Abstract
Phospholipase Cγ1 (PLCγ1) is an important signaling effector of T cell receptor (TCR). To investigate the role of PLCγ1 in T cell biology, we generated and examined mice with T cell–specific deletion of PLCγ1. We demonstrate that PLCγ1 deficiency affects positive and negative selection, significantly reduces single-positive thymocytes and peripheral T cells, and impairs TCR-induced proliferation and cytokine production, and the activation of ERK, JNK, AP-1, NFAT, and NF-κB. Importantly, PLCγ1 deficiency impairs the development and function of FoxP3+ regulatory T cells, causing inflammatory/autoimmune symptoms. Therefore, PLCγ1 is essential for T cell development, activation, and tolerance.
During thymic T cell development, CD4+CD8+ double-positive (DP) thymocytes that express functional TCR are subjected to positive or negative selection and mature into CD4 or CD8 single-positive (SP) thymocytes (Starr et al., 2003). Thymic selection also leads to the development of FoxP3+ T regulatory (T reg) cells, which play a critical role in maintaining self-tolerance (Fontenot et al., 2003; Hori et al., 2003; Khattri et al., 2003). A major drive for thymic and mature T cell development are signals emanating from the TCR (Samelson, 2002). PLCγ1 is an essential effector molecule in TCR signal transduction, which, after activation, hydrolyzes the membrane lipid phosphatidylinositol 4,5-bisphosphate (PIP2) to generate diacylglycerol (DAG) and inositol 1,4,5-trisphosphate (IP3; Rhee, 2001). Whereas DAG activates the NF-κB and GRP–Ras–ERK pathways (Ebinu et al., 2000; Lin and Wang, 2004), IP3 mediates the elevation of Ca2+, which is essential for NFAT activation (Rao et al., 1997). The capability of PLCγ1 to regulate multiple signaling pathways and transcription factors raises considerable interest in the biological role of PLCγ1. However, embryonic lethality of PLCγ1-deficient mice precludes the analysis to determine the role of PLCγ1 in T cell biology in vivo (Ji et al., 1997). Here, we generate conditional PLCγ1-deficient mice, in which PLCγ1 deficiency is restricted to the T cell lineage. Our results demonstrate that PLCγ1 plays a critical and as yet unknown multifold role in T cell biology.
RESULTS AND DISCUSSION
Generation of PLCγ1-deficient mice
To avoid embryonic lethality caused by PLCγ1 deficiency, we modified PLCγ1 locus by “floxing” exons 2–4 of PLCγ1 (Fig. S1). The offspring that inherited the “floxed” PLCγ1 (PLCγ1fl/+) were bred with PLCγ1+/− mice (Ji et al., 1997). CD4Cre (Cre) transgene was introduced into the PLCγ1fl/− mice to mediate PLCγ1 deletion at thymic DP stage (Lee et al., 2001). PLCγ1 proteins were absent or substantially reduced in DP and SP thymocytes, and splenic T cells from Cre/PLCγ1fl/− compared with Cre/PLCγ1+/− mice (Fig. S2 A), and truncated PLCγ1 was not generated (not depicted). The residual PLCγ1 proteins in peripheral T cells of Cre/PLCγ1fl/− mice may reflect preferential survival and/or amplification of rare thymocytes that failed to delete the PLCγ1 gene. To better track deletion of the “floxed” PLCγ1, we used Rosa-26-YFP (YFP) mice (Srinivas et al., 2001). PLCγ1 expression was not observed in YFP+ thymocytes and splenic T cells (Fig. S2 B). Thus, YFP expression correlates with Cre-mediated deletion of the floxed PLCγ1 in T cells.
PLCγ1 deficiency impairs thymocyte maturation
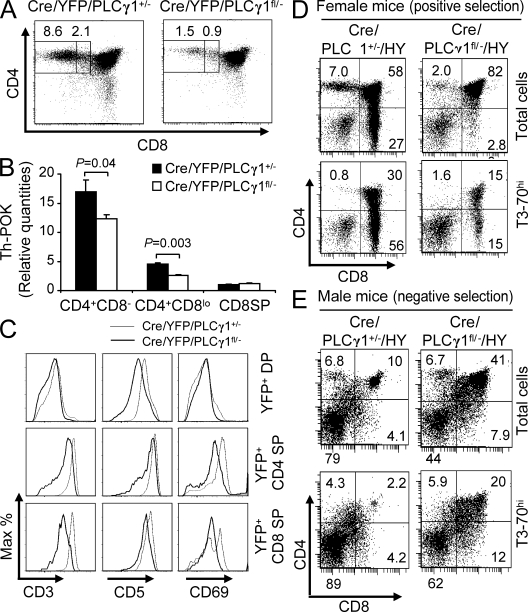
We examined T cell development in Cre/PLCγ1fl/− mice. PLCγ1+/−, PLCγ1fl/−, and Cre/PLCγ1+/− mice showed normal T cell development compared with WT mice, and thus served as experimental controls (Table I and not depicted). Total thymocyte number of Cre/PLCγ1fl/− mice was reduced compared with that of the control mice, but not statistically significant (Table I). Whereas the percentage of DN thymocytes was not significantly changed, the percentage of DP thymocytes was slightly but consistently increased in Cre/PLCγ1fl/− mice relative to controls (Table I). Moreover, there was a dramatic reduction in the percentages and absolute numbers of SP thymocytes derived from Cre/PLCγ1fl/− mice relative to controls, with CD4SP thymocytes more profoundly affected than CD8SP thymocytes (Table I). Further analysis showed that the percentage of the “bipotent” CD4+CD8loYFP+ thymocytes that differentiate into either CD4 or CD8 SP thymocytes was reduced by 55 ± 16% in Cre/YFP/PLCγ1fl/− mice compared with Cre/YFP/PLCγ1+/− mice. The percentage of CD4+CD8−YFP+ thymocytes was further reduced by 82 ± 5% (Fig. 1 A). Importantly, expression of Th-POK, the transcription factor that determines CD4 T cell commitment (He et al., 2005), was significantly lower in CD4+CD8loCD69+YFP+ and CD4+CD8−CD69+YFP+ thymocytes from Cre/YFP/PLCγ1fl/− relative to Cre/YFP/PLCγ1+/− mice (Fig. 1 B). Thus, PLCγ1 deficiency impaired the development of the bipotent CD4+CD8lo thymocytes, and further development of both CD4 and CD8 thymocytes. In addition, PLCγ1 deficiency reduced the up-regulation of Th-POK in CD4+CD8lo and CD4+CD8− thymocytes, providing one possible explanation for the greater affect on the CD4SP population. Finally, compared with Cre/YFP/PLCγ1+/− mice, Cre/YFP/PLCγ1fl/− mice lacked a small population of DP thymocytes that up-regulated CD3 and CD69 (Fig. 1 C), and had reduced CD5 up-regulation on DP thymocytes and CD3, CD5, and CD69 up-regulation on SP thymocytes (Fig. 1 C). These data are consistent with the notion that PLCγ1 deficiency reduces TCR signaling.
