHEJ and must use a largely different set of components.
Prior studies found that CSR was abrogated by Ku70- or Ku80-deficiency, leading to the common assumption that Ku70 and Ku80 are essential for end joining during CSR (Honjo et al., 2004). However, Ku70- or Ku80-deficient cells, as assayed at that time, had severe proliferation defects, which also were suggested to potentially contribute to their severe CSR impairment (Manis et al., 1998, 2002; Chaudhuri and Alt, 2004; Chaudhuri et al., 2007). Our current stimulation conditions lead to substantially increased CSR in WT cells and also appear to obviate the very severe proliferation defects previously observed in Ku-deficient B cells. Under these stimulation conditions, we confirm that CSR indeed is impaired in Ku-deficient B cells; but, on the other hand, we also find that Ku-deficient B cells are capable of undergoing substantial CSR. Indeed, Ku70- or Ku80-deficient B cells reach steady-state CSR levels that range from 25–50% those of WT B cells, which are similar in range to those found in XRCC4- or Lig4-deficient B cells. Likewise, our time course experiments confirmed that CSR in the absence of Ku or Ku plus Lig4 is quite robust; although perhaps slightly delayed kinetically as compared with that of WT B cells reminiscent of findings of CSR in Lig4-deficient CH12 cells (Han and Yu, 2008).
The spectrum of CSR junctions differs between Ku70-deficient or Ku70- plus Lig4-deficient versus Lig4- or XRCC4-deficient B cells. Thus, XRCC4- or Lig4-deficient junctions are nearly all MH mediated, whereas the junctions that occur in the absence of Ku70 or Ku70 plus Lig4, although still biased toward MH, have a substantial percentage of direct joins. These findings are consistent with findings of increased, although not to the same degree, use of MH in ISceI-mediated DSB junctions recovered from XRCC4 versus Ku-deficient Chinese hamster organ cells (Guirouilh-Barbat et al., 2004, 2007). Ku was previously thought to function in DNA repair only via C-NHEJ; thus, our finding that Ku actually promotes MH-mediated A-EJ during CSR in the absence of Ligase 4 was unexpected. The Ku heterodimer binds to ends of DSBs and then recruits other C-NHEJ repair factors (for review see Rooney et al., 2004), while also protecting the broken DNA ends from degradation. However, we find that nearly all CSR DSBs in Ku-proficient/Lig4-deficient cells are joined via MHs, which presumably requires end resection; whereas CSR DSBs in Ku plus Lig4-deficient B cells have a higher frequency of direct joins. One possible explanation for these seemingly contradictory findings would be that Ku can preferentially stabilize S region ends held by microhomologies. Alternatively, in the absence of Ku70, end-modifying polymerases, such as TdT, pol µ, or pol λ might gain access to DNA ends and perform nucleotide additions to generate “occult” end-homology (Komori et al., 1996) that cannot be distinguished from “direct” joins. In this context, Pol µ has been shown to have TdT-like activity and to add nontemplate nucleotides even in the absence of Ku, XRCC4, or Lig4 (Gu et al., 2007). A further possibility is that Lig1 and Lig3 may be differentially involved in the Ku70-dependent, MH-mediated A-EJ pathway versus the Ku70-independent pathway and, thereby, provide the end preference. In biochemical assays, Lig3 but not Lig1 performs direct end joining of oligonucleotide substrates (Chen et al., 2000; Cotner-Gohara et al., 2008). Thus, if the end preference observed in biochemical studies holds in Ku70-deficient cells, our findings might be explained by Lig3 preferentially gaining access to DNA ends in Ku70-deficient cells and Lig1 preferentially accessing ends in Ku-70 proficient, Lig4-deficient cells.
In conclusion, our studies demonstrate the existence of an A-EJ pathway that functions to generate substantial levels of chromosomal IgH CSR joins in activated B cells that lack both DSB recognition and joining components of the C-NHEJ pathway. This finding firmly demonstrates the existence of a physiologically functional A-EJ pathway that is distinct from C-NHEJ. In addition, we find that the previously described MH-based A-EJ pathway of CSR that catalyzes CSR in the absence of either XRCC4 or Lig4 is substantially influenced by Ku; thus, this pathway might be a variant of C-NHEJ that uses a different ligase; although other possibilities including a distinct, Ku dependent, A-EJ pathway can also be considered.
MATERIALS AND METHODS
Mouse strains.
