Abstract
Mutations in isocitrate dehydrogenase 1 and 2 (IDH1/2), are present in most gliomas and secondary glioblastomas, but are rare in other neoplasms. IDH1/2 mutations are heterozygous, and affect a single arginine residue. Recently, IDH1 mutations were identified in 8% of acute myelogenous leukemia (AML) patients. A glioma study revealed that IDH1 mutations cause a gain-of-function, resulting in the production and accumulation of 2-hydroxyglutarate (2-HG). Genotyping of 145 AML biopsies identified 11 IDH1 R132 mutant samples. Liquid chromatography-mass spectrometry metabolite screening revealed increased 2-HG levels in IDH1 R132 mutant cells and sera, and uncovered two IDH2 R172K mutations. IDH1/2 mutations were associated with normal karyotypes. Recombinant IDH1 R132C and IDH2 R172K proteins catalyze the novel nicotinamide adenine dinucleotide phosphate (NADPH)–dependent reduction of α-ketoglutarate (α-KG) to 2-HG. The IDH1 R132C mutation commonly found in AML reduces the affinity for isocitrate, and increases the affinity for NADPH and α-KG. This prevents the oxidative decarboxylation of isocitrate to α-KG, and facilitates the conversion of α-KG to 2-HG. IDH1/2 mutations confer an enzymatic gain of function that dramatically increases 2-HG in AML. This provides an explanation for the heterozygous acquisition of these mutations during tumorigenesis. 2-HG is a tractable metabolic biomarker of mutant IDH1/2 enzyme activity.
Isocitrate dehydrogenase 1 and 2 (IDH1 and IDH2) are NADP-dependent enzymes that catalyze the oxidative decarboxylation of isocitrate to α-ketoglutarate (α-KG) in the cytoplasm/peroxisome and mitochondrial matrix, respectively, with the concomitant production of nicotinamide adenine dinucleotide phosphate (NADPH). Their activities are distinct from the NAD-dependent enzyme IDH3, which functions in the tricarboxylic acid cycle to produce the NADH required to supply the electron transport chain.
The high-throughput sequencing of glioblastoma multiforme tumors identified a novel mutation in IDH1 that was present in 12% of tumors from glioblastoma multiforme patients (Parsons et al., 2008). Further investigation has revealed that this mutation is present in a high proportion of gliomas and secondary glioblastomas, but not in other human malignancies (Balss et al., 2008; Bleeker et al., 2009; Hartmann et al., 2009; Kang et al., 2009; Sanson et al., 2009; Watanabe et al., 2009; Yan et al., 2009). Less common mutations in IDH2 have also been identified in gliomas, and are mutually exclusive with mutations in IDH1 (Hartmann et al., 2009; Sonoda et al., 2009; Yan et al., 2009). All mutations identified to date involve a single amino acid change at arginine 132 (R132) of IDH1, or the analogous residue in IDH2 (R172). This residue is located in the active site of the enzyme and participates in isocitrate binding (Xu et al., 2004). Interestingly, all mutations are heterozygous, suggesting that alteration of R132 of IDH1 or R172 of IDH2 causes an enzymatic gain of function. Furthermore, these studies have provided evidence that the IDH1 mutation is an early event in the pathogenesis of the disease (Watanabe et al., 2009).
Recently, whole genome sequencing of a patient with acute myelogenous leukemia (AML) identified an R132 mutation in IDH1 (Mardis et al., 2009). Sequencing of additional patients revealed that IDH1 is mutated at R132, mainly to histidine and cysteine, in ∼8% of AML patients, demonstrating that this mutation is not restricted to gliomas, as previously thought (Mardis et al., 2009). More importantly, we recently reported that IDH1 R132 mutations confer a novel enzymatic activity. Surprisingly, the IDH1 R132H mutant protein was found to catalyze the NADPH-dependent reduction of α-KG to 2-hydroxyglutarate (2-HG), a rare metabolite normally present at very low levels in healthy cells and tissues (Dang et al., 2009). We set out to further investigate the role of IDH mutation in AML. Our work establishes that IDH1 R132 mutations cause production and accumulation of 2-HG in AML cells. Additionally, 2-HG screening uncovered previously unrecognized IDH2 mutations in AML. Overall, our data indicate that mutations at R132 and R172 in the active sites of IDH1 and IDH2, respectively, lead to a change in the molecular mechanism of enzyme catalysis, resulting in elevated levels of 2-HG in AML.