Table I.
Analysis of T cell populations in Cre/PLCγ1fl/− and the control mice
| Thymocytes | Splenocytes | ||||||||
| Total | DN | DP | CD4 SP | CD8 SP | Total | CD4+ | CD8+ | ||
| PLCγ1+/− (n = 6) | % | 1.5 ± 0.8 | 87 ± 2.8 | 8.5 ± 1.5 | 2.7 ± 1.2 | 22 ± 3.6 | 15 ± 2.9 | ||
| # (×106) | 216 ± 76 | 3.2 ± 2.0 | 189 ± 69 | 19 ± 8.3 | 6.4 ± 4.1 | 100 ± 46 | 22 ± 11 | 15 ± 6.6 | |
| PLCγ1fl/− (n = 6) | % | 1.1 ± 0.7 | 87 ± 4.1 | 9.4 ± 2.1 | 2.8 ± 1.5 | 18 ± 8.7 | 12 ± 5.1 | ||
| # (×106) | 238 ± 61 | 2.6 ± 1.5 | 223 ± 40 | 24 ± 6.4 | 6.9 ± 3.4 | 93 ± 42 | 17 ± 11 | 11 ± 7.6 | |
| Cre/PLCγ1+/− (n = 8) | % | 2 ± 0.8 | 87 ± 1.5 | 8.3 ± 1.5 | 2.9 ± 1.0 | 15 ± 5.3 | 12 ± 4.0 | ||
| # (×106) | 201 ± 88 | 3.6 ± 1.5 | 174 ± 72 | 17 ± 8.3 | 6.0 ± 4.0 | 118 ± 46 | 18 ± 8.6 | 13 ± 6.6 | |
| Cre/PLCγ1fl/− (n = 9) | % | 2.1 ± 1.2 | 95 ± 2.3a | 1.5 ± 0.43a | 1.2 ± 0.9b | 3.2 ± 1.5a | 5.6 ± 1.8c | ||
| # (×106) | 172 ± 81 | 3.7 ± 2.8 | 170 ± 72 | 2.7 ± 1.7c | 2.3 ± 2.0d | 77 ± 27 | 2.7 ± 2c | 4.1 ± 2.8b | |
The age of the mice analyzed was between 3 and 10 wk. Data presented are average percentage (%) or absolute number (#) of each T cell subset. The P value was calculated by comparing the percentages or absolute numbers of different T cell subsets from Cre/PLCγ1fl/− mice to those from Cre/PLCγ1+/− mice. There was no significant difference in the percentages and absolute numbers of different T cell subsets from PLCγ1+/−, PLCγ1fl/−, and Cre/PLCγ1+/− mice.
P < 0.0001.
P < 0.01.
P < 0.001.
P < 0.05.
Figure 1.
PLCγ1 deficiency impairs thymocyte maturation. (A) PLCγ1 deficiency reduced the percentages of CD4+CD8lo and CD4+CD8− thymocytes. Data represent six pairs of mice. (B) PLCγ1 deficiency reduced Th-POK expression in CD4+CD8loCD69+ and CD4+CD8−CD69+ thymocytes. Data represent two independent experiments on four Cre/YFP/PLCγ1fl/− and four Cre/YFP/PLCγ1+/− mice. (C) PLCγ1 deficiency reduced CD3, CD5, and CD69 expression levels on DP and SP thymocytes. Data represent three pairs of mice. (D) PLCγ1 deficiency blocks positive selection. CD4/CD8 expression profiles on total or T3-70hi thymocytes from female Cre/PLCγ1+/−/HY and Cre/PLCγ1fl/−/HY mice. Data represent three pairs of mice. (E) PLCγ1 deficiency impairs negative selection. CD4/CD8 expression profiles on total or T3-70hi thymocytes from male Cre/PLCγ1+/−/HY and Cre/PLCγ1fl/−/HY mice. Data represent five Cre/PLCγ1+/−/HY and six Cre/PLCγ1fl/−/HY mice.
As PLCγ1 deficiency resulted in reduction of SP thymocytes, we further examined the role of PLCγ1 in thymic positive and negative selection using HY TCR transgenic mice. Each experimental mouse carried a single copy of the HY transgene. Positive selection in female Cre/PLCγ1+/−/HY mice resulted in high CD8SP thymocyte percentages in total (20 ± 8.2%) and T3-70hi (54 ± 12%) cells (Fig. 1 D). In contrast, female Cre/PLCγ1fl/−/HY mice displayed a significant reduction in CD8SP thymocyte percentages in total (2.8 ± 0.66%, P = 0.02) and T3-70hi (19 ± 9.0%, P = 0.01) cells. Therefore, PLCγ1 deficiency blocks positive selection. In male Cre/PLCγ1+/−/HY mice, negative selection significantly reduced thymic cellularity (3.8 ± 2.0 × 106) and DP thymocyte percentages within total (6.3 ± 4.0%) and T3-70hi (2.3 ± 1.3%) cells, with an increase in DN thymocyte percentages within total (79 ± 4.0%) and T3-70hi (85 ± 6.2%) cells (Fig. 1 E). In comparison, male Cre/PLCγ1fl/−/HY mice had a markedly increased thymic cellularity (11 ± 4.6 × 106, P = 0.01) and DP thymocyte percentages within total (42 ± 11%, P < 0.001) and T3-70hi (15 ± 7.7%, P < 0.01) cells, with a reduction in DN thymocyte percentages within total (45 ± 9.8%, P < 0.001) and T3-70hi (68 ± 9.4%, P < 0.01) cells (Fig. 1 E). In addition, the CD8SP cells in the T3-70hi population was increased in male Cre/PLCγ1fl/−/HY mice (14 ± 2.6%) compared with male Cre/PLCγ1+/−/HY mice (8.7 ± 3.4%, P = 0.01; Fig. 1 E). These data indicated a failure of negative selection and possible conversion to positive selection in the absence of PLCγ1.