Ku70−/−, Lig4−/− CD21CreXrcc4c/c, and Ku80−/− HL mice have been previously described (Gu et al., 1997, Frank et al., 1998, Yan et al., 2007, Casellas et al., 1998). Ku70−/−Lig4−/−HL mice were obtained by breeding double-heterozygous Ku70+/−Lig4+/−HL mice. Animal protocols were approved by the Institutional Animal Care and Use Committee of Children’s Hospital (Boston, MA).
Splenic B cell purification and culture.
Mature B cells were isolated and enriched from the spleen and stimulated with αCD40/IL-4 to induce CSR to IgG1 and IgE as described (Yan et al., 2007). For IgG3 and IgG2b CSR, B cells were stimulated with LPS and dextran-conjugated αIgD as described (Cheng et al., 2009). For IgA CSR, B cells were stimulated with LPS (25 µg/ml), dextran-conjugated αIgD (3 ng/ml), IL-4: 10 ng/ml, IL-5: 1.5 ng/ml and TGFβ: 2 ng/ml. Cytokine sources: αCD40: eBioscience, IL-4: eBioscience, IL-5: BD dextran-conjugated αIgD: Fina BioSolutions LLC. Proliferation studies were conducted on aCD40/IL-4 stimulated splenic cells enriched with B220 magnetic beads (Miltenyi Biotec) and cells were counted every 24 h starting at 1.5 d post-enrichment to ensure that the majority of the cells in culture were B cells. Surface expression was recorded by FACS. ELISPOT and ELISA were performed as described (Yan et al., 2007) at days 4 and 6 of culture, respectively. B cell hybridoma fusions were performed at day 4 of culture and hybridomas were screened by ELISA after 1 wk of selection.
S region junction isolation and analysis.
Sμ-Sγ1 and Sμ-Sε junctions were amplified from B cells stimulated in culture with αCD40 plus IL-4 or from hybridomas derived from the above cultures with primers described previously (Yan et al., 2007) and National Center for Biotechnology Information Blast was used for sequence analysis. Calculation of statistical significance was done using the two-tailed Student T test.
Online supplemental material.
Fig. S1 shows a CSR timecourse in Ku70−/−HL and Ku80−/−HL mice. Fig. S2 shows CSR-related DNA recombination events in IgG1+ and IgE+ Ku70−/−HL and Ku70−/−Lig4−/−HL hybridomas. Fig. S3 shows IgA CSR in Ku70−/−HL B cells. Table S1 is a summary of all FACS experiments. Table S2 is a summary of all ELISA experiments. Table S3 is a summary of all hybridoma fusion data. Table S4 is a summary of all S region function data.
Acknowledgments
We thank Klaus Rajewsky, Bjoern Schwer, Monica Gostissa, and Maria-Vivienne Boboila for discussions and critical reading of the manuscript.
This work was supported by National Institutes of Health grants AI031541 and CA092625 to FWA, NIH grant AI037526 to MN and T32CA09382-26 to JHW. CB is supported by a Cancer Research Institute training grant. FWA and MN are investigators of the Howard Hughes Medical Institute.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- A-EJ
- alternative end-joining
- AID
- activation-induced cytidine deaminase
- C-NHEJ
- classical nonhomologous end-joining
- CSR
- class switch recombination
- DNA-PKcs
- DNA-protein kinase catalytic subunit
- DSB
- double-strand break
- MH
- microhomology
References
- Adachi N., Ishino T., Ishii Y., Takeda S., Koyama H. 2001. DNA ligase IV-deficient cells are more resistant to ionizing radiation in the absence of Ku70: implications for DNA double-strand break repair. Proc. Natl. Acad. Sci. USA. 98:12109–12113 10.1073/pnas.201271098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audebert M., Salles B., Calsou P. 2004. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J. Biol. Chem. 279:55117–55126 10.1074/jbc.M404524200 [DOI] [PubMed] [Google Scholar]
- Bassing C.H., Alt F.W. 2004. The cellular response to general and programmed DNA double strand breaks. DNA Repair (Amst.). 3:781–796 10.1016/j.dnarep.2004.06.001 [DOI] [PubMed] [Google Scholar]
- Boulton S.J., Jackson S.P. 1996. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 15:5093–5103 [PMC free article] [PubMed] [Google Scholar]