RESULTS AND DISCUSSION
As IDH1 is a critical enzyme in cellular metabolism, the recent report of IDH1 mutations in AML is intriguing. To investigate the role of IDH1 R132 mutations in AML, we genotyped leukemic cells obtained at initial presentation, from a series of 145 AML patients treated at the Princess Margaret Hospital with the aim of identifying mutant samples in our viable cell tissue bank. Heterozygous IDH1 R132 mutations were found in 11 (8%) of these patients (Table I). The spectrum of IDH1 mutations in AML appears to differ from that seen in central nervous system (CNS) tumors. In the CNS, the majority of mutations (80–90%) are IDH1 R132H substitutions, whereas we observe 5, 4, and 2 patients with IDH1 R132H, R132C, and R132G mutations, respectively (Table I). This is consistent with the previous report in AML (Mardis et al., 2009) and suggests that there may be functional differences among these IDH1 mutants. In four cases, leukemic cells were also available from samples taken at the time of relapse. The IDH1 mutation was retained in 4/4 of these samples (Table I). One of the AML patients harboring an IDH1 mutation had progressed from an earlier myelodysplastic syndrome (MDS). When cells from the prior MDS biopsy were genotyped, IDH1 was found to be WT. Further sequencing of a large group of well-characterized MDS samples will be necessary to determine whether IDH1 mutations are a feature of this heterogeneous disease, and whether they contribute to progression to AML. In a subset of IDH1 mutant patients (n = 8), reverse transcribed RNA was used for genotyping to assess the relative expression of mutant and WT alleles. Sequenom genotyping showed balanced allele peaks for these samples, indicating that both the WT and mutant genes are expressed. 10 established AML cell lines were also genotyped (OCI/AML-1, OCI/AML-2, OCI/AML-3, OCI/AML-4, OCI/AML-5, HL-60, MV-4-11, THP-1, K562, and KG1A) and all were IDH1 WT. This finding is consistent with the failure of other investigators to identify glioma or glioblastoma cell lines retaining the IDH1 R132 mutation after long-term culture (Bleeker et al., 2009).
Table I.
Identification of 13 AML patients bearing an IDH1 R132 or IDH2 R172 mutation
| Patient ID | Mutation | Amino acid change | FAB subtype | NPM1 and FLT3 status | Cytogenetic profile | Genotype at relapse | 2-HG level (ng/2 × 106 cells) |
| IDH1 mutations | |||||||
| 090108 | G/A | R132H | M4 | na | Normal | na | 2,090 |
| 090356 | G/A | R132H | na | na | na | na | 1,529 |
| 0034 | C/T | R132C | M5a | Normal | Normal | na | 10,285 |
| 0086 | C/G | R132G | M2 | Normal | Normal | na | 10,470 |
| 0488 | C/T | R132C | M0 | Normal | Normal | R132C | 13,822 |
| 8587 | G/A | R132H | na | Normal | Normal | na | 5,742 |
| 8665 | C/T | R132C | M1 | na | Normal | na | 7,217 |
| 8741 | G/A | R132H | M4 | NPM1 | Normal | R132H | 6,419 |
| 9544 | C/G | R132G | na | na | Normal | R132G | 4,962 |
| 747762 | G/A | R132H | M1 | NPM1 | Normal | R132H | 8,464 |
| 090148 | C/T | R132C | M1 | na | 46, xx, i(7) (p10) [20] | na | na |
| IDH2 mutations | |||||||
| 9382 | G/A | R172K | M0 | Normal | Normal | na | 19,247 |
| 0831 | G/A | R172K | M1 | Normal | Normal | na | 15,877 |
| IDH1/2 WT | |||||||
| 090239 | WT | WT | na | FLT3 | Normal | na | 112 |
| 090158 | WT | WT | M2 | na | 46, XX [15] | na | 411 |
| 090313 | WT | WT | na | na | Normal | na | 116 |
FLT3, FMS-related tyrosine kinase 3 internal tandem duplications. Na indicates that some data were not available for some patients.