PLCγ1 deficiency results in peripheral T cell lymphopenia
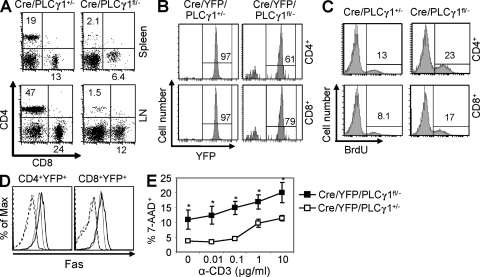
Cre/PLCγ1fl/− mice had substantial reduction in peripheral T cells (Fig. 2 A and Table I). YFP+ percentage was also substantially reduced in T cells from Cre/YFP/PLCγ1fl/− (CD4+: 57 ± 5.1%; CD8+: 78 ± 9.2%) relative to Cre/YFP/PLCγ1+/− mice (CD4+: 97 ± 1.3%, P < 0.001; CD8+: 98 ± 1.1%, P < 0.001; Fig. 2 B), suggesting that PLCγ1 deficiency caused T cell incompetence. This possibility was supported by competitive BM transplantation experiments. When cotransplanted with WT BM cells, Cre/YFP/PLCγ1fl/− BM cells, indicated by YFP expression, contributed significantly less to T cells than Cre/YFP/PLCγ1+/− BM cells, with peripheral T cells more severely affected than thymocytes (Table S1). Together, these data indicated that PLCγ1-deficient thymocytes and peripheral T cells are less competent compared with PLCγ1-sufficient T cells.
Figure 2.
PLCγ1 deficiency results in T cell lymphopenia. (A) CD4/CD8 expression profiles on spleen and lymph node T cells from Cre/PLCγ1+/− and Cre/PLCγ1fl/− mice. Data represent six pairs of mice. (B) Percentages of YFP+ cells in splenic CD4+ and CD8+ populations from Cre/YFP/PLCγ1+/− and Cre/YFP/PLCγ1fl/− mice. Data represent five pairs of mice. (C) PLCγ1-deficient peripheral T cells displayed higher rates of BrdU incorporation. Data represent three pairs of mice. (D) PLCγ1-deficient T cells showed increased Fas expression. Splenocytes from Cre/YFP/PLCγ1+/− (thin line) or Cre/YFP/PLCγ1fl/− (thick line) mice were examined for Fas expression. Dashed lines represent isotype control. Data represent four pairs of mice. (E) PLCγ1-deficient T cells are more susceptible to AICD. 7-AAD+ cells are examined in CD4+YFP+ populations. Each data point is the mean of data derived from three Cre/YFP/PLCγ1+/− or five Cre/YFP/PLCγ1fl/− mice. *, P < 0.01.
To examine the mechanism of T cell lymphopenia, we performed TUNEL and BrdU incorporation assays. T cells from Cre/PLCγ1fl/− and Cre/PLCγ1+/− mice showed comparable levels of TUNEL+ cells (unpublished data). Compared with Cre/PLCγ1+/− mice, Cre/PLCγ1fl/− mice contained higher percentages of BrdU+ cells in SP thymocytes and splenic T cells (Fig. 2 C and not depicted), which could be caused by increased cell proliferation and/or apoptosis. Further examination showed that Fas expression was increased on T cells from Cre/YFP/PLCγ1fl/− relative to Cre/YFP/PLCγ1+/− mice (Fig. 2 D), whereas FasL expression was comparable (not depicted). In addition, reactivation resulted in significantly higher cell death in CD4+ T cells from Cre/YFP/PLCγ1fl/− mice compared with Cre/YFP/PLCγ1+/− mice (Fig. 2 E), suggesting that PLCγ1-deficient T cells are more susceptible to activation-induced cell death (AICD), which is consistent with increased levels of Fas expression. Thus, increased AICD may contribute to T cell lymphopenia in PLCγ1-deficient mice.
PLCγ1 is essential for TCR-mediated proliferation and cytokine production
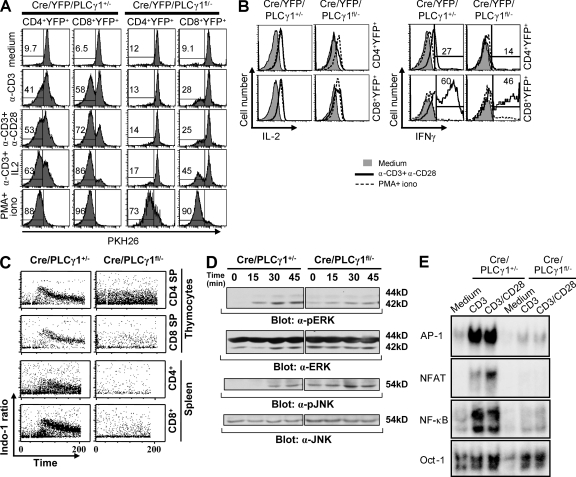
T cells from Cre/YFP/PLCγ1fl/− mice exhibited dramatic reduction of proliferation in response to stimulation with anti-CD3, anti-CD3/anti-CD28, or anti-CD3/IL-2 compared with those from Cre/YFP/PLCγ1+/− mice, and this reduction was largely restored by PMA/ionomycin stimulation (Fig. 3 A). Therefore, PLCγ1 is critical for TCR-mediated proliferation. In addition, compared with those from Cre/YFP/PLCγ1+/− mice, T cells from Cre/YFP/PLCγ1fl/− mice (n = 3) displayed significant reduction in anti-CD3/anti-CD28–induced IL-2 production by CD4+ (Cre/YFP/PLCγ1+/−: mean fluorescence intensity (MFI) = 1741 ± 80; Cre/YFP/PLCγ1fl/−: MFI = 899 ± 98, P < 0.01) and CD8+ (Cre/YFP/PLCγ1+/−: MFI = 1685 ± 69; Cre/YFP/PLCγ1fl/−: MFI = 1102 ± 119, P < 0.01) T cells (Fig. 3 B). The percentages of IFN-γ+ T cells from Cre/YFP/PLCγ1fl/− mice after anti-CD3/anti-CD28 stimulation were also reduced, but not significant in either CD4+ (Cre/YFP/PLCγ1+/−: 25 ± 16%; Cre/YFP/PLCγ1fl/−: 15 ± 2.5%, P = 0.3) or CD8+ (Cre/YFP/PLCγ1+/−: 54 ± 6.0%; Cre/YFP/PLCγ1fl/−: 38 ± 15%, P = 0.1) T cells (Fig. 3 B). Without differentiation, IL-4 was not detected in CD4+ T cells, regardless of PLCγ1 expression (unpublished data). Therefore, whereas PLCγ1 deficiency significantly reduces IL-2 production, it only slightly reduces IFN-γ production by T cells.