- Callén E., Jankovic M., Wong N., Zha S., Chen H.T., Difilippantonio S., Di Virgilio M., Heidkamp G., Alt F.W., Nussenzweig A., Nussenzweig M. 2009. Essential role for DNA-PKcs in DNA double-strand break repair and apoptosis in ATM-deficient lymphocytes. Mol. Cell. 34:285–297 10.1016/j.molcel.2009.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casellas R., Nussenzweig A., Wuerffel R., Pelanda R., Reichlin A., Suh H., Qin X.F., Besmer E., Kenter A., Rajewsky K., Nussenzweig M.C. 1998. Ku80 is required for immunoglobulin isotype switching. EMBO J. 17:2404–2411 10.1093/emboj/17.8.2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri J., Alt F.W. 2004. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat. Rev. Immunol. 4:541–552 10.1038/nri1395 [DOI] [PubMed] [Google Scholar]
- Chaudhuri J., Basu U., Zarrin A., Yan C., Franco S., Perlot T., Vuong B., Wang J., Phan R.T., Datta A., et al. 2007. Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Adv. Immunol. 94:157–214 10.1016/S0065-2776(06)94006-1 [DOI] [PubMed] [Google Scholar]
- Chen L., Trujillo K., Sung P., Tomkinson A.E. 2000. Interactions of the DNA ligase IV-XRCC4 complex with DNA ends and the DNA-dependent protein kinase. J. Biol. Chem. 275:26196–26205 10.1074/jbc.M000491200 [DOI] [PubMed] [Google Scholar]
- Cheng H.L., Vuong B.Q., Basu U., Franklin A., Schwer B., Astarita J., Phan R.T., Datta A., Manis J., Alt F.W., Chaudhuri J. 2009. Integrity of the AID serine-38 phosphorylation site is critical for class switch recombination and somatic hypermutation in mice. Proc. Natl. Acad. Sci. USA. 106:2717–2722 10.1073/pnas.0812304106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corneo B., Wendland R.L., Deriano L., Cui X., Klein I.A., Wong S.Y., Arnal S., Holub A.J., Weller G.R., Pancake B.A., et al. 2007. Rag mutations reveal robust alternative end joining. Nature. 449:483–486 10.1038/nature06168 [DOI] [PubMed] [Google Scholar]
- Cotner-Gohara E., Kim I.K., Tomkinson A.E., Ellenberger T. 2008. Two DNA-binding and nick recognition modules in human DNA ligase III. J. Biol. Chem. 283:10764–10772 10.1074/jbc.M708175200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Guo X., Ferguson D.O., Chang S. 2009. Multiple roles for MRE11 at uncapped telomeres. Nature. 60:914–918 10.1038/nature08196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deriano L., Stracker T.H., Baker A., Petrini J.H., Roth D.B. 2009. Roles for NBS1 in alternative nonhomologous end-joining of V(D)J recombination intermediates. Mol. Cell. 34:13–25 10.1016/j.molcel.2009.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkelmann M., Spehalski E., Stoneham T., Buis J., Wu Y., Sekiguchi J.M., Ferguson D.O. 2009. Multiple functions of MRN in end-joining pathways during isotype class switching. Nat. Struct. Mol. Biol. 16:808–813 10.1038/nsmb.1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco S., Murphy M.M., Li G., Borjeson T., Boboila C., Alt F.W. 2008. DNA-PKcs and Artemis function in the end-joining phase of immunoglobulin heavy chain class switch recombination. J. Exp. Med. 205:557–564 10.1084/jem.20080044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank K.M., Sekiguchi J.M., Seidl K.J., Swat W., Rathbun G.A., Cheng H.L., Davidson L., Kangaloo L., Alt F.W. 1998. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature. 396:173–177 10.1038/24172 [DOI] [PubMed] [Google Scholar]
- Gilfillan S., Dierich A., Lemeur M., Benoist C., Mathis D. 1993. Mice lacking TdT: mature animals with an immature lymphocyte repertoire. Science. 261:1175–1178 10.1126/science.8356452 [DOI] [PubMed] [Google Scholar]