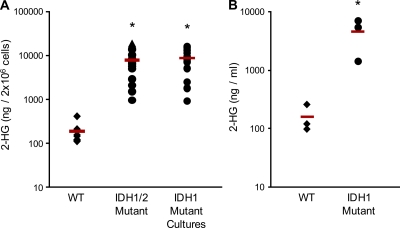
Because gliomas carrying IDH1 mutations accumulate high levels of 2-HG (Dang et al., 2009), we measured 2-HG in this set of AML samples. Levels of 2-HG were ∼50-fold higher in samples harboring an IDH1 R132 mutation (Table I, Fig. 1 A, and Table S2). 2-HG was also elevated in the sera of patients with IDH1 R132 mutant AML (Fig. 1 B). There was no relationship between the specific amino acid substitution at residue 132 and the level of 2-HG.
Figure 1.
IDH1/2 mutant AML cells and sera have increased levels of 2-HG. (A) Extracts from IDH1/2 WT (n = 10) and IDH1/2 mutant (n = 16) patient leukemia cells obtained at presentation and relapse, and IDH1 R132 mutant leukemia cells grown in culture for 14 d (n = 14) were analyzed by LC-MS to measure levels of 2-HG. (B) 2-HG was measured in sera of patients with IDH1 WT or IDH1 R132 mutant leukemia. In A and B, each point represents an individual patient sample. Diamonds represent WT, circles represent IDH1 mutants, and triangles represent IDH2 mutants. Horizontal bars indicate the mean. * indicates a statistically significant difference relative to WT patient cells (P < 0.05).
Interestingly, two IDH1 WT samples also showed high levels of 2-HG (Table I). Mutation of arginine 172 in IDH2 has been previously reported in gliomas (Ducray et al., 2009; Hartmann et al., 2009; Sonoda et al., 2009; Yan et al., 2009), but not in AML. The high 2-HG concentration prompted sequencing of the IDH2 gene in these two AML samples, and IDH2 R172K mutations were identified in both samples (Table I). This is the first report of IDH2 R172K mutations in AML. Subsequently, all samples were screened for IDH2 mutations, but no further mutations were identified.
Evaluation of the clinical characteristics of patients with or without IDH1/2 mutations revealed a significant association between IDH1/2 mutations and normal karyotype (P = 0.05), but no other differences (Table S1). Notably, there was no difference in treatment response for a subgroup of patients who received consistent treatment (n = 136). These findings are consistent with the initial study identifying IDH1 mutations in AML (Mardis et al., 2009).
Unlike in glioma, where IDH1 mutant patients have improved outcome, there does not seem to be an effect of IDH1/2 mutation status on most clinical characteristics or treatment response in AML. However, the studies reported to date are likely not sufficiently powered to adequately address these questions. Further studies using larger numbers of patients will be required to determine whether IDH1 mutation status will be a clinically useful marker. Regardless of its effect on outcome, the finding that 4/4 patients retained the mutation upon relapse suggests that it is stable through the course of disease progression, and may be clinically useful as a marker of minimal residual disease, as has been shown for nucleophosmin 1 (NPM1) mutations (Schnittger et al., 2009).
To extend these findings, panels of AML cells from WT and IDH1 mutant patients were cultured in vitro. There was no difference in the growth rates or viability of the IDH1 R132 mutant and WT cells, with both groups showing high variability in their ability to proliferate in culture, as is characteristic of primary AML cells (Fig. S1). There was no relationship between 2-HG levels in the IDH1 R132 mutant cells and their growth rate or viability in culture. After 14 d in culture, the mutant AML cells retained their IDH1 R132 mutations (11/11), and continued to accumulate high levels of 2-HG (Fig. 1 A), further confirming that IDH1 R132 mutations lead to the production and accumulation of 2-HG in AML cells.