Figure 3.
The effect of PLCγ1 deficiency on proliferation, cytokine production, and TCR signaling. (A) PLCγ1 deficiency impairs TCR-mediated proliferation. (B) The effect of PLCγ1 deficiency on IL-2 and IFN-γ production. (C) PLCγ1 deficiency impairs TCR-mediated Ca2+ mobilization in both SP thymocytes and splenic T cells. (D) PLCγ1 deficiency impairs TCR-mediated ERK and JNK activation. (E) PLCγ1 deficiency impairs TCR-mediated AP-1, NFAT, and NF-κB activation. All data shown are representative of three independent experiments. The numbers shown in B represent IFN-γ+ T cells from one pair of mice after anti-CD3/anti-CD28 stimulation.
PLCγ1 deficiency impairs TCR signaling
We further examined the effect of PLCγ1 deficiency on TCR-mediated signaling. Compared with Cre/PLCγ1+/− mice, T cells from Cre/PLCγ1fl/− mice displayed markedly reduced TCR-induced Ca2+ flux, but higher basal Ca2+ level in the CD4SP thymocytes (Fig. 3 C). Although basal level activities of ERK and JNK were high in T cells from Cre/PLCγ1fl/− mice, TCR-mediated activation of these kinases was reduced compared with those from Cre/PLCγ1+/− mice (Fig. 3 D). Moreover, regardless of CD28 stimulation, TCR-induced activation of transcription factors NFAT, NF-κB, and AP-1 was severely impaired in T cells from Cre/PLCγ1fl/− relative to Cre/PLCγ1+/− mice (Fig. 3 E). Collectively, these data demonstrate that PLCγ1 plays a central role in TCR-mediated activation of multiple signaling pathways.
PLCγ1-deficient mice develop inflammatory/autoimmune disease
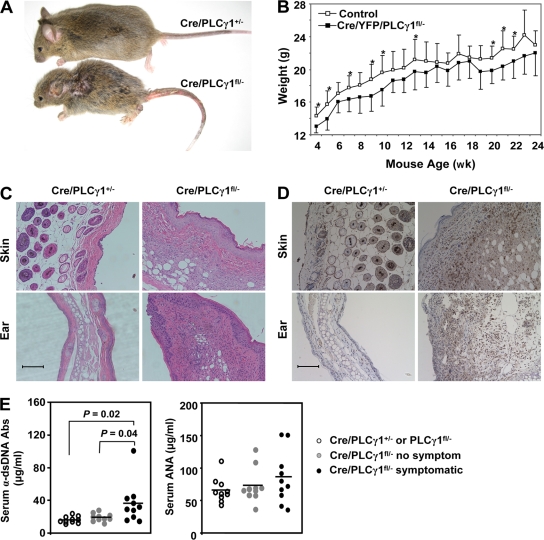
From 2–3 wk of age, most Cre/PLCγ1fl/− mice were apparently runt (Fig. 4 A), with reduced weight in both male and female mice compared with littermate control mice (Fig. 4 B and not depicted). One third of Cre/PLCγ1fl/− mice developed variable visible symptoms, including alopecia, dermatitis, and rectal prolapse (symptomatic mice), with the latter two symptoms more frequent. Whereas none of the littermate control mice exhibited significant histological abnormalities, a majority of symptomatic Cre/PLCγ1fl/− mice showed infiltration of mononuclear cells into variable tissues, particularly skin and ear (Fig. 4 C). Although Cre/PLCγ1fl/− mice were severely T cell lymphopenic, the skin and ear were infiltrated by CD3+ T cells, which was not observed in the corresponding areas of the littermate control mice (Fig. 4 D). Serum anti–double-stranded DNA antibodies were significantly higher in symptomatic Cre/PLCγ1fl/− relative to nonsymptomatic Cre/PLCγ1fl/− or control mice (Fig. 4 E). Nevertheless, serum antinuclear antibodies were comparable (Fig. 4 E). The experimental mice analyzed were backcrossed with C57BL/6 mice for three generations. We have since determined that Cre/PLCγ1fl/− mice backcrossed with C57BL/6 mice for eight generations displayed the same phenotypes (unpublished data). Although we cannot entirely exclude background gene effects, we can conclude that the inflammatory/autoimmune syndrome in Cre/PLCγ1fl/− mice is attributable to PLCγ1 deficiency.
Figure 4.
Cre/PLCγ1fl/− mice develop inflammatory/autoimmune disease. (A) Cre/PLCγ1fl/− mice were smaller than littermate control mice. A pair of 6-wk-old littermates is shown. (B) PLCγ1 deficiency reduced weight gain in Cre/PLCγ1fl/− mice. Each point represents the mean weight of 7 to 14 female mice. *: P < 0.05. (C) Infiltration of inflammatory cells in the skin and ear (H&E, x200) of Cre/PLCγ1fl/− mice. Bar, 100 µm. (D) Infiltration of CD3+ T cells in the skin and ear (x200) of Cre/PLCγ1fl/− mice. The slides were analyzed by immunohistochemistry and examined by Nikon Eclipse E600. Bar, 100 µm. (E) Levels of α-double-stranded DNA antibodies and antinuclear antibodies in the serum of symptomatic Cre/PLCγ1fl/− mice compared with the indicated control mice.