- Gu Y., Jin S., Gao Y., Weaver D.T., Alt F.W. 1997. Ku70-deficient embryonic stem cells have increased ionizing radiosensitivity, defective DNA end-binding activity, and inability to support V(D)J recombination. Proc. Natl. Acad. Sci. USA. 94:8076–8081 10.1073/pnas.94.15.8076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J., Lu H., Tippin B., Shimazaki N., Goodman M.F., Lieber M.R. 2007. XRCC4:DNA ligase IV can ligate incompatible DNA ends and can ligate across gaps. EMBO J. 26:1010–1023 10.1038/sj.emboj.7601559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guirouilh-Barbat J., Huck S., Bertrand P., Pirzio L., Desmaze C., Sabatier L., Lopez B.S. 2004. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol. Cell. 14:611–623 10.1016/j.molcel.2004.05.008 [DOI] [PubMed] [Google Scholar]
- Guirouilh-Barbat J., Rass E., Plo I., Bertrand P., Lopez B.S. 2007. Defects in XRCC4 and KU80 differentially affect the joining of distal nonhomologous ends. Proc. Natl. Acad. Sci. USA. 104:20902–20907 10.1073/pnas.0708541104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L., Yu K. 2008. Altered kinetics of nonhomologous end joining and class switch recombination in ligase IV—deficient B cells. J. Exp. Med. 205:2745–2753 10.1084/jem.20081623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honjo T., Muramatsu M., Fagarasan S. 2004. AID: how does it aid antibody diversity? Immunity. 20:659–668 10.1016/j.immuni.2004.05.011 [DOI] [PubMed] [Google Scholar]
- Kabotyanski E.B., Gomelsky L., Han J.O., Stamato T.D., Roth D.B. 1998. Double-strand break repair in Ku86- and XRCC4-deficient cells. Nucleic Acids Res. 26:5333–5342 10.1093/nar/26.23.5333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanjawala Z.E., Adachi N., Irvine R.A., Oh E.K., Shibata D., Schwarz K., Hsieh C.L., Lieber M.R. 2002. The embryonic lethality in DNA ligase IV-deficient mice is rescued by deletion of Ku: implications for unifying the heterogeneous phenotypes of NHEJ mutants. DNA Repair (Amst.). 1:1017–1026 10.1016/S1568-7864(02)00151-9 [DOI] [PubMed] [Google Scholar]
- Komori T., Okada A., Stewart V., Alt F.W. 1993. Lack of N regions in antigen receptor variable region genes of TdT-deficient lymphocytes. Science. 261:1171–1175 10.1126/science.8356451 [DOI] [PubMed] [Google Scholar]
- Komori T., Pricop L., Hatakeyama A., Bona C.A., Alt F.W. 1996. Repertoires of antigen receptors in Tdt congenitally deficient mice. Int. Rev. Immunol. 13:317–325 10.3109/08830189609061755 [DOI] [PubMed] [Google Scholar]
- Li Z., Otevrel T., Gao Y., Cheng H.L., Seed B., Stamato T.D., Taccioli G.E., Alt F.W. 1995. The XRCC4 gene encodes a novel protein involved in DNA double-strand break repair and V(D)J recombination. Cell. 83:1079–1089 10.1016/0092-8674(95)90135-3 [DOI] [PubMed] [Google Scholar]
- Lieber M.R., Lu H., Gu J., Schwarz K. 2008. Flexibility in the order of action and in the enzymology of the nuclease, polymerases, and ligase of vertebrate non-homologous DNA end joining: relevance to cancer, aging, and the immune system. Cell Res. 18:125–133 10.1038/cr.2007.108 [DOI] [PubMed] [Google Scholar]
- Manis J.P., Gu Y., Lansford R., Sonoda E., Ferrini R., Davidson L., Rajewsky K., Alt F.W. 1998. Ku70 is required for late B cell development and immunoglobulin heavy chain class switching. J. Exp. Med. 187:2081–2089 10.1084/jem.187.12.2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manis J.P., Dudley D., Kaylor L., Alt F.W. 2002. IgH class switch recombination to IgG1 in DNA-PKcs-deficient B cells. Immunity. 16:607–617 10.1016/S1074-7613(02)00306-0 [DOI] [PubMed] [Google Scholar]
- McVey M., Lee S.E. 2008. MMEJ repair of double-strand breaks (director’s cut): deleted sequences and alternative endings. Trends Genet. 24:529–538 10.1016/j.tig.2008.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelanda R., Schwers S., Sonoda E., Torres R.M., Nemazee D., Rajewsky K. 1997. Receptor editing in a transgenic mouse model: site, efficiency, and role in B cell tolerance and antibody diversification. Immunity. 7:765–775 10.1016/S1074-7613(00)80395-7 [DOI] [PubMed] [Google Scholar]
- Rass E., Grabarz A., Plo I., Gautier J., Bertrand P., Lopez B.S. 2009. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat. Struct. Mol. Biol. 16:819–824 10.1038/nsmb.1641 [DOI] [PubMed] [Google Scholar]