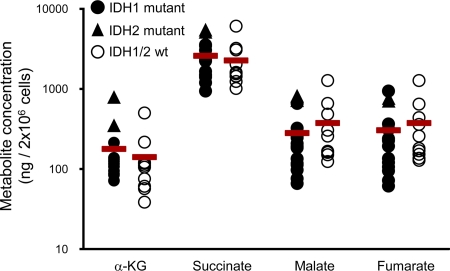
To investigate the effect of IDH1/2 mutations on the concentration of cellular metabolites proximal to the IDH reaction, α-KG, succinate, malate, and fumarate levels were measured in AML cells with IDH1/2 mutations and in a set of WT AML cells matched for AML subtype and cytogenetic profile. None of the metabolites were found to be greatly altered in the IDH1 mutants compared with the IDH1 WT cells (Fig. 2 and Table S2). Of note, the mean level of α-KG was not altered in the IDH1/2 mutant AML cells, suggesting that the mutation does not decrease the concentration of this metabolite as has been previously hypothesized (Zhao et al., 2009). These findings are consistent with measurements made in glioma tissue (Dang et al., 2009), and indicate that IDH1 mutations do not cause large changes in the cellular concentrations of these metabolites.
Figure 2.
IDH1/2 mutant AML cells do not display altered levels of central carbon metabolites. Extracts from leukemia cells of AML patients carrying an IDH1/2 mutant allele (mutant; n = 16) or WT (n = 10) obtained at initial presentation and relapse were assayed by LC-MS for levels of α-KG, succinate, malate, and fumarate. Each point represents an individual patient sample. Open circles represent WT, closed circles represent IDH1 mutants, and triangles represent IDH2 mutants. Horizontal bars represent the mean. There were no statistically significant differences between the WT and IDH1/2 mutant AML samples.
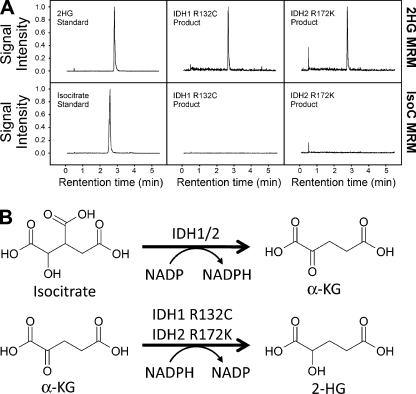
To confirm that the R132C mutation of IDH1, and the R172K mutation of IDH2 confer a novel enzymatic activity that produces 2-HG, recombinant mutant enzymes were assayed for the NADPH-dependent reduction of α-KG. When samples were analyzed by liquid chromatography mass spectrometry (LC-MS) upon completion of the enzyme assay, 2-HG was identified as the end product for both the IDH1 R132C and IDH2 R172K mutant enzymes (Fig. 3 A). No isocitrate was detectable by LC-MS, indicating that 2-HG is the sole product of this reaction (Fig. 3 A). This observation held true even when the reductive reaction was performed in buffer containing NaHCO3 saturated with CO2 (unpublished data), which would be expected to favor the formation of isocitrate via the canonical reverse reaction.
Figure 3.
Recombinant IDH1 R132C and IDH2 R172K produce 2-HG. (A) LC-MS analysis of in vitro reactions using recombinant IDH1 R132C and IDH2 R172K confirms that 2-HG and not isocitrate is the end product of the mutant enzyme reactions. Reactions were performed in triplicate in each of two independent experiments; typical chromatograms are presented. (B) The WT IDH1 enzyme catalyzes the oxidative decarboxylation of isocitrate to α-KG, with the concomitant reduction of NADP to NADPH. The IDH1 R132C and IDH2 R172K mutants reduce α-KG to 2-HG while oxidizing NADPH to NADP. These are referred to in the text as the “forward” and “partial reverse” reactions, respectively.
As discussed above, a large proportion of IDH1 mutant patients in AML have an IDH1 R132C mutation (Table I; Mardis et al., 2009). To biochemically characterize mutant IDH1 R132C, the enzymatic properties of recombinant R132C protein were assessed in vitro. Kinetic analyses showed that the R132C substitution severely impairs the oxidative decarboxylation of isocitrate to α-KG, with a significant decrease in kcat, even though the affinity for the cofactor NADP+ remains essentially unchanged (Table II). However, unlike the R132H mutant enzyme described previously (Dang et al., 2009), the R132C mutation leads to a dramatic loss of affinity for isocitrate (KM), and a drop in net isocitrate metabolism efficiency (kcat/KM) of more than six orders of magnitude (Table II). This suggests a potential difference in the substrate-level regulation of enzyme activity in the context of AML. Although substitution of cysteine at R132 inactivates the canonical conversion of isocitrate to α-KG, the IDH1 R132C mutant enzyme acquires the ability to catalyze the reduction of α-KG to 2-HG in an NADPH dependent manner (Fig. 3 B). This reductive reaction of mutant IDH1 R132C is highly efficient (kcat/KM) compared with the WT enzyme because of the considerable increase in binding affinity of both the NADPH and α-KG substrates (KM; Table II).