PLCγ1 deficiency impairs T reg cell development and function
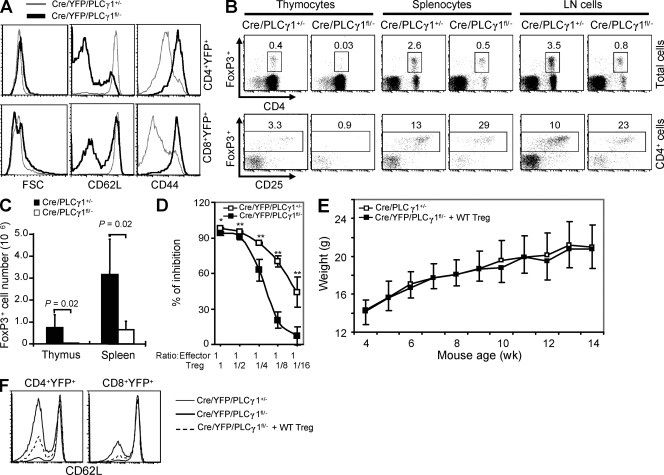
T cells in Cre/YFP/PLCγ1fl/− mice showed activated phenotype, including larger cell sizes, decreased CD62L, and increased CD44 expression compared with those in Cre/YFP/PLCγ1+/− mice (Fig. 5 A). T cell lymphopenia and the activated phenotype of PLCγ1-deficient T cells were largely corrected in mice receiving Cre/YFP/PLCγ1fl/− and WT BM cells (Fig. S3). Thus, T cell lymphopenia may contribute to the activated T cell phenotype in PLCγ1-deficient mice. Alternatively, given that the symptoms of Cre/PLCγ1fl/− mice are reminiscent of mice deficient in FoxP3+ T reg cells (Fontenot et al., 2003; Hori et al., 2003; Khattri et al., 2003), the autoimmune phenotype may be consequent to impaired T reg cells. The percentage and absolute number of T reg cells were substantially reduced in thymus in Cre/PLCγ1fl/− relative to Cre/PLCγ1+/− mice (Fig. 5, B and C). Although T reg cell percentage was reduced in lymphocytes in the spleen and lymph nodes of Cre/PLCγ1fl/− relative to Cre/PLCγ1+/− mice, it was substantially increased in CD4+ T cells (Fig. 5 B). Nevertheless, T reg cell number was dramatically reduced in the spleen from Cre/PLCγ1fl/− relative to Cre/PLCγ1+/− mice (Fig. 5 C). We performed suppression assays to determine T reg cell suppressive functions. Compared with those from Cre/YFP/PLCγ1+/− mice, YFP+ T reg cells from Cre/YFP/PLCγ1fl/− mice had reduced ability to suppress naive T cell proliferation (Fig. 5 D). Furthermore, we transferred purified WT T reg cells into 10 10-d-old Cre/YFP/PLCγ1fl/− mice to determine whether impaired T reg cells contributed to the disease phenotype in PLCγ1-deficient mice. None of them developed symptoms such as alopecia, dermatitis, and rectal prolapse. These mice gained weight at a rate similar to WT mice (Fig. 5 E). WT T reg cells also partially corrected the activated phenotype of Cre/YFP/PLCγ1fl/− T cells, as CD62L expression was increased (Fig. 5 F). Collectively, PLCγ1 deficiency impaired T reg cell development and functions, which may contribute to the inflammatory/autoimmune disease in the PLCγ1-deficient mice.
Figure 5.
PLCγ1 deficiency impairs T reg cell development and function. (A) PLCγ1-deficient T cells display activated phenotype. Data are representative of four pairs of mice. (B) Percentages of FoxP3+ T reg cells in total and CD4-gated lymphocytes in the thymus, spleen, and lymph node cells. Data represent five pairs of mice. (C) CD4+FoxP3+ cell numbers were reduced in thymus and spleen of Cre/PLCγ1fl/− mice. Data represent five pairs of mice. (D) PLCγ1 deficiency impairs the inhibitory functions of T reg cells. Error bars represent the standard deviation of triplicate measurements of each data point. *: P < 0.05, **: P < 0.01. Data are representative of two independent experiments. (E) WT T reg cell reconstitution restores normal weight gain in Cre/PLCγ1fl/− mice. Each point represents the mean weight of 7 to 14 female Cre/PLCγ1+/− mice or 6 female Cre/PLCγ1fl/− mice that received WT T reg cells. (F) WT T reg cell reconstitution restores high CD62L expression on PLCγ1-deficient T cells. Data represent three independent experiments.
The development of inflammatory/autoimmunity in Cre/PLCγ1fl/− mice could be attributed to a couple of possibilities. First, impairment of thymic negative selection allows autoreactive thymocytes that would normally be eliminated, to survive and mature into periphery and become pathogenic T cells. To determine whether PLCγ1 deficiency altered the T cell TCR repertoire, we performed spectratyping analysis of 18 Vβ subfamilies (Maślanka et al., 1995) on cDNA isolated from Cre/YFP/PLCγ1fl/− and Cre/YFP/PLCγ1+/− thymocytes. The CD4SPYFP+ cells from either of the mice showed a Gaussian distribution of CDR3 length commensurate with normal selection (unpublished data). Interestingly, the CDR3 length profiles of CD8SPYFP+ cells from Cre/YFP/PLCγ1fl/− mice differed from the Gaussian profile seen in the control PLCγ1+/− mice for several subfamilies examined (unpublished data). Thus, while PLCγ1 deficiency affects the naive CD8 repertoire, the effect, if any, on naive CD4 repertoire involves more subtle changes that do not alter the overall distribution of CDR3 lengths. Second, impaired function of T regs in Cre/PLCγ1fl/− mice also contribute to the autoimmune phenotype, as shown in mice deficient in T reg cells (Fontenot et al., 2003; Hori et al., 2003; Khattri et al., 2003; Lin et al., 2007). Consistently, knock-in mice homozygous for a single tyrosine mutation in LAT (Y136F), which impairs PLCγ1 activation, develop signs of autoimmune disease (Aguado et al., 2002; Sommers et al., 2002), which is attributed to impaired negative selection and T reg cells (Sommers et al., 2005; Koonpaew et al., 2006). Despite the similarities between LAT (Y136F) knock-in and Cre/PLCγ1fl/− mice, these two mutant mice display several significant differences. First, lymphoproliferative disorder, particularly of CD4+ T cells in LAT (Y136F) knock-in mice was in contrast to the severe lymphopenia, particularly of CD4+ T cells in Cre/PLCγ1fl/− mice. Second, CD4+ T cells from LAT (Y136F) knock-in mice exhibit substantial IL-4 production without in vitro differentiation, whereas those from Cre/PLCγ1fl/− mice do not produce IL-4. Lastly, T cells derived from LAT (Y136F) knock-in mice have normal ERK activation, whereas those from Cre/PLCγ1fl/− mice have markedly impaired ERK activation. These differences emphasize that Y136F mutation in LAT is not equivalent to the deletion of PLCγ1. Cre/PLCγ1fl/− mice are unique in uncovering the biological function of PLCγ1 in T cells. The impaired function of PLCγ1-deficient T reg cells is likely attributed to defective TCR-mediated Ca2+ flux and NFAT activation, because T cells deficient for STIM1 and STIM2 also displayed impairment of T reg functions (Oh-Hora et al., 2008). In addition, T reg function is mediated by an interaction between NFAT and FOXP3, disruption of which substantially inhibited T reg suppressive function (Wu et al., 2006).