- Reina-San-Martin B., Difilippantonio S., Hanitsch L., Masilamani R.F., Nussenzweig A., Nussenzweig M.C. 2003. H2AX is required for recombination between immunoglobulin switch regions but not for intra-switch region recombination or somatic hypermutation. J. Exp. Med. 197:1767–1778 10.1084/jem.20030569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney S., Chaudhuri J., Alt F.W. 2004. The role of the non-homologous end-joining pathway in lymphocyte development. Immunol. Rev. 200:115–131 10.1111/j.0105-2896.2004.00165.x [DOI] [PubMed] [Google Scholar]
- Rooney S., Alt F.W., Sekiguchi J., Manis J.P. 2005. Artemis-independent functions of DNA-dependent protein kinase in Ig heavy chain class switch recombination and development. Proc. Natl. Acad. Sci. USA. 102:2471–2475 10.1073/pnas.0409857102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth D.B. 2002. Amplifying mechanisms of lymphomagenesis. Mol. Cell. 10:1–2 10.1016/S1097-2765(02)00573-7 [DOI] [PubMed] [Google Scholar]
- Rush J.S., Liu M., Odegard V.H., Unniraman S., Schatz D.G. 2005. Expression of activation-induced cytidine deaminase is regulated by cell division, providing a mechanistic basis for division-linked class switch recombination. Proc. Natl. Acad. Sci. USA. 102:13242–13247 10.1073/pnas.0502779102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda E., Pewzner-Jung Y., Schwers S., Taki S., Jung S., Eilat D., Rajewsky K. 1997. B cell development under the condition of allelic inclusion. Immunity. 6:225–233 10.1016/S1074-7613(00)80325-8 [DOI] [PubMed] [Google Scholar]
- Soulas-Sprauel P., Le Guyader G., Rivera-Munoz P., Abramowski V., Olivier-Martin C., Goujet-Zalc C., Charneau P., de Villartay J.P. 2007. Role for DNA repair factor XRCC4 in immunoglobulin class switch recombination. J. Exp. Med. 204:1717–1727 10.1084/jem.20070255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taccioli G.E., Gottlieb T.M., Blunt T., Priestley A., Demengeot J., Mizuta R., Lehmann A.R., Alt F.W., Jackson S.P., Jeggo P.A. 1994. Ku80: product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science. 265:1442–1445 10.1126/science.8073286 [DOI] [PubMed] [Google Scholar]
- Wang H., Perrault A.R., Takeda Y., Qin W., Wang H., Iliakis G. 2003. Biochemical evidence for Ku-independent backup pathways of NHEJ. Nucleic Acids Res. 31:5377–5388 10.1093/nar/gkg728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.H., Alt F.W., Gostissa M., Datta A., Murphy M., Alimzhanov M.B., Coakley K.M., Rajewsky K., Manis J.P., Yan C.T. 2008. Oncogenic transformation in the absence of Xrcc4 targets peripheral B cells that have undergone editing and switching. J. Exp. Med. 205:3079–3090 10.1084/jem.20082271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.H., Gostissa M., Yan C.T., Goff P., Hickernell T., Hansen E., Difilippantonio S., Wesemann D.R., Zarrin A.A., Rajewsky K., et al. 2009. Mechanisms promoting translocations in editing and switching peripheral B cells. Nature. 460:231–236 10.1038/nature08159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie A., Kwok A., Scully R. 2009. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat. Struct. Mol. Biol. 16:814–818 10.1038/nsmb.1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C.T., Boboila C., Souza E.K., Franco S., Hickernell T.R., Murphy M., Gumaste S., Geyer M., Zarrin A.A., Manis J.P., et al. 2007. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 449:478–482 10.1038/nature06020 [DOI] [PubMed] [Google Scholar]
- Zha S., Boboila C., Alt F.W. 2009. Mre11: roles in DNA repair beyond homologous recombination. Nat. Struct. Mol. Biol. 16:798–800 10.1038/nsmb0809-798 [DOI] [PubMed] [Google Scholar]
- Zhu C., Mills K.D., Ferguson D.O., Lee C., Manis J., Fleming J., Gao Y., Morton C.C., Alt F.W. 2002. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 109:811–821 10.1016/S0092-8674(02)00770-5 [DOI] [PubMed] [Google Scholar]