Table II.
Kinetic parameters of the IDH1 R132C mutant enzyme
| Oxidative (→NADPH) | WT | R132C |
| KM,NADP+ (µM) | 49 | 21 |
| KM,isocitrate (µM) | 57 | 8.7 × 104 |
| KM,MgCl2 (µM) | 29 | 4.5 × 102 |
| Ki,αKG (µM) | 6.1 × 102 | 61 |
| kcat (s−1) | 1.3 × 105 | 7.1 × 102 |
| kcat /KM,isoc(M−1.s−1) | 2.3 × 109 | 8.2 × 103 |
| Reductive (→ NADP+) | WT | R132C |
| KM,NADPH (µM) | na | 0.3 |
| KM,αKG (µM) | na | 295 |
| kcat (s−1) | ∼7 | 5.5 × 102 |
Na indicates no measureable activity.
There is a group of rare, inherited, neurometabolic disorders called 2-hydroxyglutaric acidurias, in which 2-HG levels are dramatically increased in the CNS and sera of patients (Rzem et al., 2007; Struys, 2006). These diseases are caused by homozygous loss of function mutations in the 2-HG dehydrogenase enzymes responsible for converting 2-HG back into α-KG. Although the phenotypes are diverse, some of these patients are at higher risk of developing brain tumors. Whether 2-HG contributes directly to tumor development is unknown. The existence of these diseases raises the possibility that mutations in genes other than IDH1 could result in an increase in 2-HG in tumors. 2-HG–based metabolite screening as performed in this study might facilitate rapid and sensitive identification of these mutations.
The physiological effects of elevated 2-HG are poorly understood. It has been reported that 2-HG can increase levels of reactive oxygen species, which may drive tumor progression (Latini et al., 2003a,b). Alternatively, because of its high degree of structural similarity to α-KG, 2-HG may bind and inhibit specific α-KG–dependent enzymes such as the prolyl hydroxylases that control the stability of the hypoxia-inducible factor transcription factors. This could explain the finding that hypoxia-inducible factor target genes have been reported to be elevated in IDH1 mutant glioma tissue (Zhao et al., 2009), whereas levels of α-KG are not reduced in AML or glioma tumor cells harboring IDH1 mutations.
The surprising finding that the IDH1 R132 mutant protein acquires a novel enzymatic function provides a new explanation for the heterozygous acquisition of this mutation during tumorigenesis (Dang et al., 2009). We have shown that the IDH1 R132C mutant commonly found in AML has the ability to reduce α-KG to 2-HG in an NADPH-dependent manner, and that the usually scarce α-KG derivative 2-HG is dramatically elevated in leukemia cells and sera of patients carrying IDH1 mutations. Our previous work has also shown high levels of 2-HG in glioma tissues harboring IDH1 R132 mutations, suggesting that this is a consistent feature of IDH1 R132 mutant cells and tissues (Dang et al., 2009). In addition, we identified two IDH2 R172K mutations and corresponding elevated 2-HG levels for the first time in AML, and showed directly that IDH2 R132K mutant protein produces 2-HG in vitro. The observation that the mutation of IDH2 arginine 172 leads to production of 2-HG in AML represents an example of metabolic convergence. Mutations in the analogous arginine residue in the active sites of the two enzymes leads to the same biochemical gain of function. Further functional and mechanistic work will be required to understand the underlying biology driving the acquisition of these mutations, and to determine whether mutants of IDH1 R132 and IDH2 R172 may be useful therapeutic targets.
MATERIALS AND METHODS
Patients and clinical data.