PLCγ1 deficiency affects the development of CD4+ T cells more profoundly than that of CD8+ T cells, resulting in lower CD4/CD8 ratio in both the thymus and periphery of Cre/PLCγ1fl/− mice relative to controls, reminiscent of mice lacking both Itk and Rlk (Broussard et al., 2006). Nevertheless, the CD8SP thymocytes in PLCγ-deficient mice appeared not to be the innate type CD8+ T cells (CD122+) developed in the Itk and Rlk double-deficient mice, as they were CD122− (unpublished data). In addition, percentages of CD4+ T cells were not reduced in the Itk and Rlk double-deficient mice (Broussard et al., 2006). It is possible that CD4+ and CD8+ cells display a different degree of PLCγ1 dependence. It is proposed in the kinetic signaling model that proper TCR signaling at the thymic DP stage results in positive selection and a reduction in the intensity of CD8, leading to an appearance of the bipotent CD4+CD8lo thymocytes (Singer, 2002). Extended TCR signaling in the CD4+CD8lo thymocytes, a sign of MHC class II recognition, results in Th-POK expression, which in turn leads to differentiation into CD4 lineage. In contrast, attenuated TCR signaling as a result of CD8 expression loss, denoting MHC class I recognition, leads to CD8 T cell development. Our data showed that PLCγ1 is required for CD4+CD8lo thymocyte development as well as Th-POK expression in CD4+CD8lo and CD4+CD8− thymocytes. Consequently, PLCγ1 deficiency impairs both CD4 and CD8 development with CD4 lineage more affected.
The reason for elevated basal calcium and MAPK activity is not clear. PLCγ1-deficient T cells had activated phenotype, which may account for elevated basal Ca2+ and MAPK activity. Our study showed that both lymphopenia and impaired T reg cells may contribute to the activated T cell phenotype. In addition, Cbl-b up-regulation was severely impaired in PLCγ1-deficient T cells (unpublished data). Several studies have shown that Cbl-b plays an important role in down-regulating TCR expression and peripheral T cell activation (Bachmaier et al., 2000; Chiang et al., 2000; Naramura et al., 2002). In addition, a previous study showed that CD5 deficiency enhances PLCγ1 activation (Tarakhovsky et al., 1995). Thus, impaired CD5 up-regulation in PLCγ1-deficient T cells could enhance the activation of PLCγ2, which is involved in TCR signal transduction. It is possible that enhanced basal level Ca2+ and Erk activation may be the consequence of peripheral lymphopenia, impaired T reg cell number/functions, and defective CD5 and Cbl up-regulation.
In conclusion, we observed that PLCγ1 deficiency severely impairs TCR-induced activation of multiple signaling molecules and transcription factors, and unveiled a crucial role of PLCγ1 in T cell development, activation, and tolerance.
MATERIALS AND METHODS
Mice.
HY TCR and CD4Cre transgenic mice were purchased from Taconic. PLCγ1+/− and Rosa-26-YFP mice were generously provided by G. Carpenter (Vanderbilt University, Nashville, TN; Ji et al., 1997) and F. Costantini (Columbia University, New York, NY; Srinivas et al., 2001), respectively. PLCγ1-deficient and control mice were originally on a mixed 129xC57BL/6 background. The experimental mice were backcrossed onto the C57BL/6 background for three generations. Mice analyzed were between 6 and 12 wk old unless otherwise stated. Mice were maintained in the Biological Resource Center at the Medical College of Wisconsin (MCW). All animal protocols were approved by the MCW Institutional Animal Care and Use Committee.
Cell proliferation and intracellular cytokine analysis.
Splenocytes were labeled with PKH Fluorescent Cell Linker Dye (PKH26; Sigma-Aldrich) according to the manufacturer’s recommendation and stimulated for 72 h before analysis for cell proliferation. For intracellular cytokine staining, total splenocytes (1.5 × 106/ml) were stimulated for 36 h, followed by 6 h of monensin treatment. The cells were then harvested and stained with α-CD4 and α-CD8 antibodies, followed by intracellular staining for IL-2, IL-4, and IFN-γ according to manufacture’s recommendation (eBioscience).
Real-time PCR analysis of Th-POK expression.
CD4+CD8loCD69+YFP+ and CD4+CD8−CD69+YFP+ thymocytes were purified from Cre/YFP/PLCγ1fl/− and Cre/YFP/PLCγ1+/− mice and total RNA was extracted. Th-POK expression was examined by real-time PCR as previously described (He et al., 2005).
Calcium flux analysis.
Thymocytes or splenocytes (2 × 106) were loaded with indo-1AM in 1 ml PBS (2% FBS + 10 µg/ml indo-1AM) for 30 min. The cells were then incubated with FITC-α-CD4 + PE-α-CD8 + biotin-α-CD3 (20 µg/ml) in 200 µl PBS (2% FBS) for 15 min. After washing, the cells were resuspended in 1 ml medium and run on a LSRII (BD). Baseline data were collected for 30 s, streptavidin (Thermo Fisher Scientific) was added to a final concentration of 8 µg/ml to cross-link the TCR, and data were then collected by the LSRII for another 9 min.