Peripheral blood and bone marrow were collected from AML patients at the time of diagnosis and at relapse, after informed consent and approval from the Research Ethics Board of the University Health Network. Sample identification numbers are unique, anonymous identifiers generated by the leukemia bank at the Princess Margaret Hospital, and are independent of true patient ID numbers. The cells were separated by ficoll hypaque centrifugation, and stored at −150°C as described previously (Miyauchi et al., 1987). Patient sera were stored at −80°C. Cytogenetics and molecular testing were performed in the diagnostic laboratory of the University Health Network (Toronto, Canada). A subgroup of patients (n = 132) was given consistent treatment using a standard induction and consolidation chemotherapy regimen consisting of daunorubicin and cytarabine.
IDH1 and IDH2 genotyping.
DNA was extracted from leukemic cells and cell lines using the Puregene kit (QIAGEN). For a subset of samples (n = 96), RNA was extracted using a Qiagen RNeasy kit, and reverse transcribed into cDNA for genotyping. IDH1 and IDH2 genotype was determined at the Analytical Genetics Technology Centre at the University Health Network using a Sequenom MassARRAY platform (Sequenom). Positive results were confirmed by direct sequencing.
Cell lines.
AML cell lines and 5637 cells were obtained from the laboratory of M. Minden (Ontario Cancer Institute, Toronto, Canada). Primary AML cells were cultured in α-MEM media with 20% fetal bovine serum and 10% 5637 cell-conditioned media as previously described (Miyauchi et al., 1987). Growth curves were generated by counting viable cells as assessed by trypan blue exclusion on a Vi-CELL automated cell counter (Beckman Coulter).
Expression/purification of IDH1 and IDH2 proteins.
The human IDH1 cDNA (GenBank/EMBL/DDBJ accession no. NM_005896) and IDH2 cDNA (GenBank/EMBL/DDBJ accession no. NM_002168) were purchased from OriGene Technologies. IDH1 (full length) and IDH2 (residues 40–452) were subcloned into vector pET41a (EMD Biosciences) to enable the E. coli expression of C-terminal His8-tagged proteins. Site-directed mutagenesis was performed using the QuikChange Lightning kit (Stratagene) to change C394 to T in IDH1 and G515 to A in IDH2, resulting in the R132C and R172K mutations, respectively. WT and mutant IDH1 proteins were expressed in and purified from the E. coli Rosetta (DE3) strain (Invitrogen). Overexpression of IDH2 protein was accomplished by co-transfection of plasmids encoding respective IDH2 clones and pG-KJE8–expressing chaperone proteins (Nishihara et al., 1998, 2000).
IDH1/2 activity assays.
Enzymatic activity was assessed by following the change in NADPH absorbance at 340 nm in an SFM-400 stopped-flow spectrophotometer (Biological) in the presence of isocitrate and NADP (forward reaction) or α-KG and NADPH (reverse reaction) in standard reaction buffer (150 mM NaCl, 20 mM Tris-Cl, pH 7.5, 10 mM MgCl2, and 0.03% [wt/vol] bovine serum albumin). To measure kinetic parameters, sufficient enzyme was added to give a linear reaction for 1–5 s. Enzymatic binding constants were determined using curve-fitting algorithms to standard kinetic models with SigmaPlot software (Systat Software). For determination of kcat, enzyme was incubated with 5X Km of substrate and cofactor. An extinction coefficient of 6,200 M−1 cm−1 was used for NADPH.
2-HG and metabolite analysis.
Metabolites were extracted from cells and sera using 80% aqueous methanol, as previously described (Lu et al., 2006). For cell extraction, 2 × 106 cells were suspended in −80°C 80% methanol. For serum extraction, 1 ml of serum was mixed with 4 ml −80°C methanol. All extracts were spun at 13,000 rpm at 4°C to remove precipitate, dried at room temperature, and stored at −80°C. Metabolite levels were determined by ion-paired reverse-phase LC coupled to negative mode electrospray triple-quadrupole mass spectrometry using multiple reaction monitoring, and integrated elution peaks were compared with metabolite standard curves for absolute quantification (Dang et al., 2009).
Statistical analysis.
Fisher’s exact test was used to test for differences in categorical variables between IDH1/2 WT and IDH1/2 mutant patients. One-way analysis of variance followed by a Student’s t test with correction for multiple comparisons was used to test for differences in IDH1 activity and metabolite concentrations. Differences with P < 0.05 were considered significant.
Online supplemental material.
Fig. S1 shows growth of IDH1 R132 mutant cells in vitro. Table S1 shows characteristics of IDH1/2 mutant and WT patients. Table S2 shows metabolite concentrations in individual IDH1/2 mutant and WT AML cells. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20092506/DC1.
Acknowledgments
We thank Shaohui Wang at ChemPartner for assistance with biochemical experiments. We also thank Katherine Yen, Kevin Marks, Francisco Salituro, Jeffrey Sauders, Craig Thompson, and Lewis Cantley for discussion and comments on the manuscript.
Tak Mak is supported by grants from the Canadian Institutes of health Research, the Canadian Cancer Society, the Terry Fox Foundation, and the Leukemia and Lymphoma Society.
Stefan Gross, Edward M. Driggers, Mark A. Bittinger, Hyun Gyung Jang, Shenfang Jin, David P. Shenkein, Shinsan M. Su, Lenny Dang, and Valeria Fantin disclose financial interests, as employees of Agios Pharmaceuticals. Tak Mak owns stock options in Agios Pharmaceuticals. The authors have no further conflicts of interest.
Footnotes
Abbreviations used:
- 2-HG
- 2-hydroxyglutarate
- α-KG
- α-ketoglutarate
- AML
- acute myelogenous leukemia
- CNS
- central nervous system
- IDH1/2
- isocitrate dehydrogenase 1 and 2
- LC-MS
- liquid chromatography mass spectrometry
- MDS
- myelodysplastic syndrome
- NADPH
- nicotinamide adenine dinucleotide phosphate
- NPM1
- nucleophosmin 1
References
- Balss J., Meyer J., Mueller W., Korshunov A., Hartmann C., von Deimling A. 2008. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 116:597–602 10.1007/s00401-008-0455-2 [DOI] [PubMed] [Google Scholar]
- Bleeker F.E., Lamba S., Leenstra S., Troost D., Hulsebos T., Vandertop W.P., Frattini M., Molinari F., Knowles M., Cerrato A., et al. 2009. IDH1 mutations at residue p.R132 (IDH1(R132)) occur frequently in high-grade gliomas but not in other solid tumors. Hum. Mutat. 30:7–11 10.1002/humu.20937 [DOI] [PubMed] [Google Scholar]
- Dang L., White D.W., Gross S., Bennett B.D., Bittinger M.A., Driggers E.M., Fantin V.R., Jang H.G., Jin S., Keenan M.C., et al. 2009. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 462:739–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducray F., Marie Y., Sanson M. 2009. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 360:2248. [PubMed] [Google Scholar]
- Hartmann C., Meyer J., Balss J., Capper D., Mueller W., Christians A., Felsberg J., Wolter M., Mawrin C., Wick W., et al. 2009. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol. 118:469–474 10.1007/s00401-009-0561-9 [DOI] [PubMed] [Google Scholar]
- Kang M.R., Kim M.S., Oh J.E., Kim Y.R., Song S.Y., Seo S.I., Lee J.Y., Yoo N.J., Lee S.H. 2009. Mutational analysis of IDH1 codon 132 in glioblastomas and other common cancers. Int. J. Cancer. 125:353–355 10.1002/ijc.24379 [DOI] [PubMed] [Google Scholar]
- Latini A., Scussiato K., Rosa R.B., Leipnitz G., Llesuy S., Belló-Klein A., Dutra-Filho C.S., Wajner M. 2003a. Induction of oxidative stress by L-2-hydroxyglutaric acid in rat brain. J. Neurosci. Res. 74:103–110 10.1002/jnr.10735 [DOI] [PubMed] [Google Scholar]
- Latini A., Scussiato K., Rosa R.B., Llesuy S., Belló-Klein A., Dutra-Filho C.S., Wajner M. 2003b. D-2-hydroxyglutaric acid induces oxidative stress in cerebral cortex of young rats. Eur. J. Neurosci. 17:2017–2022 10.1046/j.1460-9568.2003.02639.x [DOI] [PubMed] [Google Scholar]
- Lu W., Kimball E., Rabinowitz J.D. 2006. A high-performance liquid chromatography-tandem mass spectrometry method for quantitation of nitrogen-containing intracellular metabolites. J. Am. Soc. Mass Spectrom. 17:37–50 10.1016/j.jasms.2005.09.001 [DOI] [PubMed] [Google Scholar]