BrdU incorporation assay.
Mice were injected i.p. with 1 mg BrdU in 200 µl PBS at 12-h intervals for 5 d. Splenocytes were then prepared for analysis. The BrdU staining was performed according to the manufacturer’s instructions (BD).
AICD.
3 × 106 purified CD4+ peripheral T cells were stimulated with plate-bound α-CD3 (5 µg/ml α-CD3 coated) in 1 ml of T cell medium for 2 d, and the cells were collected and cultured at 3 × 106 cells/ml in T cell medium in the presence of 30 U/ml IL-2 for an additional day. The cells were then collected and further cultured at 106 cells/ml in media alone or stimulated with plate-bound α-CD3 (0.01, 0.1, 1, and 5 µg/ml α-CD3 coated) for 1 d. 24 h later, the cells were collected and stained for CD4 and CD8 and resuspended in PBS with 7-AAD (1 µg/ml) and analyzed by FACS.
T reg suppression assay.
Naive CD4+ T cells (CD4+CD25−, 2 × 104) pooled from spleen and lymph node were stimulated with 2 µg/ml α-CD3 in the presence of 8 × 104 irradiated (3000Rad) splenocytes in 200 µl culture medium in a round-bottomed plate. Indicated portions of FACS-sorted CD4+CD25+YFP+ T reg cells from the spleen and lymph nodes of Cre/YFP/PLCγ1+/− or Cre/YFP/PLCγ1fl/− mice were added at the beginning of the culture. After 48 h of culture, the cells were pulsed and harvested as previously described (Zeng et al., 2008).
T reg transfer experiment.
WT T reg cells were sorted based on CD4 and GFP expression from Foxp3EGFP mice, in which a bicistronic locus encoding both Foxp3 and EGFP was under the control of the Foxp3 promoter (Haribhai et al., 2007). Each 10-d-old Cre/YFP/PLCγ1fl/− pup received 5 × 105 purified WT T reg cells through i.p. injection and were weighed and observed for 3 mo afterward.
Statistical analysis.
All P values were calculated according to two-tailed Student’s t test analysis. Datasets are presented as mean ± SD.
Online supplemental material.
Fig. S1 describes strategies for T-lineage–specific deletion of PLCγ1. Fig. S2 shows PLCγ1 protein levels in T cells derived from Cre/PLCγ1fl/− and Cre/YFP/PLCγ1fl/− and control mice. Fig. S3 demonstrates that the activated phenotype is not an intrinsic phenotype of the PLCγ1-deficient T cells. Table S1 demonstrates that PLCγ1-deficient T cells are less competitive than WT T cells in the periphery. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20090880/DC1.
Acknowledgments
We would like to thank T.L. Heil for editing of the manuscript, and H. Albert and C. Reinbold for technical assistance.
This work was supported in part by National Institutes of Health (NIH) RO1 AI052327 (R. Wen), NIH U19 AI062627 pilot grant (R. Wen), American Cancer Society Pilot Project Grant IRG-86-004 (R. Wen), NIH RO1 HL073284 (D.W.), NIH RO1 AI079087 (D. Wang), and Scholar Award from the Leukemic and Lymphoma Society (D. Wang).
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- AICD
- activation-induced cell death
- DAG
- diacylglycerol
- DP
- double-positive
- IP3
- inositol 1,4,5-trisphosphate
- MFI
- mean fluorescence intensity
- PIP2
- phosphatidylinositol 4,5-bisphosphate
- SP
- single-positive
References
- Aguado E., Richelme S., Nuñez-Cruz S., Miazek A., Mura A.M., Richelme M., Guo X.J., Sainty D., He H.T., Malissen B., Malissen M. 2002. Induction of T helper type 2 immunity by a point mutation in the LAT adaptor. Science. 296:2036–2040 10.1126/science.1069057 [DOI] [PubMed] [Google Scholar]
- Bachmaier K., Krawczyk C., Kozieradzki I., Kong Y.Y., Sasaki T., Oliveira-dos-Santos A., Mariathasan S., Bouchard D., Wakeham A., Itie A., et al. 2000. Negative regulation of lymphocyte activation and autoimmunity by the molecular adaptor Cbl-b. Nature. 403:211–216 10.1038/35003228 [DOI] [PubMed] [Google Scholar]
- Broussard C., Fleischacker C., Fleischecker C., Horai R., Chetana M., Venegas A.M., Sharp L.L., Hedrick S.M., Fowlkes B.J., Schwartzberg P.L. 2006. Altered development of CD8+ T cell lineages in mice deficient for the Tec kinases Itk and Rlk. Immunity. 25:93–104 10.1016/j.immuni.2006.05.011 [DOI] [PubMed] [Google Scholar]
- Chiang Y.J., Kole H.K., Brown K., Naramura M., Fukuhara S., Hu R.J., Jang I.K., Gutkind J.S., Shevach E., Gu H. 2000. Cbl-b regulates the CD28 dependence of T-cell activation. Nature. 403:216–220 10.1038/35003235 [DOI] [PubMed] [Google Scholar]
- Ebinu J.O., Stang S.L., Teixeira C., Bottorff D.A., Hooton J., Blumberg P.M., Barry M., Bleakley R.C., Ostergaard H.L., Stone J.C. 2000. RasGRP links T-cell receptor signaling to Ras. Blood. 95:3199–3203 [PubMed] [Google Scholar]
- Fontenot J.D., Gavin M.A., Rudensky A.Y. 2003. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 4:330–336 10.1038/ni904 [DOI] [PubMed] [Google Scholar]
- Haribhai D., Lin W., Relland L.M., Truong N., Williams C.B., Chatila T.A. 2007. Regulatory T cells dynamically control the primary immune response to foreign antigen. J. Immunol. 178:2961–2972 [DOI] [PubMed] [Google Scholar]
- He X., He X., Dave V.P., Zhang Y., Hua X., Nicolas E., Xu W., Roe B.A., Kappes D.J. 2005. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 433:826–833 10.1038/nature03338 [DOI] [PubMed] [Google Scholar]