- Mardis E.R., Ding L., Dooling D.J., Larson D.E., McLellan M.D., Chen K., Koboldt D.C., Fulton R.S., Delehaunty K.D., McGrath S.D., et al. 2009. Recurring mutations found by sequencing an acute myeloid leukemia genome. N. Engl. J. Med. 361:1058–1066 10.1056/NEJMoa0903840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyauchi J., Kelleher C.A., Yang Y.C., Wong G.G., Clark S.C., Minden M.D., Minkin S., McCulloch E.A. 1987. The effects of three recombinant growth factors, IL-3, GM-CSF, and G-CSF, on the blast cells of acute myeloblastic leukemia maintained in short-term suspension culture. Blood. 70:657–663 [PubMed] [Google Scholar]
- Nishihara K., Kanemori M., Kitagawa M., Yanagi H., Yura T. 1998. Chaperone coexpression plasmids: differential and synergistic roles of DnaK-DnaJ-GrpE and GroEL-GroES in assisting folding of an allergen of Japanese cedar pollen, Cryj2, in Escherichia coli. Appl. Environ. Microbiol. 64:1694–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara K., Kanemori M., Yanagi H., Yura T. 2000. Overexpression of trigger factor prevents aggregation of recombinant proteins in Escherichia coli. Appl. Environ. Microbiol. 66:884–889 10.1128/AEM.66.3.884-889.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons D.W., Jones S., Zhang X., Lin J.C., Leary R.J., Angenendt P., Mankoo P., Carter H., Siu I.M., Gallia G.L., et al. 2008. An integrated genomic analysis of human glioblastoma multiforme. Science. 321:1807–1812 10.1126/science.1164382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzem R., Vincent M.F., Van Schaftingen E., Veiga-da-Cunha M. 2007. L-2-hydroxyglutaric aciduria, a defect of metabolite repair. J. Inherit. Metab. Dis. 30:681–689 10.1007/s10545-007-0487-0 [DOI] [PubMed] [Google Scholar]
- Sanson M., Marie Y., Paris S., Idbaih A., Laffaire J., Ducray F., El Hallani S., Boisselier B., Mokhtari K., Hoang-Xuan K., Delattre J.Y. 2009. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J. Clin. Oncol. 27:4150–4154 10.1200/JCO.2009.21.9832 [DOI] [PubMed] [Google Scholar]
- Schnittger S., Kern W., Tschulik C., Weiss T., Dicker F., Falini B., Haferlach C., Haferlach T. 2009. Minimal residual disease levels assessed by NPM1 mutation-specific RQ-PCR provide important prognostic information in AML. Blood. 114:2220–2231 10.1182/blood-2009-03-213389 [DOI] [PubMed] [Google Scholar]
- Sonoda Y., Kumabe T., Nakamura T., Saito R., Kanamori M., Yamashita Y., Suzuki H., Tominaga T. 2009. Analysis of IDH1 and IDH2 mutations in Japanese glioma patients. Cancer Sci. 100:1996–1998 10.1111/j.1349-7006.2009.01270.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struys E.A. 2006. D-2-Hydroxyglutaric aciduria: unravelling the biochemical pathway and the genetic defect. J. Inherit. Metab. Dis. 29:21–29 10.1007/s10545-006-0317-9 [DOI] [PubMed] [Google Scholar]
- Watanabe T., Nobusawa S., Kleihues P., Ohgaki H. 2009. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am. J. Pathol. 174:1149–1153 10.2353/ajpath.2009.080958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Zhao J., Xu Z., Peng B., Huang Q., Arnold E., Ding J. 2004. Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity. J. Biol. Chem. 279:33946–33957 10.1074/jbc.M404298200 [DOI] [PubMed] [Google Scholar]
- Yan H., Parsons D.W., Jin G., McLendon R., Rasheed B.A., Yuan W., Kos I., Batinic-Haberle I., Jones S., Riggins G.J., et al. 2009. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 360:765–773 10.1056/NEJMoa0808710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Lin Y., Xu W., Jiang W., Zha Z., Wang P., Yu W., Li Z., Gong L., Peng Y., et al. 2009. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. 324:261–265 10.1126/science.1170944 [DOI] [PMC free article] [PubMed] [Google Scholar]