- Hori S., Nomura T., Sakaguchi S. 2003. Control of regulatory T cell development by the transcription factor Foxp3. Science. 299:1057–1061 10.1126/science.1079490 [DOI] [PubMed] [Google Scholar]
- Ji Q.S., Winnier G.E., Niswender K.D., Horstman D., Wisdom R., Magnuson M.A., Carpenter G. 1997. Essential role of the tyrosine kinase substrate phospholipase C-gamma1 in mammalian growth and development. Proc. Natl. Acad. Sci. USA. 94:2999–3003 10.1073/pnas.94.7.2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khattri R., Cox T., Yasayko S.A., Ramsdell F. 2003. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat. Immunol. 4:337–342 10.1038/ni909 [DOI] [PubMed] [Google Scholar]
- Koonpaew S., Shen S., Flowers L., Zhang W. 2006. LAT-mediated signaling in CD4+CD25+ regulatory T cell development. J. Exp. Med. 203:119–129 10.1084/jem.20050903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P.P., Fitzpatrick D.R., Beard C., Jessup H.K., Lehar S., Makar K.W., Pérez-Melgosa M., Sweetser M.T., Schlissel M.S., Nguyen S., et al. 2001. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 15:763–774 10.1016/S1074-7613(01)00227-8 [DOI] [PubMed] [Google Scholar]
- Lin X., Wang D. 2004. The roles of CARMA1, Bcl10, and MALT1 in antigen receptor signaling. Semin. Immunol. 16:429–435 10.1016/j.smim.2004.08.022 [DOI] [PubMed] [Google Scholar]
- Lin W., Haribhai D., Relland L.M., Truong N., Carlson M.R., Williams C.B., Chatila T.A. 2007. Regulatory T cell development in the absence of functional Foxp3. Nat. Immunol. 8:359–368 10.1038/ni1445 [DOI] [PubMed] [Google Scholar]
- Maślanka K., Piatek T., Gorski J., Yassai M., Gorski J. 1995. Molecular analysis of T cell repertoires. Spectratypes generated by multiplex polymerase chain reaction and evaluated by radioactivity or fluorescence. Hum. Immunol. 44:28–34 10.1016/0198-8859(95)00056-A [DOI] [PubMed] [Google Scholar]
- Naramura M., Jang I.K., Kole H., Huang F., Haines D., Gu H. 2002. c-Cbl and Cbl-b regulate T cell responsiveness by promoting ligand-induced TCR down-modulation. Nat. Immunol. 3:1192–1199 10.1038/ni855 [DOI] [PubMed] [Google Scholar]
- Oh-Hora M., Yamashita M., Hogan P.G., Sharma S., Lamperti E., Chung W., Prakriya M., Feske S., Rao A. 2008. Dual functions for the endoplasmic reticulum calcium sensors STIM1 and STIM2 in T cell activation and tolerance. Nat. Immunol. 9:432–443 10.1038/ni1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao A., Luo C., Hogan P.G. 1997. Transcription factors of the NFAT family: regulation and function. Annu. Rev. Immunol. 15:707–747 10.1146/annurev.immunol.15.1.707 [DOI] [PubMed] [Google Scholar]
- Rhee S.G. 2001. Regulation of phosphoinositide-specific phospholipase C. Annu. Rev. Biochem. 70:281–312 10.1146/annurev.biochem.70.1.281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samelson L.E. 2002. Signal transduction mediated by the T cell antigen receptor: the role of adapter proteins. Annu. Rev. Immunol. 20:371–394 10.1146/annurev.immunol.20.092601.111357 [DOI] [PubMed] [Google Scholar]
- Singer A. 2002. New perspectives on a developmental dilemma: the kinetic signaling model and the importance of signal duration for the CD4/CD8 lineage decision. Curr. Opin. Immunol. 14:207–215 10.1016/S0952-7915(02)00323-0 [DOI] [PubMed] [Google Scholar]
- Sommers C.L., Park C.-S., Lee J., Feng C., Fuller C.L., Grinberg A., Hildebrand J.A., Lacaná E., Menon R.K., Shores E.W., et al. 2002. A LAT mutation that inhibits T cell development yet induces lymphoproliferation. Science. 296:2040–2043 10.1126/science.1069066 [DOI] [PubMed] [Google Scholar]
- Sommers C.L., Lee J., Steiner K.L., Gurson J.M., Depersis C.L., El-Khoury D., Fuller C.L., Shores E.W., Love P.E., Samelson L.E. 2005. Mutation of the phospholipase C-γ1–binding site of LAT affects both positive and negative thymocyte selection. J. Exp. Med. 201:1125–1134 10.1084/jem.20041869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S., Watanabe T., Lin C.S., William C.M., Tanabe Y., Jessell T.M., Costantini F. 2001. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1:4 10.1186/1471-213X-1-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.K., Jameson S.C., Hogquist K.A. 2003. Positive and negative selection of T cells. Annu. Rev. Immunol. 21:139–176 10.1146/annurev.immunol.21.120601.141107 [DOI] [PubMed] [Google Scholar]
- Tarakhovsky A., Kanner S.B., Hombach J., Ledbetter J.A., Müller W., Killeen N., Rajewsky K. 1995. A role for CD5 in TCR-mediated signal transduction and thymocyte selection. Science. 269:535–537 10.1126/science.7542801 [DOI] [PubMed] [Google Scholar]
- Wu Y., Borde M., Heissmeyer V., Feuerer M., Lapan A.D., Stroud J.C., Bates D.L., Guo L., Han A., Ziegler S.F., et al. 2006. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell. 126:375–387 10.1016/j.cell.2006.05.042 [DOI] [PubMed] [Google Scholar]
- Zeng H., Chen Y., Yu M., Xue L., Gao X., Morris S.W., Wang D., Wen R. 2008. T cell receptor-mediated activation of CD4+CD44hi T cells bypasses Bcl10: an implication of differential NF-kappaB dependence of naïve and memory T cells during T cell receptor-mediated responses. J. Biol. Chem. 283:24392–24399 10.1074/jbc.M802344200 [DOI] [PMC free article] [PubMed] [Google Scholar]